Abstract
The INK4A and the INK4B genes map to chromosome 9p21, an area frequently deleted in bladder neoplasms. In addition to the p16 protein, the INK4A encodes for a second product, termed p19ARF. We analyzed tissues from 121 patients with initial Ta and T1 tumors. Deletions of the INK4A gene were observed in 17 of 121 (14.1%) cases. Point mutations were identified in 2 of 64 (3.1%) tumors. The INK4A-exon 1β and the INK4B gene were codeleted with INK4A in all of the homozygously deleted cases analyzed. The p16 promoter underwent de novo methylation in 7 of 47 (14.9%) evaluable cases. The p16-positive phenotype was observed in 18 of 56 (32%) evaluable cases. p16 negative phenotype correlated with deletion and methylation status. A statistically significant association between INK4A homozygous deletions and tumor size was observed (P = 0.003). Patients bearing tumors with INK4A homozygous deletions had a lower recurrence-free survival (P = 0.040) than those with wild type INK4A. In conclusion, deletions and methylation of the INK4A gene occur frequently in superficial bladder tumors. However, only those deletions that affect both the p16 and the p19ARF, deregulating both the pRb and p53 pathways, correlated with clinicopathological parameters of worse prognosis.
The discovery of proteins that bind to and inhibit the catalytic activity of complexes of cyclins and cyclin-dependent kinases (Cdk) has identified kinase inhibition as an intrinsic component of the stringent control imposed during cell cycle transitions. 1,2 This new family of negative regulators has been designated as cyclin-dependent kinase inhibitory (CKI) genes. The inhibitors identified so far can be subdivided into two groups, based on their sequence homology and functional similarities: INK4 and KIP family members. 1,2 The INK4A and INK4B gene map to the short arm of chromosome 9 (9p21), where they are found in tandem spanning a region of approximately 80 kb. The INK4A gene was initially described as encoding a 148-aa protein of Mr 15,845, termed p16. 3-5 The INK4B gene encodes a protein of 137 aa with Mr 14,700, known as p15. 6 The p16 and p15 proteins form binary complexes exclusively with Cdk4 and Cdk6, inhibiting their function and, by doing so, inhibiting the phosphorylation of the retinoblastoma protein (pRB) during the G1 phase of the cell cycle. Additional complexity results from the presence of a second INK4A product, which has been termed p19ARF (ARF is the acronym for alternative reading frame). 7-10 The p19ARF has been recently shown to interact with mdm2 and to block mdm2-induced p53 degradation and transactivational silencing. 11,12
Strong functional and genetic evidence supports an important role for INK4A as a tumor suppressor gene in a variety of tumors. The INK4A is altered in many tumor-derived cell lines and primary tumors, including bladder transitional cell carcinomas. 13-17 In addition, germ line mutations of the INK4A gene are frequently found in patients with familial melanoma and pancreatic adenocarcinoma. 18-20 More recently, independent studies reported the targeted deletion of the Ink4a and the specific p19ARF exon 1β loci in murine models. 21,22 Both Ink4a- and p19ARF-deficient mice were viable but developed spontaneous tumors at an early age. 21,22
We have previously reported that deletions of INK4A and INK4B genes are found frequently in bladder tumors. 15 The present study was conducted to further determine the frequency and potential clinical significance of detecting INK4A and INK4B mutations in superficial bladder cancer. We also assessed potential alterations affecting INK4A exon 1β. Methylation of the 5′ CpG island, located in the promoter region of p16, has been reported to be a common mechanism of p16 inactivation in certain neoplasms. 23,24 Therefore, we investigated the presence of de novo methylation in the promoter region of p16. We also assessed p16 phenotype in normal and tumor tissues by means of immunohistochemical assays employing a p16-specific monoclonal antibody.
Materials and Methods
Tissue and Patient Characteristics
The patients included in this study had newly diagnosed superficial bladder tumors and were admitted for the first transurethral resection between September, 1990 and April, 1992 in hospitals of the province of Quebec, Canada. Criteria for patient eligibility included presence of histologically confirmed Ta or T1 transitional cell carcinoma, as previously described. 25 The deletion and mutational status of the INK4A and INK4B genes were initially assessed on a group of 64 paired normal and tumor DNA samples. The analysis of the methylation condition of the p16 promoter was then analyzed on cases with nondeleted INK4A alleles (47 of 55 available cases), and tissue sections from 58 cases were also available for immunohistochemical assays. We then analyzed INK4A gene losses on 57 additional cases of paired normal and tumor DNA samples. The median follow-up period was 38.5 months (range, 2.1–51.7 months, n = 120). No follow-up was available in one of the cases. Ninety-six lesions were classified as superficial papillary tumors (Ta), and 23 lesions as invasive tumors into the lamina propria (T1). Thirty-eight tumors were classified as grade 1, 72 as grade 2, and 11 as grade 3. In 68 cases the tumor size was <3 cm, and in 47 cases it was >3 cm. In all cases with multiple tumors, the size of the largest tumor was the one reported. Eighty-six cases had single tumors and 35 cases presented multiple tumors. Samples were embedded in a cryopreservative solution (OCT Compound, Miles Laboratories, Elkhart, IN), snap-frozen in isopentane precooled in liquid nitrogen, and stored at −70°C. Representative hematoxylin-eosin-stained sections of each frozen block were examined microscopically to confirm the presence of tumor, and only lesions with >90% neoplastic cells were included in the study. Normal DNA samples from peripheral blood were obtained for all patients.
DNA Extraction
Tumor and normal DNA was extracted by methods previously described. 26 Briefly, tumor DNA was extracted by proteinase K digestion from consecutive 20-μm frozen tissue sections or cell suspension. For the normal DNA, 200 μl of blood were resuspended with 20 mmol/L Tris, 5 mmol/L EDTA (pH 8.0), and NP40 0.5%, vortexed, and incubated at room temperature for 20 minutes. After centrifuging, the pellet was resuspended with 10 μl of sodium dodecyl sulfate (10%) and 10 μl of proteinase K (2.5 mg/ml) and incubated at 65°C for 2 hours. The DNA was precipitated with 100 μl NH4Ac 7.5 mol/L and 500 μl ethanol (100%) at −20°C, and resuspended in 20 mmol/L Tris and 1 mmol/L EDTA (pH 8.0).
Polymerase Chain Reaction-Single Stranded Conformation Polymorphism (PCR-SSCP) and Multiplex PCR Analyses
PCR-SSCP assays were performed in a subset of the tumors following protocols previously described. 15 The primer sets used to amplify exons 1α (one fragment) and 2 (three overlapping fragments) of the INK4A gene were published by Hussussian et al. 18 For the exon 1β of the INK4A gene and the INK4B gene, the following primer pairs were used: 1) INK4A, exon 1β (fragment 1, 439 bp): 5′ TCC CAG TCT GCA GTT AAG G 3′ (forward) and 5′ GTC TAA GTC GTT GTA ACC CG 3′ (reverse); 2) INK4A, exon 1β (fragment 2, 160 bp): 5′ AAC ATG GTG CGC AGG TTC 3′ (forward) and 5′ AGT AGC ATC AGC ACG AGG G 3′ (reverse); 3) INK4B, exon 1: 5′ ATT ATC CGG GCC GCT GCG C 3‘ (forward) and 5′ TGC CGG CGA GGC CCT GG 3′ (reverse); 4) INK4B, exon 2 set A: 5′ CCC GGC CGG CAT CTC CCA TA 3′ (forward) and 5′ ACC ACC AGC GTG TCC AGG AA 3′ (reverse); 5) INK4B, exon 2 set B: 5′ CAC CCG ACC GGT GCA TGA TG 3′ (forward) and 5′ GTG GGC GGC TGG GGA ACC TG 3′ (reverse). DNA was amplified with 28 cycles of PCR using a thermal cycler (Perkin-Elmer Cetus, Foster City, CA). Briefly, the PCR reactions were performed in 10-μl volumes containing 80–100 ng of template DNA, 2.2 μCi of [α32P]dCTP (Dupont NEN Research Products, Boston, MA) or [α33P]dCTP (Amersham, Arlington Heights, IL), 3 mmol/L MgCl2, 100 μmol/L deoxynucleoside triphosphates, 3% DMSO, 0.6 U of TaqI polymerase, and 1× PCR buffer (Promega, Madison, WI). Annealing temperatures ranged from 55–65°C. For the SSCP analysis of the exon 1β (fragment 1) of the p19ARF, the PCR product was digested with NarI and EheI restriction enzymes for 2 hours at 37°C. In general, both digested and nondigested products were analyzed by SSCP. The PCR products were denatured and loaded onto a nondenaturing 8% polyacrylamide gel containing 10% glycerol and subjected to electrophoresis at room temperature for 12–16 hours at 10–12 W. After electrophoresis, the gels were dried and exposed to X-ray film at −70°C for 4–16 hours. In those cases in which mobility shifts were identified by PCR-SSCP, DNA was amplified and sequenced with the Sequenase PCR product sequencing kit (Amersham, Cleveland, OH).
An independent DNA amplification was performed for the comparative multiplex PCR assay. 27 A simultaneous amplification of genomic DNA was performed using two sets of primers, one to the target gene sequence under study and the other to an internal control gene sequence. The following sets of primers were used as internal controls for DNA quality and loading: 1) GADPH gene: 5′ TGG TAT CGT GGA AGG ACT CAT GAC 3′ (forward) and 5′ ATG CCA GTG AGC TTC CCG TTC AGC 3′ (reverse) (PCR product 189 bp); 2) ANDRR gene: 5′ GTG CGC GAA GTG ATC CAG AA 3′ (forward) and 5′ TCT GGG ACG CAA CCT CTC TC 3′ (PCR product 296 bp) (reverse); 3) D9S196 (260 bp): Genome Data Base Accession ID 62901. Each PCR reaction tube contained 50–100 ng of genomic DNA, 1× PCR buffer (Promega), 3.2 mmol/L MgCl2, 130 μmol/L dNTP, 5% DMSO, 0.4 μmol/L of each target and control set of primers, 0.5 U Taq Polymerase (Promega), and 1 μCi of [α33P]dCTP. Samples were preheated at 95°C for 5 minutes and amplified for 25 cycles with annealing temperatures ranging from 55–63°C, followed by an extension at 72°C for 10 minutes. PCR products were run in nondenaturing 8% acrylamide gels at 40–45 W for 3–4 hours. Gels were dried and exposed to sensitive film and to a phosphoimage plate. The sensitized plate was scanned by a phosphoimager (Bas 1000-Mac, Bio Imaging System Fujix, Fuji). The presence of INK4A or INK4B specific fragments was expressed as the ratio [target-band signal]/[control-band signal]. 27 For the INK4A-exon 1β, samples were also analyzed using primers directed to a smaller exon 1β fragment (fragment 2, see above) to rule out false negative results due to partial DNA degradation. To establish potential allelic losses in the tumor DNA samples, tumor DNA samples lacking INK4A (p16) and INK4B genes by Southern blot analysis were used as control DNA, validating the quantitative nature of the multiplex PCR method. Briefly, varying mixtures of tumor DNA and normal genomic DNA were coamplified. These tumor-to-normal DNA mixtures represented a range of the target content, varying from 0% (tumor sample control) to 100% (normal DNA counterpart). Samples presenting <25% of the control signal were considered homozygously deleted, and those presenting <65% as hemizygously deleted for the INK4A/B-specific fragments.
Antibodies and Immunohistochemistry
An avidin-biotin immunoperoxidase assay was performed on formalin-fixed, paraffin-embedded tissue sections. Deparaffinized sections were treated with 1% H2O2 to block endogenous peroxidase activity. Sections were subsequently immersed in boiling 0.01% citric acid (pH 6.0) in a microwave oven for 15 minutes to enhance antigen retrieval, allowed to cool, and incubated with 10% normal horse serum (Organon Teknika, West Chester, PA), to block nonspecific tissue immunoreactivities. A well characterized antibody to p16 (Ab-1, Oncogene Research Products, Cambridge, MA; 2 μg/ml final concentration) was then incubated overnight at 4°C. Biotinylated horse anti-mouse IgG antibodies (Vector Laboratories, Burlingame, CA, 1:500 final dilution) were used as the secondary reagent. This was followed by avidin-biotin immunoperoxidase complexes (Vector Laboratories, 1:25) for 30 minutes. Diaminobenzidine (Sigma Chemical, St. Louis, MO) was used as the final chromogen and hematoxylin was used as the nuclear counterstain. Sections were counterstained with hematoxylin. An esophageal adenocarcinoma case overexpressing p16 was always included as a positive control. Immunoreactivities were classified as a continuum of data (undetectable levels or 0% to 100% homogenous staining).
Analysis of Methylation
The methylation status of the 5′ CpG island in the promoter region of the p16 gene was determined with the CpG Wiz p16 Methylation kit (Oncor, Gaithersburg, MD), based on the method developed by Herman et al, 28 and following manufacturer’s protocol. Briefly, 0.5–1 μg of DNA was denatured with 3 mol/L of sodium hydroxide at 50°C for 10 minutes and then treated with sodium bisulfite. After completion of the DNA modification, the DNA was purified by precipitation. The dissolved DNA was amplified by PCR using primers specific for the methylated (M) or unmethylated (U) sequences. Two to 3 μl of template (corresponding to treated DNA, positive control for methylated DNA, positive control for unmethylated sequences, and distilled water as negative control) were amplified in presence of 1× Universal PCR buffer, 2.5 mmol/L dNTP mix, U or M primers, and AmpliTaq Gold (Perkin-Elmer), under the following conditions: preheating at 95°C for 12 minutes, followed by 35 cycles (95°C for 45 seconds, 66°C for 45 seconds, and 72°C for 1 minute). The PCR product was analyzed on a 2% agarose gel. The DNA methylation was determined by the presence of a 145-bp fragment in those samples amplified with the M primers.
Statistical Methods
Statistical analysis was performed to evaluate possible associations between INK4A/B deletions, prognostic variables measured at diagnosis, and disease recurrence. Results of the immunohistochemical analysis were analyzed as a continuous variable. A two-tailed Fisher’s exact test 29 was used to assess the associations between INK4A and INK4B status and clinicopathological parameters. The latter included tumor stage (Ta versus T1), tumor grade (G1 versus G2/G3), tumor size (≤3 cm versus >3 cm), and number of tumors (one versus multiple). Tumor recurrence was defined as a new tumor detected during follow-up cystoscopy. In the analysis of disease recurrence, patients who recurred were classified as failures, whereas patients who did not or were lost to follow-up during the study period were coded as censored. The follow-up time was calculated as the time elapsed between diagnosis by (transurethral resection) and first recurrence, or the date of the last cytoscopic evaluation. Differences in recurrence-free survival between the two groups were evaluated with survival curves using the Kaplan-Meier method 30 and the log rank test. 31 All statistical procedures were performed using SAS software. 32 All reported P values are two-sided. Proportional hazards analysis was used to obtain maximum likelihood estimates of relative risks (RR) and their 95% confidence intervals in a multivariate analysis. 33,34
Results
Tables 1 and 2 ▶ ▶ summarize the clinicopathological parameters at the time of diagnosis, correlating these data with the status of the INK4A gene, as well as the results from the univariate and multivariate analyses. Figures 1, 2, and 3 ▶ ▶ ▶ illustrate alterations occurring at the INK4A and INK4B genes, as detected by the comparative multiplex PCR, PCR-SSCP and sequencing, methylation-specific PCR, and immunohistochemistry.
Table 1.
Association between Deletions of INK4A and Clinicopathological Variables
| Variable | INK4A deletions | P value (Fisher’s test) | |||
|---|---|---|---|---|---|
| no | yes | ||||
| n | (%) | n | (%) | ||
| Tumor stage | |||||
| Ta | 85 | (88.5) | 11 | (11.5) | 1.00 |
| T1 | 21 | (91.3) | 2 | (8.7) | |
| Tumor grade | |||||
| G1 | 37 | (97.4) | 1 | (2.6) | 0.06 |
| G2/3 | 71 | (85.5) | 12 | (14.5) | |
| Tumor size | |||||
| <3 cm | 66 | (97.1) | 2 | (2.9) | 3.0× 10−3 |
| ≥3 cm | 37 | (78.7) | 10 | (21.3) | |
| Number of tumors | |||||
| 1 | 77 | (89.5) | 9 | (10.5) | 1.00 |
| multiple | 31 | (88.6) | 4 | (11.4) |
Table 2.
Results of Univariate and Multivariate Analyses for Prediction of Disease Recurrence
| Analysis type | Risk ratio | 95% Confidence interval | P value | |
|---|---|---|---|---|
| Lower | Upper | |||
| Univariate analysis | ||||
| INK4A deletion (no vs. yes) | 1.93 | 1.02 | 3.66 | 0.04 |
| Tumor stage (Ta vs. T1) | 1.03 | 0.58 | 1.84 | 0.91 |
| Tumor grade (G1 vs. G2/3) | 1.49 | 0.91 | 2.45 | 0.11 |
| Tumor size (<3 cm vs. ≥3 cm) | 1.73 | 1.10 | 2.72 | 0.02 |
| Number of tumors (1 vs. multiple) | 1.76 | 1.11 | 2.79 | 0.02 |
| Multivariate analysis | ||||
| INK4A deletions | 1.58 | 0.77 | 3.26 | 0.22 |
| Tumor stage | 0.86 | 0.45 | 1.63 | 0.64 |
| Tumor grade | 1.48 | 0.85 | 2.59 | 0.17 |
| Tumor size | 1.39 | 0.84 | 2.29 | 0.20 |
| Number of tumors | 1.85 | 1.13 | 3.02 | 0.01 |
Figure 1.
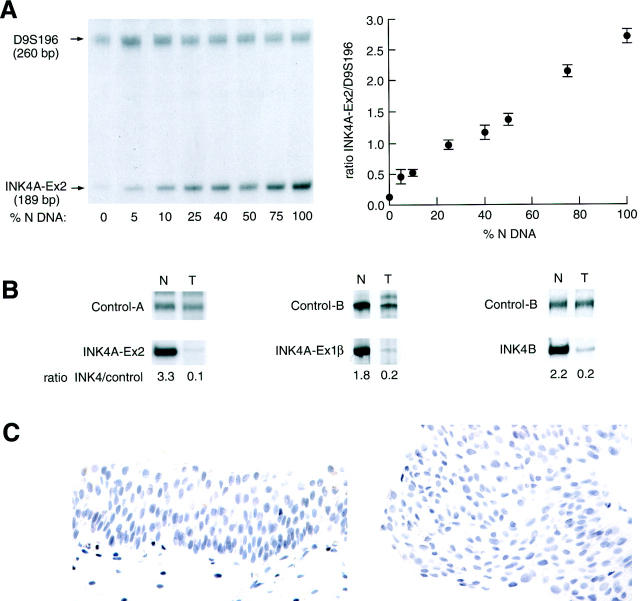
Deletion of the INK4A and the INK4B genes in superficial bladder tumors. Panel A illustrates the standardization of the assay by incremental amplification of INK4A-exon 2 with an increasing normal DNA target. The control curve was constructed with the relative (INK4A/control) ratios and the amount of normal DNA included in each sample. The vertical bars indicate the deviation of the duplicate-values. Panel B shows proportional increment in the amount of INK4A (exons 2 and 1β) and INK4B (exon 1), expressed as the ratio (INK4A or INK4B/control). Glyceraldehyde phosphate dehydrogenase androgen receptor, (ANDRR), and D9S196 were used as internal controls for DNA quality and loading. Panel C shows absence of p16 immunoreactivities in normal urothelium (left) and tumor cells (right), corresponding to an INK4A deleted case.
Figure 2.
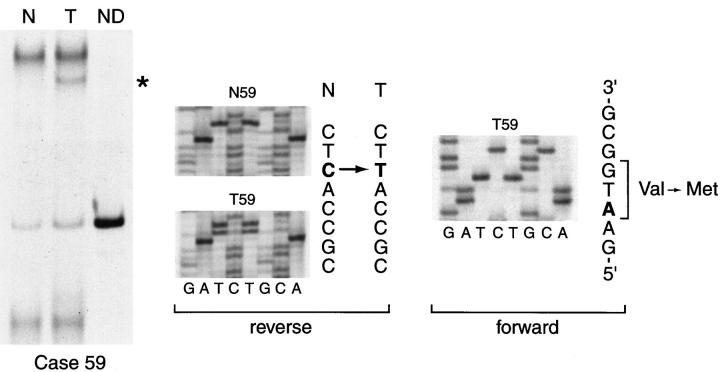
INK4A point mutations in superficial bladder tumor. The left panel illustrates the abnormal INK4A band pattern identified in the tumor DNA by PCR-SSCP (case 59). Direct sequencing results of the PCR products obtained from normal and tumor DNA from the same case are shown in the middle and right panels, corresponding to the reverse and forward sequences, respectively. The single base substitution (C → T, arrow) produces the mutation (Val→Met), as indicated in the forward sequence. N, normal DNA; T, tumor DNA; ND, nondenatured.
Figure 3.
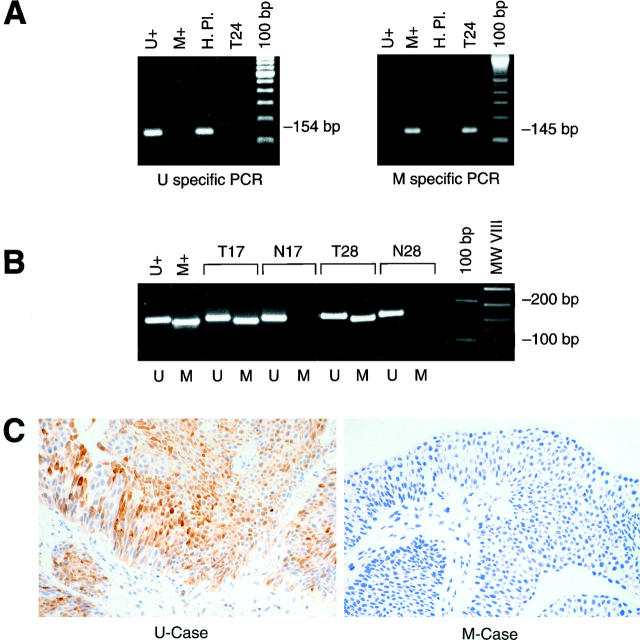
Methylation of the promoter region of INK4A/p16 in superficial bladder tumors. Panel A illustrates the amplification of unmethylated (U+) and methylated (M+) controls by a methylation-specific PCR assay. The specific band sizes are indicated. Panel B shows the results from the amplification of the DNA in two cases (cases 17 and 28). Note the presence of the methylated specific PCR product in the tumor DNA. Panel C shows representative patterns of p16 expression in an unmethylated (left) and methylated (right) bladder tumors. Note the strong nuclear staining in the unmethylated tumor. U, unmethylated; M, methylated; H Pl, human placental DNA; T24, bladder tumor-derived cell line DNA; T, tumor DNA; N, normal DNA.
We found that 17 of 121 (14.1%) cases had INK4A exon 2 losses, including 13 homozygous (ie, loss of both alleles) and 4 heterozygous (ie, loss of one allele) deletions (Figure 1) ▶ . Among the 9 homozygously deleted cases available for further analysis of INK4A exon 1β and INK4B gene, we found that all cases had a concomitant deletion of these loci (Figure 1) ▶ . Among the INK4A heterozygous cases, one case presented simultaneous deletion of the INK4B gene, two cases presented no additional alterations, and in one case the status of the INK4B gene could not be evaluated. In addition, one case showed deletion of the INK4A exon1β with no involvement of the surrounding loci (data not shown).
Two of 64 (3.1%) cases evaluated by PCR-SSCP showed extra bands or mobility band shifts in tumor DNA. Direct sequencing of normal and shifted bands demonstrated point mutations that affected both reading frames (p16 and p19ARF) of the INK4A gene. In one case, the change consisted of a transition, GAT→ AAT (Asp→Asn), that affected codon 108 of the p16. In the p19ARF this mutation involved codon 121, CGA→ CAA (Arg→ Gln) (data not shown). The second mutation was also a transition, GTG→ ATG (Val→ Met), that affected codon 59 of the p16 (Case 59, Figure 2 ▶ ). In the p19ARF this nucleotide change affected codon 72, AGA→ AAT (Ser→ Asn). This case also displayed a deletion of the contralateral INK4A allele. In addition, mobility shifts were detected in INK4A gene in both normal and tumor DNA in 4 cases corresponding to a polymorphism at codon 148 (Ala→Thr). In 11 cases we observed a transversion (C→ A) at nucleotide (−27) of the intron 1 of the INK4B gene, corresponding also to a polymorphism at that site. In cases where no signal was obtained after amplification by PCR, homozygous deletions of the INK4A and the INK4B genes were confirmed by comparative multiplex PCR.
Samples presenting one or more alleles of the INK4A gene were analyzed for the methylation status of the 5′CpG island of p16. Methylated and unmethylated control DNAs showed the corresponding fragment (Figure 3A) ▶ . Seven of 47 (14.8%) informative tumors revealed the presence of a methylation-specific fragment (Figure 3B) ▶ . In cases without the methylation-specific band, an unmethylated fragment was present, indicating the existence of good quality bisulfite-modified DNA. Therefore, the results were informative and the absence of methylated bands reflected absence of methylation in the p16 promoter.
Immunohistochemical evaluation of p16 expression was performed recording the estimated percentage of tumor cells that showed nuclear staining as continuum data. The immunohistochemical analysis was done in a blinded fashion, without knowledge of the molecular data or clinicopathological information. Normal urothelium did not show detectable nuclear or cytoplasmic p16 expression (Figure 1C) ▶ . However, different patterns were observed in tumors (Figures 1C and 3C) ▶ ▶ . Undetectable p16 levels (0% immunoreactive tumor cells) were observed in 24 cases; 1 to 20%, in 14 cases; 21 to 70%, in 15 cases; and 71 to 100%, in 3 cases. None of the cases with homozygous INK4A deletions had detectable p16 expression (Figure 1C) ▶ . Similarly, those cases that had a methylated p16 promoter did not present detectable p16 immunostaining (Figure 3C) ▶ . However, certain unmethylated cases revealed p16 nuclear overexpression (Figure 3C) ▶ .
Overall, 19 of 55 (34.6%) cases in which the deletion, point mutation, and methylation analyses were informative presented one or more alterations in the INK4A and INK4B genes. Among the tumors with homozygous deletions of the INK4A gene, 11 of 13 (84.6%) cases corresponded to Ta lesions, while the 2 (15.4%) remaining cases were T1 lesions. A significant correlation between homozygous deletions of the INK4A gene and certain clinicopathological parameters was observed. Tumors with INK4A deletions had larger diameters: 10 of 47 (21.3%) tumors >3 cm versus 2 of 68 (2.9%) tumors ≤3 cm (P = 0.003). In addition, 12 of 13 INK4A deleted cases corresponded to high grade tumors and, although this association was statistically non-significant (P = 0.061), it revealed a trend.
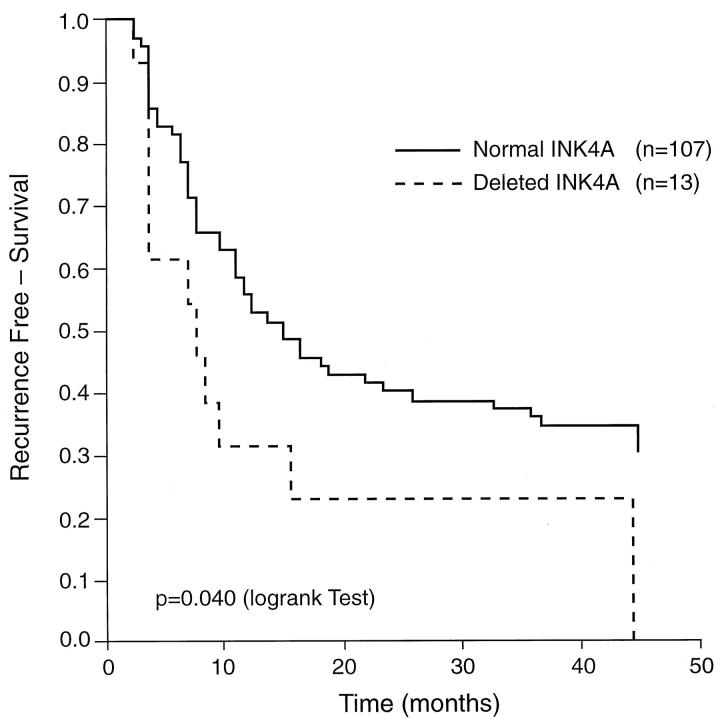
In the prospective analysis, patients with homozygous deletion of the INK4A gene had greater risk of recurrence than those with normal genotype (RR = 1.95, 95% CI = 1.02–3.66) (Table 2) ▶ . The comparison of the recurrence-free survival data of patients with normal or deleted INK4A alleles is presented in Figure 4 ▶ . The median time to survival was 7.1 months for the group of patients without INK4A deletions and 6.4 months for the group of patients with deletions. Tumor size and multiplicity were the only additional prognostic variables that affected the risk of tumor recurrence. In the multivariate analysis of risk factors by the proportional hazards model, the relative risks for recurrence were 1.6 (95% CI: 0.77–3.26, P = 0.216) for INK4A deletions and 1.9 (95% CI: 1.13–3.02, P = 0.014) for the number of tumors, after controlling for stage, grade, and tumor size. The number of patients in this analysis was 112, due to missing values for certain prognostic variables in nine cases. No correlation was found between INK4A deletions and number of tumors. No associations were observed between alterations of the INK4A, INK4B gene, and the clinicopathological parameters at the time of diagnosis (stage, grade, number of tumors, diameter) or recurrence. Only two cases presented disease progression during the follow-up period: one at 35.9 months and another at 39.5 months.
Figure 4.
Effect of the INK4A gene deletions on the recurrence-free survival in patients with superficial bladder tumors. Disease-specific survival was evaluated using the Kaplan-Meier method and curves were compared with the log rank test. The probability of recurrence-free survival was significantly higher in patients with nondeleted INK4A than in those with homozygous INK4A deletions (P = 0.04).
Discussion
The INK4A gene is deleted in a variety of primary tumors, including bladder carcinomas. 14-17 In a previous study, it was revealed that the area of deletion often comprises both the INK4A and the INK4B genes and that there is a correlation between deletions of the INK4A gene and low stage bladder tumors. 15 We observed that papillary superficial tumors (Ta) and tumors invading the lamina propria (T1) had INK4A deletions more frequently than muscle invasive tumors. 15 The INK4A gene encodes, through the utilization of exon 1α or exon 1β, two proteins involved in cell cycle arrest: p16 and p19ARF. 7-10 The present study was undertaken to analyze the status of INK4A and INK4B genes in a cohort of 121 patients first diagnosed with superficial bladder tumors.
We found deletions in 17 of 121 (14.1%) cases, most corresponding to homozygous deletions of the INK4A gene (n = 13). These 13 cases were also deleted at the telomeric exon 1β and INK4B loci. Among the four cases with heterozygous INK4A deletions, only one case showed LOH of INK4B gene. In this study we identified only two cases in which the INK4A gene had tumor-specific point mutations. This low frequency of point mutations has also been found in other studies. 14,15,35 It is of interest that in one case (case 59) the inactivation of the INK4A gene followed the “two hit” model, displaying deletion of one allele and a point mutation of the contralateral allele. 36
In addition to tumor-specific mutations, p16 can be inactivated by de novo methylation of the CpG dinucleotides located in the promoter region. 23,24 We found de novo methylation in 14.8% of the evaluable cases. The percentage of cases with this alteration is lower than that reported by other investigators. 24 This difference in results could be due to several factors. Certain previous studies reported using methylation-sensitive restriction enzymes. Although this method is simple, an incomplete digestion can be interpreted as a false positive result. The method used in the present study is more precise, because it is based on the amplification of methylated or unmethylated DNA sequences. In addition, it is possible that advanced stage lesions (T2-T4) have an increased methylation rate of p16. To further evaluate the expression of p16, we conducted immunohistochemical analysis using a monoclonal antibody specific to exon 1α. Normal uroepithelial cells revealed undetectable p16 levels. However, 12 of 24 tumors in which p16 was not detected had normal p16 alleles. The remaining 12 cases had either homozygous deletions (n = 7) or p16 methylation (n = 5). Even though other groups have reported that absence of positive staining reflects p16 inactivation, 16 it appears that the negative phenotype in bladder cancer may reflect the normal physiological levels. Other studies have reported that p16 overexpression is associated with pRB inactivation, probably reflecting accumulation of otherwise inactive p16 products. 16,37,38
The comparative analysis of INK4A alterations and clinicopathological parameters revealed that homozygous deletions were the only mutations significantly associated with poor prognosis, including outcome. Known clinicopathological parameters associated with poor outcome include detection of multiple tumors and tumors presenting with large diameter. Specifically, homozygous deletions of the INK4A gene were significantly associated with tumors displaying large diameter (>3 cm). In this setting, loss of functional p16 appears to participate in an unrestricted cell growth. In addition, we observed a significant association between INK4A deletions and a greater risk of recurrence. After controlling for tumor stage, tumor grade, tumor size, and number of tumors, the risk of recurrence among patients with INK4A deletions was 60% higher than that of patients without INK4A deletions (RR = 1.58, 95% CI: 0.77–3.26).
An unusual genetic complexity is found on 9p21, where the INK4A and the INK4B genes encode three proteins involved in cell cycle arrest: p16, p15, and p19ARF. 3-10 The p16 and p15 proteins form binary complexes exclusively with Cdk4 and Cdk6, inhibiting their function and, by doing so, inhibiting pRB phosphorylation during G1. 3-6,39 It has been recently reported that p19ARF interacts with mdm2 and blocks the mdm2-induced p53 degradation and transactivational silencing. 11,12 Thus, the INK4A gene encodes two products impacting on the two most critical tumor suppressor pathways controlling neoplasia, p16 through pRB and p19ARF preventing neutralization of p53 by mdm2. 11,12,22 The pRB pathway could be linearly depicted as p16/CycD1/Cdk4/pRb/E2F1, and it appears to be involved in most tumors. 35 The p53 pathway includes p19ARF/mdm2/p53/p21-bax, and it has been also documented to be altered in most neoplasms. 40-42 The mechanistic basis for the inactivation of both pathways during cellular transformation stems, in part, from the deactivation of a p53-dependent cell suicide program that would normally be brought about as a response to unchecked cellular proliferation resulting from pRB deficiency. This cooperative effect of pRB and p53 has been documented in cultured cells as well as primary tumors. 43-45 Homozygous deletions of the INK4A gene disrupt these pathways and, even though not functionally equivalent to direct inactivation of p53 and pRB, could generate tumors with an aggressive behavior. Data from this study support this hypothesis, because patients with superficial bladder tumors harboring homozygous deletions of the INK4A gene had a lower recurrence-free survival rate. It is of interest that other INK4A alterations, such as point mutations or de novo methylation, did not impact on clinical outcome. Although homozygous deletions of the INK4A completely inactivate both p16 and p19ARF proteins, other mutations or altered expression probably have an impact on a single protein product. Similarly, INK4B deletions do not seem to play a key role in the biology of the disease. However, based on functional differences, such as the activation by TGFβ of p15 but not p16, 6 codeletion of INK4A and INK4B genes could provide an additional selective growth.
In conclusion, the present study reports that de novo methylation of the p16 promoter region and deletions of the INK4A/B genes are common alterations in superficial bladder tumors. Whereas point mutations and methylation affect one protein product, the complete allelic loss of the INK4A includes both p16 and p19ARF, affecting the pRb and p53 suppressor pathways. The significant association of homozygous INK4A gene deletions with tumors of large size, as well as increased risk of disease recurrence, suggests that they may provide prognostic molecular parameters for the evaluation of patients affected with superficial bladder neoplasms.
Footnotes
Address reprint requests to Carlos Cordon-Cardo, M.D., Ph.D., Department of Pathology, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10021. E-mail: cordon-c@mskcc.org.
Supported in part by National Cancer Institute grants NCI-CA-47538 (to C. C. C.) and NCI-CA-47526 (to Y. F.) as part of the Cooperative Network for Evaluation of Markers of Urinary Bladder Cancer.
References
- 1.Hunter T, Pines J: Cyclins and cancer. II. Cyclin D and CDK inhibitors come of age. Cell 1994, 79:573-582 [DOI] [PubMed] [Google Scholar]
- 2.Cordon-Cardo C: Mutation of cell cycle regulators: biological and clinical implications for human neoplasias. Am J Pathol 1995, 147:545-560 [PMC free article] [PubMed] [Google Scholar]
- 3.Serrano M, Hannon GJ, Beach D: A new regulatory motif in cell cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993, 366:704-707 [DOI] [PubMed] [Google Scholar]
- 4.Kamb A, Gruis NA, Weaver Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, 3rd, Johnson BE, Skolnick MH: A cell cycle regulator potentially involved in genesis of many tumor types. Science 1994, 264:436-440 [DOI] [PubMed] [Google Scholar]
- 5.Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA: Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 1994, 368:753-756 [DOI] [PubMed] [Google Scholar]
- 6.Hannon GJ, Beach D: p15 INK4B is a potential effector of TGFβ induced cell cycle arrest. Nature 1994, 371:257-261 [DOI] [PubMed] [Google Scholar]
- 7.Quelle DE, Zindy F, Ashmun RA, Sherr CJ: Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 1995, 83:993-1000 [DOI] [PubMed] [Google Scholar]
- 8.Duro D, Bernard O, Della Valle V, Berger R, Larsen CJ: A new type of p16INK4/MTS1 gene transcript expressed in B-cell malignancies. Oncogene 1995, 11:21-29 [PubMed] [Google Scholar]
- 9.Mao L, Merlo A, Bedi G, Shapiro GI, Edwards CD, Rollins BJ, Sidransky D: A novel p16INK4A transcript. Cancer Res 1995, 55:2995-2997 [PubMed] [Google Scholar]
- 10.Stone S, Jiang P, Dayananth P, Tavtigian SV, Katcher H, Parry D, Peters G, Kamb A: Complex structure and regulation of the P16 (MTS1) locus. Cancer Res 1995, 55:2988-2994 [PubMed] [Google Scholar]
- 11.Pomerantz J, Schrieber-Agus N, Liegoeis N, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho RA: The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell 1998, 92:713-723 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Xiong Y, Yarbrough WG: ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 1998, 92:725-734 [DOI] [PubMed] [Google Scholar]
- 13.Jen J, Harper JW, Bigner SH, Bigner DD, Papadopoulos N, Markowitz S, Willson JK, Kinzler KW, Vogelstein B: Deletion of p16 and p15 genes in brain tumors. Cancer Res 1994, 54:6353-6358 [PubMed] [Google Scholar]
- 14.Spruck CH, III, Gonzalez-Zulueta M, Shibata A, Simoneau AR, Lin MF, Gonzales F, Tsai YC, Jones PA: p16 gene in uncultured tumours. Nature 1994, 370:183-184 [DOI] [PubMed] [Google Scholar]
- 15.Orlow I, Lacombe L, Hannon GJ, Serrano M, Pellicer I, Dalbagni G, Reuter VE, Zhang Z-F, Beach D, Cordon-Cardo C: Deletion of the p16 and p15 genes in human bladder tumors. J Natl Cancer Inst 1995, 87:1524-1529 [DOI] [PubMed] [Google Scholar]
- 16.Reed AL, Califano J, Cairns P, Westra WH, Jones RM, Koch W, Ahrendt S, Eby Y, Sewell D, Nawroz H, Bartek J, Sidransky D: High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res 1996, 56:3630-3633 [PubMed] [Google Scholar]
- 17.Washimi O, Nagatake M, Osada H, Ueda R, Koshikawa T, Seki T, Takahashi T, Takahashi T: In vivo ocurrence of p16 (MTS1) and p15 (MTS2) alterations preferentially in non-small cell lung cancers. Cancer Res 1995, 55:514-517 [PubMed] [Google Scholar]
- 18.Hussussian CJ, Struewing JP, Goldstein AM, Higgins PA, Ally DS, Sheahan MD, Clark WH, Jr, Tucker MA, Dracopoli NC: Germline p16 mutations in familial melanoma. Nat Genet 1994, 8:15-21 [DOI] [PubMed] [Google Scholar]
- 19.Gruis NA, van der Velden PA, Sandkuijl LA, Prins DE, Weaver-Feldhaus J, Kamb A, Bergman W, Frants RR: Homozygotes for CDKN2 (p16) germline mutation in Dutch familial melanoma kindreds. Nat Genet 1995, 10:351-353 [DOI] [PubMed] [Google Scholar]
- 20.Goldstein AM, Fraser MC, Struewing JP, Hussussian CJ, Ranade K, Zametkin DP, Fontaine LS, Organic SM, Dracopoli NC, Clark WH, Jr, Tucker MA: Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N Engl J Med 1995, 333:970-974 [DOI] [PubMed] [Google Scholar]
- 21.Serrano M, Lee H.-W, Chin L, Cordon-Cardo C, Beach D, DePinho R: Role of the INK4a locus in tumor suppression and cell mortality. Cell 1996, 85:27-37 [DOI] [PubMed] [Google Scholar]
- 22.Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Scherr CJ: Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 1997, 91:649-659 [DOI] [PubMed] [Google Scholar]
- 23.González-Zulueta M, Bender CM, Yang AS, Nguyen T, Beart RW, Van Tornout JM, Jones PA: Methylation of the 5′ CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res 1995, 55:4531-4535 [PubMed] [Google Scholar]
- 24.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger P, Baylin SB, Sidransky D: 5′ CpG island methylation is associated with transcriptional silencing of the tumor suppressor p16/CDKN2/Mts1 in human cancers. Nat Med 1995, 1:686-692 [DOI] [PubMed] [Google Scholar]
- 25.Allard P, Fradet Y, Tetu B, Bernard P, : Quebec Urology Research Group: Tumor-associated antigens as prognostic factors for recurrence in 382 patients with primary transitional cell carcinoma of the bladder. Clin Cancer Res 1995, 1:1195-1202 [PubMed] [Google Scholar]
- 26.LaRue H, Simoneau M, Fradet Y: Human papillomavirus in transitional cell carcinoma of the urinary bladder. Clin Cancer Res 1995, 1:435-440 [PubMed] [Google Scholar]
- 27.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, Horner JW, II, DePinho RA: Cooperative effects of Ink4a and ras in melanoma susceptibility in vivo. Genes Dev 1997, 11:2822-2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB: Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996, 93:9821-9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metha CR, Patel NR: A network algorithm for performing Fisher’s Exact test r × c contingency tables. J Am Stat Assoc 1983, 78:427-434 [Google Scholar]
- 30.Kaplan EL, Meier P: Nonparametric estimation from incomplete observations. J Am Stat Assoc 1958, 53:457-481 [Google Scholar]
- 31.Peto R, Pike MC, Armitage P, Breslow NE, Cox DR, Howard SV: Design and analysis of randomized trials requiring prolonged observation of each patient. Br J Cancer 1977, 35:1-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.SAS/STAT users guide, version 6. Cary, NC, SAS Institute, Inc., 1990
- 33.Cox DR: Regression models and life tables. J R Stat Soc B 1972, 34:187-220 [Google Scholar]
- 34.Cox DR: Partial likelihood. Biometrika 1975, 62:269-276 [Google Scholar]
- 35.Liggett WH, Jr, Sidransky D: Role of the p16 tumor suppressor gene in cancer. J Clin Oncol 1998, 16:1197-1206 [DOI] [PubMed] [Google Scholar]
- 36.Knudson AG, Jr: Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA 1971, 68:820-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G: Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol 1996, 16:859-867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kratzke RA, Greatens TM, Rubins JB, Maddaus MA, Niewoehner DE, Niehans GA, Geradts J: Rb and p16INK4a expression in resected non-small cell lung tumors. Cancer 1996, 56:3415-3420 [PubMed] [Google Scholar]
- 39.Lukas J, Parry D, Aagaard L, Mann DJ, Bartkova J, Strauss M, Peters G, Bartek J: Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature 1995, 375:503-506 [DOI] [PubMed] [Google Scholar]
- 40.Lonardo F, Ueda T, Huvos AG, Healey J, Ladanyi M: p53 and MDM2 alterations in osteosarcomas: correlation with clinicopathologic features and proliferative rate. Cancer 1997, 79:1541-1547 [PubMed] [Google Scholar]
- 41.Sarkis AS, Dalbagni G, Cordon-Cardo C, Zhang ZF, Sheinfeld J, Fair WR, Herr HW, Reuter VE: Nuclear overexpression of p53 protein in transitional cell bladder carcinoma: a marker for disease progression. J Natl Cancer Inst 1993, 85:53-59 [DOI] [PubMed] [Google Scholar]
- 42.Lianes P, Orlow I, Zhang ZF, Oliva MR, Sarkis AS, Reuter VE, Cordon-Cardo C: Altered patterns of MDM2 and TP53 expression in human bladder cancer. J Natl Cancer Inst 1994, 86:1325-1330 [DOI] [PubMed] [Google Scholar]
- 43.Williams BO, Remington L, Albert DM, Mukai S, Bronson RT, Jacks T: Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat Gen 1994, 7:480-484 [DOI] [PubMed] [Google Scholar]
- 44.Cordon-Cardo C, Zhang Z-F, Dalbagni G, Drobnjak M, Charytonowicz E, Hu S-X, Xu H-J, Reuter VE, Benedict WF: Cooperative effects of p53 and pRB alterations in primary superficial bladder tumors. Cancer Res 1997, 57:1217-1221 [PubMed] [Google Scholar]
- 45.Cote RJ, Dunn MD, Chatterjee SJ, Stein JP, Shi SR, Tran QC, Hu SX, Xu HJ, Groshen S, Taylor CR, Skinner DG, Benedict WF: Elevated and absent pRb expression is associated with bladder cancer progression and has cooperative effects with p53. Cancer Res 1998, 58:1090-1094 [PubMed] [Google Scholar]