Abstract
Plant shoot development depends on the perpetuation of a group of undifferentiated cells in the shoot apical meristem (SAM). In the Petunia mutant hairy meristem (ham), shoot meristems differentiate postembryonically as continuations of the subtending stem. HAM encodes a putative transcription factor of the GRAS family, which acts non-cell-autonomously from L3-derived tissue of lateral organ primordia and stem provasculature. HAM acts in parallel with TERMINATOR (PhWUSCHEL) and is required for continued cellular response to TERMINATOR and SHOOTMERISTEMLESS (PhSTM). This reveals a novel mechanism by which signals from differentiating tissues extrinsically control stem cell fate in the shoot apex.
Keywords: Meristem, differentiation, stem cell, Petunia, HAM, WUS
Plants differ from animals in that they continuously form new organs (stems, leaves, or flowers) during postembryonic shoot development. This depends on the perpetuation of a shoot apical meristem (SAM) at the very summit of the growth axis. Within the SAM, a few stem cells are specified whose daughters give rise to the entire aerial plant body (for review, see Weigel and Jürgens 2002). A primary question in plant development is how the SAM is maintained and how stem cells are specified.
A major signaling pathway that controls stem cell identity is mediated by the homeobox gene WUSCHEL (WUS). WUS is critical for SAM maintenance as judged by mutation (Laux et al. 1996) or by genetically programmed elimination during flower development (Lenhard et al. 2001; Lohmann et al. 2001). WUS expression is under negative feedback control of one of its target genes, CLAVATA3 (CLV3), which keeps stem cell maintenance and differentiation in dynamic equilibrium (Brand et al. 2000; Schoof et al. 2000). Other major factors that control SAM maintenance are the homeobox genes of the KNOTTED (KNOX) family. One of these, SHOOTMERISTEMLESS (STM), is thought to distinguish the SAM from lateral organs (Byrne et al. 2000). Ectopic KNOX expression can induce adventitious SAMs at sites that normally are fully differentiated (e.g., see Sinha et al. 1993).
Factors like WUS, STM, and CLV are all expressed within undifferentiated cells of the SAM and are meristem intrinsic. There is evidence, however, that SAM development depends on extrinsic activities present in differentiating tissues. For example, SAM function can be affected by failures in lateral organ development or in the establishment of their ad-abaxial polarities (Waites et al. 1998; Lynn et al. 1999). Clearly, the SAM is intricately linked to its differentiating environment by non-cell-autonomous control systems, the extent and molecular mechanisms of which are important issues.
Here, we describe the GRAS gene HAIRY MERISTEM (HAM) of Petunia. HAM mediates a signal from lateral organ primordia and stem provasculature that is essential and specific for maintaining the SAM. This defines a novel pathway that links stem cell perpetuation to differentiation.
Results and discussion
HAIRY MERISTEM is required for meristem maintenance
The recessive hairy meristem (ham-B4281) mutation was found in a screen for meristem defects in a population of ∼60,000 dTph1 insertions in Petunia (Koes et al. 1995). All ham mutants (100%, n = 60, stable allele hamFT-7; Fig. 3B, below) ceased organ formation during vegetative growth after initiating a variable number of leaves (9.9 ± 3.9, n = 60), whereas wild-type plants produced 17.9 ± 1.2 (n = 20) leaves before transition to flowering (Fig. 1A,C). Terminating vegetative SAMs developed a differentiated epidermis with trichomes (Fig. 1E–G), a feature normally found only on leaf primordia and subapical stem. ham axillary meristems differentiated likewise (data not shown) but without any organ formation. If axillary shoots did grow out, they arose exclusively from early vegetative nodes and terminated after several leaves. Occasionally, ham mutants developed inflorescence nodes from adventitious shoots, as judged by a nearly opposite pair of bracts, and their apices terminated with trichomes on a differentiated epidermis (Fig. 1H,I). Flowers were rare and produced fewer, although normal, floral organs per whorl [3–5 sepals (s), 3–5 petals (p), 1–3 stamens (st), 0 carpels (c), n = 10] than wild type (invariably 5s, 5p, 5st, 2c; Fig. 1B,D). Intact ham flowers have never been observed. Floral meristems terminated as flat structures (Fig. 1J) at the cost of inner whorls.
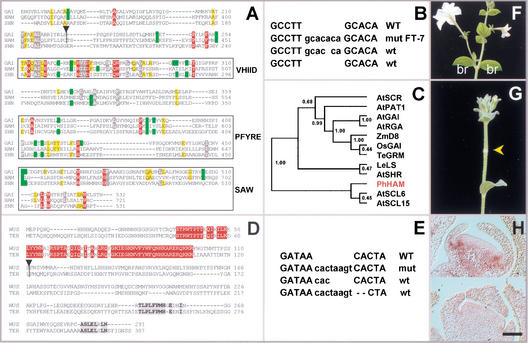
Figure 3.
Cloning and structure of HAM and TER. (A) Protein sequence alignment of the C-terminal portion of HAM with SHR and GAI. Absolutely conserved positions are red, and conserved residues are grey (>90%), yellow (>80%), or green (>70%), based on alignments of 12 GRAS sequences as in the cladogram of C. Atypical residues at conserved positions are not colored. VHIID, PFYRE, and SAW are domains as defined in Pysh et al. (1999). Triangle = dTph1 insertion in ham-B4281. (B) ham excision alleles. WT = wild-type sequence flanking the insertion in ham-B4281, wt = footprints restoring HAM function, mut = mutant footprint allele hamFT-7. (C) Tree produced by neighbor joining (ClustalG software) showing the similarities of 12 GRAS sequences. Numbers indicate bootstrap frequencies of each branchpoint in the cladogram. GenBank accession nos. AtGAI (CAA75492), AtSHR (NP195480), AtPAT1 (AAF73237), AtRGA (CAA75493), LeLS (AAD05242), AtSCL6 (NP191926), AtSCR (AAB06318), AtSCL15 (NP191622), ZmD8 (AAL10319), TeGRM (CAB51555), OsGAI (BAA90749), and PhHAM (AF481952). (D) Full protein alignment of TER (GenBank accession no. AF481951) and WUS (GenBank accession no. CAA09986). Conservation is given on the basis of these two orthologs only. Red residues indicate positions in the homeobox, and grey residues denote blocks of conspicuous colinearity. (E) As in B but for ter-B1382. (F) Wild-type Petunia inflorescence producing two bracts (br) and a flower per node. (G) HAM cosuppressor showing a node without bracts and flower (arrow). (H) In situ hybridization of HAM RNA on wild-type (upper) and cosuppressed (lower) floral meristems. Wild type shows a signal in the initiating petal primordia. The cosuppressor lacks this signal. Bar, 50 μm.
Figure 1.
Phenotype of ham mutants. (A) Wild-type W138 Petunia. (B) Wild-type W138 flower with the internal whorls of organs. Five stamens (yellow arrow) surround two fused carpels (green arrow). (C) ham-B4281 plant, terminated during vegetative rosette growth. (D) ham mutant flower lacking two stamens and both carpels. (E) Wild-type vegetative apex. p3, p2, and p1 = leaf primordia in order of decreasing age; m = meristem. (F) Vegetative ham apex shortly after termination. Primordium initiation has ceased. The meristem displays ectopic trichomes (arrows). (G) As in F, 2 wk after termination, the central dome has increased in size and is covered with trichomes. (H) Wild-type inflorescence apex. fm = floral meristem, im = inflorescence meristem, br = bract, se = sepal. (I) ham inflorescence apex with ectopic trichomes. br = last initiated bracts. (J) ham floral meristem, showing termination after initiation of three stamens (st). In place of carpels, a flat apex is visible with a small outgrowth (arrow). Bars, 100 μm.
Before termination, ham plants could not be distinguished from wild type by shoot morphology, leaf histology, or root growth (data not shown). Thus, HAM is specific for postembryonic maintenance of all shoot and floral meristems, reflecting a shared and essential property. The differentiation of epidermis with trichomes on the SAM is a unique feature of ham, pointing toward a developmental mechanism that has hitherto not been uncovered.
ham meristems differentiate as stem
The wild-type vegetative SAM contains two outer cell layers (L1 and L2; Fig. 2A) with cell divisions predominantly in the anticlinal plane, leading to a gain in surface area. The subepidermal L2 layer shows more frequent periclinal division in the SAM's periphery at sites of leaf initiation (Fig. 2A), which are marked by disappearance of PhSTM expression (Fig. 2B). PhSTM was isolated in this study as a class-1 KNOX gene, most similar to tobacco NTH15 (Nishimura et al. 1999; data not shown). Its expression pattern is typical for STM-like genes (e.g., see Long et al. 1996).
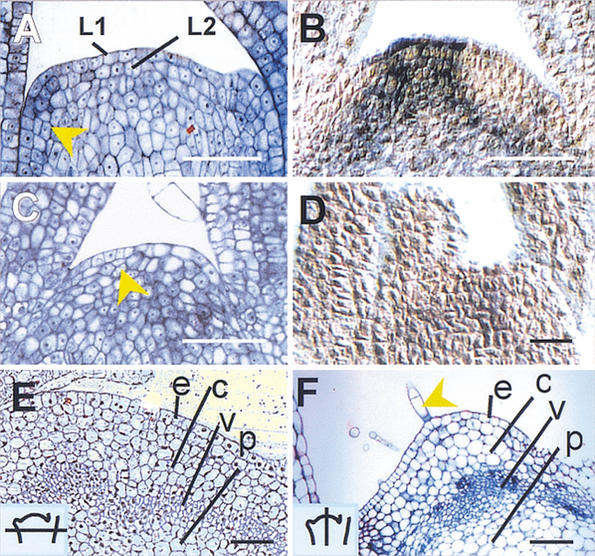
Figure 2.
Histology of ham apices. (A) Wild-type vegetative meristem. The arrow indicates a periclinal division in the L2 layer of an initiating leaf primordium. (B) In situ localization of PhSTM transcript in a wild-type vegetative apex. The signal (blue) is excluded from the leaf primordia. (C) ham vegetative apex showing a cessation of organ initiation and periclinal division in the central zone (arrow). (D) In situ localization of PhSTM transcript in a ham apex shortly after termination. (E) Transverse section of developing stem, just below a wild-type meristem (section schematized, inset). e = epidermis, c = cortex, v = vasculature, p = pith. (F) Older ham apex in longitudinal section showing a layered structure of differentiated tissue. e, c, v, p as in E. Arrow = trichome. Bars, 50 μm.
In ham seedlings with 2–4 leaves, no obvious histological deviations from wild type could be observed in the SAM (data not shown), consistent with the general absence of a ham phenotype at this stage. At termination, ham was best recognized by the absence of new leaf primordia (Fig. 2C). In L2, periclinal divisions in the central zone of the SAM were observed more frequently (Fig. 2C), and PhSTM expression was weak, although its pattern was normal (Fig. 2D). Two weeks after overt termination, ham meristems showed differentiated cell types of epidermis (trichomes), several layers of highly vacuolated subepidermal cells, and vascular differentiation (Fig. 2F). This histological structure is strikingly similar to the radial pattern of stem tissue underneath a wild-type SAM: epidermis, cortex, vasculature, and pith (Fig. 2E). At this stage, PhSTM expression could no longer be detected (data not shown).
On the basis of these data and the idea that leaf initiation requires local auxin maxima whereas stem develops by default (Reinhardt et al. 2000), we conclude that ham meristems differentiate as continuations of the stem. More importantly, PhSTM expression continues for some time after meristem termination, indicating that cells at the ham apex lose the ability to respond to PhSTM but not the potential to express it in a normal pattern.
HAM encodes a GRAS protein
ham-B4281 was genetically unstable. We differentially displayed dTph1 transposon insertions using selective inverse PCR (siPCR; see Materials and Methods). A single siPCR product fully cosegregated with the mutation (data not shown). The ham phenotype reverted to wild type whenever excision of dTph1 restored the translational reading frame (Fig. 3B). This shows that the siPCR product is part of HAM.
HAM cDNA contained a 721 amino acids open reading frame with strong homology with the GRAS family (Fig. 3A; Pysh et al. 1999). The homology extended over all the conserved residues of this family, which are located primarily in the C-terminal half of the protein. GRAS proteins involved in gibberellin signal transduction share a functional amino acid motif in the N-terminal half (DELLA; Dill et al. 2001). HAM lacks this motif. Comparative sequence analysis of a variety of GRAS proteins, ranging from gibberellin signaling proteins to root patterning factors, indicated that HAM falls into a distinct group with the putative Arabidopsis proteins AtSCL6 and AtSCL15 (SCARECROW-like; Fig. 3C). Bootstrap analysis strongly supported this group (frequency 1.00, 1000 samplings), making AtSCL6 and AtSCL15 good candidates for HAM orthologs. The dTph1 element was inserted 413 amino acids downstream from the start codon in the coding region of HAM (Fig. 3A), probably resulting in a null phenotype.
Conclusive evidence for the identity of HAM was obtained by cosuppression, using the full cDNA expressed in the sense orientation from the 35S promoter in transgenic plants. One cosuppression line was selected on the basis of absence of endogenous HAM expression (Fig. 3H) and analyzed in detail. Although vegetative development was mostly normal, axillary shoots typically terminated in hairy meristems (data not shown). Interestingly, 52% of inflorescence nodes (n = 138, 14 plants) skipped one or more organs, resuming organ initiation in the node that followed (Fig. 3F,G). This is probably a weak ham phenotype, with SAM cells differentiating into stem before acquiring organ identity. The recovery of these SAMs indicates that some self-maintaining properties of the plant's stem cell population can compensate for a partial loss of HAM function.
HAM acts non-cell-autonomously from lateral organ primordia and stem provasculature
RT-PCR detected HAM cDNA in all shoot tips and in roots but not in expanding leaves (data not shown). HAM expression was analyzed in detail by RNA in situ hybridization. In vegetative apices, HAM was expressed in deeper layers of the meristem at the presumptive site of organ initiation (Fig. 4A), as well as in the developing stem vasculature (Fig. 4B). In transverse sections, a signal was obtained in the developing primordia at least until P6 (Fig. 4C). HAM was expressed strongly in the L3-derived ground tissues in the inner part of the primordia and weakly in the main vascular bundle of older primordia (Fig. 4C). Beneath the vegetative meristem, HAM was observed in a ring that corresponds to the provasculature of the stem (Fig. 4D). In the inflorescence and floral meristems, similar HAM expression was detected consistently in all organ primordia and all floral whorls (Fig. 4E), as well as in provascular tissue (Fig. 4E). Figure 4F summarizes this pattern for a vegetative meristem.
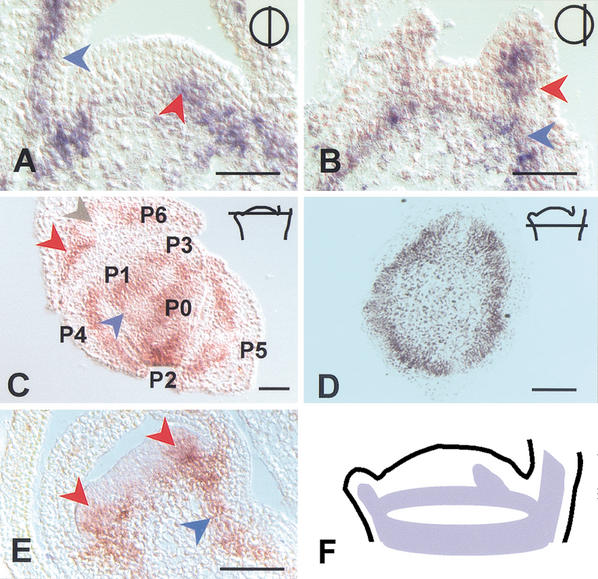
Figure 4.
Expression pattern of HAM. (A) In situ localization of HAM transcript in a near median (top right inset) longitudinal section through a vegetative apex. Signal is in the developing primordia (blue arrow) and at the presumptive position of a newly initiating primordium (red arrow). (B) As in A, with a section located more peripherally (top right inset). The signal is seen in a developing primordium (red arrow), as well as in a ring-shaped pattern that corresponds to the developing stem vasculature (blue arrow). (C) As in A, but in a transverse section. The position of the section is indicated in the top right inset. The signal is observed in the inner ground tissues of the primordia (red arrow) and is weaker in the main vascular bundle of older primordia (grey arrow). The blue arrow indicates HAM expression in a ring-shaped pattern that merges with primordia P1 and P0 and corresponds to provascular tissue of the differentiating stem. P6, P5, and so forth indicate the consecutive order of primordium initiation with decreasing age. (D) As in C, but at a position just below the meristem (indicated in top right inset). HAM expression is seen as a ring that corresponds to the provasculature of the stem. (E) HAM localization during development of the floral meristem, as exemplified for initiating petal primordia. Expression is observed in inner cell layers at the site of petal initiation (red arrows) and in subtending provascular tissue of the developing pedicel (blue arrow). (F) Schematic representation of the HAM expression pattern as exemplified for a wild-type vegetative meristem. Bar, 50 μm.
These data indicate that HAM expression from the L3 layer of the SAM may be sufficient for function in the overlying layers L1 and L2. To test this, we analyzed the HAM:ham segregation ratios for four independent revertant branches on unstable ham-B4281 homozygotes. These branches developed wild-type flowers indeterminately. Within-flower self-fertilizations from three branches gave rise to nearly normal Mendelian 3:1 (HAM:ham) patterns. However, one revertant branch did not segregate a single wild-type progeny among 80 siblings derived from the eighth and tenth flower on the branch. As gametes are derived mostly from L2 (e.g., see Stewart and Burk 1970), the wild-type phenotype of this branch was conferred by cells that were not related clonally to L2. Given the expression pattern of HAM, this branch was most likely a periclinal chimera with a revertant L3. Taken together, this essentially shows a non-cell-autonomous mode of action of HAM. Because revertant branches did not influence the mutant phenotype in other parts of the same plant, the signal must be short range.
HAM acts in parallel with TERMINATOR
ham has some important similarities with wuschel mutants of Arabidopsis (the meristem is not maintained in both). To investigate the relationships between HAM and WUS, we isolated the Petunia WUS ortholog from a mutant terminator (ter-B1382). Like wus, ter ceased shoot development after the two first true leaves (Fig. 5A,D), continuously reinitiating ectopic leaves and defective meristems from flat apices (stop-and-go growth; Fig. 5E,F). This led to increasingly bushy plants that rarely flowered. Occasional flowers had fewer floral organs per concentric whorl (data not shown), strongly resembling wus mutants (Laux et al. 1996). A single siPCR product cosegregated with ter (data not shown). The predicted TER protein was highly similar to WUS (Fig. 3D), mainly in the homeobox and with conspicuous blocks of homology in the C terminus. ter could be reverted to wild type whenever excision of dTph1 restored the open reading frame (Fig. 3E). TER gene expression patterns were identical to WUS (Fig. 5C). dTph1 was inserted 12 amino acids downstream from the homeobox, probably resulting in a null mutation. We refer to TER as PhWUS and to its mutation as ter.
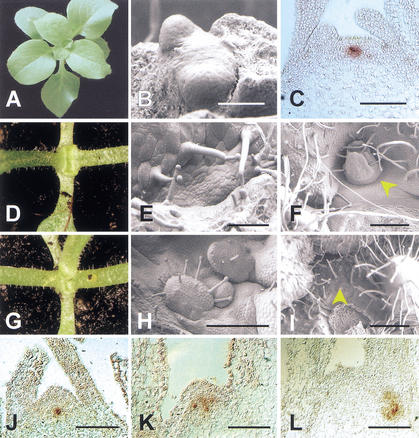
Figure 5.
Relations between ham and ter. (A) Wild-type Petunia during vegetative rosette growth. (B) Wild-type shoot apical meristem (SAM) with the first two true leaf primordia. Cotyledons have been removed. (C) In situ localization of TER (PhWUS) transcripts in a wild-type vegetative apex. (D) ter-B1382 seedling. Growth has ceased after production of the first two true leaves. (E) ter seedling apex after initiation of the two first leaves. A flat, disorganized structure replaces the SAM. (F) ter apex with an ectopic meristem (arrow, stop-and-go growth). (G) ter ham double mutant seedling. An additional leaf, compared with D, occurred with a low frequency in ter single mutants as well. (H) ter ham double mutant seedling apex. Initiation of ectopic leaves is observed (stop-and-go). (I) ter ham double mutant apex on an older plant. The SAM displays a trichome covered surface characteristic for ham single mutants. (J) In situ localization of PhWUS transcripts in a ham mutant apex shortly after termination. The signal is essentially normal. (K) In situ localization of PhWUS transcripts in a later ham mutant apex. Expression occurs in a disorganized pattern. (L) As in K. PhWUS expression in the main apex has disappeared. In the axillary position, expression is disorganized and deeply internal. Bars: B, 25 μm; C,E,J, 50 μm; F,I, 200 μm; H,K,L, 100 μm.
During early vegetative growth, ter ham double mutants showed an initial stop-and-go growth characteristic for ter but subsequently started to display ham phenotypes (Fig. 5F,I). We compared the structure of 15 meristems of both ter single mutants and ter ham double mutants in mature plants. In ter single mutants, we found ectopic leaves and meristems on a flat apex in 13 of 15 cases (Fig. 5F). In contrast, in ter ham double mutants, the typical ham phenotype of a trichome-covered surface was found in 10 of 15 cases (Fig. 5I). In the other five cases, the surface was flat and lacked trichomes but showed no sign of resumption (data not shown). Thus, in early seedling stage ter ham double mutants closely resemble ter, but at later stages they behave like ham. In situ hybridization on ham single mutants showed that, at termination, PhWUS expression was essentially normal (Fig. 5J). In later stages, we observed no expression (Fig. 5L) or a patchy pattern (Fig. 5K). PhWUS expression in axillary ham meristems did the same (Fig. 5L).
We conclude that specification and maintenance of stem cells by HAM and WUS are largely parallel processes, although maintenance of spatially correct PhWUS expression indirectly depends on HAM. Like PhSTM, PhWUS expression continues for some time after meristem termination. Thus, cells in a differentiating ham shoot apex lose the ability to respond to PhWUS activity but not the potential to express it.
Role of HAM in shoot meristem and stem cell maintenance
By acting from L3 of organ primordia and provasculature of the stem, HAM affects communication among differentiating and nondifferentiating cells of the SAM. HAM is likely to signal cell fate decisions in the shoot apex, promoting the undifferentiated state as a distinct cellular identity. Judging from the nonpleiotropic nature of the ham mutation, this may be the single most important function of HAM. It might work in a similar way as its homolog SHORTROOT (SHR) of Arabidopsis, which controls cell fate by intercellular movement (Nakajima et al. 2001).
The antidifferentiation function of HAM is distinct from that of STM and WUS. The loss of responsiveness of apical cells to PhSTM and PhWUS in ham apices establishes a hitherto unknown cellular precondition on which these homeobox factors must act. We propose that, by means of HAM, cells that exit the meristematic domain and initiate differentiation actively maintain a field of uncommitted apical cells, protecting against a default path of development into stem. In the presence of a factor like WUS, these apical cells may acquire stem cell identity (Schoof et al. 2000). In the absence of STM, conversely, they may develop into lateral organs (e.g., see Byrne et al. 2000).
In root and shoot systems of plants, the differentiation and division patterns of stem cell daughters are directed by positional information from surrounding cells (Stewart and Burk 1970; van den Berg et al. 1995). In the root, initial cells for radial cell files differentiate like their maturing neighbors unless inhibited by the quiescent center (van den Berg et al. 1997). Our data indicate that, also in the shoot, stem cell daughters differentiate according to a prepattern present in more mature (stem-) tissue, and that maintenance of stem cell activity depends on extrinsic antidifferentiation factor(s). Whereas the molecular identities of antidifferentiation activities in the root are unknown, HAM is a likely candidate for those of the shoot.
HAM adds a new level of complexity to the developmental interactions in the meristem by showing a specific, essential role of differentiating cells in maintaining the undifferentiated state. Its biological meaning would be the coupling of stem cell fate to differentiation, effectively assuring indeterminate growth potential by a positive feedback mechanism on tissue level.
Materials and methods
Transposon mutagenesis and mutant construction
For transposon mutagenesis, ∼2000 M1 plants (Petunia hybrida strain W138) from 600 different families were self-fertilized, and M2 families were inspected visually for developmental abnormalities. Mutants were verified in segregation studies and manually cross-pollinated for double mutant construction. ham ter double mutants were identified on the basis of their dTph1 insertion genotype using PCR. All plants were grown in a greenhouse under standard conditions.
Structural analysis and histology
Scanning electron microscopy was performed on fresh plant material with Hitachi S-3500N equipment. Histological sections were made by fixing tissue in 4% formaldehyde in 50-mM sodium phosphate buffer (pH = 7.2), dehydrating through an ethanol series, embedding in Epon (Fluka), sectioning to 3 μm, and staining with 0.1% toluidine blue. All sections shown were selected for near-median plane, unless indicated otherwise. For sections, only stable ham mutants were used that carried a translational frameshift allele (hamFT-7; see Fig. 3B).
Gene isolation and analysis
Full-length PhSTM (GenBank accession no. AY112704) and HAM (GenBank accession no. AF481952) cDNAs were isolated by filter screening of a ZAPII cDNA library of young inflorescence shoot tips (gift of R. Koes). PhSTM: heterologous probe from tomato LeT6 (Janssen et al. 1998, gift of N. Sinha). HAM: siPCR fragment as probe. The full coding region of PhWUS (GenBank accession no. AF481951) was amplified and sequenced with primers ter5′F (5′-GCACGAGGCCATATTCTCTTCA CT-3′) and ter3′R (5′-CCATGGTCCAAAGATACGTAGTAC-3′) from the above cDNA library.
For selective iPCR (siPCR), genomic DNA was digested with MboI, self-ligated with T4 ligase, and relinearized with SspI. Such templates were selectively preamplified using dTph1-specific primers out4 (5′-GAACGGTTGTCCTCTTGAACC-3′) and out6 (5′-GTGGCAGCCCA GATTCGATCnn-3′). Out6 flanks an MboI site in dTph1 and carries any of 16 possible combinations of 3′-terminal bases (NN) extending into plant DNA. Preamplifications were reamplified with out6 + NN primers and a nested dTph1-out1 primer (5′-GGGAATTCGCTCCGCCCCTG-3′). Products were separated on 5% native polyacrylamide gels and stained with SYBR Gold (Molecular Probes, Inc.). Candidates were cut from the gel, eluted in water, reamplified, and sequenced. A detailed protocol is available from the authors.
To analyze footprint alleles for ham and ter, genomic DNA was isolated from revertant branches on homozygous insertion mutants or from their progeny. DNAs were PCR amplified with ham3 (5′-CAAGGAG GCTCTGCAGTTGC-3′) and ham4 (5′-CTTCAGAAAAGGACTTATA AGCATCC-3′) or terF (5′-GAAAAGGCTTATTGCTGCTGCTACC-3′) and terR (5′-CCTAAAGCTCTTCTCCATAGCTAAAG-3′). Products were separated on 5% polyacrylamide gels, and bands with small size increases relative to wild type were eluted, reamplified, and sequenced.
Expression analyses
RT-PCR was performed on first strand cDNA using gene-specific primers for the 3′ untranslated region of HAM cDNA (HAM3′F, 5′-AGCTTTAT CTAATGAAGCCAGAAGAGG-3′; HAM3′R, 5′-ACAGGGAAATGAG ACAAGATACATCAC-3′). In situ hybridization was performed using DIG-labeled antisense riboprobes according to the protocol at http//www.wisc.edu/genetics/CATG/barton/protocols.html. PhWUS and PhSTM probes were from full-length cDNAs. HAM probes were either full-length cDNA or a 976-bp probe from the nonconserved 5′ part of the cDNA (corresponding to amino acids 1–325). Both HAM probes gave identical results.
Transgenics
The full HAM coding region was amplified with Pfu DNA polymerase and primers hamATG (5′-ATCTAGAGTTTAAGATGATTGCAATAC C-3′) and hamTGA (5′-AAGATCTCTCCTCAGCACCTCCAAGTC-3′), digested with BglII and XbaI, and inserted in the sense orientation into a pBI121 (Clontech)-derived vector to replace the GUS cassette. Transformation was into P. hybrida W115 using Agrobacterium tumefaciens LBA4404.
Acknowledgments
We are very grateful to Ronald Koes for providing the W138 line and invaluable input. We thank John Bowman, Thomas Laux, Didier Reinhardt, Pia Stieger, and Jan Traas for commenting on the manuscript; Bruno Hungerbühler for assistance with TER footprints and STM clones; Pia Malnoe for greenhouse support; and members of the C.K. Lab for continuous discussion and general help. This work was supported by the Swiss National Science Foundation (grant 31.55540.98) and the National Center for Competence in Research (NCCR Plant Survival).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cris.kuhlemeier@ips.unibe.ch; FAX 41-31-332-2059.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.230702.
References
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsison a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- Byrne M, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408:967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- Dill A, Jung H-S, Sun T-P. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci. 2001;98:14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BJ, Williams A, Chen JJ, Mathern J, Hake S, Sinha N. Isolation and characterisation of two knotted-like homeobox genes from tomato. Plant Mol Biol. 1998;36:417–425. doi: 10.1023/a:1005925508579. [DOI] [PubMed] [Google Scholar]
- Koes RE, van Houwelingen A, Mur L, Spelt C, Quattrocchio F, Wing J, Oppedijk B, Ahmed S, Maes T, Gerats T, et al. Targeted gene inactivation in Petuniaby PCR based selection of transposon insertion mutants. Proc Natl Acad Sci. 1995;92:8149–8153. doi: 10.1073/pnas.92.18.8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KFX, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jürgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch M, Parcy F, Simon R, Weigel D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Long JA, Moan E, Medford J, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey P. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Nishimura A, Tamaoki M, Sato Y, Matsuoka M. The expression of tobacco knotted1-type class1 homeobox genes corresponds to regions predicted by the cytohistological zonation model. Plant J. 1999;18:337–347. doi: 10.1046/j.1365-313x.1999.00457.x. [DOI] [PubMed] [Google Scholar]
- Pysh L, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey P. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKEgenes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12:507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHELgenes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Sinha N, Williams RE, Hake S. Overexpression of the maize homeobox gene KNOTTED-1causes a switch from determinate to indeterminate cell fates. Genes & Dev. 1993;7:787–795. doi: 10.1101/gad.7.5.787. [DOI] [PubMed] [Google Scholar]
- Stewart RN, Burk LG. Independence of tissues derived from apical layers in ontogeny of the tobacco leaf and ovary. Amer J Bot. 1970;57:1010–1016. [Google Scholar]
- van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. Cell fate in the Arabidopsisroot meristem is determined by directional signalling. Nature. 1995;378:62–65. doi: 10.1038/378062a0. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendricks G, Weisbeek P, Scheres B. Short range control of cell differentiation in the Arabidopsisroot meristem. Nature. 1997;378:62–65. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- Waites R, Selvadurai HRN, Oliver I, Hudson A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell. 1998;93:779–789. doi: 10.1016/s0092-8674(00)81439-7. [DOI] [PubMed] [Google Scholar]
- Weigel D, Jürgens G. Stem cells that make stems. Nature. 2002;415:751–754. doi: 10.1038/415751a. [DOI] [PubMed] [Google Scholar]