Abstract
Hibernomas are benign tumors of brown fat, frequently characterized by aberrations of chromosome band 11q13. In this study, the chromosome 11 changes in five hibernomas were analyzed in detail by metaphase fluorescence in situ hybridization. In all cases, complex rearrangements leading to loss of chromosome 11 material were found. Deletions were present not only in those chromosomes that were shown to be rearranged by G-banding, but in four cases also in the ostensibly normal homologues, resulting in homozygous loss of several loci. Among these, the gene for multiple endocrine neoplasia type I (MEN1) was most frequently deleted. In addition to the MEN1 deletions, heterozygous loss of a second region, approximately 3 Mb distal to MEN1, was found in all five cases, adding to previous evidence for a second tumor suppressor locus in 11q13.
Hibernomas are rare, benign tumors simulating brown adipose tissue in their histological appearance. 1 They primarily occur in the scapular, interscapular, and mediastinal regions of young adults and are cytogenetically characterized by translocations involving chromosome band 11q13, together with few or no other changes. 2-5 The aim of this study was to characterize in detail the 11q13 rearrangements in hibernoma by physical mapping of translocation breakpoints, using metaphase fluorescence in situ hybridization (FISH) analysis. Yeast and P1 artificial chromosome (YAC and PAC) probes were used for megabase-level breakpoint mapping, followed by hybridizations with cosmid clone probes for a more refined characterization.
Materials and Methods
Material from five tumors was obtained for cytogenetic analysis directly after resection (Table 1) ▶ . The histopathological criteria used for classification of hibernoma have previously been described 4 ; two of the cases were reviewed by the CHAMP study group. 6 Culturing, harvesting, and chromosome banding were essentially as previously described. 7 The cultures were harvested within 2 weeks and subcultured once or twice. Wright’s stain was used for G-banding, and karyotypic descriptions were done according to ISCN (1995). 8 The G-band karyotypes of cases 4 and 5 have previously been published by Dal Cin et al 2 and Mertens et al, 4 respectively. FISH on metaphase chromosomes was according to standard protocols. 7 Biotin- and digoxigenin-labeled whole chromosome painting probes (wcp) were purchased from Cambio (Cambridge) and Oncor (Gaithersburg, MD). Centromeric probes (cen) for chromosomes 5 and 11 were prepared from the α satellite clones pG-A16 and pHS53, obtained from Dr. T. Hulsebos (Amsterdam) and the American Type Culture Collection (Rockville, MD), respectively, and the 11q subtelomeric probe 2072C1 was kindly provided by Dr. L. Kearney (Oxford). YAC probes for 11q13 and the PAC clone 838L14 (Figure 1) ▶ , containing PYGM, GCK, and MEN1, 9 were provided by the Centre d’Etude du Polymorphisme Humain (Paris) and by D. Kedra (Stockholm), respectively. Human DNA was amplified from YAC clones by interAlu polymerase chain reaction. Cosmid probes for PLCB3, PYGM, PPP1A, and MLK3 (cSOM163–8, cCL15, cCLP1A, and cMLK-3, respectively) 10 were kindly provided by Dr. C. Larsson (Stockholm). GCK, MEN1, D11S1783, POLA, D11S4933, and SEA were detected with the cosmid clones 115E10, 10B11, 116B6, 33B5, 48D12, and 57H7, respectively. 11 Posthybridization washing was carried out in 0.4 or 1 × SSC for 2 minutes at 72°C. Counterstaining was obtained with diamidino phenylindole. For each hybridization, 4–15 metaphase cells were analyzed. Chromosomes were identified by the diamidino phenylindole banding patterns, complemented by cen or wcp hybridizations when necessary. Hybridizations to aberrant metaphase cells were only considered informative if signals were present on both chromosomes 11 in the cells with a constitutional chromosome complement found in the same preparation.
Table 1.
Patient Data and Karyotypes Based on G-Banding and FISH Analyses
| Case no. | Age/sex | Location | Karyotype* |
|---|---|---|---|
| 1 | 22 /M | thigh | 46,XY,del(11)(q13.3q13.3),der(11)t(11;17)(q13.3;p13),der(17)t(11;17) (q13.4;p13)/46,XY |
| 2 | 58 /F | shoulder | 46,XX,der(9)t(9;11)(p21;q13.4),del(11)(q13.3q13.4),der(11)t(9;11)(p21;q13.4)/46,XX |
| 3 | 41 /M | shoulder | 45,XY,der(13;14)(q10;q10)c/45,idem,der(2)t(2;11)(p11;q13.3),del(11)(q13.3q13.4),der(11)del(11)(q13.3q13.3)t(11;15)(q13.3;q22),der(15)t(2;15)(p11;q22) |
| 4 | 42 /M | buttock | 46,XY,der(1)ins(1;11)(p36;q13.3q13.?5)del(11)(q13.3q13.4)t(1;2)(p36;q33),der(2)ins(2;11)(q33;q13.3q13.4)t(2;5)(q33;q22),der(5)t(5;11)(q22;q13.?5),del(11)(q13.3q13.3),der(11) del(11)(q13.3q13.3)del(11)(q13.4)/46,idem,t(8;10)(q24;q22)/46,XY |
| 5 | 47 /M | buttock | 46,XY,der(5)t(5;11)(q11;q13.4),der(11)del(11)(p11)del(11)(q13.3q13.3)dup(11) (q13.3q13.4)t(5;11)(q11;q13.4),der(11)del(11)(q13.3q13.4)del(11)(q?)dup(11) (q13.4q?)/46,XY |
M, male; F, female.
*Subbands were derived from hybridization results according to integrated maps in Guru et al11; designations in bold type are those used in the text.
Figure 1.
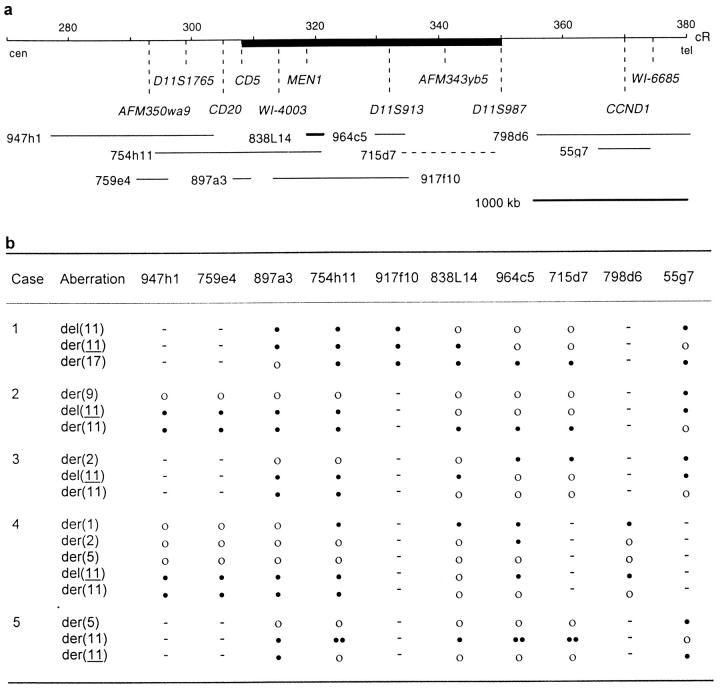
a: Physical map of 11q13. Human YAC and PAC clone inserts are represented by thin and thick lines, respectively, sized according to the 1000-kb bar; the broken line indicates that the insert size is unknown. YAC sizes, centiRay (cR) distances, and locations of genes and sequence-tagged sites are according to the Whitehead Institute/MIT Center for Genome Research (http://www-genome.wi.mit.edu/). The region commonly rearranged in the five cases is marked by a black bar. b: Results of hybridizations with the YAC and PAC probes. Abnormal chromosomes are according to Table 1 ▶ . Symbols: −, not determined; •, presence of one signal; ••, presence of two signals; ○, absence of signal.
Results
Chromosome Banding
Clonal rearrangements involving chromosome band 11q13 were detected by G-banding in all tumors (Table 1) ▶ . Cases 1 and 2 exhibited reciprocal translocations, t(11;17) and t(9;11), respectively, as the sole anomalies (Figure 2a) ▶ . In case 3, with a constitutional Robertsonian 13;14 translocation, a t(2;15;11) was found, and in case 4 there were two abnormal clones, one with a t(1;11;5;2) as the sole change and a second one with t(8;10) in addition. All of these translocations were seemingly balanced. In case 5, complex rearrangements involving chromosomes 5 and 11 were seen (Figure 2b) ▶ . FISH analyses revealed additional genetic recombinations in all cases. The karyotypes based on the combined G-banding and FISH findings are presented in Table 1 ▶ .
Figure 2.
Representative FISH and G-band images. a: Case 1. Top row: G-banding shows a seemingly balanced 11;17 translocation; second row: 897a3 (green) hybridizes to the del(11) and the der(11), whereas hybridization with 754h11 (red) results in signals on the del(11) and the der(17), as well as the der(11), indicating a breakpoint in the region covered by the YAC; third row: the der(11) and the der(17) are positive with both wcp11 (green) and 838L14 (red), but the del(11)is only wcp11 positive, indicating a cryptic deletion of the region covered by the single-copy probe; fourth row: hybridization with the cosmid probes for PLCB3 (green) and PPP1A (red), showing a red signal only on the del(11), indicating a heterozygous deletion. b: Case 5. Top row: Complex rearrangements of chromosomes 5 and 11 shown by G-banding; second row: wcps 5 (red) and 11 (green) also reveal rearrangement of the der(5), which is similar to a normal chromosome 11 by G-banding; third row: the subtelomeric probe for 11q (red) hybridizes to the der(5) and the der(11); fourth row: 964c5 (red) gives two signals on the der(11), and 897a3 (green) one signal each on the derivative chromosomes 11. c: Case 4. A normal and an aberrant metaphase cell from the same slide hybridized with cen11 (green) and the cosmid probe for MEN1 (red) show strong signals from both probes on chromosomes 11 in the normal cell (top), but signals only from the centromeric probe in the tumor cell (bottom), although the detection threshold is set very low for this cell, as indicated by the prominent red background.
Chromosome Painting, Centromeric and Subtelomeric Probes
Hybridizations with wcp11 confirmed the translocations detected by G-banding in cases 1, 2, and 3 (Figure 2a) ▶ . In case 4, material from chromosome 11 was found in all chromosomes involved in the four-way translocation: apart from the derivative chromosomes 5 and 11, it was also found in the derivative chromosomes 1 and 2, where weak, interstitial signals located at positions corresponding to the translocation breakpoints were seen. In case 5, dual-color cohybridizations of wcp5 together with wcp11 and cen5 together with the subtelomeric probe for 11q revealed complex rearrangements of both chromosomes 11 (Figure 2b) ▶ . The der(11) was completely painted with wcp11, had the chromosome 11 centromere, but showed no signal with the 11q subtelomeric probe, whereas a minor part of the der(11) was positive with wcp11, cen11, and the 11q subtelomeric probe.
YAC and PAC Probes for 11q13
In cases 1–4, the YAC 897a3 hybridized proximal to the translocation breakpoints in the derivative chromosomes 11 and gave signals on the homologues not involved in the translocations (Figure 2a) ▶ . The same results were obtained by hybridizations with 947h1 and 759e4 in cases 2 and 4. Similarly, in case 5, signals from 897a3 were retained on the two derivative chromosomes 11 (Figure 2b) ▶ . In cases 2 and 3, 754h11 was present only on the chromosome 11 homologues. However, in case 1, the signals from the probes 754h11 and 917f10 were split by the t(11;17). In case 4, 754h11 hybridized to der(1), del(11), and der(11), and, in case 5, both signals seen were present only on the der(11). The PAC 838L14 was also split between the translocation partners in case 1 but gave no signal on the del(11). In the remaining cases, only one signal from this probe was detected in aberrant metaphase cells, which was present on the derivative chromosomes 11 in cases 2 and 5, the del(11) in case 3, and the der(1) in case 4. The YACs 964c5 and 715d7 also yielded only one signal per metaphase cell in cases 1–3. In case 4, 964c5 was split between the derivative chromosomes 1 and 2, and was present on del(11), whereas in case 5, two signals were seen on the der(11). The distal 11q13 probes, 798d6 and 55g7, hybridized telomeric to the breakpoints in all cases. Taken together these results indicated that the 11q13 rearrangements clustered to a region covered by 754h11, 917f10, 838L14, 964c5, and 715d7, approximately corresponding to the interval between CD5 and D11S987.
Cosmid Probes
In aberrant metaphase cells, two signals from the cosmid probe for PLCB3, located approximately 3 Mb distal to CD5, were present in cases 1 and 3 (Figure 2c) ▶ , whereas only one signal could be observed in cases 2, 4, and 5 (Figure 3) ▶ . The probe for PYGM yielded one signal in cells from cases 2–5 and no signals in cells from case 1, whereas the GCK probe gave one signal in cases 2, 3, and 5, but none in cases 1 and 4. At hybridization with the probe for MEN1, signals were present only in case 2, where the probe hybridized to the der(11) (Figure 2c) ▶ . The probes for D11S1783, POLA, D11S4933, and SEA yielded one signal each in aberrant cells from cases 1–3. Because of shortage of material, analysis with these probes could not be performed in cases 4 and 5. However, the MLK3 probe, located between D11S4933 and SEA, gave one signal in cases 1–3, but two signals in cases 4 and 5; in case 5, both signals were located on the der(11), partly corroborating the duplications observed by YAC probe hybridizations. In all cases, hybridizations with the probe for PPP1A resulted in only one signal per aberrant cell.
Figure 3.
FISH results for the 11q13 region between PLCB3 and D11S987. The absence or presence of cosmid signals for individual loci is indicated by open and filled ellipses, respectively. The chromosomes are referred to according to their designations in Table 1 ▶ . The positions of genes and sequence-tagged sites are according to Guru et al 11 and references therein.
Discussion
Essentially, the pattern of cytogenetic aberrations detected by chromosome banding techniques in hibernoma parallels the spectrum of rearrangements seen in lipoma, ie, one chromosome segment, 11q13 in hibernoma and 12q13–15 in lipoma, recombining with a variety of other segments. 12 In lipoma, these rearrangements frequently lead to the formation of chimeric genes involving the HMGIC gene in 12q15. In this study, the breakpoints in hibernoma were mapped by molecular cytogenetic techniques. The results indicate that the molecular events differ distinctly from those in lipoma. Complex rearrangements of chromosome 11, leading to loss of chromosome material, were found in all five cases investigated. Deletions were present not only in those chromosomes that were shown to be rearranged by G-banding, but also, in four cases, in the ostensibly normal homologues. In these tumors, large deletions of up to 4 Mb were observed in one homologue, whereas the other one showed less extensive losses (Figure 3) ▶ . The whole segment between PYGM and PPP1A was heterozygously lost in three cases, whereas two cases exhibited discontinuous, heterozygous deletions of the PLCB3-MEN1 and the PPP1A regions. Such concomitant deletions in both homologues resulted in homozygous loss of PYGM in one case, GCK in two cases, and MEN1 in four cases. The most commonly deleted segment included MEN1 and was flanked by GCK and D11S1783, spanning a physical distance of approximately 100 kb. 11 To our knowledge, MEN1 is the only gene localized to this area so far.
Germline mutations of the MEN1 tumor suppressor gene underlie the multiple endocrine neoplasia type I (MEN1) syndrome, an autosomal, dominantly inherited disorder with parathyroid hyperplasia, pancreatic islet cell tumors, anterior pituitary adenomas, and other neuroendocrine neoplasms. 13 Frequent loss of heterozygosity in the MEN1 region has been demonstrated in these tumors, in hereditary as well as in sporadic cases. 14 Recent findings, however, have indicated that loss of an additional 11q13 region is implicated in the formation of MEN1-associated and sporadic endocrine neoplasms. 15,16 This region was mapped between the markers D11S4907/D11S4908 and D11S987, 16 coinciding with the area around PPP1A, which was shown to be heterozygously deleted, independently of MEN1 losses, in all five hibernomas investigated here. Thus the present study not only demonstrates the presence of MEN1 deletions in yet another tumor type, but also provides further evidence of a second tumor suppressor locus, distal to MEN1 in 11q13.
The finding of a high frequency of MEN1 deletions in hibernoma touches on the question of the histogenesis of this tumor. Morphologically, hibernomas are highly similar to brown adipose tissue, and it is known that the thermogenic activity as well as the proliferation of brown adipocytes are tightly regulated by hormones such as aldosterone and melatonin in a fashion similar to that of the endocrine tissues that become neoplastic in MEN1 patients. 17,18 In addition, electron microscopic studies have shown that hibernomas are capable of secretory activity. 19 However, if endocrine tumors and hibernomas develop along identical pathogenetic pathways, one would expect hibernomas to be part of the MEN1 syndrome. This is not the case, however, although lipomas, angiofibromas, and other mesenchymal tumors are occasionally observed in MEN1 patients. 20 Furthermore, the cytogenetic pattern of hibernomas differs considerably from that observed in MEN1-associated tumors: whereas hibernomas show only a small number of changes with consistent involvement of 11q13, the few endocrine tumors analyzed so far have had mostly complex karyotypes with a number of structural and numerical changes, rarely including any chromosome 11 aberrations. 12 Thus, although hibernomas and MEN1-asssociated endocrine tumors may share some ultrastructural and pathogenetic features, including homozygous loss of MEN1 at an early stage, it is probable that these entities subsequently progress along separate routes of tumorigenesis. This is further stressed by the fact that the benign tumors in MEN1 patients may transform into malignancy, 13 whereas hibernomas rarely, if ever, metastasize. 1
Apart from hibernomas, recurrent aberrations of 11q13 have been described in a number of other sporadically occurring solid tumors, eg, lipomas, astrocytomas, neurofibrosarcomas, adenocarcinomas of the breast and kidney, oropharyngeal squamous cell carcinomas, and extragonadal germ cell tumors. 12 In extragonadal germ cell tumors, the translocation breakpoints have been mapped to the segment between PLCB3 and MLK3, 21 closely corresponding to the region shown to be rearranged or lost in this study. The molecular genetic consequences, including possible deletions in the MEN1 region, of unbalanced and seemingly balanced 11q13 changes thus appear to be of great interest for future investigations in a broad spectrum of neoplasms.
Acknowledgments
We are grateful to Prof. Settara C. Chandrasekharappa, National Human Genome Research Institute, National Institutes of Health, for critical reading and for providing cosmid clones for the MEN1 region.
Footnotes
Address reprint requests to David Gisselsson, Department of Clinical Genetics, University Hospital, S-221 85 Lund, Sweden. E-mail: david.gisselsson@klingen.lu.se.
Supported by the Swedish Cancer Society, the IngaBritt and Arne Lundberg Research Foundation, and the John and Augusta Persson Foundation for Scientific Medical Research.
References
- 1.Enzinger FM, Weiss SW: Soft Tissue Tumors, ed 2. St. Louis, Mosby, 1995
- 2.Dal Cin P, Van Damme B, Hoogmartens M, Van Den Berghe H: Chromosome changes in a case of hibernoma. Genes Chromosom Cancer 1992, 5:178-180 [DOI] [PubMed] [Google Scholar]
- 3.Meloni AM, Spanier SS, Bush CH, Stone JF, Sandberg AA: Involvement of 10q22 and 11q13 in hibernoma. Cancer Genet Cytogenet 1994, 72:59-64 [DOI] [PubMed] [Google Scholar]
- 4.Mertens F, Rydholm A, Brosjö O, Willén H, Mitelman F, Mandahl N: Hibernomas are characterized by rearrangements of chromosome bands 11q13–21. Int J Cancer 1994, 58:503-505 [DOI] [PubMed] [Google Scholar]
- 5.Mrózek K, Karakousis CP, Bloomfield CD: Band 11q13 is nonrandomly rearranged in hibernomas. Genes Chromosom Cancer 1994, 9:145-147 [DOI] [PubMed] [Google Scholar]
- 6.Fletcher CDM, Akerman M, Dal Cin P, de Wever I, Mandahl N, Mertens F, Mitelman F, Rosai J, Rydholm A, Sciot R, Tallini G, van den Berghe H, van de Ven W, Vanni R, Willen H: Correlation between clinicopathological features and karyotype in lipomatous tumors. A report of 178 cases from the chromosomes and morphology (CHAMP) collaborative study group. Am J Pathol 1996, 148:623-630 [PMC free article] [PubMed] [Google Scholar]
- 7.Gisselsson D, Höglund M, Mertens F, Mitelman F, Mandahl N: Chromosomal organization of amplified chromosome 12 sequences in mesenchymal tumors. Genes Chromosom Cancer 1998, 23:203-212 [DOI] [PubMed] [Google Scholar]
- 8.ISCN (1995): An International System for Human Cytogenetic Nomenclature. Edited by F. Mitelman. Basel, S. Karger, 1995
- 9.Kedra D, Seroussi E, Fransson I, Trifunovic J, Clark M, Lagercrantz J, Blennow E, Mehlin H, Dumanski J: The germinal center kinase gene and a novel CDC25-like gene are located in the vicinity of the PYGM gene in 11q13. Hum Genet 1997, 100:611-619 [DOI] [PubMed] [Google Scholar]
- 10.Lemmens I, Merregaert J, Van de Ven WJM, Kas K, Zhang CX, Giraud S, Wautot V, Buisson N, De Witte K, Salandre J, Lenoir G, Calender A, Parente F, Quincey D, Courseaux A, Carle GF, Gaudray P, De Wit MJ, Lips CJM, Höppener JWM, Khodaei S, Grant AL, Weber G, Kytölä S, Teh BT, Farnebo F, Grimmond S, Phelan C, Larsson C, Forbes SA, Bassett JHD, Pannett AAJ, Thakker RV: Construction of a 1.2-Mb sequence-ready contig of chromosome 11q13 encompassing the multiple endocrine neoplasia type 1 (MEN1) gene. Genomics 1997, 44:94-100 [DOI] [PubMed] [Google Scholar]
- 11.Guru SC, Olufemi S-E, Manickam P, Cummings C, Gieser LM, Pike BL, Bittner ML, Jiang Y, Chinault AC, Nowak NJ, Brzozowska A, Crabtree JS, Wang Y, Roe BA, Weisemann JM, Boguski MS, Agarwal SK, Burns AL, Spiegel AM, Marx SJ, Flejter WL, de Jong PJ, Collins FS, Chandrasekharappa SC: A 2.8-Mb clone contig of the multiple endocrine neoplasia type 1 (MEN1) region at 11q13. Genomics 1997, 42:436-445 [DOI] [PubMed] [Google Scholar]
- 12.Mitelman F: Catalog of Chromosome Aberrations in Cancer, ed 6. New York, Wiley-Liss, 1998
- 13.Miller JA, Norton JA: Multiple endocrine neoplasia. Pollock RE eds. Surgical Oncology. 1997, :pp 213-225 Kluwer Academic Publishers, Dordrecht [Google Scholar]
- 14.Debelenko LV, Zhuang Z, Emmert-Buck MR, Chandrasekharappa SC, Manickam P, Guru SC, Marx SJ, Skarulis MC, Spiegel AM, Collins FS, Jensen RT, Liotta LA, Lubensky IA: Allelic deletions on chromosome 11q13 in multiple endocrine neoplasia type 1-associated and sporadic gastrinomas and pancreatic endocrine tumors. Cancer Res 1997, 57:2238-2243 [PubMed] [Google Scholar]
- 15.Iida A, Blake K, Tunny T, Klemm S, Stowasser M, Hayward N, Gordon R, Nakamura Y, Imai T: Allelic losses on chromosome band 11q13 in aldosterone-producing adrenal tumors. Genes Chromosom Cancer 1995, 12:73-75 [DOI] [PubMed] [Google Scholar]
- 16.Chakrabarti R, Srivatsan ES, Wood TF, Eubanks PJ, Ebrahimi SA, Gatti RA, Passaro EJ, Sawicki MP: Deletion mapping of endocrine tumors localizes a second tumor suppressor gene in chromosome band 11q13. Genes Chromosom Cancer 1998, 22:130-137 [DOI] [PubMed] [Google Scholar]
- 17.Zennaro M-C, Le Menuet D, Viengchareun S, Walker F, Ricquier D, Lombés M: Hibernoma development in transgenic mice identifies brown adipose tissue as a novel target of aldosterone action. J Clin Invest 1998, 101:1254-1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haldmeier G, Hoffmann K: Melatonin stimulates the growth of brown adipose tissue. Nature 1974, 247:224-225 [DOI] [PubMed] [Google Scholar]
- 19.Allegra SR, Gmuer C, O’Leary GP: Endocrine activity in a large hibernoma. Hum Pathol 1983, 14:1044-1052 [DOI] [PubMed] [Google Scholar]
- 20.Darling TN, Skarulis MC, Steinberg SM, Marx SJ, Spiegel AM, Turner M: Multiple facial angiofibromas and collagenomas in patients with multiple endocrine neoplasia type 1. Acta Dermatol 1997, 133:853-857 [PubMed] [Google Scholar]
- 21.Sinke RJ, Olde Weghuis D, Suijkerbuijk RF, Tanigami A, Nakamura Y, Larsson C, Weber G, de Jong B, Oosterhuis JW, Molenaar WM, Guerts van Kessel A: Molecular characterization of a recurring complex chromosomal translocation in two human extragonadal germ cell tumors. Cancer Genet Cytogenet 1994, 73:11-16 [DOI] [PubMed] [Google Scholar]