Abstract
The NPM-MLF1 fusion protein is expressed in blasts from patients with myelodysplasia/acute myeloid leukemia (MDS/AML) containing the t(3;5) chromosomal rearrangement. Nucleophosmin (NPM), a previously characterized nucleolar phosphoprotein, contributes to two other fusion proteins found in lympho-hematopoietic malignancies, anaplastic large cell lymphoma (NPM-ALK) and acute promyelocytic leukemia (NPM-RARα). By contrast, the function of the carboxy-terminal fusion partner, myelodysplasia/myeloid leukemia factor 1 (MLF1), is unknown. To aid in understanding normal MLF1 function, we isolated the murine cDNA, determined the chromosomal localization of Mlf1, and defined its tissue expression by in situ hybridization. Mlf1 was highly similar to its human homologue (86% and 84% identical nucleotide and amino acid sequence, respectively) and mapped to the central region of chromosome 3, within a segment lacking known mouse mutations. Mlf1 tissue distribution was restricted during both development and postnatal life, with high levels present only in skeletal, cardiac, and selected smooth muscle, gonadal tissues, and rare epithelial tissues including the nasal mucosa and the ependyma/choroid plexus in the brain. Mlf1 transcripts were undetectable in the lympho-hematopoietic organs of both the embryonic and adult mouse, suggesting that NPM-MLF1 contributes to the genesis of MDS/AML in part by enforcing the ectopic overexpression of MLF1 within hematopoietic tissues.
The myelodysplastic syndromes (MDS) are a group of clonal hematopoietic disorders that are defined by the abnormal morphological appearance of multiple blood cell lineages in the bone marrow and by the occurrence of cytopenias in the peripheral blood. 1 Classically, the deficiency of mature cells in the peripheral blood occurs despite increased proliferation of the nonlymphoid cells in the bone marrow, due to abnormal differentiation together with increased apoptosis. 2,3 Consistent with the presence of dysplastic features in cells of the myelomonocytic, erythroid, and/or megakaryocytic lineages, the pathogenic mutations of MDS are thought to occur in pluripotent hematopoietic stem cells. 4,5 Progression of MDS to acute myeloid leukemia (AML) is common and is thought to represent the final stage of a disease process characterized by the gradual accumulation of a series of mutations over a prolonged latency period. 1,6 Although notable exceptions exist, such as RAS gene mutations in approximately 25% of MDS cases, 7 EVI-1 activation by 3q26 rearrangements in about 3–5% of MDS/AML cases, 8 and TEL-PDGFRβ fusion by the t(5;12) in the rare chronic myelomonocytic leukemia (CMML) subtype of MDS, 9 little is known about the involvement of specific oncogenes in the pathogenesis of MDS leading to AML. To aid in the understanding of the genetic mechanisms that result in MDS/AML, our laboratory recently characterized the t(3;5) chromosomal rearrangement, 10 demonstrating that this translocation juxtaposes sequences of nucleophosmin (NPM, also known as numatrin or B23) on chromosome 5 with those of a novel chromosome 3 gene whose product, myelodysplasia/myeloid leukemia factor 1 (MLF1), lacks homology to previously characterized proteins.
NPM is a ubiquitously expressed phosphoprotein that can shuttle back and forth between the nucleus and the cytoplasm, transporting ribosomal RNA-containing proteins (ribonucleoproteins) for their release at the maturing ribosomes. 11-13 Interestingly, two other fusion proteins contain portions of NPM, namely NPM-ALK (anaplastic lymphoma kinase) 14 and NPM-RARα (retinoic acid receptor-α) 15 in anaplastic large cell lymphoma and acute promyelocytic leukemia, respectively. The NPM component of these fusion proteins contributes to their nuclear localization and confers on them the ability to form oligomers, 16 whereas the NPM fusion partners appear to be essential in determining the disease phenotype of lymphoma (ALK), AML (RARα), or MDS/AML (MLF1). In contrast to the well-characterized RARα and ALK gene products, little is known about the function of MLF1.
Previous work from this laboratory has shown that MLF1 is a novel 31-kd cytoplasmic protein that acquires nucleolar localization when fused to NPM. 10 Database analysis of MLF1 did not reveal known functional motifs, but sequence homology with an ubiquitously expressed protein of unknown function, MLF2, was identified. 17 In hematopoietic cell lines, MLF1 expression is variable and inconclusive, with some leukemic cell lines expressing the gene (eg, K562, HEL, and kg1) and others not (eg, HL60 and U937). 10 However, MLF1 (also referred to as HLS7) expression was found by representational display analysis to be activated in a cell line induced to undergo lineage switching from an erythroid to a monoblastic phenotype 18 and in CD34+ human bone marrow cells, 19 implying a potential role for the gene in normal hematopoiesis.
To better define the function of MLF1 in both hematopoietic cells and nonhematopoietic tissues, we now report the cloning of the murine homologue Mlf1, together with the determination of its chromosomal localization and tissue expression pattern. Whereas strikingly high levels of expression were found in certain tissues, including striated skeletal and heart muscle, lympho-hematopoietic organs showed no detectable Mlf1 signal, suggesting a role for the ectopic overexpression of MLF1 in the pathogenesis of MDS/AML.
Materials and Methods
cDNA Cloning
A 1-kb HindIII restriction fragment containing the entire coding sequence of the human MLF1 cDNA 10 was labeled with [α-32P]dCTP by the random oligonucleotide primer method (Rediprime, Amersham, Little Chalfont, UK) and used to screen a cDNA library prepared from mouse testis mRNA (Uni-ZAP XR Library, catalog no. 937308, Stratagene, La Jolla, CA) under low-stringency hybridization conditions [5 × SSC, 40% formamide, 0.08% each of bovine serum albumin, polyvinylpyrollidone, and Ficoll, 1% sodium dodecyl sulfate (SDS), 10 mmol/L Tris-HCl, pH 7.5, 40 μg/ml salmon sperm DNA; 42°C overnight]. Double-stranded DNA templates were sequenced using BigDye chemistry and Taq sequencing methods, as recommended by the manufacturer (Perkin-Elmer/Applied Biosystems, Norwalk, CT). Samples were electrophoresed and analyzed on PE/ABI 373 and 377 DNA sequencers. Contig assembly was performed using Staden’s X-windows software, and the consensus sequence was analyzed using Wisconsin Package v. 9.1 software (Genetics Computer Group, Inc., Madison, WI) and various WWW resources.
Mlf1 Chromosomal Mapping: Interspecific Mouse Backcross Mapping
Interspecific backcross progeny were generated by mating (C57BL/6J × Mus spretus) F1 females and C57BL/6J males as described. 20 A total of 205 N2 mice were used to map the Mlf1 locus. DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, Southern blot transfer, and hybridization were performed essentially as described. 21 All blots were prepared with Hybond-N+ nylon membrane (Amersham). The probe, an 879-bp EcoRI/XhoI fragment containing the mouse Mlf1 cDNA, was labeled with [α-32P]dCTP using a random primed labeling kit (Stratagene) and washing was done to a final stringency of 0.5 × SSCP/0.1% SDS at 65°C. Fragments of 9.4, 5.1, 4.7, 3.9, and 0.5 kb were detected in TaqI-digested C57BL/6J DNA and fragments of 8.8, 4.7, 3.2, and 0.5 kb were detected in TaqI-digested M. spretus DNA. The presence or absence of the 8.8- and 3.2-kb TaqI M. spretus-specific fragments, which cosegregated, was followed in the backcross mice. A description of the probes and restriction fragment length polymorphisms for the loci linked to Mlf1 including Mme, Fgg, and Ntrk1 has been reported previously. 22,23 Recombination distances were calculated using Map Manager, version 2.6.5. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
Northern Blot Analysis
A commercially available Northern blot prepared with polyA+ RNA from various mouse tissues (Clontech, Palo Alto, CA) was prehybridized at 68°C in ExpressHyb Solution (Clontech) for 30 minutes, then hybridized for 1 hour under the same conditions with the 879-bp EcoRI/XhoI Mlf1 cDNA, labeled by random priming with [α-32P]dCTP (Rediprime, Amersham). After washing for 40 minutes at room temperature in 2× SSC/0.05% SDS and 40 minutes at 50°C in 0.1× SSC/0.1% SDS, autoradiography was performed overnight at −80°C. The Mlf1 probe was removed by following the manufacturer’s recommendations for stripping and complete removal of the signal was documented by autoradiography. The blot was then rehybridized with a 2.0-kb human β-actin cDNA probe (supplied by the manufacturer), which strongly cross-hybridizes with mouse β-actin.
In Situ Hybridization
In situ hybridizations were performed as previously described. 24 The 879 bp EcoRI/XhoI mouse Mlf1 cDNA, which encompasses the entire coding region of the gene, was cloned into pBluescript SK+. The vector was linearized with either EcoRI (T7;antisense) or XhoI (T3;sense) before riboprobe synthesis.
Results and Discussion
cDNA Cloning and Sequence Analysis of Mlf1
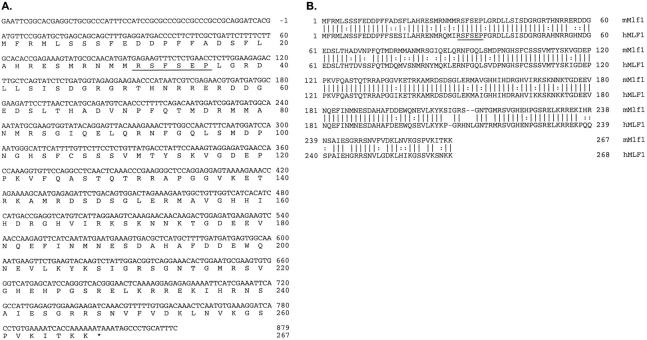
To isolate the mouse homologue of MLF1, a mouse testis cDNA library was screened using the human cDNA 10 as a probe under low-stringency hybridization conditions. Nine partially overlapping clones with insert sizes ranging from 0.9 to 1.7 kb were obtained. The 801-bp Mlf1 coding sequence (Figure 1A) ▶ was 86% identical at the nucleotide level to the human MLF1 coding sequence and the deduced 267-amino acid sequence of mouse Mlf1 showed 84% identity to its 268-amino acid human homologue (Figure 1B) ▶ . Motif searches using the Mlf1 amino acid sequence revealed that residues 31–36 (RSFSEP) match one of the two recently described motifs (RSXpSXP, RXY/FXpSXP) 25,26 that mediate phosphoserine-dependent binding of proteins such as Raf, BAD, and Cbl to the 14-3–3 family of signal transduction proteins. 27-36 No other potential functional motifs were identified in this analysis.
Figure 1.
A: Nucleotide sequence of the mouse Mlf1 cDNA with translation of the open reading frame. B: Comparison of the deduced amino acid sequence of mouse Mlf1 with human MLF1. The mouse sequence is 86% and 84% identical to the human MLF1 nucleotide and deduced amino acid sequence, respectively. The 14-3–3 binding motif (RSFpSEP) found in the two proteins is underlined. The GenBank accession number for mouse Mlf1 is AF100171.
Chromosomal Localization of Mlf1
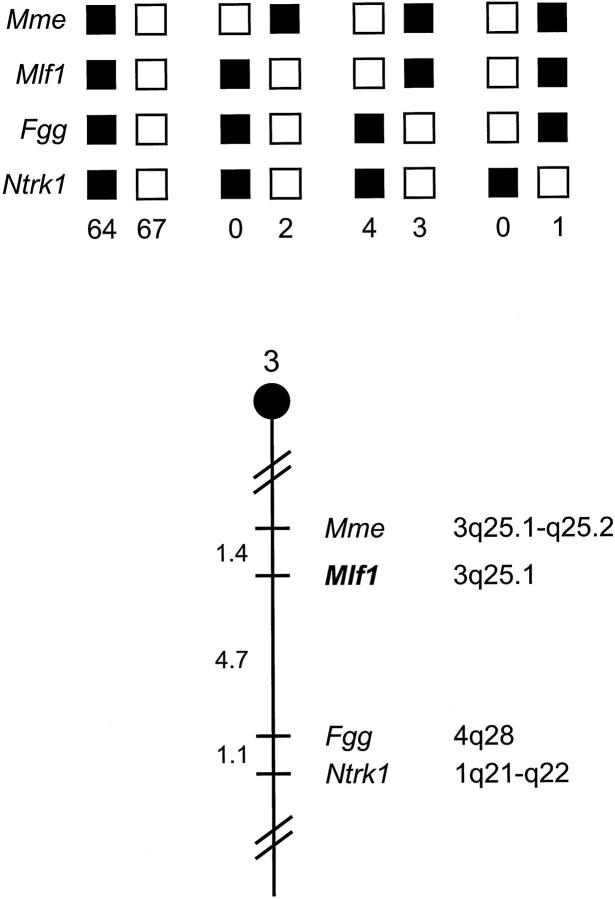
To determine whether previously described mouse disease phenotypes are associated with alterations of the genomic region encompassing Mlf1, the chromosomal localization of the gene was determined by interspecific backcross analysis using progeny derived from matings of [(C57BL/6J × M. spretus)F1 × C57BL/6J] mice (Figure 2) ▶ . This interspecific backcross mapping panel has been typed for over 2700 loci that are well distributed among all of the autosomes, as well as the X chromosome. 20 C57BL/6J and M. spretus DNAs were digested with several enzymes and analyzed by Southern blot hybridization for informative restriction fragment length polymorphisms using the mouse Mlf1 cDNA probe. The 8.8- and 3.2-kb TaqI M. spretus restriction fragment length polymorphisms were used to follow the segregation of the Mlf1 locus in backcross mice. The mapping results indicated that Mlf1 is located in the central region of mouse chromosome 3, linked to Mme, Fgg, and Ntrk1. Although 141 mice were analyzed for every marker and are shown in the segregation analysis (Figure 2) ▶ , up to 191 mice were typed for some pairs of markers. Each locus was analyzed in pairwise combinations for recombination frequencies using the additional data. The ratios of the total number of mice exhibiting recombinant chromosomes to the total number of mice analyzed for each pair of loci and the most likely gene order are: centromere–Mme–2/144–Mlf1–9/191–Fgg–2/191–Ntrk1. The recombination frequencies [expressed as genetic distances in centiMorgans (cM) ± the SE] are: Mme–1.4 ± 1.0–Mlf1–4.7 ± 1.5–Fgg–1.1 ± 0.7–Ntrk1.
Figure 2.
Mlf1 maps in the central region of mouse chromosome 3. Mlf1 was localized to mouse chromosome 3 by interspecific backcross analysis. The segregation patterns of Mlf1 and flanking genes in 141 backcross animals that were typed for all loci are shown at the top of the figure. For individual pairs of loci, more than 141 animals were typed (see text). Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J × M. spretus) F1 parent. The shaded boxes represent the presence of a C57BL/6J allele and white boxes represent the presence of a M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 3 linkage map showing the location of Mlf1 in relation to linked genes is shown at the bottom of the figure. Recombination distances between loci in (centiMorgans) are shown to the left of the chromosome and the positions of the loci in human chromosomes are shown to the right. References for the human map positions of the loci cited in this study can be obtained from GDB (Genome Data Base), a computerized database of human linkage information maintained by The William H. Welch Medical Library of The Johns Hopkins University (Baltimore, MD).
We have compared our interspecific map of chromosome 3 with a composite mouse linkage map that reports the map location of many uncloned mouse mutations (provided from the Mouse Genome Database, a computerized database maintained at The Jackson Laboratory, Bar Harbor, ME). Mlf1 mapped in a region of the composite map that lacks mouse mutations with a phenotype that might be expected for an alteration in this locus (data not shown).
The central portion of mouse chromosome 3 shares regions of homology with human chromosomes 3q, 4q, and 1q. The placement of Mlf1 in this interval is in good agreement with the human mapping studies that have localized MLF1 to 3q25.1. 37
Tissue Distribution of Mlf1 Expression
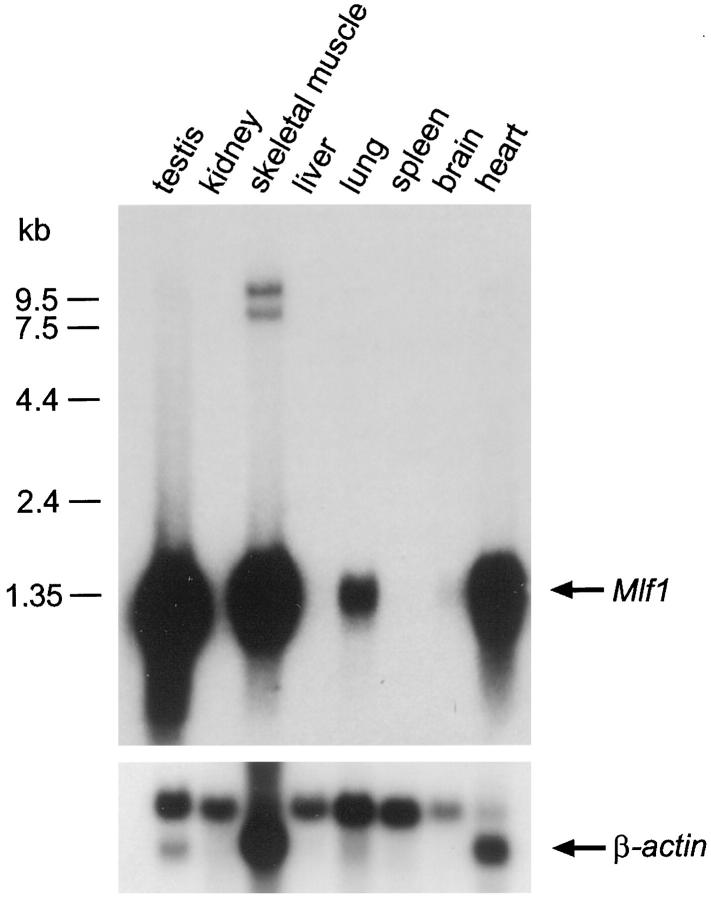
To help gain insight into normal Mlf1 function, we determined the expression pattern of the gene by both Northern blotting and in situ hybridization. Northern blot analysis of polyA+ RNA prepared from various mouse tissues demonstrated a relatively restricted expression pattern (Figure 3) ▶ . Strikingly high levels of Mlf1 mRNA were found in striated muscle tissue of both skeletal and cardiac origin and in testis. Skeletal and heart muscle also expressed two larger RNA species, approximately 7.5 and 9.5 kb in size, which hybridized with the Mlf cDNA probe, in addition to the predominant 1.3-kb Mlf1 transcript. These larger transcripts were not detected in any other tissues and their significance is unknown. High levels of Mlf1 mRNA were also present in the lung, whereas polyA+ RNA from brain tissue showed only a very weak signal. Mlf1 mRNA was not detected in spleen, liver, or kidney tissues.
Figure 3.
Tissue-specific expression of Mlf1 during embryonic and postnatal life. Northern blot analysis of Mlf1 expression. The mouse Mlf1 cDNA was hybridized to approximately 2 μg polyA+ RNA prepared from the mouse tissues indicated. After stripping of the signal, the membrane was rehybridized with a β-actin cDNA to control for RNA loading.
To further refine our analysis of Mlf1 expression, we next performed in situ hybridization experiments using sections of mouse embryos of embryonic ages 10, 12, 14, 15, 16, and 19 days, and of newborn and adult mice. Consistent with our Northern hybridization results, a strikingly high level of Mlf1 expression was found in both cardiac and skeletal muscle. Mlf1 transcripts were detected in both the atrial and ventricular myocardium as early as embryonic day 12 (Figure 4A) ▶ and became even more prominent in these tissues in the newborn mouse (Figure 4E) ▶ . Mlf1 was also strongly expressed in non-cardiac striated muscle tissue, such as the paraspinal muscles (Figure 4D) ▶ . Smooth muscle tissues also expressed Mlf1, although a more heterogeneous pattern of expression was evident. The vascular smooth muscle layers of the aorta were negative (Figure 4D) ▶ , but expression could be detected in the wall of the pulmonary veins in the newborn mouse (Figure 4E) ▶ . Similarly, the smooth muscle layer of the esophageal wall showed a significant level of Mlf1 expression (Figure 4E) ▶ . In contrast, the strong signal found in the bronchial wall of the newborn mouse was clearly of mucosal origin (Figure 4E) ▶ . Within the epithelial lining of the airways, Mlf1 expression was also detected during development, specifically in the nasopharynx of the day 14 embryo (Figure 4B) ▶ . The two other sites of significant Mlf1 expression were in germinal cells and the central nervous system. Mlf1 mRNA was not only detected in the ovary of the developing mouse (Figure 4C) ▶ , but also was expressed at high level in the germ cells of the adult testis (Figure 4G) ▶ . Within the central nervous system, Mlf1 expression was confined to the ependymal lining of the ventricles and the choroid plexus (Figure 4F) ▶ . Thus, Mlf1 expression occurred in a specific pattern during development and adult life.
Figure 4.
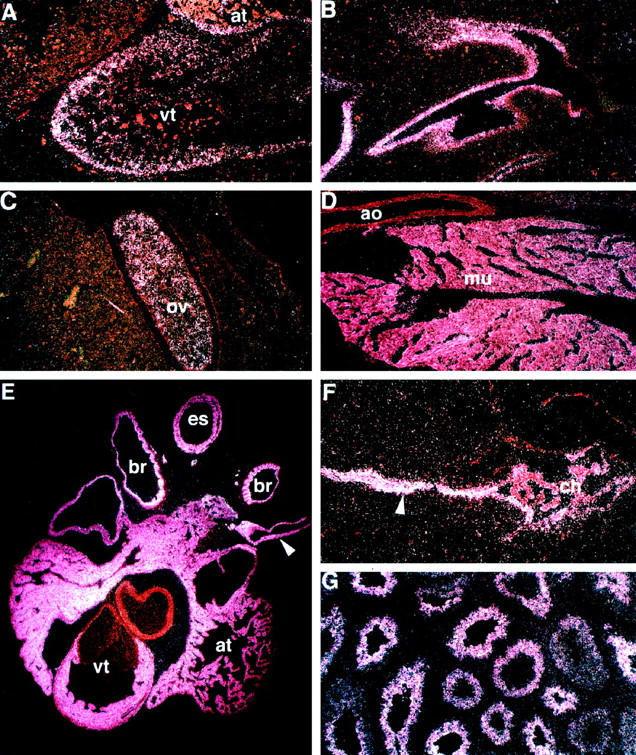
Mlf1 in situ hybridization. Panel A shows a dark field image illustrating the distribution of the Mlf1 mRNA in the developing heart of a day 12 mouse embryo. The bright white grains indicate the localization of the Mlf1-specific transcripts in the myocardial cells of the ventricle (vt) and atrium (at). Panel B is a section through the nasal cavity region of a day 14 embryo. The expression is limited to the primitive mucosal epithelium. Panel C is a section through the developing ovary (ov) of a day 14 embryo, showing a strong signal in the germinal cell population. Panel D illustrates a section through the paraspinal muscles of a newborn mouse; there is strong signal in the skeletal muscle tissue (mu) but no signal in adjacent structures such as the aorta (ao). Panel E shows the heart of a newborn mouse. The myocardial cells of the ventricle (vt) and atrium (at), as well as the smooth muscle cell layer in the pulmonary veins (arrowhead), show strong signal. There is also high-level Mlf1 expression in the mucosal lining cells of the bronchial airways (br) and in the smooth muscle layer of the esophagus (es). Panel F is a section through a newborn brain, demonstrating expression in the ependymal lining (arrowhead) and the choroid plexus (ch). In panel G, the testis of a mature mouse is shown, illustrating Mlf1 expression in the spermatogenetic cell layers. A-G, dark field illumination. Original magnifications, A-D, F, and G, ×100; E, ×40.
Our data demonstrate that the MLF1 sequence is very highly conserved between mouse and man, including the presence of a consensus 14-3–3 binding motif 25,26 in both proteins. The 14-3–3 protein family (which is comprised of multiple isoforms) has been demonstrated to interact with a large number of diverse cellular proteins (including Raf, 33-35 BAD, 29,36 p53, 38 Cbl, 32 Cdc25, 39,40 BCR and BCR-ABL, 41,42 protein kinase C, 43 and certain MEK kinases, 44 among others) and appears to modulate signal transduction through several pathways as a result. 27,30,31 MLF1 physically associates with 14-3–3 proteins, as well as certain apoptotic control proteins. 45 (X Wu, Y Sun, and SW Morris, manuscript in preparation). Thus, the ectopic overexpression of the protein in hematopoietic cells that occurs due to NPM-MLF1 could theoretically dysregulate both growth control and cell death signaling pathways. The ability to effect such pleomorphic responses may be a requirement of a gene involved in the genesis of MDS, given that the currently accepted MDS pathogenesis models suggest that the affected pluripotent stem cell clone contains a genetic hit that enhances or permits the proliferation of immature cells but interferes with the maturation and increases the apoptotic death of these cells as they attempt to differentiate in the bone marrow. 2,3 These events result in the clinical paradox of variable peripheral blood cytopenias in the presence of a normo- or hypercellular marrow, because the increased proliferation of cells is offset by an equally increased rate of intramedullary apoptosis. 46-48 In MDS patients who progress to acute leukemia, additional genetic events presumably occur that inhibit the apoptotic response and lead to autonomous growth by a leukemic clone.
Mlf1 expression was strikingly absent from spleen, thymus, and bone marrow of the adult mouse as well as embryonic sites of lympho-hematopoiesis such as yolk sac, the aorta-gonad-mesonephros region, and fetal liver (data not shown). Low-level expression of Mlf1 in small but functionally important subsets of hematopoietic cells or short-lived expression during a particular stage of hematopoiesis, however, cannot be ruled out unequivocally. The distinctly high levels of Mlf1 observed in cardiac and skeletal muscle, as well as in some sites of smooth muscle tissue, raise the intriguing possibility that Mlf1 plays a role in the differentiation and function of these tissues. Unfortunately, the chromosomal position of Mlf1 does not coincide with the location of known mouse mutations that might assist in elucidating the normal developmental function of the gene in muscle or other tissues in which we have identified expression. Definitive answers concerning normal Mlf1 function will require the analysis of animals lacking the gene, which is currently in progress.
Acknowledgments
We thank Xiaoli Cui, Bart Jones, Mary Barnstead, Pam Groen, Kathy Saalfeld, and Lisa Artmayer for excellent technical help, Alicia Emly for photographic assistance, and Doris Dodson for manuscript preparation.
Footnotes
Address reprint requests to Stephan W. Morris, M. D., St. Jude Children’s Research Hospital, Department of Experimental Oncology, Room 5024, Thomas Tower, 332 N. Lauderdale, Memphis, TN 38105-2794. E-mail: steve.morris@stjude.org.
Supported by the Pediatric Scientist Development Program through a grant from St. Jude Children’s Research Hospital (supporting J. K. H.), the American Heart Association, Ohio-West Virginia affiliate (SW-96–43) (to D. P. W.), the Department of Health and Human Services under contract with ABL (to N. A. J., N. G. C., and D.J.G.), National Cancer Institute grants CA 76301 (to S. W. M.) and Cancer Center Support (CORE) grant CA 27165, and by the American Lebanese Syrian Associated Charities, St. Jude Children’s Research Hospital.
References
- 1.Dunbar CE, Nienhuis AW: Myelodysplastic syndromes. Handin RI Lux SE Stossel TP eds. Blood Principles & Practice of Hematology. 1995, :pp 377-414 JB Lippincott Company, Philadelphia [Google Scholar]
- 2.Parker JE, Mufti GJ: Ineffective haemopoiesis and apoptosis in myelodysplastic syndromes. Br J Haematol 1998, 101:220-230 [DOI] [PubMed] [Google Scholar]
- 3.Raza A, Mundle S, Shetty V, Alvi S, Chopra H, Span L, Parcharidou A, Dar S, Venugopal P, Borok R, Gezer S, Showel J, Loew J, Robin E, Rifkin S, Alston D, Hernandez B, Shah R, Kaizer H, Gregory S, Preisler H: A paradigm shift in myelodysplastic syndromes. Leukemia 1996, 10:1648-1652 [PubMed] [Google Scholar]
- 4.Janssen JW, Buschle M, Layton M, Drexler HG, Lyons J, Van den Berghe H, Heimpel H, Kubanek B, Kleihauer E, Mufti GJ, Bartram CR: Clonal analysis of myelodysplastic syndromes: evidence of multipotent stem cell origin. Blood 1989, 73:248-254 [PubMed] [Google Scholar]
- 5.Tsukamoto N, Morita K, Maehara T, Okamoto K, Karasawa M, Omine M, Naruse T: Clonality in myelodysplastic syndromes: demonstration of pluripotent stem cell origin using X-linked restriction fragment length polymorphisms. Br J Haematology 1993, 83:589-594 [DOI] [PubMed] [Google Scholar]
- 6.Head DR: Revised classification of acute myeloid leukemia. Leukemia 1996, 10:1826-1831 [PubMed] [Google Scholar]
- 7.Bartram CR: Molecular genetic aspects of myelodysplastic syndromes. Semin Hematol 1996, 33:139-149 [PubMed] [Google Scholar]
- 8.Lopingco MC, Perkins AS: Molecular analysis of Evi1, a zinc finger oncogene involved in myeloid leukemia. Curr Top Microbiol Immunol 1996, 211:211-222 [DOI] [PubMed] [Google Scholar]
- 9.Golub TR, Barker GF, Lovett M, Gilliland DG: Fusion of PDGF receptor β to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 1994, 77:307-316 [DOI] [PubMed] [Google Scholar]
- 10.Yoneda-Kato N, Look AT, Kirstein MN, Valentine MB, Raimondi SC, Cohen KJ, Carroll AJ, Morris SW: The t(3;5)(q25.1;q34) of myelodysplastic syndrome and acute myeloid leukemia produces a novel fusion gene, NPM-MLF1. Oncogene 1996, 12:265-275 [PubMed] [Google Scholar]
- 11.Borer RA, Lehner CF, Eppenberger HM, Nigg EA: Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 1989, 56:379-390 [DOI] [PubMed] [Google Scholar]
- 12.Chan W-Y, Liu QR, Borjigin J, Busch H, Rennert OM, Tease LA, Chan P-K: Characterization of the cDNA encoding human nucleophosmin and studies of its role in normal and abnormal growth. Biochemistry 1989, 28:1033-1039 [DOI] [PubMed] [Google Scholar]
- 13.Schmidt-Zachmann MS, Franke WW: cDNA cloning and amino acid sequence determination of a major constituent protein of mammalian nucleoli: correspondence of the nucleoplasmin-related protein NO38 to mammalian protein B23. Chromosoma 1988, 96:417-426 [DOI] [PubMed] [Google Scholar]
- 14.Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT: Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 1994, 263:1281-1284 [DOI] [PubMed] [Google Scholar]
- 15.Redner RL, Rush EA, Faas S, Rudert WA, Corey SJ: The t(5;17) variant of acute promyelocytic leukemia expresses a nucleophosmin-retinoic acid receptor fusion. Blood 1996, 87:882-886 [PubMed] [Google Scholar]
- 16.Bischof D, Pulford K, Mason DY, Morris SW: Role of the nucleophosmin (NPM) portion of the non-Hodgkin’s lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis. Mol Cell Biol 1997, 17:2312-2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuefer MU, Look AT, Williams DC, Valentine V, Naeve CW, Behm FG, Mullersman JE, Yoneda-Kato N, Montgomery K, Kucherlapati R, Morris SW: cDNA cloning, tissue distribution, and chromosomal localization of myelodysplasia/myeloid leukemia factor 2 (MLF2). Genomics 1996, 35:392-396 [DOI] [PubMed] [Google Scholar]
- 18.Williams JH, Daly LN, Ingley E, Beaumont JG, Tilbrook PA, Tsai S, Klinken SP: Characterization of novel genes associated with a hemopoietic lineage switch. Blood 1998, 92:475(abstr. no. 1961) [Google Scholar]
- 19.Matsumoto N, Yoneda-Kato N, Yamamoto Y, Kishimoto Y, Fukuhara S: Myelodysplasia/myeloid leukemia factor 1, MLF1, as a new marker of prognosis, and leukemia progression from myelodysplastic syndrome. Blood 1997, 90:201(abstr. no. 885) [Google Scholar]
- 20.Copeland NG, Jenkins NA: Development and applications of a molecular genetic linkage map of the mouse genome. Trends Genet 1991, 7:113-118 [DOI] [PubMed] [Google Scholar]
- 21.Jenkins NA, Copeland NG, Taylor BA, Lee BK: Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virology 1982, 43:26-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rattner A, Hsieh JC, Smallwood PM, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J: A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc Natl Acad Sci USA 1997, 94:2859-2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tessarollo L, Tsoulfas P, Martin-Zanca D, Gilbert DJ, Jenkins NA, Copeland NG, Parada LF: trkC, a receptor for neurotrophin-3, is widely expressed in the developing nervous system and in non-neuronal tissues. Development 1993, 118:463-475[published erratum appears in Development 1993, 118:following 1384] [DOI] [PubMed] [Google Scholar]
- 24.Morris SW, Naeve C, Mathew P, James PL, Kirstein MN, Cui X, Witte DP: ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin’s lymphoma, encodes a neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK). Oncogene 1997, 14:2175-2188 [DOI] [PubMed] [Google Scholar]
- 25.Muslin AJ, Tanner JW, Allen PM, Shaw AS: Interaction of 14-3–3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 1996, 84:889-897 [DOI] [PubMed] [Google Scholar]
- 26.Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC: The structural basis for 14-3–3: phosphopeptide binding specificity. Cell 1997, 91:961-971 [DOI] [PubMed] [Google Scholar]
- 27.Aitken A, Jones D, Soneji Y, Howell S: 14-3–3 proteins: biological function and domain structure. Biochem Soc Trans 1995, 605–611 [DOI] [PubMed]
- 28.Bonnefoy-Berard N, Liu YC, von Willebrand M, Sung A, Elly C, Mustelin T, Yoshida H, Ishizaka K, Altman A: Inhibition of phosphatidylinositol 3-kinase activity by association with 14-3–3 proteins in T cells. Proc Natl Acad Sci USA 1995, 92:10142-10146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu SY, Kaipia A, Zhu L, Hsuan JJ: Interference of BAD (Bcl-xL/Bcl-2-associated death promoter)-induced apoptosis in mammalian cells by 14-3–3 isoforms and P11. Mol Endocrinol 1997, 11:1858-1867 [DOI] [PubMed] [Google Scholar]
- 30.Morrison D: 14-3–3: modulators of signaling proteins? Science 1994, 266:56-57 [DOI] [PubMed] [Google Scholar]
- 31.Reuther GW, Pendergast AM: The roles of 14-3–3 proteins in signal transduction. Vitam Horm 1996, 52:149-175 [DOI] [PubMed] [Google Scholar]
- 32.Robertson H, Langdon WY, Thien CBF, Bowtell DDL: A c-Cbl yeast two hybrid screen reveals interactions with 14-3–3 isoforms, and cytoskeletal components. Biochem Biophys Res Comm 1997, 240:46-50 [DOI] [PubMed] [Google Scholar]
- 33.Roy S, McPherson RA, Apolloni A, Yan J, Lane A, Clyde-Smith J, Hancock JF: 14-3–3 facilitates Ras-dependent Raf-1 activation in vitro, and in vivo. Mol Cell Biol 1998, 18:3947-3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorson JA, Yu LWK, Hsu AL, Shih NY, Graves PR, Tanner JW, Allen PM, Piwnica-Worms H, Shaw AS: 14-3–3 proteins are required for maintenance of Raf-1 phosphorylation, and kinase activity. Mol Cell Biol 1998, 18:5229-5238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tzivion G, Luo Z, Avruch J: A dimeric 14-3–3 protein is an essential cofactor for Raf kinase activity. Nature 1998, 394:88-92 [DOI] [PubMed] [Google Scholar]
- 36.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ: Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3–3 not BCL-XL. Cell 1996, 87:619-628 [DOI] [PubMed] [Google Scholar]
- 37.Raimondi SC, Dube ID, Valentine MB, Mirro J, Jr, Watt HJ, Larson RA, Bitter MA, Le Beau MM, Rowley JD: Clinicopathologic manifestations and breakpoints of the t(3;5) in patients with acute nonlymphocytic leukemia. Leukemia 1989, 3:42-47 [PubMed] [Google Scholar]
- 38.Waterman MJF, Stavridi ES, Waterman JLF, Halazonetis TD: ATM-dependent activation of p53 involves dephosphorylation and association with 14-3–3 proteins. Nat Genet 1998, 19:175-178 [DOI] [PubMed] [Google Scholar]
- 39.Conklin DS, Galaktionov K, Beach D: 14-3–3 proteins associate with cdc25 phosphatases. Proc Natl Acad Sci USA 1995, 92:7892-7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H: Mitotic and G2 checkpoint control: regulation of 14-3–3 protein binding by phosphorylation of Cdc25C on serine-216. Science 1997, 277:1501-1505 [DOI] [PubMed] [Google Scholar]
- 41.Braselmann S, McCormick F: BCR and RAF form a complex in vivo via 14-3–3 proteins. EMBO J 1995, 14:4939-4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reuther GW, Fu H, Cripe LD, Collier RJ, Pendergast AM: Association of the protein kinases c-Bcr and Bcr-Abl with proteins of the 14-3–3 family. Science 1994, 266:129-133 [DOI] [PubMed] [Google Scholar]
- 43.Meller N, Liu YC, Collins TL, Bonnefoy-Berard N, Baier G, Isakov N, Altman A: Direct interaction between protein kinase Cθ (PKCθ) and 14-3–3τ in T cells: 14-3–3 overexpression results in inhibition of PKCθ translocation and function. Mol Cell Biol 1996, 16:5782-5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fanger GR, Widmann C, Porter AC, Sather S, Johnson GL, Vaillancourt RR: 14-3–3 proteins interact with specific MEK kinases. J Biol Chem 1998, 273:3483-3476 [DOI] [PubMed] [Google Scholar]
- 45.Suzuki Y, Demoliere C, Kitamura D, Takeshita H, Deuschle U, Watanabe T: HAX-1, a novel intracellular protein, localized on mitochondria, directly associates with HS1, a substrate of Src family tyrosine kinases. J Immunol 1997, 158:2736-2744 [PubMed] [Google Scholar]
- 46.Bincoletto C, Saad STO, Soares da Silva E, Queiroz MLS: Autonomous proliferation and bcl-2 expression involving haematopoietic cells in patients with myelodysplastic syndrome. Br J Cancer 1998, 78:621-624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajapaksa R, Ginzton N, Rott LS, Greenberg PL: Altered oncoprotein expression and apoptosis in myelodysplastic syndrome marrow cells. Blood 1996, 88:4275-4287 [PubMed] [Google Scholar]
- 48.Raza A, Gezer S, Mundle S, Gao XZ, Alvi S, Borok R, Rifkin S, Iftikhar A, Shetty V, Parcharidou A, Loew J, Marcus B, Khan Z, Chaney C, Showel J, Gregory S, Preisler H: Apoptosis in bone marrow biopsy samples involving stromal and hematopoietic cells in 50 patients with myelodysplastic syndromes. Blood 1995, 86:268-276 [PubMed] [Google Scholar]