Abstract
The differential diagnosis of neuroblastoma from other small round-cell tumors of childhood, although clinically of great importance, is sometimes difficult due to the almost indistinguishable appearance of such tumors by conventional microscopy. Because neuroblastomas are characterized by the synthesis of catecholamines, we investigated the possibility that expression of genes involved in this pathway could serve as a molecular marker for this disease. A reverse transcriptase polymerase chain reaction assay was used to analyze expression of tyrosine hydroxylase and dopa decarboxylase in 84 pediatric malignancies including 55 neuroblastomas, 6 Ewing’s sarcomas/primitive neuroectodermal tumors, 7 lymphomas, 6 leukemias, 2 rhabdomyosarcomas, 6 osteosarcomas, and 2 phaeochromocytomas. Of the 55 neuroblastoma samples analyzed, 54 expressed clearly detectable levels of both genes. The one sample that did not express either of the genes was rediagnosed both clinically and by molecular genetic analysis as a Ewing’s sarcoma. Of the 29 non-neuroblastoma tumor samples examined, the only tumor samples that expressed clearly detectable levels of both tyrosine hydroxylase and dopa decarboxylase were phaeochromocytomas. Like neuroblastomas, these tumors are characterized by high levels of catecholamines. These findings suggest that expression of genes involved in catecholamine biosynthesis may be useful for differentiating neuroblastoma from other small round-cell tumors of childhood.
Neuroblastoma is the most common solid tumor of early childhood. Although patients with localized disease have a favorable prognosis, the majority of children with neuroblastoma present with metastases and have a poor outcome despite intensive multimodal therapy. 1 The accurate diagnosis of this disease and other pediatric malignancies has become increasingly important with the continued development of treatments tailored to specific tumor types, and the resultant improvement in survival rates. 2 Neuroblastoma, together with lymphoma, osteosarcoma, Ewing’s family of tumors, rhabdomyosarcoma, and lymphoblastic leukemia, all belong to a group of undifferentiated pediatric malignancies known as the small round-cell tumors of childhood. In some instances, the differential diagnosis of this group of tumors can prove difficult, 2 due to the fact that they share morphological similarities that can make them indistinguishable by conventional light microscopy.
The accurate diagnosis of small round-cell tumors can in some cases be facilitated by cytogenetic and, more recently, by molecular biological analysis. Thus, for example, the Ewing’s family of tumors, consisting of Ewing’s sarcoma and primitive neuroectodermal tumors (PNET), is characterized by the genetic abnormality of a chromosomal translocation at t(11;22) in the majority of cases and the less common t(21;22) in a small number of cases. 3,4 Recent molecular advances have allowed for the PCR-based detection of such translocations. 3,5 However, many of the small round-cell tumors of childhood, including neuroblastoma, do not have consistent molecular genetic abnormalities amenable to either cytogenetic or DNA analysis. Because neuroblastomas are characterized by the secretion of catecholamines, we have investigated the possibility of employing expression of genes involved in the catecholamine biosynthetic pathway as potential molecular markers for this disease. The results demonstrated that coexpression of two genes, tyrosine hydroxylase and dopa decarboxylase, appears to be highly specific for neuroblastoma and suggest that these markers may aid in distinguishing neuroblastoma from other small round-cell tumors of childhood.
Materials and Methods
Tumor Samples
Samples of 55 primary neuroblastoma tumors from untreated patients, obtained either from the Neuroblastoma Tumor Bank of the U. S. Pediatric Oncology Group (Memphis, TN), or from the Sydney Children’s Hospital, Sydney, Australia, and representing all clinical stages, have been described previously. 6 The 29 non-neuroblastoma tumor samples were obtained at diagnosis from patients presenting at the Sydney Children’s Hospital included 2 phaeochromocytomas, 6 Ewing’s sarcomas/PNETs, 7 lymphomas, 6 leukemias, 2 rhabdomyosarcomas, and 6 osteosarcomas. All samples were taken during the course of the patients’ routine management.
Analysis of Gene Expression by Polymerase Chain Reaction
Total cellular RNA was isolated from frozen tumor tissue as previously described. 7 High quality intact RNA was routinely obtained from over 95% of tumors processed. Complementary DNA (cDNA) was synthesized from 2-μg aliquots of RNA with random hexanucleotide primers and Moloney murine virus reverse transcriptase. 8 Aliquots of cDNA corresponding to 50 ng of RNA were amplified in a well-established reverse transcriptase polymerase chain reaction (RT-PCR) assay, 9 which involved co-amplification of the target gene sequence (tyrosine hydroxylase or dopa decarboxylase, respectively) with a control sequence (β2-microglobulin), for 30 cycles using gene-specific oligonucleotide primers. For amplification of dopa decarboxylase, the forward and reverse primers, respectively, were 5′-GGGGACCACAACATGCTGCTC-3′ and 5′-CCACTCCATTCAGAAGGTGCC-3′. The primers for β2-microglobulin have previously been described. 8 Following an initial denaturation step of 3 minutes at 94°C, each cycle consisted of denaturation at 94°C for 45 seconds, primer annealing at 55°C for 45 seconds, and primer extension at 72°C for 90 seconds. Where indicated, PCR amplification of dopa decarboxylase was performed in the absence of β2-microglobulin primers for 35 cycles under the same PCR conditions. Due to the high guanine and cytosine content of the tyrosine hydroxylase gene, an annealing temperature of 72°C was used for PCR reactions amplifying this gene. The forward and reverse primers for tyrosine hydroxylase were 5′-CCCTGACCTGGACTTGGACCACCC-3′ and 5′-TCTCCTCGGCGGTGTACTCCACAC-3′, respectively. β2-microglobulin gene-specific primers suitable for amplification at this elevated annealing temperature were 5′-ATGTCTCGCTCCGTGGCCTTAGCTG-3′ and 5′-TCCATTCTCTGCTGGATGACGTGAG-3′ for the forward and reverse primers, respectively. Following electrophoresis on 12% polyacrylamide gels and staining with ethidium bromide, PCR products were visualized and photographed under ultraviolet transillumination. In all cases, analyses were performed in triplicate.
RT-PCR analysis for the t(11;22) translocation specific to the Ewing’s family of tumors was performed essentially as described. 10 Briefly, cDNA was synthesized from 1 μg of total RNA using random hexanucleotide primers and the entire mixture was subjected to PCR amplification with t(11;22) translocation-specific primers. 3 Following an initial denaturation step of 10 minutes at 96°C, each cycle consisted of denaturation at 96°C for 30 seconds, primer annealing at 65°C for 1 minute, and primer extension at 75°C for 1 minute for a total of 40 cycles. PCR products were subjected to agarose gel electrophoresis and photographed.
Results
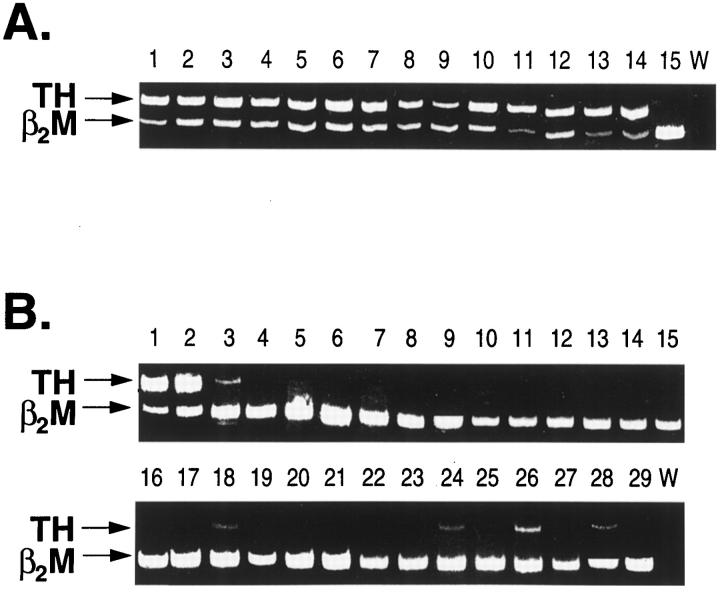
To determine the frequency and specificity of tyrosine hydroxylase and dopa decarboxylase gene transcripts in neuroblastoma and other pediatric malignancies, expression of these target genes was evaluated in 84 tumor samples using a competitive RT-PCR assay. Included were 55 neuroblastomas, 6 Ewing’s sarcomas/PNETs, 7 lymphomas, 6 leukemias, 6 osteosarcomas, 2 rhabdomyosarcomas, and 2 phaeochromocytomas. The phaeochromocytomas were included as positive controls, because this tumor type is also characterized by high levels of catecholamine secretion. 11 After RT-PCR, clearly detectable tyrosine hydroxylase gene expression was evident in 54 of 55 neuroblastoma specimens, as shown by a representative gel in Figure 1A ▶ . For the remaining tumor sample (Panel A, lane 15; tumor no. AG), however, no expression of this gene could be discerned. In contrast to this result, tyrosine hydroxylase gene expression was undetectable in 22 of the 29 non-neuroblastoma tumor specimens (Figure 1B) ▶ . Of the seven specimens in which any tyrosine hydroxylase expression was evident, only the phaeochromocytoma positive controls displayed levels of expression comparable to those of neuroblastoma. The remaining five specimens, comprising one Ewing’s tumor, one leukemia, two osteosarcomas, and one rhabdomyosarcoma, displayed very low levels of tyrosine hydroxylase gene expression, as evidenced by the tyrosine hydroxylase PCR product being considerably weaker than the control β2-microglobulin product in each case (Figure 1B) ▶ . No tyrosine hydroxylase expression could be detected in any of the lymphomas.
Figure 1.
Expression of tyrosine hydroxylase (TH) in pediatric tumors following competitive RT-PCR analysis as described in the Materials and Methods. A: Tyrosine hydroxylase gene expression in primary neuroblastoma tumors of stage I (lanes 1–2), stage II (lanes 3–5), stage III (lanes 6–9), stage IV (lanes 10–13), stage IVs (lane 14) and tumor no. AG (lane 15). B: Tyrosine hydroxylase gene expression in phaeochromocytomas (lanes 1–2), Ewing’s family of tumors (lanes 3–8), lymphomas (lanes 9–15), leukemias (lanes 16–21), osteosarcomas (lanes 22–27) and rhabdomyosarcomas (lanes 28–29). W, water control; β2M, β2-microglobulin.
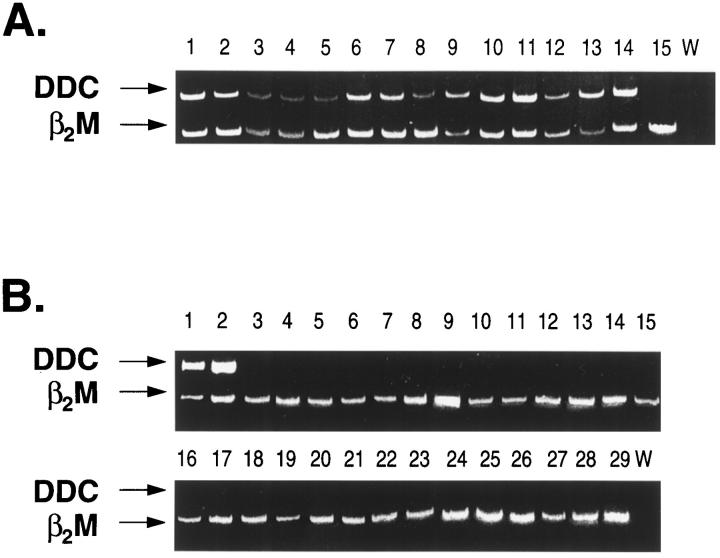
Variable but clearly detectable expression of the dopa decarboxylase gene was similarly evident in the 54 neuroblastoma specimens which had been positive for tyrosine hydroxylase expression (Figure 2A) ▶ . The one sample which had failed to express tyrosine hydroxylase (tumor no. AG) also showed no dopa decarboxylase expression (Panel A, lane 15). The complete absence of dopa decarboxylase expression from this sample was confirmed by conducting PCR amplification of the target gene in the absence of competing β2-microglobulin gene primers. Even under these more stringent conditions, no PCR product for dopa decarboxylase was evident (data not shown). In contrast, apart from the two phaeochromocytoma controls, expression of dopa decarboxylase was not observed in any of the other 27 non-neuroblastoma tumor samples examined (Figure 2B) ▶ .
Figure 2.
Expression of dopa decarboxylase (DDC) in pediatric tumors following competitive RT-PCR analysis. The legend to this figure is the same as Figure 1 ▶ .
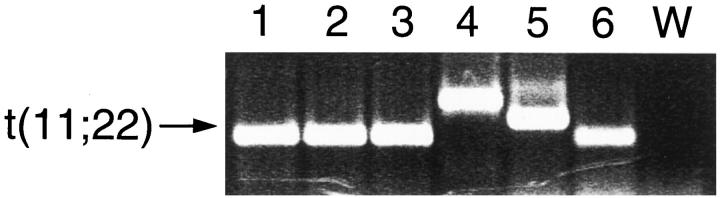
Further investigation of tumor no. AG, which had failed to express either tyrosine hydroxylase or dopa decarboxylase, revealed that the patient had independently been clinically rediagnosed as having Ewing’s sarcoma. To confirm this clinical diagnosis, RT-PCR analysis was performed using PCR primers which amplify the t(11;22) hybrid transcript believed to be specific for Ewing’s family of tumors. 3 Following amplification, a positive PCR product was obtained from this tumor sample (Figure 3 ▶ , lane 6), consistent with the positive result obtained from the other Ewing’s sarcoma samples included in this study. The differing sizes of the products reflect differences in the length of the hybrid transcripts, which can vary depending on the location of the translocation breakpoints. 3,5 Sequencing of the PCR product from tumor no. AG confirmed a Ewing’s-specific transcript (data not shown).
Figure 3.
RT-PCR analysis of the Ewing’s t(11;22) transcript. Lanes 1–5: Five cytogenetically confirmed Ewing’s sarcoma specimens. Lane 6: Tumor no. AG. W, water control.
Discussion
The increased survival rates observed over recent decades in pediatric malignancies have resulted largely from the development and streamlining of specific therapies for specific tumor types. 2 Accordingly, accurate diagnosis has become increasingly important in order to achieve optimal treatment. Despite improvements in molecular genetic techniques, differential diagnosis can still present a difficulty, particularly among the small round-cell tumors of childhood. For neuroblastoma, there have been reports of this disease being misdiagnosed as acute leukemia; often the correct diagnosis is made only postmortem. 12,13 Similarly, Kessler et al 14 reported 5 cases of intrarenal neuroblastoma that were originally misdiagnosed as Wilms’ tumor, despite the fact that Wilms’ tumor is not included in the small round-cell tumors of childhood. Thus, there is a continuing need for the development of methods to improve the differential diagnosis of these malignancies. The present findings suggest that among the small round-cell tumors of childhood, coexpression of genes involved in the catecholamine biosynthetic pathway is a molecular characteristic specific to neuroblastoma.
Tyrosine hydroxylase is the first and rate-limiting enzyme in the catecholamine biosynthetic pathway, thus suggesting a requirement for tight regulatory control of the gene encoding this enzyme. This indeed appears to be the case, since the use of tyrosine hydroxylase expression as a specific marker of residual neuroblastoma cells has been demonstrated in numerous studies involving the detection of circulating tumor cells in patient’s peripheral blood and bone marrow samples. 15-19 In the present study, the expression of tyrosine hydroxylase in the non-neuroblastoma tumor specimens was at very low levels, suggesting that it is unlikely to play any significant biological role. Such low-level expression might be due either to contamination of the tumor sample by surrounding tissue or to the phenomenon known as illegitimate transcription. 20 Illegitimate transcription is the expression of otherwise tissue-specific genes in any cell type, with the detection of such expression resulting from the increased sensitivity afforded by PCR. 20,21 Interestingly, of the two genes investigated in the present study, expression of dopa decarboxylase demonstrated greater specificity for neuroblastoma than did expression of tyrosine hydroxylase. Using coexpression of two target genes as a specific marker of neuroblastoma affords increased confidence in the accuracy of the differential diagnosis. In this regard, Hoon et al 22 have recently reported that RT-PCR analysis performed with more than one target gene provides a more sensitive and reliable means of detecting residual melanoma than assays using only a single target gene.
A number of chromosomal abnormalities have proven useful in the characterization of individual pediatric disease types, as has been demonstrated in the present context by the use of the t(11;22) translocation for the diagnosis of Ewing’s sarcoma. Similarly, for rhabdomyosarcoma, recent studies have shown that the t(2;13) and t(1;13) translocations are valuable in the diagnosis of alveolar rhabdomyosarcoma, 23,24 whereas a number of genetic rearrangements occurring in leukemia have also proven useful as markers for the detection of that disease. 25,26 For neuroblastoma, although aggressive tumors are often characterized by deletions of chromosome 1p and amplification of the N-myc oncogene, these abnormalities occur, on average, in 30% or less of tumors. 27,28 In contrast, the present results suggest that coexpression of tyrosine hydroxylase and dopa decarboxylase is ubiquitous in neuroblastoma and hence a useful molecular diagnostic marker for this malignancy.
As molecular techniques become increasingly valuable in the diagnosis and detection of childhood malignancies, their limitations are also becoming evident. For example, recent reports have identified the t(11;22) translocation in confirmed cases of rhabdomyosarcoma 29 as well as neuroblastoma, 10 indicating that it is possible for tumor markers to be present in more than one disease. Such findings highlight the need for a panel of molecular markers to maximize the likelihood of making the correct differential diagnosis. The results presented here suggest that expression of the tyrosine hydroxylase and dopa decarboxylase genes may prove a valuable aid in the differential diagnosis of neuroblastoma from other small round-cell tumors of childhood.
Acknowledgments
We thank the Neuroblastoma Biology Subcommittee of the Pediatric Oncology Group for the review and approval of this research project and for the samples of neuroblastoma tumors provided for analysis.
Footnotes
Address reprint requests to A/Prof. M. D. Norris, Children’s Cancer Institute Australia for Medical Research, High Street (P.O. Box 81), Randwick, New South Wales, Australia, 2031.
Supported by grants to M. D. N., M. H., and G. M. M. from the National Health and Medical Research Council of Australia and the New South Wales Cancer Council. J. G. is the recipient of an Australian Postgraduate Award.
Children’s Cancer Institute Australia for Medical Research is affiliated with the University of New South Wales and Sydney Children’s Hospital.
References
- 1.Brodeur GM, Castleberry RP: Neuroblastoma. 3rd Edition. Pizzo PA Poplack DG eds. Principles and Practice of Pediatric Oncology, 1997, :pp 761-797 Lippincott-Raven, Philadelphia [Google Scholar]
- 2.McManus AP, Grusterson BA, Pinkerton CR, Shipley JM: The molecular pathology of small round-cell tumours: relevance to diagnosis, prognosis and classification. J Pathol 1996, 178:116-121 [DOI] [PubMed] [Google Scholar]
- 3.Delattre O, Zucman J, Melot T, Garau XS, Zucker J-M, Lenoir GM, Ambros PF, Sheer D, Turc-Carel C, Triche TJ, Aurias A, Thomas G: The Ewing family of tumors: a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N Engl J Med 1994, 331:294-299 [DOI] [PubMed] [Google Scholar]
- 4.Peter M, Couturier J, Pacquement H, Michon J, Thomas G, Magdelenat H, Delattre O: A new member of the ETS family fused to EWS in Ewing tumors. Oncogene 1997, 14:1159-1164 [DOI] [PubMed] [Google Scholar]
- 5.Zucman J, Delattre O, Desmaze C, Plougastel B, Joubert I, Melot T, Peter M, De Jong P, Rouleau G, Aurias A, Thomas G: Cloning and characterization of the Ewing’s sarcoma and peripheral neuroepithelioma t(11: 22) translocation breakpoints. Genes Chromosomes Cancer 1992, 5:271-277 [DOI] [PubMed] [Google Scholar]
- 6.Norris MD, Bordow SB, Marshall GM, Haber PS, Cohn SL, Haber M: Expression of the gene for multidrug resistance-associated protein and outcome in patients with neuroblastoma. N Engl J Med 1996, 334:231-238 [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N: Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987, 162:156-159 [DOI] [PubMed] [Google Scholar]
- 8.Noonan K, Beck C, Holtzmayer T, Chin JE, Wunder JS, Andrulis IL, Gazdar AF, Willman CL, Griffith B, Von Hoff DD, Roninson IB: Quantitative analysis of MDR1 (multidrug resistance) gene expression in human tumors by polymerase chain reaction. Proc Natl Acad Sci USA 1990, 87:7160-7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bordow SB, Haber M, Madafiglio J, Cheung B, Marshall GM, Norris MD: Expression of the multidrug resistance-associated protein (MRP) gene correlates with amplification and overexpression of the N-myc oncogene in childhood neuroblastoma. Cancer Res 1994, 54:5036-5040 [PubMed] [Google Scholar]
- 10.Burchill SA, Wheeldon J, Cullinane C, Lewis IJ: EWS-FLI1 fusion transcripts identified in patients with typical neuroblastoma. Eur J Cancer 1997, 33:239-243 [DOI] [PubMed] [Google Scholar]
- 11.Chrousos GP: Endocrine Tumors. 2nd Edition. Pizzo PA Poplack DG eds. Principles and Practice of Pediatric Oncology, 1993, :pp 889-912 Lippincott-Raven, Philadelphia [Google Scholar]
- 12.Boyd JE, Parmley RT, Langevin A-M, Saldivar VA: Neuroblastoma presenting as acute monoblastic leukemia. J Pediatr Hematol Oncol 1996, 18:206-212 [DOI] [PubMed] [Google Scholar]
- 13.Slamovits TL, Rosen CE, Suhrland MJ: Neuroblastoma presenting as acute lymphoblastic leukemia but correctly diagnosed after orbital fine-needle aspiration biopsy. J Clin Neuro-Ophthalmol 1991, 11:158-161 [PubMed] [Google Scholar]
- 14.Kessler OJ, Siegel JF, Brock WA: Intrarenal neuroblastoma masquerading as Wilms tumor. Urology 1998, 51:313-316 [DOI] [PubMed] [Google Scholar]
- 15.Naito H, Kuzumaki N, Uchino J, Kobayashi R, Shikano T, Ishikawa Y, Matsumoto S: Detection of tyrosine hydroxylase mRNA and minimal neuroblastoma cells by the reverse transcription-polymerase chain reaction. Eur J Cancer 1991, 27:762-765 [DOI] [PubMed] [Google Scholar]
- 16.Burchill SA, Bradbury FM, Smith B, Lewis IJ, Selby P: Neuroblastoma cell detection by reverse transcriptase-polymerase chain reaction (RT-PCR) for tyrosine hydroxylase mRNA. Int J Cancer 1994, 57:671-675 [DOI] [PubMed] [Google Scholar]
- 17.Burchill SA, Bradbury FM, Selby P, Lewis IJ: Early clinical evaluation of neuroblastoma cell detection by reverse transcriptase-polymerase chain reaction (RT-PCR) for tyrosine hydroxylase mRNA. Eur J Cancer 1995, 31A:553-556 [DOI] [PubMed] [Google Scholar]
- 18.Miyajima Y, Kato K, Numata S-I, Kudo K, Horibe K: Detection of neuroblastoma cells in bone marrow and peripheral blood at diagnosis by the reverse transcriptase-polymerase chain reaction for tyrosine hydroxylase mRNA. Cancer 1995, 75:2757-2761 [DOI] [PubMed] [Google Scholar]
- 19.Gilbert J, Norris MD, Marshall GM, Haber M: Low specificity of PGP9.5 expression for detection of micrometastatic neuroblastoma. Brit J Cancer 1996, 75:1779-1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chelly J, Concordet J-P, Kaplan J-C, Kahn A: Illegitimate transcription: transcription of any gene in any cell type. Proc Natl Acad Sci USA 1989, 86:2617-2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarkar G, Sommer SS: Access to a messenger RNA sequence or its protein product is not limited by tissue or species specificity. Science 1989, 244:331-334 [DOI] [PubMed] [Google Scholar]
- 22.Hoon DSB, Wang Y, Dale PS, Conrad AJ, Schmid P, Garrison D, Kuo C, Foshag LJ, Nizze AJ, Morton DL: Detection of occult melanoma cells in blood with a multiple-marker polymerase chain reaction assay. J Clin Oncol 1995, 13:2109-2116 [DOI] [PubMed] [Google Scholar]
- 23.Arden KC, Anderson MJ, Finckenstein FG, Czekay S, Cavanee WK: Detection of the t(2: 13) chromosomal translocation in alveolar rhabdomyosarcoma using the reverse transcriptase-polymerase chain reaction. Genes Chromosomes Cancer 1996, 16:254-260 [DOI] [PubMed] [Google Scholar]
- 24.Anderson J, Renshaw J, McManus A, Carter R, Mitchell C, Adams S, Pritchard-Jones K: Amplification of the t(2;13) and t(1;13) translocations of alveolar rhabdomyosarcoma in small formalin-fixed biopsies using a modified reverse transcriptase polymerase chain reaction. Am J Pathol 1997, 150:477-482 [PMC free article] [PubMed] [Google Scholar]
- 25.Maurer J, Janssen JW, Thiel E, van Denderen J, Ludwig W-D, Aydemir U, Heinze B, Fonatsch C, Harbott J, Reiter A, Riehm H, Hoelzer D, Bartram CR: Detection of chimeric BCR-ABL genes in acute lymphoblastic leukaemia by the polymerase chain reaction. Lancet 1991, 337:1055-1058 [DOI] [PubMed] [Google Scholar]
- 26.Chang K-S, Lu J, Wang G, Trujillo JM, Estey E, Cork A, Chu D-T, Freireich EJ, Strass SA: The t(15;17) breakpoint in acute promyelocytic leukemia cluster within two different sites of the myl gene: targets for the detection of minimal residual disease by the polymerase chain reaction. Blood 1992, 79:554-558 [PubMed] [Google Scholar]
- 27.Brodeur GM: Neuroblastoma: clinical significance of genetic abnormalities. Cancer Surv 1990, 9:673-688 [PubMed] [Google Scholar]
- 28.Schwab M, Praml C, Amler LC: Genomic instability in 1p and human malignancies. Genes Chromosomes Cancer 1996, 16:211-229 [DOI] [PubMed] [Google Scholar]
- 29.Thorner P, Squire J, Chilton-MacNeill S, Marrano P, Bayani J, Malkin D, Greenberg M, Lorenzana A, Zielenska M: Is the EWS/FLI-1 fusion transcript specific for Ewing sarcoma and peripheral primitive neuroectodermal tumor? Am J Pathol 1996, 148:1125-1138 [PMC free article] [PubMed] [Google Scholar]