Abstract
The outcome of patients with renal cell carcinoma is limited by the development of metastasis after nephrectomy. To evaluate the genetic basis underlying metastatic progression of human renal cell carcinoma in vivo, we performed a comparative genomic hybridization analysis in 32 clear-cell renal-cell carcinoma metastases. The most common losses involved chromosomes 3p (25%), 4q (28%), 6q (28%), 8p (31%), and 9p (47%). The most common gains were detected at 17q (31%) and Xq (28%). There was one high-level gene amplification at chromosome 11q22–23. The mean number of aberrations in lymph node (4.8 ± 2.8) and lung metastases (6.2 ± 4.0) was lower than in other hematogenous metastases (11.5 ± 8.7, P < 0.05), suggesting that hematogenous dissemination is linked to an acquisition of complex genomic alterations. As genetic differences between primary tumors and metastases give information on genetic changes that have contributed to the metastatic process, relative DNA sequence copy number changes in 19 matched tumor pairs were compared. Genomic changes, which frequently occurred in metastases but not in the corresponding primary tumor were losses of 8p and 9p and gains of 17q and Xq. An abnormal function of genes in these regions may contribute to the metastatic process. According to a statistical analysis of shared genetic changes in matched tumor pairs, a high probability of a common clonal progenitor was found in 11 of 19 patients (58%). Six metastases (32%) were genetically almost completely different from the primary, suggesting that detection of genomic alterations in primary tumors gives only a restricted view of the biological properties of metastatic renal cell carcinoma.
Prognosis of patients with renal cell carcinoma (RCC) is limited by the development of metastases. Five-year survival rates range from 50% to 85% in patients with organ-confined renal cancer (stage I and stage II). In contrast, less than one-third of patients with regional lymph node metastases survive 5 years. All but 5% to 10% of those patients with hematogenous metastases die within 5 years after diagnosis. 1 The most common sites of distant metastasis are lung, liver, and bones, but metastases can also develop at any other site. The metastatic behavior of RCC is often bizarre and unpredictable 2-4 .
Radiotherapy, chemotherapy, or hormonal therapy have little or no effect on metastatic RCC. Gene therapy with retroviral vector-mediated lymphokine gene transfer gave promising results in preclinical studies and is under further investigation. 5 The metastatic cells represent the prime targets of cancer therapy. However, little is known about genetic changes with importance for the development of RCC metastases. 6-9 Chromosome 9p losses in primary RCC were associated with short metastasis-free survival. 10,11 Expression of the epidermal growth factor receptor gene 12 and the p53 gene 13 were associated with metastatic disease.
Cancer is a genetically heterogeneous disease. Multiple clones of malignant cells are frequently detected by standard cytogenetics, by fluorescence in situ hybridization (FISH), and by flow cytometry. 14-17 Animal studies suggest that the metastatic proportions of different cell clones may be related to the metastatic process. 18,19 Therefore, chromosomal alterations responsible for metastasis may be present only in cell subpopulations of the primary tumor, which may not be detectable by molecular analyses. However, chromosomal alterations with relevance for the metastatic process should be enriched in tissue samples from metastases. Analyzing genetic changes in the metastases rather than in the more commonly targeted primary tumors could shed new light onto the molecular mechanisms of the metastatic process. Studies comparing chromosomal changes in metastases with those found in the corresponding primary tumors in the same patient would be informative in revealing differences between primary and metastatic lesions and thereby pinpointing genetic events that could have predisposed to metastatic dissemination.
Very little is known about genetic changes present in RCC metastases. Given the fact that RCC metastases may develop years or even decades after the removal of the primary tumor, genetic evolution certainly takes place within metastases. Analysis of primary tumors and their corresponding metastases allows one to assess the extent to which primary and metastatic cell clones are different from one another. A recently developed mathematical model allows testing of the statistical probability of a common clonal progenitor in primary tumors and their metastases. 20
To search for cytogenetic events related to metastases, 32 RCC metastases and 19 corresponding primary tumors were screened by comparative genomic hybridization (CGH). CGH allows the detection of all clonal DNA-sequence copy number aberrations (>10 MB) across the entire genome. 21 The results implicated several genomic regions that might carry genes involved in metastasis.
Materials and Methods
Patients
The tumor specimens were obtained from the archives of the Institute for Pathology, University Basel. Twenty-seven patients with metastatic RCC were selected for this study according to the following criteria: 1) histologically representative metastatic specimens containing at least 75% tumor cells and having no necrosis were available, 2) the primary tumor was a histologically proven clear-cell RCC, and 3) the patient had not received any chemotherapy or radiotherapy before the diagnosis of metastases. Tissue was obtained from surgically resected metastases (n = 17) and from autopsies (n = 15). The anatomical distribution of metastatic sites was as follows: lymph node (n = 6), lung (n = 13), pancreas (n = 2), liver (n = 1), bone (n = 2), brain (n = 3), pelvis or abdominal cavity (n = 2), and soft tissue (n = 2). Tissue of different metastatic sites was obtained in three patients. Tissue of the corresponding primary tumor was available in 19 patients.
Comparative Genomic Hybridization
Tumor DNA was extracted from 8 frozen and 24 formalin-fixed and paraffin-embedded metastases and from 8 frozen and 11 formalin-fixed and paraffin-embedded primary tumors. Specimens were trimmed to ensure a minimum of 75% tumor cells in the sample. Tissue preparation and DNA extraction was as described. 22,23 One microgram of tumor DNA was nick translated by using a commercial kit (BioNick kit, Life Technologies, Gaithersburg, MD) and Spectrum Green-dUTPs (Vysis, Downers Grove, IL) for direct labeling of tumor DNA. Spectrum-Red-labeled normal reference DNA (Vysis) was used for co-hybridization.
CGH and Digital Image Analysis
The hybridization mixture consisted of 200 ng of Spectrum-Green-labeled tumor DNA, 200 ng of Spectrum-Red-labeled normal reference DNA, and 20 μg of Cot-1 DNA (GIBCO, Gaithersburg, MD) dissolved in 10 μl of hybridization buffer (50% formamide, 10% dextran sulfate, 2X SSC, pH 7.0). Hybridization, image acquisition, image analysis, and control experiments were exactly as described. 22,23 At least four observations per autosome and two observations per sex chromosome were included in each analysis. Each CGH experiment included a tumor cell line (Spectrum Green MPE-600, Vysis) with known aberrations (positive control) and a hybridization of two differentially labeled sex mismatched normal DNAs to each other (negative control). A gain of DNA sequences was assumed at chromosomal regions where the hybridization resulted in a tumor to normal ratio >1.20. Over-representations were considered amplifications when the fluorescence ratio values exceeded 1.5 in a subregion of a chromosome arm. A loss of DNA sequences was presumed where the tumor-to-normal ratio was <0.80. To define an aberration it was additionally required that the first SD was above (gain) or below (deletion) 1.00. As some false aberrations were detected in normal tissues at 1p, 16p, 19, and 22, these G-C-rich regions, known to produce false positive results by CGH, were excluded from all analyses. 24
VHL Deletion by Microsatellite Analysis
The von Hippel-Lindau gene (VHL) on chromosome 3p25.5 is the strongest tumor suppressor gene candidate in RCC because somatic VHL gene mutations are present in ∼50% of RCCs 25 with 3p losses. Microsatellite analysis was used to detect VHL deletion in primary tumors and its metastases from 18 patients, where normal tissue was also available. The same tumor DNA specimens were used for CGH and microsatellite analysis. Normal DNA, extracted either from surrounding normal tissue or from liver tissue obtained at autopsy, was used in the loss of heterozygosity (LOH) analysis.
LOH analysis was performed using the polymorphic microsatellite markers D3S1350 26 and D3S1038, 27 mapping to the VHL region on chromosome 3p25-p26. Microsatellites were amplified from 50 to 150 ng of genomic DNA using 6 pmol of the corresponding primer pairs, with one primer carrying an IRD-41 label, in polymerase chain reaction (PCR) buffer (Perkin Elmer, Norwalk, CT) with 200 mmol/L each of dATP, dCTP, dGTP, and dTTP and 1.25 U of Taq DNA polymerase (Perkin Elmer) in a total reaction volume of 25 μl. PCR was performed on an Eppendorf Mastercycler 5330 using the following conditions: denaturation at 95°C for 5 minutes, 35 cycles with 95°C for 20 seconds, 62°C (D3S1350) or 56°C (D3S1038) for 20 seconds, and 72°C for 50 seconds, followed by a final extension step of 5 minutes at 72°C.
PCR products were separated on 5% LongRanger gels (FMC BioProducts, Denmark), and the band densities were measured using a LICOR DNA sequencer 4000 (MWG Biotech) and RFLPscan software (Scanalytics, CSPI). The percent integrated optical density values of the bands representing the two alleles were compared with each other and with those obtained from normal tissue. LOH was scored if the decrease in optical density between one band and the corresponding allele in normal tissue was >60%.
Statistics
Statistical differences in the prevalence of the most common gains and losses between the primary and metastatic tumors were analyzed using Fisher’s exact test.
The degree of clonal relationship (CR) between primary tumors and metastases by CGH was based on a probabilistic model developed by a mathematician (A. A. Schäffer). 20 Intuitively, the primary tumors and metastases should have a high probability to be clonally related if they share a set of gains and losses not likely to be shared at random. The following probabilistic model was developed to quantify this intuition.
1. Let a1, a2, a3, and so forth be the specific abnormalities. The probability that ai occurs is p(ai) = number of occurrences of ai/number of tumors.
2. P = set of aberrations in the primary tumor; M = set of aberrations in the metastasis. For any patient, the events in common are P ∩ M = c1, c2, c3,… ck.
3. p(ci) is the probability of the event (gain or loss) ci in the data set. The probability that the shared events occurred independently in the two tumors of the same patient is p(c1) × p(c2) × p(ck) = X. Thus, when the tumors have no events in common, the product defining X has no terms; it is standard in probability theory that such an empty product is defined as 1. The standard definition makes sense here because the events in common, the probability of a CR is estimated as 1 − X. Thus, when the tumors have no events in common, the probability of a CR is estimated as 0. Gains and losses were classified by chromosome arm, and the analysis ignored the band intervals.
Results
Genetic Alterations in RCC Metastases
There was a high number of genetic changes in RCC metastases. The mean number (±SD) of changes per specimen was 8.1 ± 6.7 (median, 7; range, 1 to 29). The mean number of losses per tumor was 4.6 ± 4.5 (median, 3.5; range, 0 to 19), the mean number of gain per tumor 3.6 ± 3.4 (median, 3; range, 0 to 18). The most common gains were seen at 8q (22%), 17q (31%), 20q, 21q (22% each), and Xq (28%). There was one high-level gene amplification at chromosome 11q22–23. The most common losses involved 2q (22%), 3p (25%), 4q (28%), 6q (28%), 8p (31%), 9p (47%), and 10q, 13q, and 18q (22% each).
Primary sites of metastatic dissemination in patients with RCC are lungs and lymph nodes. 2 In an effort to evaluate whether the pattern of metastatic dissemination of cancer is associated with a specific chromosomal alteration, we analyzed lymph node metastases, lung metastases, and other metastatic sites as groups. The number of aberrations in lung metastases (6.2 ± 4.0) was significantly lower than in other hematogenous metastases (11.5 ± 8.7, P < 0.05). Also, the number of aberrations in lymph node metastases (4.8 ± 2.8) was lower than in hematogenous metastases (P < 0.05), excluding lung metastases. The number of aberrations was not different in lung and lymph node metastases (P = 0.6). The most frequent aberrations were also tested for an association with the metastatic site. No significant differences were found between specific aberrations and location of metastasis.
Pairwise Analysis of Primary Tumors and Their Metastases
The most common losses in 19 primary tumors involved 3p (63%), 4q (26%), 6q (21%), and 9p (26%). Gains were frequently detected at 7p (21%) and 16q (32%). None of the genetic changes in primary tumors had a frequency difference with the metastases significant at the P < 0.05 level when evaluated as a group. Closest to a significant difference were 3p losses. Chromosomal losses on 3p occurred in 12 of 19 primary tumors (63%) but in only 8 of 32 metastases (25%).
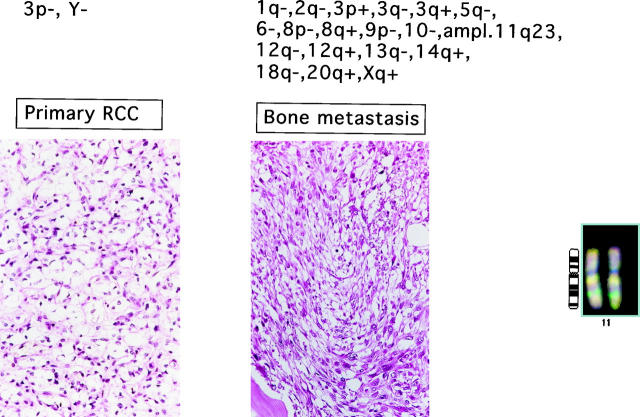
The pairwise analysis revealed that primary tumors and their corresponding metastases were never identical. The degree of clonal relationship between the primary and metastatic cell clones varied substantially from one patient to another. For example, several tumors and their metastases shared three to four genetic changes and had only one different aberration. At the other end of the spectrum, there was a metastasis with 23 genetic changes, and only two of them could be discovered in the corresponding primary tumor. One example for a tumor pair without shared genetic changes is shown in Figure 1 ▶ . In 14 patients, the metastases had more genetic aberrations than the corresponding primary tumor, whereas in 5 patients, fewer genetic alterations were found in the metastases. Genomic changes, which frequently occurred in metastases but not in the corresponding primary tumor, included 8p− (26%), 9p−, 17q+ (21% each), 21q+ (26%), and Xq+ (21%).
Figure 1.
Example of genetic aberrations in primary tumor and bone metastasis (femur). Gene amplification on 11q22–23 is shown on the right.
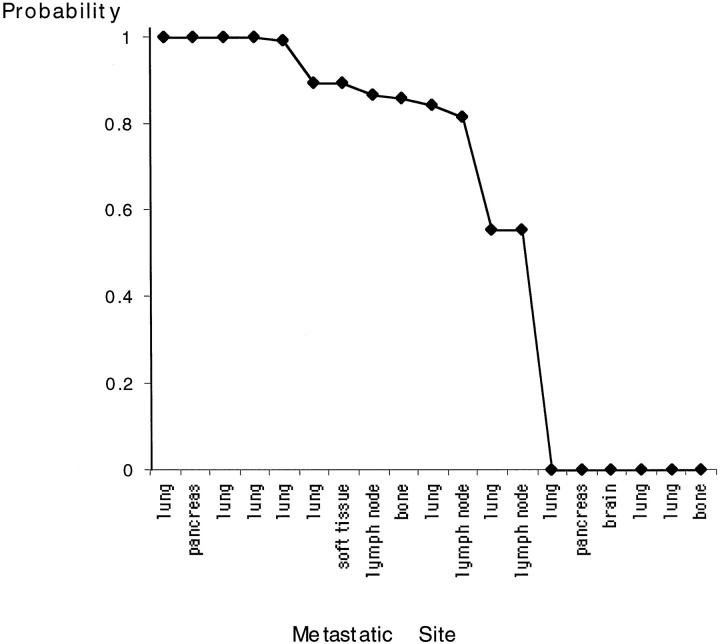
The number of shared genetic changes is a rough estimate of the degree of clonal relationship between primary tumor and metastasis. Commonly occurring genetic changes are likely to be shared between two specimens, whereas more infrequent changes provide strong evidence for a common clonal progenitor. A mathematical model was developed to more accurately quantify the degree of clonal relationship by estimating the probability that shared genetic changes in the paired specimens are not likely to be shared by chance alone. 20 According to this statistical analysis, 11 of the 19 paired specimens had a high probability (>0.8) for being clonally related. In seven of these cases, the probability was ≥0.9 (Figure 2) ▶ . In six patients (32%), the genetic changes were completely different between the primary and metastatic lesions, so that no probability of a common clonal progenitor emerged. In two cases, the few shared genetic changes between the two specimens were likely to be attributable to chance alone. No significant relationships were found in the clinical and pathological parameters and the degree of clonal relationship.
Figure 2.
Statistical probability of clonal relationship between primary tumor and metastasis of different locations.
CGH of Metastases at Different Locations
Metastases from different anatomical sites were available from three patients. The genetic composition of these metastases was never identical (Table 1) ▶ . In one patient, the lymph node metastasis had seven, the lung metastasis three, and a metastasis in the diaphragm had nine aberrations. Lymph node and lung metastases shared one (18q+), whereas lymph node and diaphragm metastasis shared four aberrations (8p−, 9p−, 10q, and 15q−). Compared with the primary tumor, additional alterations were detected in all three metastatic locations. Another patient with metastasis to lung and peritoneum showed 10 DNA-sequence copy number changes in the lung and 17 in the peritoneum. Both metastases shared seven aberrations (1p−, 6q−, 7p−, 8p−, 8q+, 11p−, and12q+). The peritoneal tumor revealed additional aberrations (1q+, 3p−, 5p+, 7q+, 11q+, 14q−, 15q−, 17p+, 17q+, and 20p−) that were not present in the lung. Brain and lung metastases of a third patient had no identical alterations. The number of aberrations in these metastases was 6 and 11 per tumor.
Table 1.
CGH of Metastases at Different Locations
| Patient 1 | Patient 2 | Patient 3 | |
|---|---|---|---|
| Primary tumor | 3p−, 4q−, 6q−, 9p−, 11q+, 18q−, | ||
| Lymph node | 8p−, 9p−, 9q−, 10q−, 15q−, 17p+, 17q+ | ||
| Lung | 5q+, 17q+, 18q+ | 1p−, 6q−, 7p−, 8p−, 8q+, 11p−, 12q+, 21q+, Xp−, Xq−, | 2q−, 3p−, 4p−, 4q−, 6q−, 9p−, 13q−, 17p+, Xp+, Xq+, Y− |
| Diaphragm | 4p−, 5q+, 8p−, 9p−, 9q−, 10q−, 12q+, 13q+, 15q− | ||
| Peritoneum | 1p−, 1q+, 3p−, 5p+, 6q−, 7p−, 7q+, 8p−, 8q+, 11p−, 11q+, 12q+, 14q−, 15q−, 17p+, 17q+, 20p− | ||
| Brain | 8p−, 2p+, 5p+, 5q+, 15q+, 17q+ |
VHL Deletion in Primary Tumors and Their Metastases
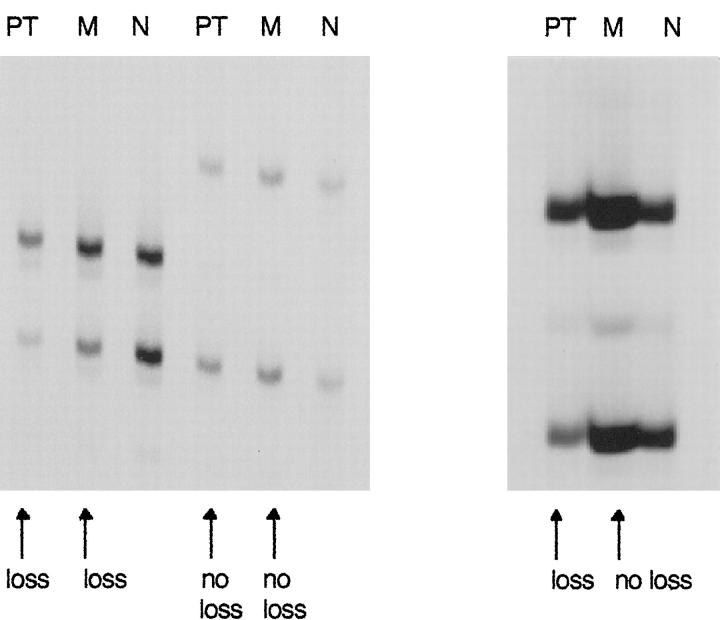
It has been suggested that chromosome 3p deletions are involved in initiation of clear-cell RCC. 28,29 If VHL loss is an early event in RCC carcinogenesis, LOH of VHL should be present in both primary tumor and metastasis. To further investigate the unexpected result of more 3p losses in primary tumors than in corresponding metastases, loss of heterozygosity (LOH) of VHL was evaluated, because microsatellite analysis is more sensitive than CGH. We analyzed LOH for VHL in all patients with paired specimens (Table 2) ▶ . One patient was excluded because no normal tissue was available. Thirteen patients were informative (heterozygous) for the VHL locus. The correspondence of 3p alterations in primary tumors and metastases was higher in the LOH analysis than in the CGH analysis. Seven primary tumors (54%) showed LOH for VHL. In metastases, LOH for VHL occurred in six cases (46%). There was only one tumor with a VHL deletion in the primary tumor but not in the corresponding metastasis (Figure 3) ▶ . Two additional metastases (11%) had microsatellite instability at the VHL locus.
Table 2.
LOH for VHL in 18 Matched Pairs of Primary RCC and Their Metastases
| Patient | Primary | Metastasis |
|---|---|---|
| 1 | LOH | LOH |
| 2 | n.i. | n.i. |
| 3 | LOH | LOH |
| 4 | − | − |
| 5 | LOH | LOH |
| 6 | n.i. | n.i. |
| 7 | LOH | LOH |
| 8 | LOH | LOH |
| 9 | − | − |
| 10 | − | MIN |
| 11 | LOH | LOH |
| 12 | − | − |
| 13 | n.i. | n.i. |
| 14 | − | − |
| 15 | LOH | − |
| 16 | n.i. | n.i. |
| 17 | n.a. | n.a. |
| 18 | − | MIN |
| 19 | n.i. | n.i. |
LOH, loss of heterozygosity; −, no loss of heterozygosity; n.i., not informative; MIN, microsatellite instability; n.a., no normal tissue available.
Figure 3.
Microsatellite analysis of three tumor pairs for LOH of the VHL gene in RCC. Left: LOH of VHL in primary tumor and metastasis. PT, primary tumor; M, metastasis; N, normal tissue. Middle: Primary tumor and metastasis without LOH of VHL. Right: LOH for VHL detected in primary tumor but not in metastasis.
Discussion
In this study, CGH was used to analyze the genetic basis underlying metastatic progression of RCC. It showed that metastases are genetically highly complex. The number of genetic aberrations detected in metastases (8.1 per tumor) was clearly higher than the number previously found in primary clear-cell (4.2 per tumor) RCC. 10 This is consistent with the theory that RCC progression from nonmetastatic primary tumors to metastasis is driven by an accumulation of genetic changes. 30 Several previous studies have suggested that a high number of CGH aberrations goes along with dedifferentiation and poor prognosis in RCC and other malignancies. 10,31,32
Hematogenous spread of RCC may occur 1) via the vena cava through the right heart to the lungs, 2) retrograde to pelvic structures, or 3) by paravertebral veins (Batson’s plexus). 3 After passing through the pulmonary circulation, tumor cells spread to multiple sites. It has been suggested that dissemination of RCC occurs stepwise from the lungs to downstream organs. 2 Our finding that lung metastases have lower numbers of aberrations than other hematogenous metastases is consistent with the hypothesis that progression from the lung to other locations is associated with an accumulation of genetic changes. However, our results on multiple metastases in other organs (eg, brain) indicate that in some cases lung metastases and metastases in other organs may be genetically very different, suggesting that there is no linear relationship between the metastases. Instead, it might be that different metastases originate from different primary tumor clones. One might also speculate that only few specific genetic changes in the primary tumors are necessary for metastasis whereas more complex genetic aberrations may originate in the metastasis itself.
The CGH findings from those patients with paired specimens allowed us to study the clonal relationship between primary tumors and their metastases. Although most samples showed a high degree of concordance between primary tumors and metastases, some metastases had no or little evidence of clonal relationship. One explanation for this finding could be the selection pressure of the tissue growth environment, for example, immune surveillance and hypoxia. Growth conditions may differ at varying metastatic sites, 33 giving growth advantage to different cell clones in primary tumors and metastases.
The absence of shared genetic changes by CGH also includes the possibility that a clonal relationship was missed because of genetic heterogeneity within the primary tumor. Previous flow cytometric analyses have illustrated marked intratumor heterogeneity in RCC. 34-37 However, cytogenetic studies of different renal tumor samples of primary RCC suggest that besides different clonal abnormalities shared cytogenetic abnormalities are present. 38 In a previous CGH analysis of a sarcomatoid and an epithelioid tumor area within one tumor, 31 we found six identical aberrations, also suggesting that despite genetic heterogeneity evidence for a clonal relationship can be found even in morphologically heterogeneous primary tumors.
Interestingly, there were 5 of 19 metastases with fewer alterations than in the corresponding primary tumor. This is consistent with previous cytogenetic findings in breast cancer, indicating that simultaneous lymph node metastases are genetically less complex than the primary breast carcinoma, possibly because only one or few of the multiple clones in the primary tumor metastasize. 39
There are few data available relating specific genetic events to the metastatic behavior in RCC. To identify genetic changes that are linked to metastasis, we have investigated pairs of primary tumors and metastases. We assume that cell clones with metastasis-specific genetic changes may represent minor cell populations in the primary tumor that are not detectable by CGH. However, chromosomal alterations with relevance for the metastatic process should be enriched in the metastatic cells. Therefore, chromosomal changes might be especially relevant for the metastatic process if they are identified in metastases but not in the corresponding primary tumors.
Losses of chromosome 8p and 9p and gains of 17q, 21q, and Xq were frequently found in metastases, but not in matching primary tumors. A disturbed function of genes in these regions of the genome may therefore contribute to the metastatic process. Suggested tumor suppressor genes on 9p include p16 (CDKN2) and p15 (MTS2), all mapped to 9p21. 40,41 Inactivation of CDKN2 by homozygous deletion, rearrangement, or point mutation was found to be rare in primary RCC. 42 However, Kinoshita et al detected a hemi- or homozygous deletion of CDKN2 in three of five RCC metastases, 11 suggesting that a gene in the 9p21–22 region could be linked to RCC metastasis. Interestingly, acquisition of p16 and p15 gene mutations takes place during metastases of non-small-cell lung cancer. 43 An association between chromosome 17 gains and lymph node metastasis has been demonstrated in breast cancer, 44 suggesting that oncogenes on chromosome 17 might play a role for the metastatic process. Previous studies have shown that amplification and overexpression of the HER-2/neu gene on chromosome 17q21–22 is of minor importance in the oncogenesis of RCC. 45 Chromosome Xq gains have been rarely detected in solid tumors, except prostate cancer and mediastinal B-cell lymphoma. 46,47 Previously, it was shown that gains of chromosome Xq were enriched in cells of of breast cancer metastases, suggesting that tumors with Xq gain have a higher risk to metastasize. 20
Remarkably, 3p losses were detected in 12 of 19 primary tumors (63%) but in only 8 of 32 metastases (25%). This finding is completely consistent with previous results of Gronwald et al, detecting 3p losses in 71% of primary tumors but in only 20% of metastases by CGH. 9 This finding suggested that tumor cell populations without relative copy number alterations on 3p may have a growth advantage during metastasis. Using fluorescence in situ hybridization (FISH), Gronwald et al detected 3p losses in most cells of the primary tumors but only in a minority of the metastatic depots. They hypothesized that a second copy of 3p can be regained as a result of mitotic reduplication of the remaining allele of 3p. The hypothesis of mitotic recombination in some RCC metastases is consistent with the results of our microsatellite analysis, which revealed in all but one tumor pair no difference in heterozygosity of the VHL gene. Such a duplication of the remaining allele could provide a growth advantage by an elevated expression of a putative oncogene on 3p. The observations of high-level amplifications at 3p in bladder (3p24) 22 and breast cancer (3p14) 21 are arguments for the existence of at least one putative oncogene on 3p. Interestingly, a loss of 3p in the primary tumor and balanced state in the metastatic tumor was also observed previously in a case of breast carcinoma. 32
High-level amplifications may highlight locations of dominant oncogenes involved in tumor progression. In this study, a high-level amplification at 11q22–23 was found in 1 of 32 metastases of clear-cell RCC. In contrast to the matched primary tumor, this metastasis showed large areas with sarcomatoid differentiation. As we have previously detected an amplification also at 11q22–23 in the sarcomatoid component of a chromophobe RCC, 31 it is tempting to speculate that activation of oncogenes at these loci might have contributed to sarcomatoid transformation. 11q22–23 amplifications have been rarely detected in solid tumors. They were found in malignant mesothelioma, glioma, osteosarcoma, malignant fibrous histiocytoma, and lung and cervical cancer (for review see Ref. 48 ). Several potential oncogenes exist on 11q22–23, including the human proto-oncogene v-cbl, the HRX proto-oncogene, and the VACM-1 gene. VACM-1 belongs to a family of genes involved in cell cycle regulation. The 11q22–23 region also includes genes of the matrix metalloproteinase family. Importantly, previous studies suggested that production of metalloproteinase-2 increases the metastatic potential of RCC cell lines. 49,50
In summary, the present study shows that multiple, genetically almost completely different clones exist in various tumor locations of one and the same patient. The considerable genetic heterogeneity between primary RCC and metastases at various locations may underlie its poor responsiveness to therapy and explains why biomarkers of prognosis measured exclusively in primary tumors give a restricted view of the biological potential of RCC.
Acknowledgments
We thank C. Egenter, M. Mirlacher, H. Novotny, H. Ogier, M. Storz, and the staff of the Institute for Pathology, University Basel, for their technical support.
Footnotes
Address reprint requests to Dr. Holger Moch, Institut für Pathologie der Universität Basel, Schönbeinstrasse 40, CH-4003 Basel, Switzerland. E-mail: moch@ubaclu.unibas.ch.
Supported by the Swiss National Science Foundation (31–50752.97), Krebsforschung Schweiz (KFS 367-9-1996), and Basler Krebsliga (3/97).
H. Bissig and J. Richter contributed equally to this work.
References
- 1.Robson C, Churchill B, Anderson W: The results of radical nephrectomy for renal cell carcinoma. J Urol 1969, 101:297-303 [DOI] [PubMed] [Google Scholar]
- 2.Weiss L, Harlos JP, Torhorst J, Gunthard B, Hartveit F, Svendsen E, Huang WL, Grundmann E, Eder M, Zwicknagl M: Metastatic patterns of renal carcinoma: an analysis of 687 necropsies. J Cancer Res Clin Oncol 1988, 114:605-612 [DOI] [PubMed] [Google Scholar]
- 3.Bennington J, Beckwith J: Tumors of the Kidney, Renal Pelvis and Ureter. 1975. Armed Forces Institute of Pathology, Washington, DC,
- 4.Johnsen JA, Hellsten S: Lymphatogenous spread of renal cell carcinoma: an autopsy study. J Urol 1997, 157:450-453 [PubMed] [Google Scholar]
- 5.Gastl G, Finstad CL, Guarini A, Bosl G, Gilboa E, Bander NH, Gansbacher B: Retroviral vector-mediated lymphokine gene transfer into human renal cancer cells. Cancer Res 1992, 52:6229-6236 [PubMed] [Google Scholar]
- 6.Trent JM, Stanisic T, Olson S: Cytogenetic analysis of urologic malignancies: study of tumor colony forming cells and premature chromosome condensation. J Urol 1984, 131:146-151 [DOI] [PubMed] [Google Scholar]
- 7.Sato M, Hattori T, Nishimura T, Akimoto M: Characterization of primary and metastatic cell lines established from a patient with renal cell carcinoma. Nippon Hinyokika Gakkai Zasshi 1993, 84:650-655 [DOI] [PubMed] [Google Scholar]
- 8.Peier AM, Meloni AM, Sandberg AA, Leong SP, Carroll PR: Cytogenetic findings in a metastatic renal cell carcinoma. Cancer Genet Cytogenet 1995, 80:168-169 [DOI] [PubMed] [Google Scholar]
- 9.Gronwald J, Störkel S, Holtgreve-Grez H, Hadaczek P, Brinkschmidt C, Jauch A, Lubinski J, Cremer T: Comparison of DNA gains and losses in primary renal clear cell carcinomas and metastatic sites: importance of 1q and 3p copy number changes in metastatic events. Cancer Res 1997, 57:481-487 [PubMed] [Google Scholar]
- 10.Moch H, Presti JC, Jr, Sauter G, Buchholz N, Jordan P, Mihatsch MJ, Waldman FM: Genetic aberrations detected by comparative genomic hybridization are associated with clinical outcome in renal cell carcinoma. Cancer Res 1996, 56:27-30 [PubMed] [Google Scholar]
- 11.Kinoshita H, Yamada H, Ogawa O, Kakehi Y, Osaka M, Nakamura E, Mishina M, Habuchi T, Takahashi R, Sugiyama T, Yoshida O: Contribution of chromosome 9p21–22 deletion to the progression of human renal cell carcinoma. Jpn J Cancer Res 1995, 86:795-799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhlman D, Nguyen P, Manivel J, Zhang G, Hagen K, Fraley E, Aeppä D, Niehans G: Epidermal growth factor receptor and transforming growth factor α expression in papillary and nonpapillary renal cell carcinoma: correlation with metastatic behavior and prognosis. Clin Cancer Res 1995, 1:913-920 [PubMed] [Google Scholar]
- 13.Uhlman D, Nguyen P, Manivel J, Aeppli D, Resnick J, Fraley E, Zhang G, Niehans G: Association of immunohistochemical staining for p53 with metastatic progression and poor survival in patients with renal cell carcinoma. J Natl Cancer Inst 1994, 86:21470-147524 [DOI] [PubMed] [Google Scholar]
- 14.Sauter G, Moch H, Gasser T, Mihatsch M, Waldman F: Heterogeneity of chromosome 17 and erbB-2 gene copy number in primary and metastatic bladder cancer. Cytometry 1995, 21:40-46 [DOI] [PubMed] [Google Scholar]
- 15.Pandis N, Heim S, Bardi G, Idvall I, Mandahl N, Mitelman F: Chromosome analysis of 20 breast carcinomas: cytogenetic multiclonality and karyotypic-pathologic correlations. Genes Chromosomes & Cancer 1993, 6:51-57 [DOI] [PubMed] [Google Scholar]
- 16.Kallioniemi OP: Comparison of fresh and paraffin-embedded tissue as starting material for DNA flow cytometry and evaluation of intratumor heterogeneity. Cytometry 1988, 9:164-169 [DOI] [PubMed] [Google Scholar]
- 17.Trent J, Yang JM, Emerson J, Dalton W, McGee D, Massey K, Thompson F, Villar H: Clonal chromosome abnormalities in human breast carcinomas. II. Thirty-four cases with metastatic disease. Genes Chromosomes & Cancer 1993, 7:194-203 [DOI] [PubMed] [Google Scholar]
- 18.Waghorne C, Thomas M, Lagarde A: Genetic evidence for progressive selection and overgrowth of primary tumors by metastatic cell subpopulations. Cancer Res 1988, 48:6109-6114 [PubMed] [Google Scholar]
- 19.Bell C, Frost P, Kerbel R: Cytogenetic heterogeneity of genetically marked and metastatically competent “dominant” tumor cell clones. Cancer Genet Cytogenet 1991, 54:153-161 [DOI] [PubMed] [Google Scholar]
- 20.Kuukasjärvi T, Karhu R, Tanner M, Kähkönen M, Schäffer A, Nupponen N, Pennanen S, Kallioniemi A, Kallioniemi O, Isola J: Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res 1997, 57:1597-1604 [PubMed] [Google Scholar]
- 21.Kallioniemi A, Kallioniemi O, Sudar D, Rutovitz D, Gray J, Waldman F, Pinkel D: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258:818-821 [DOI] [PubMed] [Google Scholar]
- 22.Richter J, Jiang F, Görög J, Sartorius G, Egenter C, Gasser T, Moch H, Mihatsch M, Sauter G: Marked genetic differences between stage pTa and stage pT1 papillary bladder cancer detected by comparative genomic hybridization. Cancer Res 1997, 57:2860-2864 [PubMed] [Google Scholar]
- 23.Jiang F, Richter J, Schraml P, Bubendorf L, Gasser T, Mihatsch M, Sauter G, Moch H: Chromosomal imbalances in papillary renal cell carcinoma: genetic differences between histological subtypes. Am J Pathol 1998, 153:1467-1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kallioniemi O, Kallioniemi A, Piper J, Isola J, Waldman F, Gray J, Pinkel D: Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes & Cancer 1994, 10:231-243 [DOI] [PubMed] [Google Scholar]
- 25.Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM, Lubensky I, Duan DR, Florence C, Pozzatti R, Walther MM, Bander NH, Grossman HB, Brauch H, Pomer S, Brooks JD, Isaacs WB, Lerman MI, Zbar B, Linehan WM: Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet 1994, 7:85-90 [DOI] [PubMed] [Google Scholar]
- 26.Li H, Schmidt L, Wei MH, Hustad T, Lerman MI, Zbar B, Tory K: Three tetrameric repeat polymorphisms on human chromosome 3: D3S1349; D3S1350; D3S1351. Hum Mol Genet 1993, 2:819. [DOI] [PubMed] [Google Scholar]
- 27.Crossey PA, Maher ER, Jones MH, Richards FM, Latif F, Phipps ME, Lush M, Foster K, Tory K, Green JS, Oostra B, Yates JRW, Linehan WM, Affara NA, Lerman M, Zbar B, Nakamura Y, Ferguson-Smith MA: Genetic linkage between von Hippel-Lindau disease and three microsatellite polymorphisms refines the localisation of the VHL locus. Hum Mol Genet 1993, 2:279-282 [DOI] [PubMed] [Google Scholar]
- 28.Zbar B, Brauch H, Talmadge C, Linehan M: Loss of alleles of loci on the short arm of chromosome 3 in renal cell carcinoma. Nature 1987, 327:721-727 [DOI] [PubMed] [Google Scholar]
- 29.van den Berg A, Buys C: Involvement of multiple loci on chromosome 3 in renal cell cancer development. Genes Chromosomes & Cancer 1997, 19:59-76 [DOI] [PubMed] [Google Scholar]
- 30.Presti J, Rao H, Chen Q, Reuter V, Li F, Fair W, Jhanwar S: Histopathological, cytogenetic, and molecular characterization of renal cortical tumors. Cancer Res 1991, 51:1544-1552 [PubMed] [Google Scholar]
- 31.Jiang F, Moch H, Richter J, Gasser T, Gschwind R, Egenter C, Bubendorf L, Sauter G, Mihatsch M: Comparative genomic hybridization reveals frequent chromosome 13q and 4q losses in renal cell carcinomas with sarcomatoid transformation. J Pathol 1998, 185:382-388 [DOI] [PubMed] [Google Scholar]
- 32.Isola J, Kallioniemi O, Chu L, Fuqua S, Hilsenbeck S, Osborne K, Waldman F: Genetic aberrations detected by comparative genomic hybridization predict outcome in node-negative breast cancer. Am J Pathol 1995, 147:905-911 [PMC free article] [PubMed] [Google Scholar]
- 33.Nowell PC: The clonal evolution of tumor cell populations. Science 1976, 194:23-28 [DOI] [PubMed] [Google Scholar]
- 34.Kushima M, Kushima R, Hattori T, Tomoyoshi T: Heterogeneity and progression of renal cell carcinomas as revealed by DNA cytofluorometry and the significance of the presence of polyploid cells. Urol Res 1995, 23:381-386 [DOI] [PubMed] [Google Scholar]
- 35.Ljungberg B, Stenling R, Roos G: DNA content in renal cell carcinoma with reference to tumor heterogeneity. Cancer 1985, 56:503-508 [DOI] [PubMed] [Google Scholar]
- 36.Baretton G, Kuhlmann B, Krech R, Lohrs U: Intratumoural heterogeneity of nuclear DNA-content and proliferation in clear cell type carcinomas of the kidney. Virchows Arch B Cell Pathol 1991, 61:57-63 [DOI] [PubMed] [Google Scholar]
- 37.Ljungberg B, Nordenson I, Roos G: Cytogenetic and flow cytometric DNA analysis in renal cell carcinoma. Eur Urol 1991, 19:59-64 [DOI] [PubMed] [Google Scholar]
- 38.Nordenson I, Ljungberg B, Roos G: Chromosomes in renal carcinoma with reference to intratumor heterogeneity. Cancer Genet Cytogenet 1988, 32:35-41 [DOI] [PubMed] [Google Scholar]
- 39.Teixeira M, Bardi G, Andersen J, Heim S: Karyotypic comparisons of multiple tumorous and macroscopically normal surrounding tissue samples from patients with breast cancer. Cancer Res 1996, 56:855-859 [PubMed] [Google Scholar]
- 40.Cairns P, Mao L, Merlo A, Lee DJ, Schwab D, Eby Y, Tokino K, van der Riet P, Blaugrund JE, Sidransky D: Rates of p16 (MTS1) mutations in primary tumors with 9p loss. Science 1994, 265:415-417 [DOI] [PubMed] [Google Scholar]
- 41.Kamb A, Liu Q, Harshman K, Tavtigian S, Cordon-Cardo C, Skolnick M: Rates of p16 (MTS1) mutations in primary tumors with 9p loss. Science 1994, 264:416-417 [Google Scholar]
- 42.Cairns P, Tokino K, Eby Y, Sidransky D: Localization of tumor suppressor loci on chromosome 9 in primary human renal cell carcinomas. Cancer Res 1995, 55:224-227 [PubMed] [Google Scholar]
- 43.Okamoto A, Hussain SP, Hagiwara K, Spillare EA, Rusin MR, Demetrick DJ, Serrano M, Hannon GJ, Shiseki M, Zariwala M, Xiong Y, Beach DH, Yokota J, Harris CC: Mutations in the p16INK4/MTS1/CDKN2, p15INK4B/MTS2, and p18 genes in primary and metastatic lung cancer. Cancer Res 1995, 55:1448-1451 [PubMed] [Google Scholar]
- 44.Herrington CS, Leek RD, McGee JO: Correlation of numerical chromosome 11 and 17 imbalance with metastasis of primary breast cancer to lymph nodes. J Pathol 1995, 176:353-359 [DOI] [PubMed] [Google Scholar]
- 45.Weidner U, Peter S, Strohmeyer T, Hussnatter R, Ackermann R, Sies H: Inverse relationship of epidermal growth factor receptor and HER2/neu gene expression in human renal cell carcinoma. Cancer Res 1990, 50:4504-4509 [PubMed] [Google Scholar]
- 46.Joos S, Otano Joos MI, Ziegler S, Bruderlein S, du Manoir S, Bentz M, Moller P, Lichter P: Primary mediastinal (thymic) B-cell lymphoma is characterized by gains of chromosomal material including 9p and amplification of the REL gene. Blood 1996, 87:1571-1578 [PubMed] [Google Scholar]
- 47.Nupponen NN, Kakkola L, Koivisto P, Visakorpi T: Genetic alterations in hormone-refractory recurrent prostate carcinomas. Am J Pathol 1998, 153:141-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knuutila S, Bjorkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy ML, Tapper J, Pere H, El-Rifai W, Hemmer S, Wasenius VM, Vidgren V, Zhu Y: DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol 1998, 152:1107-1123 [PMC free article] [PubMed] [Google Scholar]
- 49.Gohji K, Nomi M, Hara I, Arakawa S, Kamidono S: Influence of cytokines and growth factors on matrix metalloproteinase-2 production and invasion of human renal cancer. Urol Res 1998, 26:33-37 [DOI] [PubMed] [Google Scholar]
- 50.Miyake H, Hara I, Yoshimura K, Eto H, Arakawa S, Wada S, Chihara K, Kamidono S: Introduction of basic fibroblast growth factor gene into mouse renal cell carcinoma cell line enhances its metastatic potential. Cancer Res 1996, 56:2440-2445 [PubMed] [Google Scholar]