Abstract
Efficient splicing of the td group I intron in vivo is dependent on the ribosome. In the absence of translation, the pre-mRNA is trapped in nonnative-splicing-incompetent conformations. Alternatively, folding of the pre-mRNA can be promoted by the RNA chaperone StpA or by the group I intron-specific splicing factor Cyt-18. To understand the mechanism of action of RNA chaperones, we probed the impact of StpA on the structure of the td intron in vivo. Our data suggest that StpA loosens tertiary interactions. The most prominent structural change was the opening of the base triples, which are involved in the correct orientation of the two major intron core domains. In line with the destabilizing activity of StpA, splicing of mutant introns with a reduced structural stability is sensitive to StpA. In contrast, Cyt-18 strengthens tertiary contacts, thereby rescuing splicing of structurally compromised td mutants in vivo. Our data provide direct evidence for protein-induced conformational changes within catalytic RNA in vivo. Whereas StpA resolves tertiary contacts enabling the RNA to refold, Cyt-18 contributes to the overall compactness of the td intron in vivo.
Keywords: Group I intron, RNA structure, in vivo splicing, DMS probing in vivo, RNA chaperone StpA, aminoacyl-tRNA-synthetase Cyt-18
Most RNAs have to fold into a defined native structure in order to fulfill their function. Because RNA supports a variety of non-Watson-Crick base pairs, it becomes difficult for a population of RNA molecules to adopt a single structure. As a consequence, RNAs tend to misfold in vitro and in vivo (Lindahl and Adams 1966; Walstrum and Uhlenbeck 1990; Woodson and Cech 1991; Pan and Woodson 1998; Semrad and Schroeder 1998; Treiber et al. 1998; Nikolcheva and Woodson 1999; Treiber and Williamson 2001). A variety of methods—hydroxyl-radical footprinting, oligonucleotide hybridization, UV melting, native gel electrophoresis, X-ray synchrotron analysis, and single molecule fluorescence measurements—were used to study the folding pathway of the Tetrahymena group I intron in vitro (Zarrinkar and Williamson 1994, 1996; Banerjee and Turner 1995; Rook et al. 1998; Sclavi et al. 1998; Treiber et al. 1998; Russell et al. 2000; Zhuang et al. 2000; Russell and Herschlag 2001). These studies lead to a model in which the folding pathways are part of a rugged free-energy landscape. This ruggedness is due to the multiple folding pathways a molecule can use to achieve the native state. These pathways are interrupted by various kinetic traps resulting from nonnative structures. In vitro this folding problem is solved by precise renaturation protocols, which include heating procedures to resolve misfolded structures. There are two possibilities how proteins can assist RNA in achieving a unique native conformation. From in vitro experiments it has been proposed that they either act as RNA chaperones by resolving nonnative conformations without recognizing specific structures or sequences (Herschlag 1995) or they bind specifically to an RNA and recognize a defined 3D structure, thereby stabilizing it through tight binding (Weeks 1997). The Escherichia coli proteins S12 and StpA can act as RNA chaperones in the folding process of intron RNA both in vitro and in vivo (Coetzee et al. 1994; Zhang et al. 1995, 1996; Clodi et al. 1999), whereas the proteins Cyt-18 and CBP2, as well as the I-AniI maturase interact specifically with group I intron RNA (Weeks and Cech 1995; Caprara et al. 1996a; Solem et al. 2002). Several other proteins have been reported to exert RNA chaperone activity in vitro, like the nucleocapsid protein of HIV NCp7, the A1 hnRNP, the cold-shock protein CspA, the host factor I protein Hfq, the yeast La protein, and the tumor suppressor protein p53 (Tsuchihashi et al. 1993; Herschlag et al. 1994; Jiang et al. 1997; Nedbal et al. 1997; Pannone et al. 1998; Clodi et al. 1999; Negroni and Buc 2000; Zhang et al. 2002).
The E. coli protein StpA has been assigned RNA chaperone activity due to its ability to accelerate trans-splicing of the T4 phage thymidylate synthase (td) group I intron in vitro, as well as to stimulate RNA annealing and RNA dissociation (Zhang et al. 1995, 1996). The stpA gene was isolated as a suppressor of the Td− phenotype of a splicing-defective td intron (Zhang and Belfort 1992). In vivo it rescues folding of the td pre-mRNA in mutants that are kinetically trapped in nonproductive conformations due to nonsense codons in the upstream exon (Semrad and Schroeder 1998; Clodi et al. 1999). StpA has been shown recently to induce strand transfer during primer extension by the HIV reverse transcriptase, suggesting that proteins with RNA chaperone activity might confer a higher genetic flexibility to retroviruses (Negroni and Buc 2000).
Here, we undertook a first approach to analyze the effect of the protein StpA on RNA structure by probing the conformation of the td intron in vivo with dimethyl sulfate (DMS). We compared the modification patterns of the td RNA in the absence and presence of the RNA chaperone StpA in vivo. Additionally, the td intron structure was probed in the absence and presence of the aminoacyl-tRNA synthetase Cyt-18, a protein that binds specifically to the core of group I introns (Caprara et al. 1996a, 2001). StpA has a major effect on junction J3/4, which is part of the base triple interactions responsible for the correct orientation of the P4/P6 domain relative to the P3/P8 domain (Michel et al. 1990; Green and Szostak 1994; Cate et al. 1996). The same residues in J3/4 have been implicated previously in the activity of Cyt-18 (Myers et al. 1996; Chen et al. 2000). We showed that both proteins affect the modification status of the adenines in J3/4, but do so in opposite ways. Our observations were further underlined by an analysis of the effect of these proteins on the conformation and splicing activity of structurally destabilized td intron mutants.
Results
Secondary structure of the td intron in vivo
DMS readily enters the E. coli cell and methylates accessible bases in RNA and DNA (Zaug and Cech 1995; Wells et al. 2000). Methylation occurs at positions essential for Watson-Crick base pairing, N1 of adenines and N3 of cytidines, and can be detected because they provoke premature stops during primer extension. The structure of the td wild-type intron has been probed recently in vivo (C. Waldsich, B. Masquida, E. Westhof, and R. Schroeder, unpubl. data). The in vivo DMS modification pattern is in good agreement with the structural model based on phylogeny and biochemical in vitro data (Michel and Westhof 1990, 1994; Jaeger et al. 1994). Of interest for the results described here is the modification status of residues in junctions J3/4 and J8/7, in loops L9 and L9.2, and in stem P3, which are all involved in tertiary interactions. These residues are weakly to moderately modified and these results are summarized in Figure 1A.
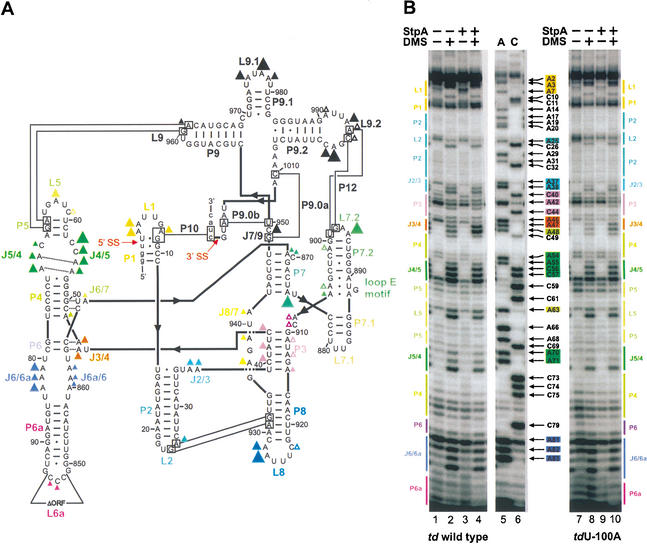
Figure 1.
In vivo DMS modification pattern of the td group I intron. (A) Structural model of the td intron based on phylogeny and biochemical data obtained in vitro (Michel and Westhof 1990; Cech et al. 1994; Leontis and Westhof 1998). Residues modified by DMS in vivo are shown as triangles. The different size of the triangles indicates the relative modification intensities. Secondary structure elements are termed P1–P10, joining regions between stems are termed J2/3–J8/7, and hairpin loops are numbered L1–L9.2. Long-range tertiary interactions are indicated by thin black lines and the base triple interactions and the sheared A-A base pairs in the P4/P5 internal loop are indicated by dashed lines. The splice sites (5′ and 3′ SS) are marked with red arrows. The deletion of the intron ORF is also indicated in loop L6a. Intron sequence is displayed in uppercase letter and exon sequences are shown as lowercase letters. The following color code is used: yellow, P1; turquoise, P2; purple, P3; green, P4/P5 domain and P7 extension; blue-purple, P6 extension; blue, P8; and black, P9 extension. (B) Intron residues of the P4/P6 domain, which are accessible to DMS, are displayed in these gels. The DMS modification pattern of the wild-type intron is shown in the left panel and of the nonsense codon mutant tdU-100A in the right panel. Boxed nucleotides correspond to positions within the intron that are modified by DMS (for the color code, see A). A and C denote the sequencing lanes.
Because the wild-type td construct splices to a high extent, the population of RNA molecules that was probed with DMS in vivo was mostly circular intron RNA. This was determined by primer extension with a primer complementary to the P2 region, which extends to the 5′/3′ junction of the circular intron and not to the upstream exon. To probe the structure of the intron in a population of molecules that are in part unspliced, td mutants containing a nonsense codon in the upstream exon (tdG-51U, tdU-100A, tdU-223A) were also subjected to in vivo DMS modification. Figure 1B shows the DMS modification pattern of the P4/P6 domain of the td wild type (lanes 1,2) and of the nonsense codon mutant tdU-100A (lanes 7,8). The DMS modification pattern of wild-type excised intron did not differ from the intron modification pattern of mutant pre-mRNAs (Fig. 1B; data not shown). This indicates that the structure of the circular intron, with exception of the splices sites, is indistinguishable from that of an intron still residing within the precursor RNA. Thus, the nonsense codon in the upstream exon does not influence the intron core structure. Nevertheless, all subsequently described experiments in which the structure of the td intron was probed in the presence of either StpA or Cyt-18 were performed both with the wild-type td construct and with at least one of the mutants containing a nonsense codon in the upstream exon. The same effects of StpA or Cyt-18 on the structure of the td intron, which will be described here, were observed for the wild-type excised intron, as well as for the nonsense codon containing mutant pre-mRNAs (Fig. 1B, lanes 3,4,9,10).
StpA rescues splicing of exonic td mutants containing nonsense codons
Efficient splicing of the td intron in vivo is dependent on the ribosome (Semrad and Schroeder 1998; Waldsich et al. 1998). Uncoupling splicing from translation by introducing nonsense codons in the upstream exon reduces splicing of these mutant pre-mRNAs compared to the wild type. In the absence of translation, the td pre-mRNA becomes trapped in nonnative splicing-incompetent conformations, resulting in a reduced splicing activity. We had shown previously that alternatively to the ribosome, proteins with RNA chaperone activity such as StpA are able to promote correct folding of the td pre-mRNA, thereby rescuing splicing of a nonsense codon mutant (Clodi et al. 1999). To test whether this effect of StpA also applies to other exonic nonsense codon mutants, we measured the splicing activity of mutants with nonsense codons 51, 100, and 223 nucleotides upstream of the 5′ splice site in the absence and presence of StpA. StpA indeed promotes splicing of all these mutants in vivo (Fig. 2A–C).
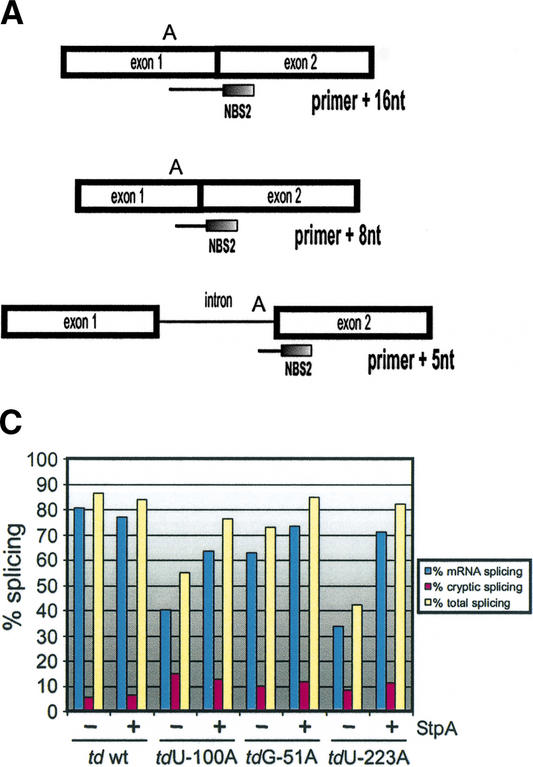
Figure 2.
Splicing activity of td mutants in the absence and presence of the RNA chaperone StpA. (A) Schematic representation of the in vivo splicing assay. Primer extension with the oligonucleotide NBS2, which hybridizes to the 5′ end of the downstream exon, terminates at the first A in the template in the presence of ddTTP as follows: primer + 16 nt corresponds to the mRNA, primer + 8 nt to the cryptic spliced RNA, and primer + 5 nt to the pre-mRNA. (B) Representative gels display the splicing activity of the td wild type and of exonic nonsense codon mutants in the presence and absence of StpA. Comparison of the mRNA extension product (primer + 16 nt) with the pre-mRNA extension product (primer + 5 nt) allows the determination of the splicing activity. The primer + 8 nt product is due to an alternative 5′ splice site, which is defined by the formation of a cryptic P1 stem also serving as a substrate for the intron core (Chandry and Belfort 1987). (C) Quantification of the individual experiments shown in B. Blue bars, % mRNA splicing; purple bars, % cryptic splicing; and yellow bars, % total splicing. (D) Western blot analysis showing the endogenous levels of StpA compared to those of cells transformed with the plasmid pSU20/StpA encoding StpA. The StpA levels are shown for the wild-type td intron and for the nonsense codon mutants, each in the absence and in the presence of an additional plasmid-encoded StpA gene. As a reference, 0.3 μg of purified StpA protein are displayed.
StpA expression levels in E. coli cells containing in addition to the endogenously expressed StpA either an empty vector or the StpA-encoding plasmid were analyzed by immunoblotting using antibodies against a C-terminal StpA peptide. An ∼10-fold increase in StpA levels was detected in cells coexpressing StpA, as compared to cells containing the vector control (Fig. 2D). However, it should be mentioned, that a weak cross-reaction of the antibody with the highly related StpA paralog H-NS cannot be precluded and may contribute to signals seen in the immunoblots. Samples corresponding to 500 μL of cells coexpressing StpA at an OD600 of 0.2 give a signal approximately equivalent to 0.2 μg of purified StpA, whereas the signal of endogenously expressed StpA corresponds to 0.02 μg StpA (Fig. 2D; data not shown). Furthermore, no fluctuation of StpA levels could be observed among samples from cells additionally containing different plasmid-encoded td alleles. Therefore, the presence of different td alleles in the cell does not influence the StpA protein amount in cells.
Opposite structural effects of StpA versus Cyt-18 on essential base triple interactions in the td intron
Although StpA is not able to rescue splicing-deficient intronic td mutants that have a single nucleotide exchange in stem P7, the Neurospora crassa aminoacyl-tRNA-synthetase Cyt-18 is known to rescue splicing of various td intron mutants and of td exonic nonsense codon mutants (Mohr et al. 1992; Myers et al. 1996; Semrad and Schroeder 1998; Clodi et al. 1999; Chen et al. 2000). Cyt-18 specifically recognizes and binds to the 3D shape of the td intron core, thereby inducing the formation of the correct structure. Thus, there is a significant difference between the effects of StpA and Cyt-18 with respect to their ability to rescue exonic and intronic td mutants.
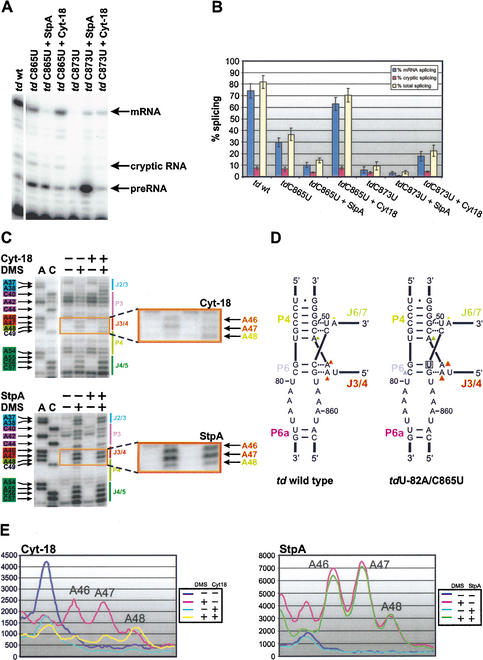
To study the differences in binding as well as in the mechanism of action of StpA and Cyt-18, we compared the DMS modification patterns of the td intron in the absence and presence of either protein in vivo (Fig. 3). The most prominent changes in the presence of either StpA or Cyt-18 are found at positions A46 and A47 in J3/4. These adenines have been shown previously to interact with stem P6, forming base triples (Michel et al. 1990; Green and Szostak 1994; Cate et al. 1996). These base triples are involved in the correct orientation of the P4/P6 domain relative to the P3/P8 domain and are crucial for binding of Cyt-18 to the intron RNA (Caprara et al. 1996a; Myers et al. 1996; Chen et al. 2000). In the absence of either protein, residues A46 and A47 are moderately modified, whereas the flanking residue A48 in P4 is always less intensely modified. Consistent with the role of Cyt-18 in stabilizing the intron structure, this protein protects residues A46 and A47 from modification (Fig. 3A, upper panel), whereas the intensity of A48 remains unchanged. In the presence of Cyt-18, the decreased modification of bases A46 and A47 compared to residue A48 suggests that the base triple interaction between P6 and J3/4 is strengthened (Fig. 3A,B, upper panels).
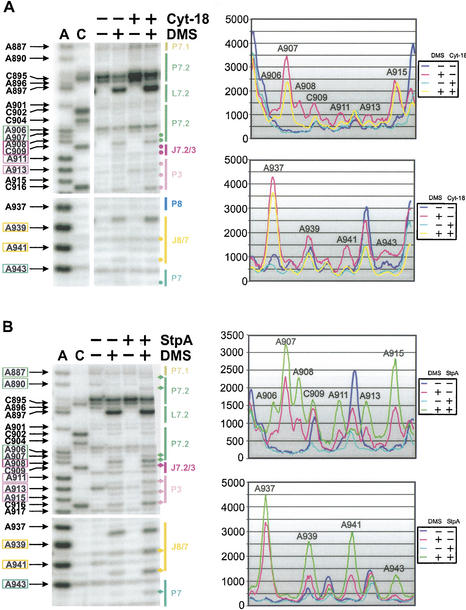
Figure 3.
StpA and Cyt-18 induce structural changes in the base triple interactions between adenines in J3/4 and stem P6. Changes in the DMS modification pattern of the td intron in vivo due to the presence of Cyt-18 is shown in the upper panel or due to the presence of StpA in the lower panel. (A) The P4/P5 domain is shown in these representative gels. Numbered nucleotides, which are highlighted by colored boxes at the left of the gel, are modified by DMS. The P4/P5 domain is marked in green and the joining segment J3/4 is highlighted in orange. The gel part boxed in orange is outlined to point out the different effect of Cyt-18 versus StpA on the residues A46 and A47 in J3/4. The sequencing lanes are labeled with A and C. In the presence of Cyt-18 the amount of td RNA is increased in the cells, as reflected by the increase of nonspecific stops in untreated samples (cf. lanes 3 and 5), as well as by the increased modification intensity of residues A55, C56, C57 and A63 in lanes 4 and 6. These differences in the td RNA amount were normalized as described in Materials and Methods. (B) PhosphorImager quantification (right panel) of the outlined gel segments in the presence of Cyt-18 or StpA. The opposite effects of these proteins on the accessibility of the two adenines in J3/4 to DMS are summarized in the middle panel.
In contrast, in the presence of StpA, modification of A46 and A47 in J3/4 is enhanced. Again the modification of A48 remains unchanged (Fig. 3A, lower left panel). Quantification of the DMS modifications confirmed the increased accessibility of residues A46 and A47 relative to nucleotide A48 (Fig. 3B, lower right panel). This increase in accessibility to DMS suggests that in the presence of StpA, the base triples are mostly unfolded. Thus, StpA loosens the base triple interactions and has the opposite effect on this structural element as Cyt-18. Comparison of the modification intensities in the presence of StpA or Cyt-18 is shown in the middle panel of Figure 3B.
StpA loosens and Cyt-18 stabilizes the overall 3D structure of the td intron
The stability of the 3D structure of the td intron is achieved by several long-range tertiary interactions (Fig. 1; Michel and Westhof 1990, 1994; Jaeger et al. 1994; Brion et al. 1999a,b). Among these essential tertiary contacts are the base triples. Formation of the base triples is crucial for proper folding of the intron; thus, opening of this interaction by StpA might also perturb other tertiary contacts. The state of other tertiary interactions was also quantified in the presence of StpA and Cyt-18 (Fig. 4A,B). The additional changes found in the presence of either protein are in line with the opposite effects of both proteins on the base triples (Fig. 6A, see below). StpA renders residues A939 and A941 in junction J8/7 and nucleotide A943 in stem P7, as well as bases A887, A890, and A906-A908, which are part of the loop E motif, slightly more accessible to DMS (Leontis and Westhof 1998; Waldsich et al. 2002). The residues A911, A913, and A915, which are part of the late-forming pseudoknot P3, are also slightly more accessible to DMS, as well as the adjacent bases in junction J3/7.2 (Fig. 4B; Michel and Westhof 1990; Sclavi et al. 1998). Similarly, positions A961 and A994-C996, which participate in the formation of or are close to long-range tertiary interactions also display an enhanced modification intensity in the presence of StpA (data not shown). In brief, StpA has a destabilizing effect on the overall 3D structure of the td intron.
Figure 4.
StpA loosens additional tertiary interactions, whereas Cyt-18 contributes to the overall compactness of the td intron. The representative gels (left panel) display the DMS modification pattern of the P7.1 and P7.2 extension and part of the stem P3, as well as the junction J8/7 in the absence and presence of Cyt-18 (A) or StpA (B). Residues that show enhanced modification in the presence of StpA or reduced accessibility to DMS in the presence of Cyt-18 are highlighted in colored boxes at the left of the gel and the specific bands are marked with diamonds (StpA) or dots (Cyt-18). The P7 extension is marked in green, stem P3 and junction J7.2/3 are highlighted in purple, and J8/7 is labeled in yellow. The corresponding PhosphorImager quantifications of these gel segments are shown in the right panels. Due to differing td RNA amounts in the presence of either protein, we again had to normalize these differences as described in Materials and Methods.
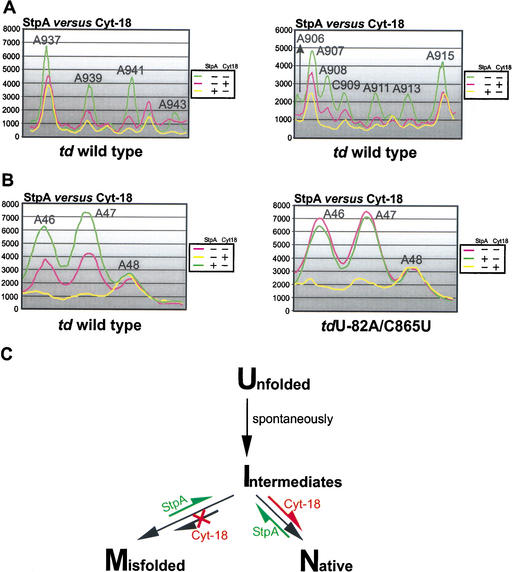
Figure 6.
Opposite effects of StpA and Cyt-18. (A) The opposite effects of these proteins on the accessibility of the residues in J8/7 (left panel) and of bases, which are close to or part of the loop E motif in the P7.1-P7.2 extension (right panel), to DMS are summarized. (B) Comparison of the accessibility to DMS of the A46–A48 in the presence and absence of either StpA and Cyt-18 in the context of the wild-type td intron (left panel) and the mutant tdU-82A/C865U (right panel). (C) A model for the mechanisms of action of the RNA chaperone StpA and the group I intron-specific splicing factor Cyt-18. The equilibrium shifts within the folding pathway induced by either StpA or Cyt-18 are marked in green or red. U, unfolded molecules, having formed the secondary but not the tertiary structure; M, misfolded molecules, secondary and tertiary structure of the intron are formed but alternative mispaired conformations at the 2D or 3D level occur within the pre-mRNA; I, intermediate molecules, secondary and part of the 3D structure are formed, but these molecules still lack the formation of long-range tertiary interactions; and N, molecules with a native 3D fold having a stable, productive structure leading to splicing competent, functional RNA.
In vitro Cyt-18 was shown to stabilize the tertiary structure of group I introns, thereby rescuing splicing of td mutant introns with a destabilized 3D fold. In vivo we observed changes in the modification pattern throughout the tertiary structure motifs. Residues A939 and A941 in junction J8/7, nucleotide A943 in stem P7, bases A911 and A913 in stem P3, and positions A906-C909 in the loop E motif are less accessible to DMS in the presence of Cyt-18 (Fig. 4A). In addition, nucleotides A961, A995, and C996 involved in and residue A994 close to long-range tertiary interactions are also further protected from modification (data not shown). Thus, the protection of A46 and A47 in J3/4 coincides with that of other residues involved in tertiary interactions, suggesting that Cyt-18 generally stabilizes the td intron core structure in vivo.
A mutant with a destabilized base triple is sensitive to StpA but is rescued by Cyt-18
For a further understanding of the effect of StpA on the base triple interactions, we weakened the P6 element by mutating C865 to U, thereby loosening the base triple interaction with J3/4 (Fig. 5A; Chen et al. 2000). This mutation decreases the splicing activity from 80% in the wild type to 37% in the mutant (Fig. 5A,B). The splicing activity of mutant tdC865U is further decreased from 37% to 15% in the presence of StpA. In contrast, Cyt-18 rescues splicing of this mutant to 70% (Fig. 5A,B). Thus, the mutant tdC865U with a destabilized 3D structure is sensitive to StpA but is rescued by Cyt-18. Noteworthy, the expression levels of endogenous StpA and plasmid-borne StpA were quantified in the presence of plasmids carrying intron mutants. The nature of the mutation does not influence the StpA expression levels, which are comparable to those in the presence of the wild-type intron (Fig. 2D; data not shown).
Figure 5.
Destabilization of a base triple interaction or of the pseudoknot P7 within the td intron core renders splicing in vivo sensitive to the RNA chaperone StpA. (A) Representative gels display the splicing activity of the wild-type td intron and the mutants tdC865U and tdC873U in the presence and absence of either Cyt-18 or StpA. (B) Quantification of at least four individual experiments with standard deviations. Blue bars, % mRNA splicing; red bars, % cryptic splicing; and yellow bars, % total splicing. (C) The DMS modification pattern of residues close to the joining segment J3/4 in the mutant tdU-82A/C865U is shown in the absence and presence of either Cyt-18 (upper panel) or StpA (lower panel). Numbered nucleotides, which are highlighted by colored boxes at the left of the gel, are modified by DMS. The P4/P5 domain is shown in green and the joining segment J3/4 is marked in orange. Part of stem P3 and junction J2/3 are indicated in blue. The gel part boxed in orange is outlined to point out the different modification status of A46 and A47. Due to differing td RNA amounts in the presence of either protein, we again had to normalize these differences as described in Materials and Methods. (D) Relative modification intensities of residues involved in the formation of base triples in the wild-type td intron (left panel) and in the mutant tdU-82A/C865U (right panel). The mutation site is marked with a gray square. Modified residues are labeled with a triangle (for color code, see Fig. 1A). The different size of the triangles correlates with the relative modification intensities. (E) PhosphorImager quantification of the outlined gel segments (shown in C) in the absence and presence of Cyt-18 (right panel) or StpA (left panel). These graphs show the modification pattern of A46 to A48 in the mutant tdU-82A/C865U and the influence of StpA and Cyt-18 on their modification intensity.
We aimed to determine whether the splicing deficiencies of this mutant correlate with the DMS modification pattern. The structural probing experiments were done with the mutant tdC865U and with a double mutant containing an additional mutation 82 nucleotides upstream of the 5′ splice site, introducing a nonsense codon in the upstream exon (Semrad and Schroeder 1998; Clodi et al. 1999). This ensures that a population of pre-mRNA molecules rather than circular intron RNA was probed. Nonsense codons within the upstream exon do not lead to changes in the DMS modification pattern within the intron core compared to the wild type. In both mutants, tdC865U and tdU-82A/C865U modification of residues A46 and A47 in J3/4 and C79 in stem P6 is significantly increased, suggesting that the base triples, as well as P6, are not formed (Fig. 5C–E). In addition, other nucleotides involved in tertiary contacts are more accessible to DMS (data not shown). These findings are consistent with the UV melting profile, which indicated that the tertiary structure is not formed in the mutant tdC865U (Chen et al. 2000). The modification intensity of adenines in J3/4 in the mutant tdU-82A/C865U is equivalent to the intensity observed in the wild-type intron in the presence of StpA (Fig. 6B). This indicates that the base triple interactions in the mutant are already opened and therefore StpA does not further increase the modification intensity. Although StpA has no further effect on the DMS modification level of this structural motif, Cyt-18 closes the base triples, protecting them from DMS modification (Fig. 5C–E). These results further show that the two proteins, Cyt-18 and StpA, have opposite mechanisms of action, indicated by opposite effects on the modification pattern of the adenines in joining region J3/4 and on the splicing activity of a structurally destabilized mutant intron.
The mutant tdC873U has a destabilized P7 element, resulting in a UV melting profile without a distinct peak for the melting of the tertiary structure and might therefore also be sensitive to StpA (Brion et al. 1999b). We performed splicing assays with this mutant in the presence and absence of StpA and could detect a decrease of the splicing activity from 10% in the absence to 4% in the presence of StpA. Cyt-18 rescues splicing of this mutant to 22% (Fig. 5A,B). We therefore suggest that td intron mutants with a destabilized tertiary fold might in general be sensitive to StpA, although mutants with alterations in other elements of tertiary structure need to be further investigated.
Discussion
RNA molecules have the tendency to be trapped in nonnative conformations, which are often almost equally stable as the native state. This is known as the RNA folding problem (Herschlag 1995; Weeks 1997; Treiber and Williamson 1999; Woodson 2000a,b; Thirumalai et al. 2001). In vitro, renaturation protocols involve heating procedures that resolve these misfolded structures. In vivo, cells have to overcome this folding problem by alternative solutions. As a consequence, the existence of proteins with RNA chaperone activity has been postulated and, indeed, RNA folding was found to be promoted by several proteins (Tsuchihashi et al. 1993; Coetzee et al. 1994; Herschlag et al. 1994; Herschlag 1995; Zhang et al. 1995; Nedbal et al. 1997; Clodi et al. 1999; Negroni and Buc 2000). It, however, is unknown which feature of the RNA signals its nonnative conformation and thus making it a target for RNA chaperones.
The effect of StpA on splicing is dependent on the structural stability of the td intron
In this work we report the first structural approach to unravel the mechanism of action of an RNA chaperone. For this purpose we determined the effect of the RNA chaperone StpA on the td group I intron in vivo using DMS as a structural probe. DMS modifications reflect the relative occupancy of interactions averaged over the population of RNA molecules and over time. We therefore interpret our results in a way that an increased accessibility of residues involved in tertiary interactions correlates with an opening of specific contacts and with a reduced compactness of the fold. The base triple interactions, as well as other tertiary contacts, are more accessible in the presence of StpA, suggesting that this protein resolves the 3D fold of the intron (Fig. 6A,B, left panel).
A major question is why StpA does not hamper the splicing activity of the wild-type td intron although it opens the base triples between stem P6 and junction J3/4. A possible explanation is that splicing or refolding of the wild-type intron is faster than the activity of StpA. In td mutants with a reduced structural stability, the thermodynamically disfavored native state probably accounts for a slower splicing rate, enabling StpA to interact with the intron prior to splicing. When StpA acts on these mutant pre-mRNAs, it further destabilizes the native conformation, resulting in an even further reduced splicing rate. In contrast, mutants with a nonsense codon in the upstream exon should in principle have a similar structural stability compared to the wild type, as indicated by the DMS modification patterns. A part of the pre-mRNA population, however, adopts a splicing-incompetent conformation with respect to folding at the splice sites (Pichler and Schroeder 2002). When StpA resolves the nonnative interactions in trapped RNAs by opening the tertiary structure, the molecules presumably get a chance to refold and to reach the native productive state, which has a fast splicing rate. We therefore conclude that the structural stability of the intron is a key determinant for the positive or negative effect of the RNA chaperone StpA on splicing of the td group I intron.
Up to now the activity of RNA chaperones was deduced from in vitro folding kinetics using RNA annealing or strand displacement assays (Coetzee et al. 1994; Zhang et al. 1995). These experiments lead to the suggestion that RNA chaperones resolve RNA structures (Herschlag 1995). Here, we present the first structural evidence that the RNA chaperone StpA might exert its activity via the resolution of tertiary interactions within the intron structure in vivo. The destabilization of the RNA structure by proteins with RNA chaperone activity is probably a general characteristic of these proteins. The nucleocapsid protein of HIV NCp7 induces structural changes in the tRNA(Lys,3), disrupting metal-ion-binding pockets and thereby unfolding the tRNA (Hargittai et al. 2001). Whether the opening of the tertiary structure by RNA chaperones is indeed a general mechanism of action of these proteins is a question that needs further analysis.
Cyt-18 contributes to the overall compactness of the td group I intron structure
The aminoacyl-tRNA-synthetase Cyt-18, which specifically recognizes and binds to the tRNA-like features of the group I intron core, causes protection of adenines in J3/4 from DMS modification (Fig. 6B). These protections could either result from a direct interaction of the protein Cyt-18 with residues A46 and A47 in J3/4 or as a consequence of the stabilization of the td 3D fold by the protein. In favor of the latter interpretation is the fact that additional residues, which are not part of the protein binding site but which participate in the formation of tertiary contacts are protected from modification by Cyt-18 (Fig. 6A). Furthermore, it has been proposed that positions N1 and N6 of adenines in J3/4 interact with stem P6 via hydrogen bonds (Chen et al. 2000). Protections specific for Cyt-18 that occur throughout the molecule indicate that Cyt-18 might contribute to the overall compactness of the td intron in vivo. Either Cyt-18 binds to the folded intron, thereby stabilizing its 3D shape, or it binds to a folding intermediate, guiding the formation of the correct intron structure. Each mechanism illustrates nicely how Cyt-18 might favor the native state of td intron mutants having a compromised tertiary fold. In favor of the second model is the fact that Cyt-18 is able to rescue splicing of kinetically trapped td mutants containing a nonsense codon in the upstream exon. If Cyt-18 guides folding of the td intron in vivo, it might prevent the formation of folding traps. Based on the analyses of Cyt-18 activity on td mutants impaired in 3D folding, an induced-fit model has been proposed for binding of Cyt-18 to the P4/P6 domain of the intron (Chen et al. 2000). Our results are in excellent agreement with data from the Lambowitz group supporting the idea that Cyt-18 guides folding of the td intron (Caprara et al. 1996b, 2001; Myers et al. 1996; Saldanha et al. 1996; Chen et al. 2000).
Opposite mechanisms of action: StpA versus Cyt-18
The difference in the mechanism of action of the two proteins is shown by their opposite effects on the tertiary structure of the td intron in vivo (Fig. 6A,B). Whereas StpA loosens, Cyt-18 contributes to the compactness of the tertiary structure. Figure 6C depicts our current view of how StpA and Cyt-18 influence the folding pathway of the td intron. At this time, it is unknown which intermediates occur during in vivo folding, but it is known, however, that misfolding of the td pre-mRNA interferes with splicing (Semrad and Schroeder 1998; Waldsich et al. 1998). We propose that StpA resolves the tertiary fold of the intron, giving the molecule another chance to fold into the native splicing competent structure. In contrast, Cyt-18 binds to folding intermediates, thereby preventing the formation of folding traps by guiding the formation of the correct native structure. Taken together our data provide the first direct evidence for protein-induced conformational changes within catalytic RNA in vivo.
Materials and methods
Plasmids and strains
The thymine deficient (thyA−) variant of E. coli strain C600 (F- thr-1 leuB6 thi-1 lacY1 supE44 rfbD1 fhuA21) was transformed with a pTZ18U derivative harboring the T4 phage-derived td construct tdP6Δ2. The intron ORF encoding an endonuclease was deleted up to L6a (Salvo et al. 1990) and the EcoRI fragment was introduced into the pTZ18U vector (USB), which carries an ampicillin resistance gene. All mutant constructs were done in the context of pTZ18U/tdP6Δ2. The mutant tdC870U was described previously in Schroeder et al. (1991). The mutant tdC873U was described previously in Belfort et al. (1987). The plasmid encoding the RNA chaperone StpA was obtained from Marlene Belfort (Wadsworth Center, New York State Department of Health, Albany, NY) and is a pSU20 derivative (Zhang et al. 1995). The plasmid encoding the Neurospora crassa tyrosyl-tRNA-synthetase Cyt-18 was obtained from Alan Lambowitz and is a pACYC184 derivative (Mohr et al. 1992).
Western blot analysis of StpA expression
E. coli cells were grown to OD600nm 0.2 and harvested and an equivalent of 500 μL of cell culture was lysed with sample buffer and loaded onto 18% SDS-PAGE. Immunoblotting was performed as described in Kramer et al. (1998). For antibody production the StpA peptide CPRAGKKRQPRPAKYKFTDVNGETK (containing an additional cystein residue at the N terminus) was coupled to keyhole limpet hemocyanin using the Imject Maleimide Activated mcKLH Kit (Pierce) and rabbits were immunized.
Cells harboring both the endogenous StpA gene and the pSU20/StpA plasmid were transformed with either the td wild type or mutants thereof as specified in Figure 2D and used for StpA expression analysis. The ratio of endogenous versus overexpressed StpA was estimated by comparing the band intensity of the cellular StpA with that of a dilution series of purified StpA protein.
In vivo DMS modification
All strains were grown in TBYE medium at 37°C (Belfort et al. 1983). Thymine was added to a final concentration of 52 mg/L when thymidylate synthase–deficient strains were used. In addition, IPTG was added to a final concentration of 1 mM because the td gene is under control of a lac-promotor. The cells were grown to an OD600nm of 0.2–0.3 and then the DMS modification was performed in vivo. First, the cells were harvested and the pellet was resuspended in a TM buffer (10 mM TRIS at pH 7, 10 mM MgCl2). DMS was then added to a final concentration of 150 mM and the samples were incubated 1 min at room temperature. To stop the DMS reaction, β-mercaptoethanol was added to a final concentration of 0.7 M. This method was adjusted from Zaug and Cech (1995). After having harvested the cells, the pellet was frozen until RNA preparation as described in Semrad and Schroeder (1998). Primer extension was used to detect the modification sites within the td RNA (von Ahsen and Noller 1993; Semrad and Schroeder 1998). The oligonucleotides used in this study are summarized as follows: tdP2–5, 5′-TTACAAGTATAAGTCACCTTA-3′; tdP6a-5, 5′-TGTAGAACCCGGGCAGTC-3′; tdL7.2–5,: 5′-TCTATCGTT TCGAGTCAC-3′; tdP9–5, 5′-ATCCAGCTGCATGTCACC-3′; Intron-Exon2, 5′-TTAAACGGTAGCATTATGTTCAG-3′. An aliquot of the RNA preparation was used for a poison primer assay to monitor RNA amounts and the in vivo splicing activity (see below).
Quantification was also performed using the PhosphorImager program ImageQuant. The analysis of the in vivo DMS modification gels sometimes required the extrapolation of the modification intensity values due to differing td RNA amounts in the presence and absence of either StpA or Cyt-18. Comparison of DMS nonspecific breaks (see gel in Fig. 3A: break 5′ to A63 or 5′ to A46), as well as of modification sites within loops (see gel in Fig. 3A; A63, A55-C57), which were taken as internal standards, was used to normalize the different td RNA amounts in the absence and presence of either protein. Furthermore, it has to be mentioned that the very 3′ terminal domain and several positions within the intron are not clearly amenable to our analysis because of strong breaks of the RNA backbone or because of premature termination of reverse transcription in the absence of DMS treatment.
In vivo splicing assay
Cells were grown as described above. After having prepared the RNA primer extension using the oligonucleotide, NBS2 was performed in the presence of ddTTP in order to terminate the extension at the first adenine in the template (see Fig. 2A). NBS2: 5′-GACGCAATATTAAACGGT-3′. The primer extension products were quantified for determining the splicing activity (Semrad and Schroeder 1998; Waldsich et al. 1998).
Acknowledgments
We thank Oliver Mayer for the generous gift of purified StpA protein and for his comments on the manuscript. We appreciate Eric Westhof's and Luc Jaeger's comments and thank them for sharing their knowledge on td intron structure. We thank Andrea Pichler for constant discussions and for critically reading the manuscript. We also acknowledge Andrea Barta, Nicolas Piganeau, Herbert Wank, Albert Weixlbaumer, Nikolai Windbichler, and Uwe von Ahsen for comments on the manuscript. This work was supported by a grant from the Austrian science foundation (FWF), project no. F1703.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL renee@gem.univie.ac.at; FAX 43-1-4277-9546.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.231302.
References
- Banerjee AR, Turner DH. The time dependence of chemical modification reveals slow steps in the folding of a group I ribozyme. Biochemistry. 1995;34:6504–6512. doi: 10.1021/bi00019a031. [DOI] [PubMed] [Google Scholar]
- Belfort M, Moelleken A, Maley GF, Maley F. Purification and properties of T4 phage thymidylate synthetase produced by the cloned gene in an amplification vector. J Biol Chem. 1983;258:2045–2051. [PubMed] [Google Scholar]
- Belfort M, Chandry PS, Pedersen-Lane J. Genetic delineation of functional components of the group I intron in the phage T4 td gene. Cold Spring Harb Symp Quant Biol. 1987;52:181–192. doi: 10.1101/sqb.1987.052.01.023. [DOI] [PubMed] [Google Scholar]
- Brion P, Michel F, Schroeder R, Westhof E. Analysis of the cooperative thermal unfolding of the td intron of bacteriophage T4. Nucleic Acids Res. 1999a;27:2494–2502. doi: 10.1093/nar/27.12.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion P, Schroeder R, Michel F, Westhof E. Influence of specific mutations on the thermal stability of the td group I intron in vitro and on its splicing efficiency in vivo: A comparative study. RNA. 1999b;5:947–958. doi: 10.1017/s1355838299990477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprara MG, Lehnert V, Lambowitz AM, Westhof E. A tyrosyl-tRNA synthetase recognizes a conserved tRNA-like structural motif in the group I intron catalytic core. Cell. 1996a;87:1135–1145. doi: 10.1016/s0092-8674(00)81807-3. [DOI] [PubMed] [Google Scholar]
- Caprara MG, Mohr G, Lambowitz AM. A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J Mol Biol. 1996b;257:512–531. doi: 10.1006/jmbi.1996.0182. [DOI] [PubMed] [Google Scholar]
- Caprara MG, Myers CA, Lambowitz AM. Interaction of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase (CYT-18 protein) with the group I intron P4-P6 domain. Thermodynamic analysis and the role of metal ions. J Mol Biol. 2001;308:165–190. doi: 10.1006/jmbi.2001.4581. [DOI] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Cech TR, Damberger SH, Gutell RR. Representation of the secondary and tertiary structure of group I introns. Nat Struct Biol. 1994;1:273–280. doi: 10.1038/nsb0594-273. [DOI] [PubMed] [Google Scholar]
- Chandry PS, Belfort M. Activation of a cryptic 5′ splice site in the upstream exon of the phage T4 td transcript: Exon context, missplicing, and mRNA deletion in a fidelity mutant. Genes & Dev. 1987;1:1028–1037. doi: 10.1101/gad.1.9.1028. [DOI] [PubMed] [Google Scholar]
- Chen X, Gutell RR, Lambowitz AM. Function of tyrosyl-tRNA synthetase in splicing group I introns: An induced-fit model for binding to the P4-P6 domain based on analysis of mutations at the junction of the P4-P6 stacked helices. J Mol Biol. 2000;301:265–283. doi: 10.1006/jmbi.2000.3963. [DOI] [PubMed] [Google Scholar]
- Clodi E, Semrad K, Schroeder R. Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J. 1999;18:3776–3782. doi: 10.1093/emboj/18.13.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee T, Herschlag D, Belfort M. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes & Dev. 1994;8:1575–1588. doi: 10.1101/gad.8.13.1575. [DOI] [PubMed] [Google Scholar]
- Green R, Szostak JW. In vitro genetic analysis of the hinge region between helical elements P5-P4-P6 and P7-P3-P8 in the sunY group I self-splicing intron. J Mol Biol. 1994;235:140–155. doi: 10.1016/s0022-2836(05)80022-1. [DOI] [PubMed] [Google Scholar]
- Hargittai MR, Mangla AT, Gorelick RJ, Musier-Forsyth K. HIV-1 nucleocapsid protein zinc finger structures induce tRNA(Lys,3) structural changes but are not critical for primer/template annealing. J Mol Biol. 2001;312:985–997. doi: 10.1006/jmbi.2001.5021. [DOI] [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger L, Michel F, Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J Mol Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Gieffers C, Holzl G, Hengstschlager M, Peters JM. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr Biol. 1998;8:1207–1210. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J Mol Biol. 1998;283:571–583. doi: 10.1006/jmbi.1998.2106. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Adams A. Native and renatured transfer ribonucleic acid. Science. 1966;152:512–514. doi: 10.1126/science.152.3721.512. [DOI] [PubMed] [Google Scholar]
- Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- ————— Slippery substrates. Nat Struct Biol. 1994;1:5–7. doi: 10.1038/nsb0194-5. [DOI] [PubMed] [Google Scholar]
- Michel F, Ellington AD, Couture S, Szostak JW. Phylogenetic and genetic evidence for base-triples in the catalytic domain of group I introns. Nature. 1990;347:578–580. doi: 10.1038/347578a0. [DOI] [PubMed] [Google Scholar]
- Mohr G, Zhang A, Gianelos JA, Belfort M, Lambowitz AM. The Neurospora CYT-18 protein suppresses defects in the phage T4 td intron by stabilizing the catalytically active structure of the intron core. Cell. 1992;69:483–494. doi: 10.1016/0092-8674(92)90449-m. [DOI] [PubMed] [Google Scholar]
- Myers CA, Wallweber GJ, Rennard R, Kemel Y, Caprara MG, Mohr G, Lambowitz AM. A tyrosyl-tRNA synthetase suppresses structural defects in the two major helical domains of the group I intron catalytic core. J Mol Biol. 1996;262:87–104. doi: 10.1006/jmbi.1996.0501. [DOI] [PubMed] [Google Scholar]
- Nedbal W, Frey M, Willemann B, Zentgraf H, Sczakiel G. Mechanistic insights into p53-promoted RNA-RNA annealing. J Mol Biol. 1997;266:677–687. doi: 10.1006/jmbi.1996.0813. [DOI] [PubMed] [Google Scholar]
- Negroni M, Buc H. Copy-choice recombination by reverse transcriptases: Reshuffling of genetic markers mediated by RNA chaperones. Proc Natl Acad Sci. 2000;97:6385–6390. doi: 10.1073/pnas.120520497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolcheva T, Woodson SA. Facilitation of group I splicing in vivo: Misfolding of the Tetrahymena IVS and the role of ribosomal RNA exons. J Mol Biol. 1999;292:557–567. doi: 10.1006/jmbi.1999.3083. [DOI] [PubMed] [Google Scholar]
- Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: Mispairing of the catalytic core. J Mol Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- Pannone BK, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: Evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Schroeder R. Folding problems of the 5′ splice-site containing P1 stem of the group I td intron: Substrate binding inhibition in vitro and mis-splicing in vivo. J Biol Chem. 2002;277:17987–17993. doi: 10.1074/jbc.M111798200. [DOI] [PubMed] [Google Scholar]
- Rook MS, Treiber DK, Williamson JR. Fast folding mutants of the Tetrahymena group I ribozyme reveal a rugged folding energy landscape. J Mol Biol. 1998;281:609–620. doi: 10.1006/jmbi.1998.1960. [DOI] [PubMed] [Google Scholar]
- Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: Commitment to form the native conformation is late in the folding pathway. J Mol Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- Russell R, Millett IS, Doniach S, Herschlag D. Small angle X-ray scattering reveals a compact intermediate in RNA folding. Nat Struct Biol. 2000;7:367–370. doi: 10.1038/75132. [DOI] [PubMed] [Google Scholar]
- Saldanha R, Ellington A, Lambowitz AM. Analysis of the CYT-18 protein binding site at the junction of stacked helices in a group I intron RNA by quantitative binding assays and in vitro selection. J Mol Biol. 1996;261:23–42. doi: 10.1006/jmbi.1996.0439. [DOI] [PubMed] [Google Scholar]
- Salvo JL, Coetzee T, Belfort M. Deletion-tolerance and trans-splicing of the bacteriophage T4 td intron. Analysis of the P6-L6a region. J Mol Biol. 1990;211:537–549. doi: 10.1016/0022-2836(90)90264-m. [DOI] [PubMed] [Google Scholar]
- Schroeder R, von Ahsen U, Belfort M. Effects of mutations of the bulged nucleotide in the conserved P7 pairing element of the phage T4 td intron on ribozyme function. Biochemistry. 1991;30:3295–3303. doi: 10.1021/bi00227a018. [DOI] [PubMed] [Google Scholar]
- Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- Semrad K, Schroeder R. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes & Dev. 1998;12:1327–1337. doi: 10.1101/gad.12.9.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solem A, Chatterjee P, Caprara M G. A novel mechanism for protein-assisted group I intron splicing. RNA. 2002;8:412–425. doi: 10.1017/s1355838202029321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalai D, Lee N, Woodson SA, Klimov D. Early events in RNA folding. Annu Rev Phys Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- ————— Beyond kinetic traps in RNA folding. Curr Opin Struct Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z, Khosla M, Herschlag D. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993;262:99–102. doi: 10.1126/science.7692597. [DOI] [PubMed] [Google Scholar]
- von Ahsen U, Noller HF. Methylation interference experiments identify bases that are essential for distinct catalytic functions of a group I ribozyme. EMBO J. 1993;12:4747–4754. doi: 10.1002/j.1460-2075.1993.tb06163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldsich C, Semrad K, Schroeder R. Neomycin B inhibits splicing of the td intron indirectly by interfering with translation and enhances missplicing in vivo. RNA. 1998;4:1653–1663. doi: 10.1017/s1355838298981444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldsich, C, Masquida, B., Westhof, E., and Schroeder, R. 2002. Monitoring intermediate folding states of the td group I intron in vivo. EMBO J. 19 (In press). [DOI] [PMC free article] [PubMed]
- Walstrum SA, Uhlenbeck OC. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry. 1990;29:10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- Weeks KM, Cech TR. Protein facilitation of group I intron splicing by assembly of the catalytic core and the 5′ splice site domain. Cell. 1995;82:221–230. doi: 10.1016/0092-8674(95)90309-7. [DOI] [PubMed] [Google Scholar]
- Wells SE, Hughes JM, Igel AH, Ares M., Jr Use of dimethyl sulfate to probe RNA structure in vivo. Meth Enzymol. 2000;318:479–493. doi: 10.1016/s0076-6879(00)18071-1. [DOI] [PubMed] [Google Scholar]
- Woodson SA. Compact but disordered states of RNA. Nat Struct Biol. 2000a;7:349–352. doi: 10.1038/75106. [DOI] [PubMed] [Google Scholar]
- ————— Recent insights on RNA folding mechanisms from catalytic RNA. Cell Mol Life Sci. 2000b;57:796–808. doi: 10.1007/s000180050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA, Cech TR. Alternative secondary structures in the 5′ exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry. 1991;30:2042–2050. doi: 10.1021/bi00222a006. [DOI] [PubMed] [Google Scholar]
- Zarrinkar PP, Williamson JR. Kinetic intermediates in RNA folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- ————— The kinetic folding pathway of the Tetrahymena ribozyme reveals possible similarities between RNA and protein folding. Nat Struct Biol. 1996;3:432–438. doi: 10.1038/nsb0596-432. [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Cech TR. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: Telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA. 1995;1:363–374. [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Belfort M. Nucleotide sequence of a newly-identified Escherichia coli gene, stpA, encoding an H-NS-like protein. Nucleic Acids Res. 1992;20:6735. doi: 10.1093/nar/20.24.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Derbyshire V, Salvo JL, Belfort M. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Rimsky S, Reaban ME, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: Regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]