Abstract
Recent studies have reported that independent adaptive radiations can lead to identical ecomorphs. Our phylogenetic analyses of nuclear and mitochondrial DNA sequences here indicate that a major radiation of ranid frogs on Madagascar produced morphological, physiological, and developmental characters that are remarkably similar to those that independently evolved on the Indian subcontinent. We demonstrate further that, in several cases, adult and larval stages each evolved sets of characters which are not only convergent between independent lineages, but also allowed both developmental stages to invade the same adaptive zone. It is likely that such covariations are produced by similar selective pressures on independent larval and adult characters rather than by genetic or functional linkage. We briefly discuss why larval/adult covariations might constitute an important evolutionary phenomenon in species for which more than one developmental stage potentially has access to multiple environmental conditions.
Ever since Darwin (1), Huxley (2), and Simpson (3), adaptive radiation (4) has been used as a major concept in evolutionary biology. In this context, convergences in morphological, ecological, and physiological characteristics are usually regarded as occasional curiosities, i.e., the exception rather than the rule. However, most features whose radiation has been studied were also the major characters used in the systematic classification of the organisms bearing them. Characters can therefore be circularly interpreted as showing lack of convergence through a phylogenetic hypothesis inferred partly from these very same characters. The development of molecular phylogenetics has contributed a major breakthrough in the study of these radiations. For example, comparisons between morphological characters and results of molecular phylogenetic analyses have demonstrated extensive and multiple convergences of ecologically specialized species (i.e., ecomorphs) of cichlid fishes (5, 6), plethodontid salamanders (7–9), Anolis lizards (10), and mammals (cf. ref. 11 and references therein).
Difficulties in detecting convergent ecomorphs without the use of molecular data are particularly pervasive in anuran amphibians because their body plan is remarkably conserved. For example, despite the fact that Archaeobatrachia and Neobatrachia diverged about 200 million years ago (mya) (12), superficial examination shows no striking morphological fixed differences between these two groups. This situation is diametrical to that, e.g., of mammals, whose orders underwent extensive remodeling of morphological and physiological character complexes in a much shorter time (10, 12, 13). Hence, most conspicuous morphological features are usually used in anuran systematics despite their possible adaptive properties.
Ranid frogs form a nearly cosmopolitan family containing roughly 1,000 species, i.e., about one-fifth of all living Amphibia. Specific morphological characters within this family have led to the taxonomic recognition of subfamilies such as Ranixalinae (with adults adapted to life in streams or on rocks, and torrential or semiterrestrial larvae), Mantellinae (a diverse group of arboreal and torrential Madagascan frogs), Rhacophorinae (tree frogs), and Tomopterninae (burrowing frogs).
Given the low dispersal abilities of amphibians over salty environments, tectonic movements, as well as sea-level changes, might have been of major importance in shaping the distribution of lineages within ranid frogs. After the Madagascar–Seychelles–Indian plate drifted from the rest of Gondwana [starting around 130 mya (14)], first Madagascar, then Seychelles disconnected about 88 mya (15) and 65 mya (16), respectively. Subsequent attachment of the Indian subcontinent to Eurasia [about 65 to 35 mya (14)] rendered dispersal possible between these two previously disjunct areas. Because tree frogs and burrowing frogs occur on both Madagascar and India, it is usually interpreted that they originated before the Madagascan and the India–Seychelles land masses separated (88 mya).
We report here on the first molecular phylogenetic analyses of mitochondrial (mt) and nuclear (nu) DNA sequences (nearly 3 kb from each of the 28 ingroup and four outgroup taxa) of all Madagascan and Asian subfamilies within Ranidae.
Materials and Methods
Species and DNA Methods.
We included members from all Asian and Madagascan ranid subfamilies as recognized by Dubois (17) (with the exception of the African subfamily Ptychadeninae, represented by a single emigrant species on Madagascar, Ptychadena mascareniensis), and all ranid subfamilies as recognized by Blommers-Schlösser (18) (Table 1). Genomic DNA was extracted from tissue samples by using standard protocols. Three mtDNA and three nuDNA fragments were PCR-amplified and sequenced (dRhodamine Cycle Sequencing; Applied Biosystems) on both strands (sequencing of complementary strands was performed on different PCR products). The target mtDNA fragments were (i) a ≈750-bp segment including portions of the 12S and 16S rRNA genes as well as the tRNAVal gene [primers for PCR and sequencing were H3296, 5′-GCTAGACCATKATGCAAAAGGTA-3′ (19); L2519, 5′-AAACTGGGATTAGATACCCCACTAT-3′ (19); 12V16A, 5′-ACAAGCGCCAGGGWAYTACGAGC-3′; and 12V16B, 5′-TTCATTGTTATTTAATCTTTCCC-3′]; (ii) a ≈550-bp segment of the 16S rRNA gene [modified 16Sar, 5′-CGCCTGTTTAYCAAAAACAT-3′ (20); and modified 16Sbr 5′-CCGGTYTGAACTCAGATCAYGT-3′] (20); and (iii) a 580-bp segment of the cytochrome b gene [CB-J-10933, 5′-TATGTTCTACCATGAGGACAAATATC-3′ (20); CytbA, 5′-CCATGAGGACAAATATCATTYTGRGG-3′; CytbB, 5′-CTTCTACTGGTTGTCCTCCGATTCA-3′; and CytbC, 5′-CTACTGGTTGTCCTCCGATTCATGT-3′]. The nuDNA fragments were (i) a 316-bp segment of exon 1 and a 175-bp segment of exon 4 of the rhodopsin gene [Rhod1A, 5′-ACCATGAACGGAACAGAAGGYCC-3′; Rhod1B, 5′-AACGGAACAGAAGGYCCMAACTT-3′; Rhod1C, 5′-CCAAGGGTAGCGAAGAARCCTTC-3′; Rhod1D, 5′-GTAGCGAAGAARCCTTCAAMGTA-3′; Rhod4A, 5′-CAAGAATCAGCCACCACCCAGAA-3′; Rhod4B, 5′-GAATCAGCCACCACCCAGAAGGA-3′; Rhod4C, 5′-TTGTTCAGCATAATGTAGATGAC-3′; and Rhod4D, 5′-AGCATAATGTAGATGACRGGGTT-3′]; and (ii) a 529- to 532-bp segment of exon 1 of the tyrosinase gene [Tyr1A, 5′-AGGTCCTCTTRAGCAAGGAATG-3′; Tyr1B, 5′-AGGTCCTCYTRAGGAAGGAATG-3′; Tyr1C, 5′-GGCAGAGGAWCRTGCCAAGATGT-3′; Tyr1D, 5′-TCCTCCGTGGGCACCCARTTCCC-3′; Tyr1E, 5′-GAGAAGAAAGAWGCTGGGCTGAG-3′; Tyr1F, 5′-TCATCTCCCGYCAYCTTCTGGAT-3′; and Tyr1G, 5′- TGCTGGGCRTCTCTCCARTCCCA-3′].
Table 1.
List of species examined in this study
| Group | Family | Subfamily (Dubois) | Subfamily (Blommers-Schlösser) | Genus | Species | Origin |
|---|---|---|---|---|---|---|
| Outgroup | Bufonidae | NA | NA | Bufo | melanostictus | India |
| Microhylidae | NA | NA | Kaloula | taprobanica | Sri Lanka | |
| Microhylidae | NA | NA | Microhyla | sp. | India | |
| Hyperoliidae | NA | NA | Hyperolius | sp. | Kenya | |
| Ingroup | Ranidae | Dicroglossinae | Raninae | Euphlyctis | cyanophlyctis | India |
| Ranidae | Dicroglossinae | Raninae | Hoplobatrachus | crassus | Sri Lanka | |
| Ranidae | Dicroglossinae | Raninae | Limnonectes (Fejervarya) | cf. limnocharis | India | |
| Ranidae | Dicroglossinae | Raninae | Limnonectes (Fejervarya) | syhadrensis | India | |
| Ranidae | Dicroglossinae | Raninae | Limnonectes (Limnonectes) | corrugatus | Sri Lanka | |
| Ranidae | Dicroglossinae | Raninae | Limnonectes (Limnonectes) | kuhlii | Vietnam | |
| Ranidae | Ranixalinae | Nyctibatrachinae | Nyctibatrachus | major | India | |
| Ranidae | Ranixalinae | Nyctibatrachinae | Nyctibatrachus | cf. aliciae | India | |
| Ranidae | Ranixalinae | Indiraninae | Indirana | sp. 1 | India | |
| Ranidae | Ranixalinae | Indiraninae | Indirana | sp. 2 | India | |
| Ranidae | Ranixalinae | Cacosterninae | Nannophrys | ceylonensis | Sri Lanka | |
| Ranidae | Raninae | Petropedetinae | Micrixalus | fuscus | India | |
| Ranidae | Raninae | Petropedetinae | Micrixalus | kottigeharensis | India | |
| Ranidae | Raninae | Raninae | Rana | temporalis | India | |
| Ranidae | Raninae | Raninae | Rana | curtipes | India | |
| Ranidae | Raninae | Raninae | Rana | temporaria | Belgium | |
| Ranidae | Rhacophorinae | Rhacophorinae | Philautus | wynaadensis | India | |
| Ranidae | Rhacophorinae | Rhacophorinae | Philautus | charius | India | |
| Ranidae | Rhacophorinae | Rhacophorinae | Philautus (Kirtixalus) | microtympanum | Sri Lanka | |
| Ranidae | Rhacophorinae | Rhacophorinae | Rhacophorus | malabaricus | India | |
| Ranidae | Rhacophorinae | Rhacophorinae | Polypedates | cruciger | Sri Lanka | |
| Ranidae | Rhacophorinae | Rhacophorinae | Aglyptodactylus | madagascariensis | Madagascar | |
| Ranidae | Rhacophorinae | Rhacophorinae | Boophis | xerophilus | Madagascar | |
| Ranidae | Rhacophorinae | Rhacophorinae | Boophis | tephraeomystax | Madagascar | |
| Ranidae | Tomopterninae | Raninae | Tomopterna (Laliostoma) | labrosa | Madagascar | |
| Ranidae | Tomopterninae | Raninae | Tomopterna (Sphaerotheca) | breviceps | Sri Lanka | |
| Ranidae | NA (Mantellidae) | Mantellinae | Mantella | madagascariensis | Madagascar | |
| Ranidae | NA (Mantellidae) | Mantellinae | Mantidactylus | cf. ulcerosus | Madagascar |
Sequences were aligned with clustal w (21), using three different sets of alignment parameters (weighted matrix and gap/extension penalties = 10/5, 8/4, and 12/6, respectively), and positions at which the three alignments differed were excluded in the subsequent analyses (22). Gaps resulting from the alignment were treated as missing data.
For each mtDNA and nuDNA fragment, we checked for possible saturation of substitution types by plotting the number of transitions (Ti) and transversions (Tv) against uncorrected pairwise distances. Saturation plots were also examined separately for first, second, and third positions of protein-coding genes.
Three data sets were analyzed: (i) mtDNA, (ii) nuDNA, and (iii) “total DNA” evidence, i.e., concatenation of the two former data sets. Partition homogeneity tests (23) indicated that the nuDNA and mtDNA data sets were not incongruent. paup* (24) was used for all phylogenetic analyses. Maximum parsimony (MP) analyses were first performed with all characters weighted equally. Stability of the cladograms was tested with the Goloboff fit criterion (25) with k = 0, 2, 4, and 8, which allows individual down-weighting of noisy characters. We also used the maximum likelihood (ML) method of phylogeny inference with the following settings (paup*): empirical nucleotide frequencies, Ti/Tv ratio, and proportion of invariable sites estimated by means of ML, Hasegawa–Kishino–Yano (HKY) model (26) with rate heterogeneity, rates for variable sites assumed to follow a γ distribution with shape parameter estimated by ML, and tree bisection–reconnection (TBR) branch-swapping. Given the high computation burden of ML analyses, these were performed while excluding some of the taxa within clades that were very well supported in MP analyses.
The stability of individual clades was estimated by computing bootstrap values (27) for MP and ML trees, and by Bremer support indices—i.e., the number of additional character transformations necessary to collapse an internal branch (28)—for MP trees. Specific alternative hypotheses were compared statistically by means of Templeton (MP) and Kishino–Hasegawa (MP and ML) tests (29).
We tried to root the trees by using outgroup taxa chosen on the basis of earlier molecular and morphological studies of interfamilial relationships in Neobatrachia. The best rooting was evaluated with rasa 2.3b [Optimal Outgroup Analysis (30)] as well as by examining saturation plots. Beside performing analyses including all outgroup taxa, we tested the influence of outgroup choice by using individual outgroup taxa separately.
Results
Sequence Variation and Saturation.
Alignment resulted in a data matrix of 2,692 unambiguously aligned characters, of which 979 (706 mtDNA + 276 nuDNA) are parsimony informative. Given their high level of divergence (37–68% uncorrected pairwise divergence) and their obvious saturation in numbers of transitions and transversions (data not shown), cytochrome b third positions were excluded from all further analyses. After removal of these positions, mtDNA contained 513 informative characters. Saturation plots for the remaining data showed a slight saturation when outgroup taxa were included, which disappeared when they were excluded. All further analyses were therefore performed without an outgroup.
Adaptive Radiation of Madagascan Ranidae.
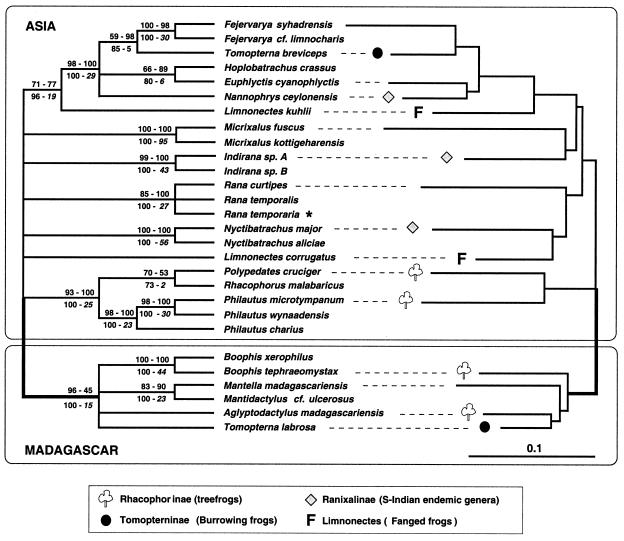
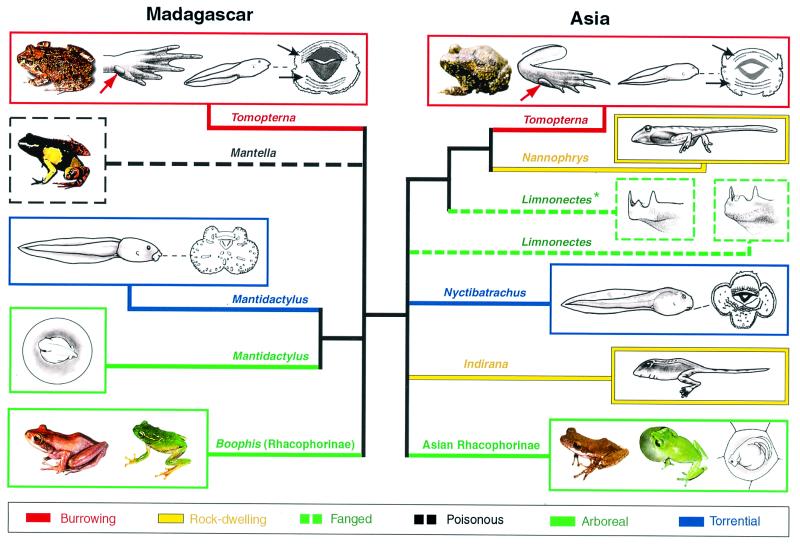
Our results do not support the traditional grouping of ranids according to ecomorphs but rather reveal that their relationships were primarily shaped by plate tectonics. Indeed, the inferred phylogeny exhibits a branch (indicated as bold in Fig. 1) separating a Madagascan from an Indian group despite the fact that identical ecomorphs are found in both groups. MP bootstrap analyses of the mtDNA, nuDNA, and mtDNA + nuDNA data sets all yielded support for that branch (cf. Fig. 1 Left). ML analyses yielded a single best tree (−ln L = 14498.13736) shown in Fig. 1 Right, and ML bootstrap analysis (400 replicates) yielded a 98% support for the branch separating Madagascan from Indian ranids. Constraining the grouping of any Indian ranid within the Madagascan group (or vice versa) in a total evidence (mtDNA + nuDNA) analysis requires a minimum of 15 additional evolutionary events (in comparison to the best tree of length, TL = 3981) under MP, and a significant (P = 0.0251) reduction in likelihood under ML. Although the exact placement of the root within the Asian group could not be unambiguously identified, rooting the best tree on any Madagascan lineage could be rejected with high statistical significance (P = 0.0069) under ML. Our analyses therefore unambiguously indicate (in agreement with figure 1 in ref. 31) that all native ranids on Madagascar form a clade (i.e., a monophyletic group) and that this lineage has, within the limits of the conserved anuran body plan, experienced a dramatic diversification in morphology, reproductive strategies, and choice of habitat. The adaptive radiation on Madagascar produced more than 120 species comprising (Figs. 1 and 2): burrowing frogs, arboreal frogs, torrential frogs, and the colorful and poisonous mantellas. Concurrently, developmental and/or larval characters greatly diversified such that nine different reproductive modes have been recognized (32). For example, the tadpoles can be surface-feeders or bottom-dwellers (both with specific mouth parts), have the ability to move between different water bodies, or go through direct “terrestrial” development in the egg up to complete metamorphosis (Fig. 2).
Figure 1.
Unrooted molecular phylogeny of Asian and Madagascan Ranidae. MP analyses of the mtDNA and nuDNA data sets both yield majority rule bootstrap consensus trees compatible with that shown on the Left. The first and second values given above the branches are bootstrap values (1,000 replicates, 50 random additions each) for the analyses of mtDNA and nuDNA data sets, respectively. The first and second values below the branches indicate bootstrap values and Bremer support, respectively, for the analysis of the combined mtDNA and nuDNA data sets. Right shows the best tree under ML. Wherever the tree is rooted, there is no possibility to make Rhacophorinae, Tomopterninae, Ranixalinae, or the genus Limnonectes monophyletic. This finding indicates multiple convergences of ecomorphs among and within Madagascar, the Indian subcontinent, and Asia. Monophyly of Madagascan Ranidae (bold branch) is strongly supported (see text for details). We included a European Rana species (indicated by an asterisk) which, as expected, clusters with Asian members of the genus.
Figure 2.
Convergences in larval and adult ecomorphs between and within Madagascan and Asian ranids. All Asian species are from the Indian subcontinent, except one (Limnonectes kuhlii, indicated by an asterisk) coming from Vietnam. Adult burrowing frogs (red boxes), beside having a toad-like general morphology and short hind limbs, exhibit a suite of characters [such as feet with a crescentic inner metatarsal tubercle (red arrows) and partly connected lateral metatarsals] that are adaptations to the fossorial zone. Their tadpoles have a general body shape and mouth parts [such as rows of keratinized teeth (black arrows)] which are of the typical ancestral aquatic type. Arboreal frogs (green boxes) exhibit similar adult ecomorphs in Madagascan and Indian Rhacophorinae. Some Asian tree frogs exhibit a development on land with complete metamorphosis in the egg, showing a remarkable convergence with some Madagascan arboreal species (Mantidactylus). Some rock-dwelling frogs (yellow boxes) have semiterrestrial larvae with stout hind-limbs, a strongly developed tail with much reduced fin membranes, and with the ventral side of the body and the spiracle tube (as well as, sometimes, the anal tube) flattened. Certain torrential frogs (blue boxes) have larvae that lack horny teeth and exhibit, around the mouth, enlarged lobes that are richly provided with papillae. Fanged frogs (dashed green boxes) have protruding fangs in the lower jaw. Although poisonous frogs do not occur in Asia, some poisonous mantellas (black dashed box) exhibit aposematic colors and are remarkably convergent with the neotropical poison arrow frogs, Dendrobatidae.
Convergences Between Madagascan and Asian Radiations.
The Indian subcontinent holds several adult and larval ecomorphs that are extremely similar to those found on Madagascar. Hence, the results of our molecular phylogenetic analyses prompt revision of how variations of the anuran body plan may have evolved. For instance, the genus Tomopterna was defined as a taxon on the basis of shared characters involved in burrowing (red boxes, Fig. 2). Recent phylogenetic analyses based on 20 morphological characters clustered African, Madagascan, and Asian Tomopterna species as a monophyletic unit (33). Our analyses support, however, that the burrowing ecomorph evolved independently in Madagascan and Asian ranids: constraining the grouping of burrowing frogs to the exclusion of all other ranids resulted in an impressive and very significant increase of tree length (+123 steps, P < 10−4) and reduction of likelihood (P < 10−4).
Similarly, and as previously suggested by Richards and Moore (31), Madagascan and Asian rhacophorine tree frogs do not form a clade (Fig. 1). These brownish to bright-green frogs are ecologically characterized by an arboreal life and have very similar adult morphologies (green boxes, Fig. 2). Grouping rhacophorines to the exclusion of all other ranids requires a significant increase in tree length (+32 steps, P = 0.0183) and reduction of likelihood (P = 0.0158).
Convergences Among Asian Groups.
Our analyses indicate that convergences also occurred among Asian groups. Fanged frogs [Limnonectes (Limnonectes)] have a remarkable discontinuous distribution: they are found from North India to New Guinea, and a single species is known from Sri Lanka, whereas they are absent from Southern India. The name of the group originates from the fact that both males and females have protruding fangs in the lower jaw (dashed green boxes, Fig. 2). Our analyses indicate that fanged frogs do not form a monophyletic group (cf. Figs. 1 and 2). Constraining this monophyly requires a significant (P = 2 × 10−4) decrease in likelihood. Furthermore, ML analyses of the separated (data not shown) and combined (Fig. 1 Right) mtDNA and nuDNA data sets all indicate that the Sri Lankan fanged frog (L. corrugatus) groups with members of the South-Indian endemic genus Nyctibatrachus (which, incidentally, also possess small odontoid processes in the lower jaw).
Within the Indian subcontinent, Nannophrys and Indirana both have semiterrestrial larvae (yellow boxes in Fig. 2) that exhibit unique adaptive morphological features, such as a strongly developed tail with much reduced fin membranes, which allow them to hop over wet rocks (34). These striking similarities led to the suggestion that this suite of characters “cannot be coincidence or convergence, but has to reflect true phylogenetic relationships” (35). Nevertheless, our analyses clearly indicate that these morphological features evolved independently in Sri Lankan and Indian ranids (cf. Figs. 1 and 2). Constraining the grouping of both taxa presenting semiterrestrial larvae to the exclusion of all other ranids requires a very significant increase of tree length (+70 steps, P < 10−4) and reduction of likelihood (P < 10−4).
Discussion
In addition to the strictly aquatic or semiaquatic habitat, four other adaptive zones can be recognized for ranid frogs: arboreal, rocky, fossorial, and torrential. This diversity indicates that various lineages of ranid frogs evolved adaptations that allowed them to shift to new adaptive zones which are comparable to those observed for plethodontid salamanders (36).
A remarkable outcome of our analyses is that convergences of adult characters were often paralleled by convergences of other characters in the corresponding larvae. For example, some Asian rhacophorines (e.g., genus Philautus) and Madagascan species of the genus Mantidactylus that are adapted to the arboreal zone (green boxes, Fig. 2), also present larvae with direct development on trees (i.e., complete metamorphosis in the egg without free-swimming tadpole stage).
Similarly, the genus Nyctibatrachus (South India) and some members of the genus Mantidactylus (Madagascar) have both adult and larval stages adapted to the torrential zone (blue boxes, Fig. 2). Their larvae experienced striking convergent evolution in the oral anatomy, i.e., they lack horny labial teeth and developed lobed lips richly provided with papillae.
As a third example, both Indirana and Nannophrys (Indian subcontinent) have an adult stage that is ecologically adapted to life on rocks at the edge of hill-streams (yellow boxes, Fig. 2). As noted above, the larvae of these species have independently acquired very similar adaptations for jumping on wet rock surfaces.
These examples indicate that larvae and adults can each evolve specializations that are associated with different sets of morphological characters but that yield adaptation toward the same habitat. This association could give the impression that these adult and larval characters are genetically or functionally linked, especially as similar covariations are observed in independent lineages. We think they are not, but rather that selection on independent larval and adult characters have yielded a similar pattern of covariation as if they were.
Such spectacular covariation between adult and larval traits is obviously not observed in all ranid frog groups, as larvae and adult stages can be adapted to different adaptive zones. For example, the adults in both lineages of the (polyphyletic, see above) genus Tomopterna became adapted to the fossorial zone, whereas their larvae retained the ancestral aquatic development. Similarly, some Asian and Madagascan Rhacophorinae (e.g., genus Polypedates and Boophis, respectively) exhibit an arboreal adult stage, whereas their larvae develop in pools. Interestingly, in Polypedates species, early larval development takes place in “foam nests” laid in trees above pools. The larvae eventually fall in the pool in which they complete their development and metamorphosis. We suggest that, in Asian Rhacophorinae, foam nests could be an obligate first step toward direct development on land or trees.
Within Amphibia, extensive adaptive radiation has been demonstrated in detail only for plethodontid salamanders (7), in which both spectacular multiple convergences of morphological characters [e.g., projectile tongues (8)] and parallel evolution toward the same adaptive zones (36) were demonstrated. Our analyses here indicate (i) major adaptive radiations in ranid frogs both in larval and adult stages, (ii) multiple convergences in each of these developmental stages, and (iii) parallel evolution of larval and adult characters such that both stages are adapted to the same habitat. Points i and ii indicate that larval characters can undergo dramatic and multiple convergences as soon as they experience selective pressures of intensity and variability that are similar to those usually experienced by adult stages. Point iii suggests that constraints in anuran amphibians might yield, convergently in independent lineages, covariation between the two developmental stages. Whether this covariation is incidental (i.e., caused by strictly independent selection on larval and adult characters) or due to a yet-unascertained mechanism of larval-dependent selection on adult characters remains to be investigated. One central question will be to assess whether the frequency of species with both stages adapted to the same habitat is higher than that expected under the null hypothesis of independent selection on larvae and adults. The possibility that developmental constraints and selective pressures (7) could reduce the “morphospace” to which both adult and larval stages have access will need to be incorporated in such an investigation.
Finally, our analyses indicate that all Madagascan Ranidae have evolved from a single lineage that underwent a major radiation. This result calls for taxonomic revision (see Appendix) and increases the support for the uniqueness of the Madagascan fauna, which, in turn, strengthens the worth and need for protection of its diversity through sound and objective management.
Acknowledgments
We thank Miguel Vences and Frank Glaw for permission to use their photographs. S. Struyf, K. Roelants, and I. Tallon assisted in the graphical work. Miguel Vences, Frank Glaw, Sanath Hettiarachchi, Madura De Silva, Amal Wijesekera, Janaka Gamachchi, Malaka Bopage, and Yves Samyn kindly provided samples. G. Lenglet and P. Jouk provided a Limnonectes kuhlii and Mantella madagascariensis, respectively. A. Caccone, P. Mardulyn, A. Meyer, G. Orti, and three anonymous reviewers provided helpful comments on earlier versions of the manuscript. We are grateful to Dave L. Swofford for giving us the opportunity to use the successive beta versions of paup*. This work was supported by grants to F.B. (Fund for Scientific Research, FWO; Onderzoeksraad VUB) and M.C.M. (Belgian National Fund for Scientific Research, FNRS; the Free University of Brussels, ULB; and the Van Buuren Fund).
Abbreviations
- mya
million years ago
- mtDNA
mitochondrial DNA
- nuDNA
nuclear DNA
- MP
maximum parsimony
- ML
maximum likelihood
Appendix
Our analyses suggest that Tomopterna (Laliostoma) labrosa is the sister species of the genus Aglyptodactylus (cf. Fig. 1 Right), a result that is compatible with the observation of several adaptations for burrowing in the recently discovered Aglyptodactylus laticeps (33). Hence, these findings indicate that Tomopterna labrosa is not closely related to other Tomopterna species and we propose to raise it to the generic rank (i.e., Laliostoma labrosa).
If further work validates the monophyly of the Madagascan group, it would prompt taxonomic revisions to conform with a phylogenetic framework (37), i.e., the most recent common ancestor of the genera Mantella, Mantidactylus, Boophis, Aglyptodactylus, and Laliostoma, and all of the descendants of that ancestor should then be grouped in a single family, for which the name Mantellidae is available.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF249001–AF24919).
References
- 1.Darwin C. On the Origin of Species by Means of Natural Selection. London: John Murray; 1859. [Google Scholar]
- 2.Huxley J. Evolution: The Modern Synthesis. New York: Harper and Brothers; 1942. [Google Scholar]
- 3.Simpson G G. Tempo and Mode in Evolution. New York: Columbia Univ. Press; 1944. [Google Scholar]
- 4.Givnish T J. In: Molecular Evolution and Adaptive Radiation. Givnish T J, Sytsma K J, editors. Cambridge, U.K.: Cambridge Univ. Press; 1997. pp. 1–54. [Google Scholar]
- 5.Rüber L, Verheyen E, Meyer A. Proc Natl Acad Sci USA. 1999;96:10230–10235. doi: 10.1073/pnas.96.18.10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kocher T D, Conroy J A, McKaye K R, Stauffer J R. Mol Phylogenet Evol. 1993;2:158–165. doi: 10.1006/mpev.1993.1016. [DOI] [PubMed] [Google Scholar]
- 7.Wake D B. Am Nat. 1991;138:543–567. [Google Scholar]
- 8.Wake D B, Larson A. Science. 1987;238:42–48. doi: 10.1126/science.238.4823.42. [DOI] [PubMed] [Google Scholar]
- 9.Jackman T R, Applebaum G, Wake D B. Mol Biol Evol. 1997;14:883–891. doi: 10.1093/oxfordjournals.molbev.a025830. [DOI] [PubMed] [Google Scholar]
- 10.Losos J B, Jackman T R, Larson A, de Queiroz K, Rodriguez-Schettino L. Science. 1998;279:2115–2118. doi: 10.1126/science.279.5359.2115. [DOI] [PubMed] [Google Scholar]
- 11.Fleagle J G, McGraw W S. Proc Natl Acad Sci USA. 1999;96:1157–1161. doi: 10.1073/pnas.96.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S, Hedges B. Nature (London) 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 13.Milinkovitch M C. Trends Ecol Evol. 1995;10:328–334. doi: 10.1016/s0169-5347(00)89120-x. [DOI] [PubMed] [Google Scholar]
- 14.Smith A G, Smith D G, Funnel B M. Atlas of Mesozoic and Cenozoic Coastlines. Cambridge, U.K.: Cambridge Univ. Press; 1994. [Google Scholar]
- 15.Storey B C. Nature (London) 1995;377:301–308. [Google Scholar]
- 16.Courtillot V, Jaupart C, Manighetti I, Tapponnier P, Besse J. Earth Planet Sci Lett. 1999;166:177–195. [Google Scholar]
- 17.Dubois A. Bull Mens Soc Linn Lyon. 1992;61:305–352. [Google Scholar]
- 18.Blommers-Schlösser R M A. Ethol Ecol Evol. 1993;5:199–218. [Google Scholar]
- 19.Richards C M, Moore W S. Mol Phylogenet Evol. 1996;5:522–532. doi: 10.1006/mpev.1996.0047. [DOI] [PubMed] [Google Scholar]
- 20.Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Ann Entomol Soc Am. 1994;87:651–701. [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatesy J, DeSalle R, Wheeler W. Mol Phylogenet Evol. 1993;2:152–157. doi: 10.1006/mpev.1993.1015. [DOI] [PubMed] [Google Scholar]
- 23.Farris J S, Källersjö M, Kluge A G, Bult C. Cladistics. 1994;10:315–319. [Google Scholar]
- 24.Swofford D. Sunderland, MA: Sinauer; 1998. , Version 4.0d64 (in progress). [Google Scholar]
- 25.Goloboff P. Cladistics. 1993;9:83–91. doi: 10.1111/j.1096-0031.1993.tb00209.x. [DOI] [PubMed] [Google Scholar]
- 26.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 28.Bremer K. Cladistics. 1994;10:295–304. [Google Scholar]
- 29.Kishino H, Hasegawa M. J Mol Evol. 1998;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 30.Lyons-Weiler J, Hoelzer G A, Tausch R J. Biol J Linn Soc. 1998;64:493–511. [Google Scholar]
- 31.Richards C M, Moore W S. Herpetol J. 1998;8:41–46. [Google Scholar]
- 32.Glaw F, Vences M. A Field Guide to the Amphibians and Reptiles of Madagascar. 2nd Ed. Köln, Germany: Vences and Glaw; 1994. [Google Scholar]
- 33.Glaw F, Vences M, Böhme W. J Zool Syst Evol Res. 1998;36:17–37. [Google Scholar]
- 34.Kirtisinghe P. Ceylon J Sci Biol Sci. 1958;1:171–176. [Google Scholar]
- 35.Dubois A. Alytes. 1987;5:7–95. [Google Scholar]
- 36.Wake D B. Mem S Calif Acad Sci. 1966;4:1–111. [Google Scholar]
- 37.Ford L S, Cannatella D C. Herpetol Monogr. 1993;7:94–117. [Google Scholar]