Abstract
The mitotic kinase Aurora A (Aur-A) is required for formation of a bipolar mitotic spindle and accurate chromosome segregation. In somatic cells, Aur-A protein and kinase activity levels peak during mitosis, and Aur-A is degraded during mitotic exit. Here, we investigated how Aur-A protein and kinase activity levels are regulated, taking advantage of the rapid synchronous cell division cycles of Xenopus eggs and cell-free systems derived from them. Aur-A kinase activity oscillates in the early embryonic cell cycles, just as in somatic cells, but Aur-A protein levels are constant, indicating that regulated activation and inactivation, instead of periodic proteolysis, is the dominant mode of Aur-A regulation in these cell cycles. Cdh1, the APC/C activator that targets many mitotic proteins for ubiquitin-dependent proteolysis during late mitosis and G1 in somatic cells, is missing in Xenopus eggs and early embryos. We find that addition of Cdh1 to egg extracts undergoing M phase exit is sufficient to induce rapid degradation of Aur-A. Aur-A contains both of the two known APC/C recognition signals, (1) a C-terminal D box similar to those required for ubiquitin-dependent destruction of cyclin B and several other mitotic proteins, and (2) an N-terminal KEN box similar to that found on cdc20, which is ubiquitinated in response to APC/CCdh1. The D box is required for Cdh1-induced destruction of Aur-A but the KEN box is not. Destruction also requires a short region in the N terminus, which contains a newly identified recognition signal, the A box. The A box is conserved in vertebrate Aur-As and contains serine 53, which is phosphorylated during M phase. Mutation of serine 53 to aspartic acid, which can mimic the effect of phosphorylation, completely blocks Cdh1-dependent destruction of Aur-A. These results suggest that dephosphorylation of serine 53 during mitotic exit could control the timing of Aur-A destruction, allowing recognition of both the A box and D box by Cdh1-activated APC/C.
Keywords: Mitosis, anaphase-promoting complex/cyclosome (APC/C), proteolysis, Xenopus eggs, D box, A box
The serine/threonine kinase Aurora A (Aur-A) is required for progression through mitosis, and overexpression of Aur-A generates aneuploidy (for review, see Bischoff and Plowman 1999; Goepfert and Brinkley 2000; Nigg 2001). Aur-A was first identified as a Drosophila mutant in which centrosomes fail to separate and cells arrest in mitosis with chromosomes arranged on circular monopolar mitotic spindles (Glover et al. 1995). In C. elegans embryos lacking Aur-A, centrosomes separate normally but then collapse and fail to assemble a bipolar spindle (Schumacher et al. 1998; Hannak et al. 2001). Thus, Aur-A is required for formation of a functional bipolar spindle and the completion of mitosis. In somatic cells, Aur-A mRNA, protein, and kinase activity levels are low during G1 and S phase, rise during G2 and mitosis, and drop during mitotic exit into G1 of the next cell cycle (Gopalan et al. 1997; Kimura et al. 1997; Bischoff et al. 1998; Farruggio et al. 1999; Honda et al. 2000; Walter et al. 2000; Crosio et al. 2002). During G2 and early prophase, Aur-A protein concentrates around centrosomes and, as mitosis progresses, is also found on spindle microtubules near the poles; Aur-A relocalizes to centrosomes during telophase and is destroyed late during mitotic exit (Gopalan et al. 1997; Kimura et al. 1997; Roghi et al. 1998; Bischoff and Plowman 1999; Giet et al. 1999; Honda et al. 2000; Crosio et al. 2002; Meraldi et al. 2002). It seems likely that localization at the spindle poles, which is mediated by the N-terminal noncatalytic domain (Giet and Prigent 2001), is essential for Aur-A's function during mitosis.
Overexpression of Aur-A results in cells that contain multiple centrosomes and multipolar spindles, and these cells quickly become aneuploid (Zhou et al. 1998; Meraldi et al. 2002). In humans, the Aur-A containing locus 20q13 is amplified in many breast and colorectal cancers (Sen et al. 1997; Bischoff et al. 1998; Zhou et al. 1998). Forced overexpression of either human or Xenopus Aur-A transforms NIH3T3 cells and promotes tumor formation in nude mice; kinase-dead forms do not (Bischoff et al. 1998; T. Andresson, L. Amundedottir, P. Leder, and J. Ruderman, in prep.). Mutations leading to defects in the timing or extent of Aur-A destruction would also lead to overexpression of Aur-A, and therefore would also be expected to be deleterious.
All Aur-A family members contain two different sequences known to be required for the destruction of other mitotic proteins: the D box and the KEN box (Glotzer et al. 1991; King et al. 1996; Pfleger et al. 2001). These sequences target specific mitotic proteins for ubiquitination by the APC/C (anaphase-promoting complex/cyclosome) and subsequent proteolysis by the proteasome (for review, see Townsley and Ruderman 1998; Zachariae 1999; Tyers and Jorgensen 2000). Two major forms of the APC/C function during mitotic exit. Cdc20-activated APC/C recognizes D box-containing proteins such as cyclin B and securin, initiating their ubiquitination during the metaphase/anaphase transition. The destruction of cyclin B is followed by inactivation of cdc2 and dephosphorylation of cdc2 targets, including the APC/C activator Cdh1. Dephosphorylated Cdh1 then binds the APC/C, programming it to ubiquitinate KEN box-containing proteins including Cdc20 and a small number of other known targets. Cdh1-activated APC/C then maintains the ubiquitination-dependent destruction of several KEN box and D box-containing proteins during the completion of mitotic exit and G1 (Kramer et al. 2000; Petersen et al. 2000; Sudo et al. 2001).
Aur-A is unusual in that it contains both KEN-like and D box-like sequences and, in fact, has three potential D boxes. Previous investigations had reported variously that removal of the N-terminal KEN sequence did not block destruction (Arlot-Bonnemains et al. 2001), that none of the D box sequences were required for the in vitro ubiquitination of Aur-A (Honda et al. 2000), that mutation of the C-terminal D box completely stabilized Aur A (Castro et al. 2002), and that removal of the C-terminal D box partially stabilized and removal of second D box completely blocked destruction (Arlot-Bonnemains et al. 2001).
We find that in eggs, as in somatic cells, Aur-A kinase activity is high in M phase and low in interphase. In striking contrast, however, Aur-A protein levels remain constant across the first several embryonic cell divisions, indicating that kinase activation and inactivation is the sole mode of regulating Aur-A activity in these cell cycles. The complete stability of Aur-A after fertilization is perfectly reproduced in egg extracts undergoing mitotic exit and progression through the first cell cycle in vitro. Addition of the APC/C activator Cdh1, which is absent from eggs and appears only much later in development (Lorca et al. 1998; Kramer et al. 2000), leads to the rapid and complete destruction of Aur-A, whereas addition of Cdc20 does not. Mutation of the C-terminal D box completely stabilized Aur-A against Cdh1-dependent destruction whereas mutation of the N-terminal K6EN box did not. Additional deletions revealed the existence of an N-terminal sequence (residues 34–68) that is absolutely required for Cdh1-dependent destruction of Aur-A and can direct the degradation of the N terminus in the absence of any conventional D box. This region includes a short sequence that is highly conserved in vertebrate Aur-As and contains serine 53, a site of mitotic phosphorylation (L. Littlepage, H. Wu, J. Deanehan, and J. Ruderman, in prep.). We refer to this region as the A box. Mutation of serine 53 to aspartic acid completely blocks destruction, suggesting that dephosphorylation of ser53 during mitotic exit is a prerequisite for Aur-A degradation.
Results
Aur-A kinase activity oscillates after fertilization, but Aur-A protein levels remain constant
To monitor Aur-A levels and kinase activity, we generated and affinity-purified antibodies against the N-terminal noncatalytic domain (1–146) of Xenopus Aur-A. This antibody is highly specific: It recognizes a single band of the correct size on blots of whole cell extracts, and shows no reactivity with bands of the sizes of Aur-B or Aur-C (Figs. 1 and 3A, below). It recognizes a single band in reticulocyte translation product programmed by Aur-A mRNA and immunoprecipitates more than 80% of Aur-A protein from egg lysates (see Supplemental Fig. 1 in Supplementary Material at http://www.genesdev.org).
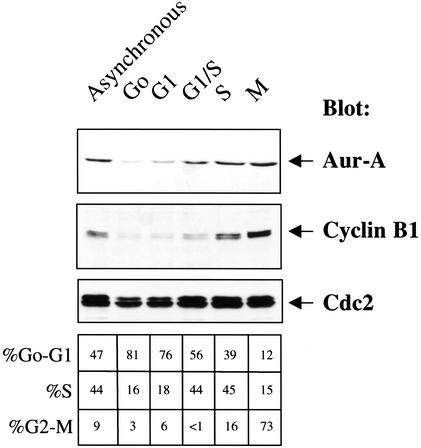
Figure 1.
Aur-A protein levels in Xenopus somatic tissue culture cells. XTC cells analyzed were synchronized as described in Materials and Methods. Fifty micrograms of protein was loaded per lane, separated by SDS-PAGE, and blotted with anti-cyclin B1, anti-cdc2 or affinity-purified Xenopus Aur-A antibodies.
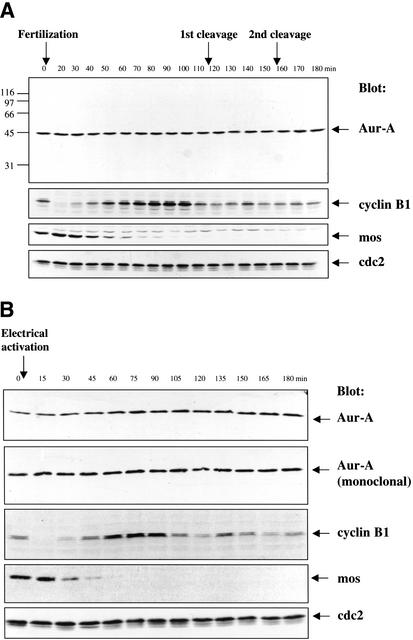
Figure 3.
Aur-A protein levels are constant following fertilization or electrical activation of eggs. (A) Fertilization. Eggs were fertilized, and samples were taken at 10-min intervals. Samples were analyzed by SDS-PAGE followed by Western blotting with the indicated antibodies. The equivalent of one embryo was loaded per lane. (B) Electrical activation. Eggs were electrically activated, and samples were taken at 15-min intervals. Samples were analyzed by SDS-PAGE followed by Western blotting with the indicated antibodies. The equivalent of one-half an embryo per time point was loaded per lane.
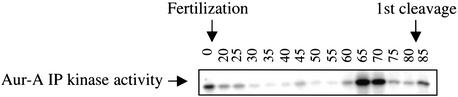
As in mammalian somatic cells, the amount of Aur-A protein peaks during mitosis (Fig. 1). To identify the pathways that regulate the periodic accumulation, activation, and destruction of Aur-A, we chose to exploit the simplified cell cycles of Xenopus eggs, where it is possible to obtain large amounts of cells that proceed through mitosis synchronously, and concentrated cytoplasmic extracts derived from these cells that faithfully reproduce many events of cell cycle progression in vitro. To determine whether Aur-A kinase activity oscillates across the first embryonic cell cycles, metaphase II-arrested eggs were fertilized and samples were taken at 10-min intervals. Aur-A was immunoprecipitated from each sample and assayed for in vitro kinase activity towards myelin basic protein. As shown in Figure 2, Aur-A kinase activity was high in eggs, dropped after fertilization, remained low during interphase, rose during mitosis 1 and dropped again at the end of mitosis 1.
Figure 2.
Aur-A kinase activity oscillates during the early embryonic cell cycle. Eggs were fertilized and collected at 10-min intervals following fertilization. Using the equivalent of five eggs, Aur-A was immunoprecipitated with affinity-purified Aur-A antibody and assayed for kinase activity towards MBP.
Surprisingly, immunoblots revealed that Aur-A protein levels were constant across the first three cycles (Fig. 3A). Several synchrony and loading controls established that this result was not due to poor synchrony or uneven loading. Cyclin B showed its characteristic destruction, resynthesis, and loss during the first cycle, Mos was degraded on schedule and failed to reappear during subsequent cycles, and cdc2 remained constant, all as expected (Watanabe et al. 1991). Identical results were obtained in numerous independent experiments.
The finding that Aur-A protein levels remained constant across the first three cell cycles differed from a previous report that Aur-A protein is degraded within 30 min of electrical activation, a treatment that mimics most events initiated by fertilization (Frank-Vaillant et al. 2000). In view of Aur-A's localization at centrosomes, its role in spindle function, and the fact that the sperm provides the centrioles that organize the first mitotic spindle (Shaver 1953; Maller et al. 1976; Masui et al. 1978), it seemed possible that the different fates of Aur-A following fertilization versus activation could be a spindle- or centrosome-dependent difference in the way Aur-A degradation is regulated. Since the postactivation disappearance of Aur-A had been obtained using a Xenopus Aur-A monoclonal antibody (Roghi et al. 1998), it seemed possible that the disappearance of the Aur-A signal on blots might reflect epitope masking during exit from the metaphase II arrest. However, as shown in Figure 3B, neither of these possibilities proved to be the explanation. Following electrical activation of eggs, Aur-A protein levels on immunoblots appeared constant when probed with either our affinity-purified polyclonal antibody or the monoclonal antibody. In addition, as shown in Figure 4, Aur-A protein levels remained constant even when ongoing protein synthesis was blocked with cycloheximide. Thus, we conclude that Aur-A protein does not undergo significant degradation, either following fertilization or parthenogenetic activation, and that Aur-A protein levels remain constant across the first few cleavage division cycles. Taken together, these results also reveal that posttranslational regulation of Aur-A's kinase activity provides the major mode of regulating its function during the embryonic cell cycles.
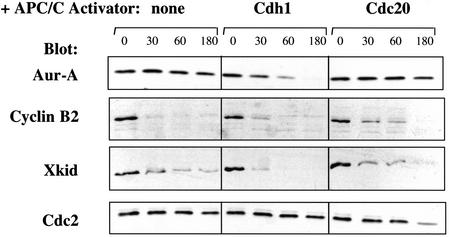
Figure 4.
The addition of Cdh1 induces destruction of Aur-A during mitotic exit. Recombinant Cdh1 or Cdc20 protein was added to CSF-arrested extracts, and calcium was added to induce M-phase exit. Samples were taken at 0, 30, 60, and 180 min after calcium addition and analyzed by SDS-PAGE followed by blotting with the indicated antibodies.
Aur-A is stable in extracts of activated eggs and degraded upon addition of the APC/C activator Cdh1
The regulated proteolysis of several proteins during mitotic exit is dependent on the presence of one of two known recognition signals, the KEN box and the D box. Aur-A contains sequences resembling both types of destruction motifs. Cdc20 is present in eggs and early embryos, but Cdh1 protein is undetectable and appears only much later in development (Lorca et al. 1998; Kramer et al. 2000; S. Rankin and M. Kirschner, pers. comm.). Because (1) Aur-A contains a KEN sequence similar to those required for Cdh1-dependent destruction of other proteins, (2) Cdh1 is not present in the early cell cycles, and (3) Aur-A is stable in the early cell cycles, we reasoned that the stability of Aur-A might be due to the absence of Cdh1. Prompted by the finding that addition of recombinant Cdh1 protein to Xenopus egg interphase extracts can induce destruction of Cdc20 and two other KEN-box proteins (Pfleger and Kirschner 2000), we investigated whether addition of Cdh1 would trigger destruction of Aur-A during M phase exit in eggs. Extracts were prepared from metaphase II-arrested eggs (CSF extracts), cycloheximide was added to block further protein synthesis, calcium was added to induce M phase exit, and the fate of endogenous Aur-A protein was followed on immunoblots. As expected (Glotzer et al. 1991; Funabiki and Murray 2000), cyclin B and another D box protein, the chromokinesin Xkid, were rapidly degraded, whereas cdc2 remained stable (Fig. 4, left panels). Addition of recombinant Cdh1 protein induced complete destruction of endogenous Aur-A (top panel), whereas addition of Cdc20 did not. This result strongly suggests that (1) Aur-A is targeted for destruction by Cdh1-activated APC/C, and (2) the lack of Cdh1 accounts for the lack of Aur-A degradation during the early embryonic cell cycles. Consistent with this idea is the recent report that recombinant Aur-A is degraded in lysates of G1-stage Xenopus somatic tissue culture cells, which are presumed to contain Cdh1 (Arlot-Bonnemains et al. 2001).
Mutation or deletion of the R378 D box stabilizes Aur-A during mitotic exit
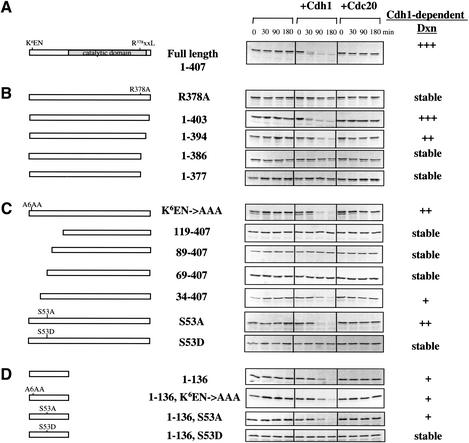
In recent work, Prigent and colleagues investigated regions required for destruction of Aur-A in somatic cells during G1 phase (Arlot-Bonnemains et al. 2001). They found that mutation or removal of the most highly conserved D box sequence, R378xxLxxVxE, blocked the disappearance of Aur-A in lysates of G1 cells, whereas deletion of the sequence K6EN did not. To determine whether the D box or KEN box is required for Cdh1-dependent destruction during mitotic exit in eggs, we used the following approach. Extracts were prepared from metaphase II-arrested eggs, radiolabeled translation product encoded by wild-type or mutant Aur-A was added in the presence or absence of Cdh1 or Cdc20, and calcium was then added to induce mitotic exit. As shown in Figure 5A, wild-type Aur-A translation product showed the same pattern of Cdh1-dependent destruction seen for endogenous Aur-A: it was stable during mitotic exit and rapidly degraded in the presence of Cdh1. Truncations that remove the C-terminal D box R378xxLxxVxE stabilized Aur-A in this assay. A single point mutation in the D box, Arg378Ala, also blocked the Cdh1-dependent destruction of Aur-A during mitotic exit (Fig. 5B).
Figure 5.
The C-terminal D box and the N-terminal region (34–68), that is, the A box, are required for Cdh1-dependent destruction of Aur-A, but the KEN box is not. CSF extracts containing cycloheximide were incubated with Cdh1 or Cdc20, as indicated, and mixed with the indicated Aur-A in vitro translation products. Calcium was added to initiate M-phase exit, and samples were taken at 0, 30, 90, and 180 min. Samples were analyzed by SDS-PAGE followed by autoradiography. (A) Full-length wild-type Aur-A. (B) C-terminal truncations and D box point mutation. (C) N-terminal mutations. (D) The N-terminal noncatalytic domain.
From these results, we conclude that (1) the R378 D box is required for destruction during both mitotic exit (Fig. 5) and G1 (Arlot-Bonnemains et al. 2001), (2) the R378 D box functions as a Cdh1 recognition site, and (3) when the R378 D box is removed, the two other less conserved D box-like sequences do not function as alternative degradation signals. Because Aur-A is stable in the early cycles, when endogenous Cdc20 is active, and remains stable even when excess Cdc20 is added, we can further conclude that (4) the R378 D box functions as a Cdh1-specific recognition signal.
Identification of the A box, a novel APC/C recognition signal
Mutation (Fig. 5C) or deletion (data not shown) of the KEN box did not block Cdh1-dependent destruction during mitotic exit in eggs. Although the KEN box itself is not required for Aur-A destruction, either during mitotic exit or G1, analysis of further truncations revealed the existence of a short region within the N terminus which is essential for Cdh1-dependent destruction (Fig. 5C). Removal of the first 33 residues slightly delayed but did not block destruction. In contrast, removal of the first 68 residues completely stabilized Aur-A against Cdh1-induced destruction. Thus, the region 34–68 contains a novel APC/CCdh1 recognition signal.
This region (Fig. 6) contains a short sequence, Q47RILGPSNVPQRV, which is highly conserved in vertebrate Aur-As. This region also includes serine 53, a residue that is phosphorylated during mitosis (see Materials and Methods). Because this sequence is found in all vertebrate Aur-As, but not in Aur-B or C, we refer to this region as the A box. Mutation of ser53 to alanine did not block destruction, but mutation to aspartic acid (a residue that can often mimic a phosphorylated ser or thr residue) completely blocked Cdh1-induced destruction (Fig. 5C). This result strongly suggests that mitotic phosphorylation of Aur-A at ser53 blocks destruction and that Cdh1-dependent destruction during mitotic exit requires dephosphorylation of serine 53. As such, this would represent the first example where recognition of a target by the APC/C is regulated by the phosphorylation status of the target protein.
Figure 6.
Sequence of the conserved Aur-A A box region, which is required for Cdh1-dependent destruction. Residues conserved between Aur-A and Aur-B in this region are highlighted. Serine 53 is indicated in bold.
Surprisingly, the entire N-terminal noncatalytic domain, residues 1–136, by itself was degraded following Cdh1 addition (Fig. 5D). A hybrid protein consisting of the entire N terminus human Aur-A fused to GFP was also degraded (R. Crane and L. Littlepage, unpubl.), indicating that this region contains a portable destruction domain. Mutating K6EN to AAA did not interfere with destruction of the N-terminal fragment, further confirming that the conserved KEN sequence is not required. In contrast, mutating ser53 to asp completely blocked Cdh1-induced destruction of the N-terminal fragment (Fig. 5C). These findings suggest that the A-box serves as the primary site regulating Aur-A destruction. The ability of the N-terminal domain by itself to undergo Cdh1-induced destruction suggests an ordered pathway for Aur-A destruction, in which dephosphorylation of Aur-A late in mitosis is followed by D box-dependent APC/CCdh1-mediated ubiquitination of the C terminus, which then permits recognition of the A box, followed by full ubiquitination and destruction of Aur-A (see Discussion).
The effects of expressing nondegradable Aur-A mutants in somatic cells are similar to those seen with wild-type or inactive Aur-A
The vast majority of maternal Aur-A protein remains stable during the early cell cycles, clearly indicating that these cycles proceed in the presence of high Aur-A protein levels. Therefore, we turned to mammalian somatic cells to investigate whether the introduction of nondegradable Aur-A would affect cell cycle progression and if the two different types of nondegradable mutants would have similar or different effects.
Xenopus wild-type and mutant constructs were subcloned into the retroviral vector pMSCV-IRES-GFP, which expresses the inserts at low levels. This vector was chosen in hopes of being able to reduce effects due simply to overexpression. NIH3T3 cells were infected with constructs encoding wild-type Aur-A, a kinase-dead mutant (K169R), Aur-A lacking the N-terminal 118 residues, or Aur-A containing the single D box point mutation, R378A. Several independently derived stable cell lines were isolated for each construct. Although the tagged Aur-A proteins were expressed at levels too low to be reliably detectable on immunoblots, expression was readily confirmed by RT-PCR (see Supplemental Fig. 2 in Supplementary Material at http://www.genesdev.org).
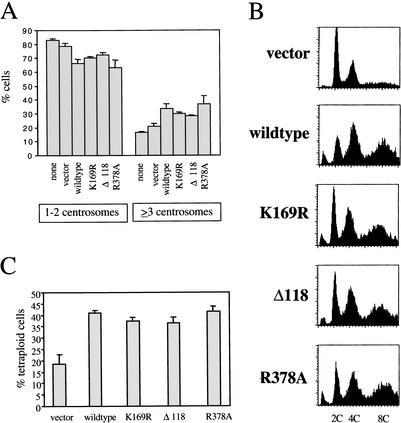
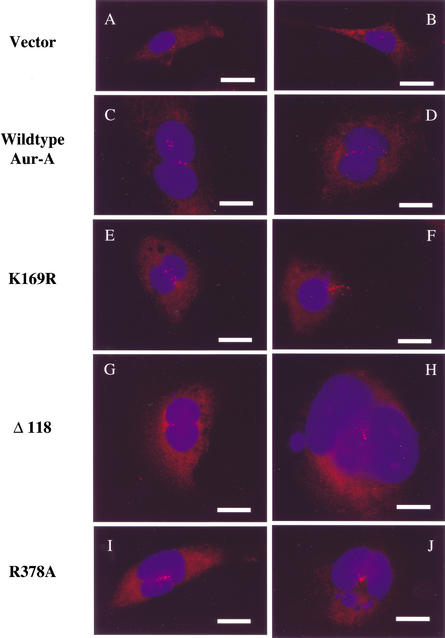
Cells were stained with γ-tubulin antibodies to monitor centrosomes and with Hoechst to reveal nuclei. GFP-positive cells were examined by fluorescence microscopy. Cells carrying wild-type Aur-A showed a modest decrease in cells with the normal number of centrosomes per cell (one or two) and a corresponding increase in cells containing three or more centrosomes (Fig. 7A), in agreement with previous work (Zhou et al. 1998; Meraldi et al. 2002). Many of those cells contained numerous centrosomes (Fig. 8). Surprisingly, the same was true of cells expressing the Aur-A K169R mutant, which has no detectable kinase activity (Andresson and Ruderman 1998). This result indicates that the kinase activity of the ectopically expressed Aur-A is not required for its ability to generate cells with abnormally high numbers of centrosomes. Cell lines stably infected with wild-type or kinase-dead Aur-A showed a substantial increase in the number of binucleate cells with 4C or greater DNA content, and almost all of those cells had multiple centrosomes (Figs. 7B and 8; data not shown). A similar study was reported very recently by Meraldi et al. 2002), where wild-type and kinase-dead Aur-A were expressed using transient transfection. By monitoring the effects shortly after transfection, those authors concluded that centrosome amplification and aneuploidy were not, as commonly assumed, the results of increased centrosome duplication during S phase but were instead the consequence of defects in mitotic exit and cytokinesis. These results indicate that the major way in which Aur-A overexpression interferes with mitotic exit is not by sustained phosphorylation of substrates but instead through noncatalytic interactions with other proteins.
Figure 7.
Expression of wild-type, kinase-dead, and nondegradable Aur-A mutants in NIH3T3 cells all result in an increase in cells containing multiple centrosomes and greater than 2N amounts of DNA. NIH3T3 cells were infected with retroviruses expressing vector alone, wild-type Aur-A, K169R, Δ118, or R378A, as indicated, and stable, GFP-expressing cell lines were isolated. (A) Aur-A expression results in increased centrosome number per cell. DNA was visualized using Hoechst (blue) and centrosomes using γ-tubulin antibody (red). Centrosome number was counted in GFP-positive cells. Graph shows compiled results from four experiments for vector, wild-type, and R378A-infected cells and three experiments for none, K169R-, and Δ118-infected cells; 200–450 cells were counted per experiment. (B) Aur-A expression results in cells with greater than 2N amounts of DNA. DNA content of GFP-positive cells was determined by FACS. Similar results were seen in three additional experiments. (C) Aur-A expression results in cells with multiple nuclei. Cells were scored for DNA content by FACS. Graph shows results from four experiments.
Figure 8.
Expression of wild-type, kinase-dead, and nondegradable Aur-A mutants in NIH3T3 cells all result in centrosome amplification and multinucleation. NIH3T3 cells stably infected with retroviruses expressing vector alone (A,B), wild-type Aur-A (C,D), K169R (E,F), Δ118 (G,H), and R378A (I,J) were stained for DNA with Hoechst (blue) and centrosomes with γ-tubulin (red). GFP-positive cells were selected for these images. Bars, 25 μm.
To test the effects of nondegradable versions of Aur-A and to ask whether these effects would be similar or different depending on the type of stabilizing mutation, we examined cell lines carrying Aur-A lacking the N terminus (Δ1–118) or mutated in the D box (R378A). Expression of either mutant led to roughly the same fraction of cells expressing the same defects seen with wild-type and kinase-dead Aur-A. Both led to an increase in cells with more than two centrosomes and 4C or greater DNA content (Fig. 7), and in many cases, cells with large numbers of centrosomes (Fig. 8). Thus, both nondegradable mutants had easily detectable effects, indistinguishable from those obtained with wild-type or kinase-dead Aur-A. This result, which was somewhat unexpected, suggests that in somatic cells, even modest increases in Aur-A are sufficient to quickly generate aneuploidy.
Discussion
The work presented here establishes four main points. First, in contrast to its periodic synthesis and destruction in the somatic cell cycles, Aur-A is maintained at constant levels during the early embryonic cell cycles, and oscillations in its kinase activity are regulated solely at the posttranslational level. Second, this stability appears to be due to the lack of the APC/C activator Cdh1, addition of which induces rapid destruction during mitotic exit. Mutation of the C-terminal D box blocks Cdh1-induced destruction, but mutation of the N-terminal KEN box does not. Third, in addition to the D box, destruction requires a newly identified APC/C recognition signal, the A box, which is located within the N-terminal noncatalytic domain. Fourth, the A-box contains a conserved residue, serine 53, a site of mitotic phosphorylation. Mutation of serine 53 to alanine does not block Cdh1-induced destruction of Aur-A but mutation to aspartic acid does, suggesting that phosphorylation might negatively regulate Aur-A destruction until the last stages of mitotic exit. The sequence surrounding serine 53 does not resemble any known kinase consensus sequence, raising obvious questions about the identity of the Aur-A serine 53 kinase and how phosphorylation at this site is regulated.
Earlier work had reported that Aur-A was degraded during mitotic exit in eggs (Frank-Vaillant et al. 2000). We find, however, that Aur-A levels are constant following fertilization and during the early embryonic cell cycles and that its stability can be attributed to a lack of endogenous Cdh1 protein in eggs and early embryos. This finding agrees with those of Castro et al. (2002), which appeared after the completion of our work. In retrospect, the stability of Aur-A protein in eggs and early embryonic cycles is not surprising in view of the fact that Aur-A mRNA is deadenylated and ceases translation following fertilization (Paris et al. 1991; for review, see Wickens et al. 2000).
Despite the presence of three D box-like sequences, in Aur-A, a single point mutation in the R378 D box is sufficient to completely block Cdh1-induced destruction during mitotic exit in eggs, in agreement with the recent results of Castro et al. (2002). In contrast, in studies where the stability of Aur-A was followed in lysates of G1 cells, mutations in both the R378 D box and a second D box were required to fully stabilize Aur-A (Arlot-Bonnemains et al. 2001). One possible explanation of this difference is that the R378 D box is the only one used during mitotic exit, as in early embryos which lack G1 phases, whereas both D boxes are required to maintain destruction during G1, an idea that is currently being tested.
Full-length Aur-A requires both an A box and a D box for its Cdh1-dependent destruction. Other examples are known of proteins, such as cdc6, the kinase Hsl1p, and some cyclin A proteins, whose APC/C-mediated destruction requires two signals (the KEN box and the D box for complete destruction; Petersen et al. 2000; Burton and Solomon 2001). In those cases, however, mutation in either sequence alone only partially stabilized those proteins (Petersen et al. 2000; Jacobs et al. 2001; Kaspar et al. 2001). Aur-A is different in that mutation of either the A box or the D box completely stabilized the protein. Taken together, these results may have indicated that efficient recognition and ubiquitination of Aur-A strictly requires both the A box and the D box. However, the N-terminal noncatalytic domain by itself, which contains the A box, is rapidly and completely degraded in response to Cdh1. This result suggests that a multistep pathway of regulated Aur-A ubiquitination may exist. One possibility is that recognition of both the A box and D box is blocked during mitosis by phosphorylation of serine 53. Dephosphorylation of serine 53 during mitotic exit could result in a conformational change that makes the D box accessible to binding and ubiquitination by the APC/C which, in turn, could result in a second conformational change that then allows recognition of the A box by the APC/C, followed by full ubiquitination and degradation. In this model, the A box in the context of the full-length protein would be blocked from APC/C recognition by the nonubiquitinated D box region and unmasked following D box-dependent ubiquitination. When expressed as a short domain, the dephosphorylated A box in the N-terminal fragment 1–136 would be accessible to recognition by the APC/CCdh1. Although this model remains to be tested, dephosphorylation of the A box as the first step is most consistent with the fact that partially ubiquitinated Aur-derivatives, either directed by the A box or the D box, were never seen to accumulate when full-length proteins were assayed. This suggests that dephosphorylation of the A box probably represents the first step.
Cyclin B provides the earliest and, even now, the best understood example of how mutation of a destruction motif affects cell cycle progression. Single point mutations in the cyclin B D box completely block its destruction during mitotic exit, resulting in constitutive activation of its partner kinase cdc2 and cell cycle arrest at anaphase onset (Glotzer et al. 1991; Holloway et al. 1993; Parry and O'Farrell 2001; Stemmann et al. 2001). Stable cyclin A also has dramatic effects, delaying or blocking progression through metaphase, depending on the experimental set-up (den Elzen and Pines 2001; Jacobs et al. 2001; Parry and O'Farrell 2001). Recent studies in which Aur-A was introduced by transient transfections, which typically lead to considerable overexpression, had shown that higher than normal levels of Aur-A result in abnormal cells but do not block cell cycle progression (Meraldi et al. 2002). However, the specific effects of nondegradable mutants were not tested. Hoping to distinguish between a simple overexpression phenotype resulting from the accumulation of nondegradable Aur-A and the effect of a nondegradable mutant, we used retroviral-mediated infection to obtain stable cell lines of early passage NIH3T3 cells expressing very low levels of Aur-A. Introduction of either type of nondegradable mutant had the same effects, with no evidence of specific arrest points. It is of course possible that live analysis of single cells carrying mutant versus wild-type Aur-A, or expression of the mutants at higher levels, will reveal important differences, and those studies are underway.
Even modest overexpression of catalytically inactive Aur-A rapidly generates aneuploid cells containing multiple centrosomes, probably through association with interacting proteins such as scaffolds, upstream activators, or downstream targets (Ferrell 2000). In contrast, Aur-A's ability to transform cells and cause tumors absolutely requires its catalytic activity (Bischoff et al. 1998; T. Andresson, L. Amundedottir, P. Leder, and J. Ruderman, in prep.). Thus, in addition to looking for interacting proteins, identifying additional downstream targets of Aur-A is of considerable interest. The finding that Aur-A kinase activity oscillates after fertilization in the face of constant protein levels indicates that, in the early embryonic cycles, periodic activation and inactivation of its kinase activity is the sole mode of regulating Aur-A function. The Xenopus egg system, which is unencumbered by the complications of ongoing Aur-A transcription, synthesis, and destruction, provides an especially favorable system for identifying the upstream regulators that control the periodic activation and inactivation of Aur-A kinase activity.
Materials and methods
Fertilization and electrical activation of eggs
Eggs were collected and fertilized as described (Heasman et al. 1991). Briefly, frogs were induced to lay eggs by injection of 500 IU of chorionic gonadotropin (Sigma, CG-10). Eggs were collected, washed in 1× MMR (100 mM NaCl, 2 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 0.1 mM EDTA, 5 mM HEPES at pH 7.8), and mixed with sperm, and fertilization was initiated by the addition of 0.1× MMR. Ten embryos were collected per time point and lysed by homogenization in 100 μL NP-40 buffer (50 mM Tris at pH 7.4, 150 mM NaCl, 0.1 mM EDTA, 1% NP-40). Samples were spun at 14,000 rpm for 5 min and cytoplasmic fractions were collected. For Aur-A, mos, cdc2, and cyclin B1 immunoblots, the equivalent of one egg was loaded per lane. Eggs were electrically activated as described (Murray 1991). Briefly, eggs were dejellied, suspended in 0.2× MMR, placed in an electrical activation chamber (Murray 1991), and pulsed with 12V of current.
Egg extracts
Concentrated “CSF” extracts were prepared from unfertilized eggs, which arrest naturally in metaphase II, as described (Murray 1991). Extracts were made in the presence of 100 μg/mL cycloheximide, aliquoted, frozen in liquid nitrogen, and stored at −80°C.
Construction of Aur-A mutants
Mutations were created by PCR using primers designed to yield desired mutants. Point mutants were introduced using a QuickChange Site-Directed Mutagenesis Kit (Stratagene). Constructs used for in vitro translation were tagged at their N terminus with the AU1 epitope (Andresson and Ruderman 1998), subcloned into the pCS2+ vector, and expressed using coupled transcription and translation reaction (TnT Kit, Promega). For expression in Sf9 cells, N-terminally histidine-tagged constructs were cloned into the pFastBacHT vectors (GIBCO-BRL Life Technologies). Constructs used for retroviral expression of AU1-tagged Aur-A mutants were inserted into pMSCV [original vector gift from Owen Witte (University of California at Los Angeles, Los Angeles, CA) and modified by Joan Brugge (Harvard Medical School, Boston, MA) for use in the Gateway Cloning System (Invitrogen Life Technologies)]. All constructs were sequenced to verify mutations and to ensure that no additional mutations were introduced. For detailed information on clone constructions, please contact the authors.
Antibodies
Rabbit antisera generated against a bacterially expressed fusion protein containing GST fused to the unique N terminus of Aur-A (1–146) was characterized previously (Mendez et al. 2000). For affinity purification, his-tagged Aur-A protein was purified from Sf9 cells (Andresson and Ruderman 1998) and attached to HiTrap affinity columns (Amersham Pharmacia Biotech) following the manufacturer's directions. Antiserum was affinity-purified according to Harlow and Lane (1988). Antibodies were eluted at low pH (100 mM glycine at pH 2.5), high pH (100 mM triethylamine at pH 11.5), and in the presence of organic solvent (50% ethylene glycol at pH 10.5). Fractions were dialyzed overnight in PBS (8 g/L NaCl, 0.2 g/L KCl, 1.44 g/L Na2HPO4, 0.24 g/L KH2PO4 at pH 7.2), and assayed by immunoblotting, and selected fractions were pooled and stored at −80°C. Affinity-purified Aur-A antibody recognized a single band on blots of whole-cell extracts of both Xenopus tissue culture cells and eggs (Figs. 1 and 3). This antibody also recognized a single band on blots of reticulocyte translation product programmed by Aur-A mRNA and was also able to immune-deplete more than 80% of Aur-A from cell extracts (Supplemental Fig. 1 in Supplementary Material at http://www.genesdev.org).
All samples were analyzed by SDS-PAGE using the standard Laemmli system (Laemmli 1970). This system gives a more accurate estimation of Aur-A protein amounts (Frank-Vaillant et al. 1999) than does the modified Anderson gel system (Anderson et al. 1973), which is more useful for distinguishing unmodified from phosphorylated forms of Aur-A (Andresson and Ruderman 1998). For immunoblotting, samples were transferred from gels onto nitrocellulose paper (Bio-Rad) using a Hoefer semi-dry transfer unit. Blots were blocked with 3% nonfat dry milk made in TBS-T (10 mM Tris at pH 7.4, 150 mM NaCl, 0.1% Tween-20) and then incubated with the following antibodies: affinity-purified Xenopus Aur-A (1:2000 dilution in 1% BSA); Xenopus Aur-A monoclonal (1:100 dilution in 1% BSA, gift from Claude Prigent, Universite de Rennes, Rennes, France; Roghi et al. 1998); Mos [1:1000 dilution of MosXe(C237) antibody; Santa Cruz Biotechnology, sc-86]; Xkid (1:2000 dilution of Xkid antibody, gift from Hironori Funabiki and Andrew Murray, Harvard University, Cambridge, MA; Funabiki and Murray 2000); Cyclin B1 (1:10,000 in 1% BSA, gift from Jim Maller, University of Colorado, Denver, CO; Taieb et al. 2001); Cyclin B2 (1:500 dilution in 4% milk, gift from Tim Hunt, Clare Hall Labs, South Mimms, UK; Hochegger et al. 2001). Blots were washed three times in TBS-T and incubated with a secondary antibody. The secondary antibody conditions used were as follows: 1:5000 dilution of horseradish peroxidase-conjugated rabbit secondary antibody (Amersham) in 3% milk, 1:2000 dilution of horseradish peroxidase-conjugated mouse secondary antibody (Amersham) in 3% milk or 1:50,000 dilution of horseradish peroxidase-conjugated anti-sheep secondary antibody (Jackson ImmunoResearch) in 2.5% donkey serum (Jackson ImmunoResearch). Blots then were washed three times in TBS-T. Antibodies were detected using enhanced chemiluminescence (ECL; Amersham Pharmacia) and visualized by autoradiography (Marsh, Blue Sensitive autoradiography film).
Aur-A and cdc2 kinase assays
Eggs were fertilized and 10 embryos were collected at each time point. Embryos were resuspended in extraction buffer (0.25M sucrose, 0.1M NaCl, 2.5 mM MgCl2, 20 mM HEPES at pH 7.6) containing 1× protease inhibitors, EDTA-free, that were resuspended in dH2O (Roche) and 10 μg/mL PMSF. Buffer was removed by pipetting, embryos were crushed by spinning at 14,000 rpm for 5 min, and the cytoplasmic fraction was collected. Samples were precleared by incubation with 20 μL protein A-sepharose beads CL-4B (Amersham Pharmacia Biotech) for 1 h at 4°C. For Aur-A, the equivalent of five embryos was incubated with 20 μL Aur-A affinity-purified antibodies plus 30 μL protein A-sepharose beads for 2 h at 4°C. Then samples were washed twice in NP-40 buffer plus 10 μg/mL PMSF and once in Aur-A kinase buffer (20 mM HEPES at pH 7.6, 10 mM MgCl2, 0.1 mM EGTA). For kinase reactions, samples were resuspended in Aur-A kinase buffer plus 0.5 mM DTT, 50 μM ATP, 10 μCi 32P-γ-ATP and 4 μg substrate myelin basic protein (MBP, GIBCO-BRL). For cdc2 kinase assays, the equivalent of one embryo was incubated with 10 μL p13Suc1 beads (Upstate Biotechnology) under the conditions used for Aur-A except that, during the final wash, the samples were washed once in H1 kinase buffer (80 mM B-glycerophosphate, 15 mM MgCl2, 20 mM EGTA) and resuspended in H1 kinase buffer plus 50 μM ATP, 5 μCi 32P-γ-ATP and 1 μg histone H1 substrate (Roche 223549).
Aur-A destruction assays
Twenty microliters CSF extract containing cycloheximide was mixed with 2.5 μL (1.5 μg) baculovirus-expressed, purified Cdh1 or Cdc20 (Pfleger and Kirschner 2000) and incubated for 5 min on ice. To monitor destruction of endogenous Aur-A, 2.5 μL of 4 mM CaCl2 was added to yield a final concentration of 0.4 mM to induce mitotic exit, and incubations were carried out at 23°C. Five microliter aliquots were taken at the indicated times and mixed with 95 μL SDS gel sample buffer. The equivalent of 0.5 μL of extract (about four eggs) was loaded per lane. To follow the stability of Aur-A mutants, these were expressed as 35S-labeled in vitro translation products that had been synthesized using the TnT coupled reticulocyte lysate system (Promega) containing 35S-labeled methionine and cysteine. One microliter translation product was added to CSF extract containing Cdh1 or Cdc20, and incubated for an additional 15 min on ice and then activated by the addition of CaCl2.
Identification of sites of mitotic phosphorylation on Aur-A
His6 tagged Aur-A was cloned into pFastBacHT (GIBCO-BRL Life Technologies) and baculovirus was isolated using the BAC-TO-BAC baculovirus expression system (GIBCO-BRL Life Technologies). Virus was amplified and Aur-A was recovered from Sf9 cells. To obtain inactive, nonphosphorylated Aur-A (Aur-AKI), proteins were expressed in the presence of serine/threonine kinase inhibitors (Calbiochem) for 4 h before harvesting. Aur-A was purified on Ni-NTA agarose beads (QIAGEN). To determine sites of mitotic phosphorylation, Aur-AKI was incubated with oocyte (interphase) extracts or with extracts prepared from metaphase II-arrested eggs. His-tagged Aur-A was recovered on nickel beads using 20 mM B-glycerophosphate, 10 mM HEPES-KOH at pH 7.7, 5 mM EGTA, 5 mM B-mercaptoethanol, 150 mM NaCl, 1% Chaps, 1 mM PMSF, and protease inhibitors (Roche protease inhibitor tablets, EDTA-free). Samples were washed 4× in pulldown buffer, boiled in sample buffer, and separated by SDS-PAGE. Coomassie-stained Aur-A bands were cut out and analyzed by mass spectrometry by William Lane at the Harvard Microchemistry Facility. Briefly, proteins were analyzed by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (μLC/MS/MS) on a Finnigan LCQ DECA quadrupole ion trap mass spectrometer. No phosphorylation sites were detected on Aur-A that had been incubated with oocyte extract. Three sites, including ser53, were identified on Aur-A samples that had been incubated with metaphase II egg extract. A full description of the other mitotic phosphorylation sites, and the sites/regions required for kinase activation and inactivation is underway (L. Littlepage, H.Wu, J. Deanehan, and J. Ruderman, in prep.).
XTC cell culture and synchronization
XTC cells (gift from Tim Mitchison, Harvard Medical School, Boston, MA) were grown at room temperature (22°C) in the dark in L-15 media diluted 3.5:1 in H2O and supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin. NIH3T3 cells (passage 2, gift from Frank McKeon, Harvard Medical School, Boston, MA) were grown at 37°C with 10% CO2 in DMEM supplemented with 10% newborn calf serum, 1% penicillin-streptomycin, and 1% glutamine. Cells were synchronized at the indicated stages as follows. For G0, cells were collected after incubation in serum-free media for 43 h. For G1, G0-arrested cells were released into serum-containing media and collected after 2 h. For G1/S, cells were incubated with media containing 2 mM thymidine for 16 h, washed with PBS, released into regular media for 12 h, and then incubated in media containing 2 mM thymidine for 15 h. S-phase cells were prepared by releasing G1/S phase cells into regular media minus thymidine and collected after 5 h. M-phase cells were prepared by incubation in media containing 2 mM thymidine for 18 h, washed in PBS, released into regular media for 2 h and incubated with 100 ng/mL nocodazole for 12 h. Mitotic cells were then collected by shake-off.
Retrovirus-mediated expression of Aur-A in NIH3T3 cells
Retroviruses were prepared by transient transfection (TransIT transfection, Mirus) of retroviral plasmids carrying the indicated Aur-A mutants into Phoenix-Eco retroviral producer cells (gift from Garry Nolan, Stanford School of Medicine, Stanford, CA; http://www.stanford.edu/group/nolan). Cells were grown at 37°C with 10% CO2 in DMEM supplemented with 10% FCS, 1% penicillin-streptomycin, and 1% glutamine. Viral supernatant was collected 48 h after the transfection, and aliquots were frozen at −80°C. To infect NIH3T3 cells on a 10-cm plate, 2 mL of virus was mixed with 2 mL of media and 5 μg/mL polybrene (Sigma, H9268) and incubated with cells for 4 h. Five milliliters of media was added, and cells were incubated overnight. Fresh media was added and cells were allowed to express virus for several days before splitting cells for analysis and for freezing stably transfected cell lines.
Immunofluorescence
NIH3T3 cells carrying the indicated Aur-A mutants were seeded onto 18 mm2 coverslips with 10,000 cells plated per coverslip. Following an overnight incubation, the samples were fixed with 3% formaldehyde prepared in PBS (GIBCO-BRL Life Technologies) for 7–10 min and washed 4× in PBS. Then samples were fixed with −20°C methanol for 5 min at −20°C, washed 4× with PBS, and blocked with 3% milk made in PBST (PBS plus 0.1% Triton X-100) for 30 min. Cells were stained for centrosomes using anti-γ tubulin (GTU-88, Sigma; 1:250 in 3% milk) for 30 min. Samples were washed 3× in PBST and stained with the secondary antibody Alexa 594 anti-mouse (A-11032, Molecular Probes; 1:2000 in 3% milk). Samples were washed twice, stained for DNA with Hoechst 33258 (1 μg/mL) for one min, washed twice in PBST and mounted using 90% glycerol in 100 mM Tris at pH 8.0. Samples were analyzed at the Nikon Imaging Center at Harvard Medical School on a Nikon E1000 Eclipse automated upright microscope using a 60×, 1.4 N.A. objective lens. Images were collected using a Hamamatsu Orca II dual-mode cooled CCD camera and processed using Imaging System software (MetaMorph).
FACS analysis
Cells from a 10-cm plate were fixed in 1% paraformaldehyde made in PBS for 1 h at 4°C. Cells were washed twice in 10 mL PBS. Cells were postfixed for at least 16 h in 70% ethanol at −20°C. Samples were washed twice in 10 mL PBS plus 1% BSA and resuspended in 400 μL PBS plus 1% BSA. Fifty microliters of propidium iodide (PI) solution (stock: 500 μg/mL PI in 3.8 × 10−2M sodium citrate at pH 7.0) and 50 μL RNase A (10 mg/mL prepared in 10 mM Tris-HCl at pH 7.5) were added to each sample and incubated at 37°C for 30 min. Cells were filtered through the lids of Falcon polystyrene round-bottom tubes with cell strainer cap (VWR 21008-948) and analyzed at the Dana Farber Cancer Institute Flow Cytometry Facility.
Acknowledgments
We are extremely grateful to our colleagues for their generous and timely gifts of antibodies, plasmid DNAs and other reagents: Tim Hunt, Jim Maller, Joan Brugge, Owen Witte, Garry Nolan, Cathie Pfleger, Marc Kirschner, Hironori Funabiki, Andrew Murray, Puck Ohi, Tim Mitchison, and Frank McKeon. We thank Cathie Pfleger and Puck Ohi for many helpful discussions, Joan Brugge for her sustained interest in this work, Thorkell Andresson and Holger Bastians for numerous contributions in the early stages of the project, and Jennifer Waters Schuler in the Harvard Medical School Nikon Imaging Center. Jim Horn in the Machine Shop rapidly provided a new electrical activation chamber at a critical time after our original one went missing. We thank all the members of the Ruderman lab for their interest, feedback, and high spirits. This work was supported by a grant from the NIH to J.V.R. and an NSF predoctoral fellowship to L.E.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ruderman@hms.harvard.edu; FAX 617-432-0555.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1007302.
References
- Anderson CW, Baum PR, Gesteland RF. Processing of adenovirus-2 induced proteins. J Virol. 1973;12:241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresson T, Ruderman JV. The kinase Eg2 is a component of the Xenopus oocyte progesterone-activated signaling pathway. EMBO J. 1998;17:5627–5637. doi: 10.1093/emboj/17.19.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlot-Bonnemains Y, Klotzbucher A, Giet R, Uzbekov R, Bihan R, Prigent C. Identification of a functional destruction box in the Xenopus laevis aurora-A kinase pEg2. FEBS Lett. 2001;508:149–152. doi: 10.1016/s0014-5793(01)03048-4. [DOI] [PubMed] [Google Scholar]
- Bischoff JR, Plowman GD. The Aurora/Ipl1p family: Regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9:454–459. doi: 10.1016/s0962-8924(99)01658-x. [DOI] [PubMed] [Google Scholar]
- Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. D box and KEN box motifs in budding yeast Hsl1p are required for APC- mediated degradation and direct binding to Cdc20p and Cdh1p. Genes & Dev. 2001;15:2381–2395. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A, Arlot-Bonnemains Y, Vigneron S, Labbe JC, Prigent C, Lorca T. APC/Fizzy-Related targets Aurora-A kinase for proteolysis. EMBO Rep. 2002;3:457–462. doi: 10.1093/embo-reports/kvf095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P. Mitotic phosphorylation of histone H3: Spatio-temporal regulation by mammalian Aurora kinases. Mol Cell Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farruggio DC, Townsley FM, Ruderman JV. Cdc20 associates with the kinase aurora2/Aik. Proc Natl Acad Sci. 1999;96:7306–7311. doi: 10.1073/pnas.96.13.7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE., Jr What do scaffold proteins really do? Sci STKE. 2000;2000:E1. doi: 10.1126/stke.2000.52.pe1. [DOI] [PubMed] [Google Scholar]
- Frank-Vaillant M, Jessus C, Ozon R, Maller JL, Haccard O. Two distinct mechanisms control the accumulation of cyclin B1 and Mos in Xenopus oocytes in response to progesterone. Mol Biol Cell. 1999;10:3279–3288. doi: 10.1091/mbc.10.10.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant M, Haccard O, Thibier C, Ozon R, Arlot-Bonnemains Y, Prigent C, Jessus C. Progesterone regulates the accumulation and the activation of Eg2 kinase in Xenopus oocytes. J Cell Sci. 2000;113:1127–1138. doi: 10.1242/jcs.113.7.1127. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Giet R, Prigent C. The non-catalytic domain of the Xenopus laevis auroraA kinase localises the protein to the centrosome. J Cell Sci. 2001;114:2095–2104. doi: 10.1242/jcs.114.11.2095. [DOI] [PubMed] [Google Scholar]
- Giet R, Uzbekov R, Kireev I, Prigent C. The Xenopus laevis centrosome aurora/Ipl1-related kinase. Biol Cell. 1999;91:461–470. [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Glover D, Leibowitz M, McLean D, Parry H. Mutations in Aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- Goepfert TM, Brinkley BR. The centrosome-associated Aurora/Ipl-like kinase family. Curr Top Dev Biol. 2000;49:331–342. doi: 10.1016/s0070-2153(99)49016-7. [DOI] [PubMed] [Google Scholar]
- Gopalan G, Chan CSM, Donovan PJ. A novel mammalian, mitotic spindle-associated kinase is related to yeast and fly chromosome segregation regulators. J Cell Biol. 1997;138:643–656. doi: 10.1083/jcb.138.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E, Kirkham M, Hyman AA, Oegema K. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J Cell Biol. 2001;155:1109–1116. doi: 10.1083/jcb.200108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Heasman J, Holwill S, Wylie CC. Fertilization of cultured Xenopus oocytes and use in studies of maternally inherited molecules. In: Kay BK, Peng HB, editors. Methods in cell biology. San Diego, CA: Academic Press; 1991. pp. 213–230. [DOI] [PubMed] [Google Scholar]
- Hochegger H, Klotzbucher A, Kirk J, Howell M, le Guellec K, Fletcher K, Duncan T, Sohail M, Hunt T. New B-type cyclin synthesis is required between meiosis I and II during Xenopus oocyte maturation. Development. 2001;128:3795–3807. doi: 10.1242/dev.128.19.3795. [DOI] [PubMed] [Google Scholar]
- Holloway SL, Glotzer M, King RW, Murray AW. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993;73:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Honda K, Mihara H, Kato Y, Yamaguchi A, Tanaka H, Yasuda H, Furukawa K, Urano T. Degradation of human aurora2 protein kinase by the anaphase-promoting complex-ubiquitin-proteasome pathway. Oncogene. 2000;19:2812–2819. doi: 10.1038/sj.onc.1203609. [DOI] [PubMed] [Google Scholar]
- Jacobs HW, Keidel E, Lehner CF. A complex degradation signal in Cyclin A required for G1 arrest, and a C-terminal region for mitosis. EMBO J. 2001;20:2376–2386. doi: 10.1093/emboj/20.10.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar M, Dienemann A, Schulze C, Sprenger F. Mitotic degradation of cyclin A is mediated by multiple and novel destruction signals. Curr Biol. 2001;11:685–690. doi: 10.1016/s0960-9822(01)00205-6. [DOI] [PubMed] [Google Scholar]
- Kimura M, Kotani S, Hattori T, Sumi N, Yoshioka T, Todokoro K, Okano Y. Cell cycle-dependent expression and spindle pole localization of a novel human protein kinase, Aik, related to Aurora of Drosophila and yeast Ipl1. J Biol Chem. 1997;272:13766–13771. doi: 10.1074/jbc.272.21.13766. [DOI] [PubMed] [Google Scholar]
- King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lorca T, Castro A, Martinez AM, Vigneron S, Morin N, Sigrist S, Lehner C, Doree M, Labbe JC. Fizzy is required for activation of the APC/cyclosome in Xenopus egg extracts. EMBO J. 1998;17:3565–3575. doi: 10.1093/emboj/17.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller J, Poccia D, Nishioka D, Kidd P, Gerhart J, Hartman H. Spindle formation and cleavage in Xenopus eggs injected with centriole-containing fractions from sperm. Exp Cell Res. 1976;99:285–294. doi: 10.1016/0014-4827(76)90585-1. [DOI] [PubMed] [Google Scholar]
- Masui Y, Forer A, Zimmerman AM. Induction of cleavage in nucleated and enucleated frog eggs by injection of isolated sea-urchin mitotic apparatus. J Cell Sci. 1978;31:117–135. doi: 10.1242/jcs.31.1.117. [DOI] [PubMed] [Google Scholar]
- Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Paris J, Le Guellec R, Couturier A, Le Guellec K, Omilli F, Camonis J, MacNeill S, Philippe M. Cloning by differential screening of a Xenopus cDNA coding for a protein highly homologous to cdc2. Proc Natl Acad Sci. 1991;88:1039–1043. doi: 10.1073/pnas.88.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DH, O'Farrell PH. The schedule of destruction of three mitotic cyclins can dictate the timing of events during exit from mitosis. Curr Biol. 2001;11:671–683. doi: 10.1016/s0960-9822(01)00204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Denchi EL, Gieffers C, Matteucci C, Peters JM, Helin K. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes & Dev. 2000;14:2330–2343. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes & Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Lee E, Kirschner MW. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes & Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roghi C, Giet R, Uzbekov R, Morin N, Chartrain I, Le Guellec R, Couturier A, Doree M, Philippe M, Prigent C. The Xenopus protein kinase pEg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. J Cell Sci. 1998;111:557–572. doi: 10.1242/jcs.111.5.557. [DOI] [PubMed] [Google Scholar]
- Schumacher JM, Ashcroft N, Donovan PJ, Golden A. A highly conserved centrosomal kinase, AIR-1, is required for accurate cell cycle progression and segregation of developmental factors in Caenorhabditis elegans embryos. Development. 1998;125:4391–4402. doi: 10.1242/dev.125.22.4391. [DOI] [PubMed] [Google Scholar]
- Sen S, Zhou H, White RA. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene. 1997;14:2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- Shaver JR. Studies on the initiation of cleavage in the frog egg. J Exp Zool. 1953;122:169–192. [Google Scholar]
- Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. Dual inhibition of sister chromatid separation at metaphase. Cell. 2001;107:715–726. doi: 10.1016/s0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- Sudo T, Ota Y, Kotani S, Nakao M, Takami Y, Takeda S, Saya H. Activation of Cdh1-dependent APC is required for G1 cell cycle arrest and DNA damage-induced G2 checkpoint in vertebrate cells. EMBO J. 2001;20:6499–6508. doi: 10.1093/emboj/20.22.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taieb F, Gross SD, Lewellyn AL, Maller JL. Activation of the anaphase-promoting complex and degradation of cyclin B is not required for progression from meiosis I to II in Xenopus oocytes. Curr Biol. 2001;11:508–513. doi: 10.1016/s0960-9822(01)00145-2. [DOI] [PubMed] [Google Scholar]
- Townsley F, Ruderman JV. Proteolytic ratchets that control progression through mitosis. Trends Cell Biol. 1998;8:238–244. doi: 10.1016/s0962-8924(98)01268-9. [DOI] [PubMed] [Google Scholar]
- Tyers M, Jorgensen P. Proteolysis and the cell cycle: With this RING I do thee destroy. Curr Opin Genet Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Walter AO, Seghezzi W, Korver W, Sheung J, Lees E. The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene. 2000;19:4906–4916. doi: 10.1038/sj.onc.1203847. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Hunt T, Ikawa Y, Sagata N. Independent inactivation of MPF and cytostatic factor (Mos) upon fertilization of Xenopus eggs. Nature. 1991;352:247–248. doi: 10.1038/352247a0. [DOI] [PubMed] [Google Scholar]
- Wickens M, Goodwin EB, Kimble J, Strickland S, Hentze MW. Translational control of developmental decisions. In: Sonenberg N, Hershey JWB, Mathews M, editors. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2000. pp. 295–370. [Google Scholar]
- Zachariae W. Progression into and out of mitosis. Curr. Opin. Cell Biol. 1999;11:708–716. doi: 10.1016/s0955-0674(99)00041-1. [DOI] [PubMed] [Google Scholar]
- Zhou H, Kuang J, Kuo W, Gray J, Sahin A, Brinkley BR, Sen S. Tumor amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Gen. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]