Abstract
Methylation of histone H3 at Lys 9 is causally linked to formation of heterochromatin and to long-term transcriptional repression. We report an unexpected pattern of H3 Lys 9 methylation occurring at a subset of inducible inflammatory genes. This pattern is characterized by relatively low constitutive levels of H3 Lys 9 methylation that are erased upon activation and restored concurrently with post-induction transcriptional repression. Changes in H3 Lys 9 methylation strongly correlate with RNA polymerase II recruitment and release. In particular, remethylation correlates with RNApolII release more strongly than does histone deacetylation. We propose that, by generating a window of time in which transcription is permitted, dynamic modulation of H3 Lys 9 methylation adds an additional regulatory level to transcriptional activation of tightly controlled inducible genes.
Keywords: histone methylation, transcriptional repression, histone methyltransferase, histone code
The N-terminal tails of core histones are the convergent substrates of several enzymes controlling transcription and replication (Berger 2001). Combination of several covalent modifications along a single histone tail is believed to generate a sort of encrypted code (histone code; Strahl and Allis 2000), which acts by providing alternative docking surfaces for proteins regulating chromatin organization, transcription, DNA replication and repair (Jenuwein and Allis 2001).
The N-terminal tails of histone H3 and H4 are methylated at several lysine and arginine residues, methylation of individual residues being causally linked to either transcriptional activation or repression (Jenuwein 2001; Zhang and Reinberg 2001). Recently, SET-domain-containing proteins of the evolutionary conserved SU(VAR)3–9 family have been shown to act as lysine-specific histone methyltransferases (HMTases; Rea et al. 2000; Jenuwein 2001). By selectively methylating H3-Lys9, SU(VAR)3–9 family HMTases generate a docking site for heterochromatin-associated HP1 proteins (Bannister et al. 2001; Lachner et al. 2001; Nakayama et al. 2001), thus linking H3 Lys 9 methylation to formation and spreading of heterochromatin. Analysis of H3 Lys 9 methylation over large chromosomal regions has shown that this modification is preferentially associated with constitutively condensed heterochromatin and with developmentally inactive genes (Litt et al. 2001; Noma et al. 2001). Mammalian Suv39h HMTases are specifically required for the induction of a specialized high-density H3 Lys 9 methylation pattern at pericentric heterochromatin, which is required to protect genome stability in both somatic and germ cells (Peters et al. 2001). Suv39h-independent H3 Lys 9 methylation is directly involved in the chromosome-wide chromatin remodelling leading to the transcriptional shutdown of the inactive X chromosome (Heard et al. 2001; Boggs et al. 2002; Peters et al. 2002).
Experimental evidence indicates that H3 Lys 9 methylation may also have a wide role in transcriptional repression of euchromatic genes. First, the retinoblastoma tumor suppressor protein recruits SUV39H1 to cell cycle genes, where it mediates H3 Lys 9 methylation and transcriptional repression (Nielsen et al. 2001). Second, a broad euchromatic and heterochromatic Suv39h-independent H3 Lys 9 methylation is observed in mammalian cells (Peters et al. 2001), consistent with the existence of several other potential SET-domain HMTases, some of which methylate H3 Lys 9 selectively in euchromatin (Ogawa et al. 2002; Schultz et al. 2002). Third, HP1 proteins bind to several euchromatic sites in Drosophila and mammalian cells (James et al. 1989; Minc et al. 2000) and interact with transcriptional corepressors (Lehming et al. 1998; Seeler et al. 1998; Nielsen et al. 1999; Ryan et al. 1999). An attractive model is that some transcriptional corepressors may act as scaffolds recruiting H3 Lys 9 HMTases and HP1 proteins to specific euchromatic sites, thus locally repressing transcription (Schultz et al. 2002). Finally, Drosophila HP1 has been demonstrated to repress selected euchromatic genes in a Su(var)3–9-dependent manner (Hwang et al. 2001).
Regarded as a stable and potentially irreversible modification (Jenuwein and Allis 2001; Zhang and Reinberg 2001), histone methylation at lysine residues is usually considered incompatible with the requirements of rapidly inducible gene expression. Recently, however, it has been reported that H3-Lys 4 methylation at a tandem array of mouse mammary tumor virus (MMTV) promoters is rapidly downregulated upon glucocorticoid hormone treatment (Ma et al. 2001). Although the significance of this finding is not clear, it suggests either that the methyl mark can be rapidly removed from histone proteins or that methylated histones can be replaced or degraded.
We therefore set out to analyze H3 Lys 9 methylation at inducible endogenous genes. Evidence from gene-targeted mice and cells lacking transcriptional repressors expressed in cells of the monocytic lineage (Toney et al. 2000; Karsunky et al. 2002) indicates that transcriptional inactivity of some inflammatory genes in unstimulated cells, as well as post-induction transcriptional shutdown, depends on the activity of specific transcriptional repressors. The possibility therefore exists that H3 Lys 9 methylation may mediate repression in this context. We found that in unstimulated cells some inducible genes are associated with nucleosomes methylated at H3 Lys 9, and that this modification is rapidly reversed upon activation and then restored.
Results and Discussion
LPS-induced gene activity in DCs
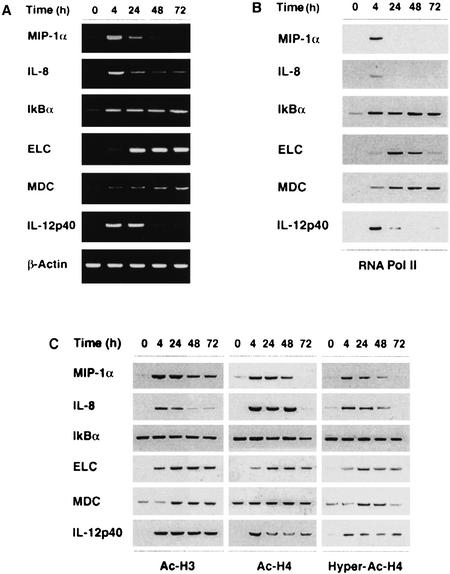
Primary human monocyte-derived dendritic cells (DCs; Sallusto and Lanzavecchia 1994) are post-mitotic, terminally differentiated cells that respond to bacterial products and inflammatory cytokines by activating transcription of several genes implicated in inflammation, chemotaxis, and T lymphocyte activation. The genes we examined are strongly induced by lipopolysaccharide (LPS) stimulation of DCs. A representative set of a larger panel of genes investigated is shown in Figure 1a. Interleukin 8 (IL-8), macrophage inflammatory protein 1α (MIP-1α), and IkBα (inhibitor of nuclear factor κ B) are induced by inflammatory stimuli in various cell types; low basal IkBα mRNA levels are detected ubiquitously. Conversely, the genes encoding the macrophage-derived chemokine (MDC), the EBV-induced molecule 1 ligand chemokine (ELC) and the p40 subunit of interleukin 12 (IL-12p40) are expressed exclusively by cells of the monocytic lineage, with DCs being the most efficient producers.
Figure 1.
Kinetics of LPS-induced gene expression in human monocyte-derived DCs. (A) Steady-state mRNA levels of LPS-induced genes were analyzed by RT-PCR. Dendritic cells were stimulated with 100 ng/ml LPS as indicated. (B) RNApolII recruitment to the same genes was analyzed by Chromatin Immunoprecipitation (ChIP) assay using an antibody directed against the RNApolII large subunit Rpb1. Immunoprecipitated DNA was analyzed by PCR using promoter-specific primers. The peak of RNApolII recruitment to the IkBα, IL-8 and MIP-1α genes occurs at earlier time points (0.5–2 h) than those shown in the figure. (C) Promoter-specific histone H3 and H4 acetylation was investigated by ChIP assays carried out with antibodies against acetylated H3 (Ac-Lys 9/Lys 14) and H4. The antibody against hyperacetylated H4 has a preference towards tetra- and tri-acetylated H4.
Chromatin immunoprecipitation (ChIP) assays carried out with an antibody recognizing the RNA polymerase II (RNApolII) large subunit Rpb1 showed transient recruitment to all genes except IkBα, where recruitment was persistent (Fig. 1b). Indeed, IkBα is directly regulated by NF-kB, which undergoes irreversible activation in LPS-stimulated DCs. Release of RNApolII from the MDC gene usually started between 48 and 72 h, with different kinetics in DCs from different donors (in this donor a slow kinetics of release was observed). With the exception of IkBα, which showed high constitutive levels of histone H3 and H4 acetylation (which were left substantially unaffected by stimulation), all of the genes tested underwent hyperacetylation at histone H3 and H4 upon LPS stimulation (Fig. 1c). The increase in H3 and H4 acetylation levels correlated with recruitment of RNApolII. Remarkably, however, both H3 and H4 hyperacetylation at all the genes tested persisted after RNApolII release. This observation cannot be accounted for by a different sensitivity of anti-polII ChIP versus anti-acetyl H4/H3 ChIPs, as their efficiency is comparable. Thus H3 and H4 acetylation levels are not predictive of transcriptional termination at any of the genes analyzed, and RNApolII release occurs independently of any extensive H3/H4 deacetylation.
Dynamic H3 Lys 9 methylation at inducible inflammatory genes
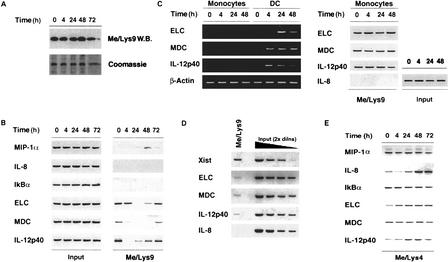
To analyze H3 Lys 9 methylation status at the genes under investigation, we used an antibody that specifically recognizes histone H3 only when methylated at Lys 9. Total H3 Lys 9 methylation (which mainly reflects heterochromatic H3 Lys 9 methylation) did not vary during LPS treatment (Fig. 2a). Using ChIP, we did not detect any H3 Lys 9 methylation above background levels at the IL-8 and IkBα promoters (Fig. 2b); a low-level H3 Lys 9 methylation was detected in several donors at the MIP-1α promoter, but only transiently and at late time points, long after transcriptional termination. The significance of this weak, late, and transient H3 Lys 9 methylation is unclear. In contrast to the genes mentioned above, ELC, MDC, and IL-12p40 promoters were found to be associated with detectable levels of H3 Lys 9 methylation in unstimulated cells. LPS stimulation determined the erasure of H3 Lys 9 methylation at all three promoters, which was followed by remethylation at later time points. Monocytes, which express very low levels of IL-12p40, ELC, and MDC mRNAs, showed similar basal levels of H3 Lys 9 methylation. However, no histone demethylation was observed following LPS treatment (Fig. 2c). Thus H3 Lys 9 demethylation strongly correlates with the high level transcriptional activity specifically observed in DCs.
Figure 2.
Dynamic changes in histone H3 Lys 9 methylation levels at the promoters of tightly regulated inducible genes. (A) DCs were stimulated with LPS as indicated and bulk levels of H3 methylated at Lys 9 were analyzed by Western blot. (B) Lys 9 methylation of H3 at the promoters of the genes under investigation was analyzed by ChIP assay. The left panel (Input) shows the relative amounts of the probed regions contained in the starting chromatin extracts. The right panel shows ChIP analysis of specific association of H3 methylated at Lys 9 with the promoters investigated. The induction of the genes we analyzed is asynchronous in LPS-stimulated DCs. It is therefore possible that the apparent gradation in H3 Lys 9 methylation changes (and in RNApolII recruitment) reflects different kinetics with which such changes occur in individual cells of the population analyzed. (C) LPS-stimulated monocytes express very low levels of ELC, MDC, and IL-12p40 mRNAs (transcripts can be detected at a higher number of PCR cycles). No detectable H3 Lys 9 demethylation was observed in these cells (right). (D) Density of H3 Lys 9 methylation at inducible euchromatic genes with respect to heterochromatic genes. Signals obtained in the Me/Lys 9 ChIP assay were compared to serial twofold dilutions of a fraction of the input. PCR was carried out at a lower number of cycles than in the experiment shown in B,C to maintain the signals obtained with differentially methylated promoters in a linear range. (E) ChIP analysis of promoter-specific H3 Lys 4 methylation at inducible genes. Promoter-specific levels of H3 Lys 4 methylation were analyzed by ChIP assay using an antibody recognizing H3 methylated at Lys 4. The negative controls for these experiments were parallel samples in which the immunoprecipitation step was carried out without antibody (data not shown). In these samples, only background levels of promoter sequences were detectable after 40 cycles of PCR.
We next compared the relative density of H3 Lys 9 methylation at the ELC, MDC, and IL-12p40 promoters with the density of H3 Lys 9 methylation at the promoters of genes known to be assembled into heterochromatin. To this aim, we analyzed H3 Lys 9 methylation at the Xist gene, which is silenced on the active X chromosome in females and on the single X chromosome in males. By comparing the ChIP signals with serial two-fold dilutions of a fraction of the input (and considering a bi-allelic methylation at inflammatory genes versus a mono-allelic methylation at the Xist gene), we estimated H3 Lys 9 methylation at inflammatory genes to be about eight-fold lower than that of Xist (Fig. 2d). However, these data should be interpreted very cautiously: since the anti-methyl H3 Lys 9 antibody has a rather low affinity towards its ligand, a difference in the density of this modification on a string of nucleosomes may have dramatic and non-linear effects on the efficiency of the immunoprecipitation. Therefore, the only conclusion we can draw from these data is that the density of this modification at euchromatic genes is lower than at heterochromatic genes. Precise quantification will have to await the generation of high affinity monoclonal antibodies. The relatively low density of H3 Lys 9 methylation at euchromatic genes may arise either from the selective modification of specific nucleosomes or from hemi-methylation of several nucleosomes. The first situation would be reminiscent of the nucleosome-selective H3 Lys 9 methylation in the cyclin E promoter (Nielsen et al. 2001).
These results indicate that H3 Lys 9 methylation is a dynamically regulated histone modification targeting specific nucleosomes in a subset of inducible genes. The behavior of the methyl mark at H3 Lys 9 is fully consistent with its known role as a repressive modification. In fact, comparison of RNApolII recruitment kinetics (Fig. 1b) with H3 Lys 9 methylation status (Fig. 2b) indicates that (1) RNApolII recruitment starts when demethylation begins; (2) the peak of RNApolII recruitment to these genes occurs when H3 Lys 9 methylation is at its lowest level; and (3) release of RNApolII temporally correlates with remethylation. MDC has the slowest remethylation kinetics among the genes tested and, in DCs from several donors, remethylation at 72 h temporarily overlaps with RNApolII occupancy. The time interval between H3 Lys 9 remethylation and RNApolII release at the MDC gene suggests that remethylation may only pose the gene for repression but is by itself not sufficient.
Overall, changes in H3 Lys 9 methylation status correlated with RNApolII persistence on genes bearing constitutive H3 Lys 9 methylation more than changes in H3/H4 acetylation status. In particular, H3 Lys 9 remethylation temporally correlated with RNApolII release, while histone deacetylation lagged behind it.
These results suggest that H3 Lys 9 demethylation and subsequent remethylation may generate a time window that is permissive for transcriptional activation.
H3 Lys 4 methylation at inducible genes
We next investigated whether inducible gene expression in this system is associated with changes in H3 Lys 4 methylation. Methylation of histone H3 at Lys 4 correlates with transcriptional activity in tetrahymena (Strahl et al. 1999), Saccharomyce pombe (Noma et al. 2001), and mammals (Heard et al. 2001; Litt et al. 2001; Boggs et al. 2002; Nishioka et al. 2002), while it mediates transcriptional repression in Saccharomyces cerevisiae (Briggs et al. 2001; Nagy et al. 2002). High levels of H3 Lys 4 methylation in S. pombe and mammalian cells are found at transcriptionally active euchromatic regions, thus suggesting that this may act as a stable and long-range mark of active chromatin; indeed, when considering large chromosomal regions, this modification strongly anti-correlates with H3 Lys 9 methylation (Heard et al. 2001; Litt et al. 2001; Boggs et al. 2002). Moreover, enforced expression of an H3 Lys 4-specific HMTase potentiates the activation function of a transcriptional activator (Nishioka et al. 2002). However, the behavior of H3 Lys 4 methylation at rapidly inducible genes is poorly defined. An antibody directed against methyl H3 Lys 4 (Briggs et al. 2001; Litt et al. 2001; Noma et al. 2001) immunoprecipitated detectable amounts of all the promoters under investigation (Fig. 2e). Control ChIPs carried out with no antibody immunoprecipitated only background amounts of the same promoters (data not shown). At the ELC, MDC, and IL-12p40 genes, H3 Lys 4 methylation therefore coexisted with the constitutive H3 Lys 9 methylation. Since the fragments analyzed have an average size of 1 kb and presumably contain several nucleosomes, it is possible that methylation of the two residues does not coexist on a single H3 molecule, as suggested by in vitro results (Wang et al. 2001; Nishioka et al. 2002). LPS stimulation did not determine dramatic changes in the amount of H3 methylated at Lys 4 that was associated with these genes, and the changes detected did not follow simple patterns. In detail, Lys 4 methylation at MIP-1α, IkBα, and MDC was substantially stable throughout the treatment. H3 Lys 4 methylation at the IL-8 promoter was stable during the first 24 hr of treatment (i.e., the time during which the gene is activated and then again reset to a basal level of activity) and increased at 48 hr (the possible role of this late increase is therefore difficult to understand, as it did not correlate with any change in transcriptional activity). In DCs from several donors (such as the one shown in Fig. 2e), the IL-12p40 promoter showed a mild increase in Lys 4 methylation (usually lagging behind Lys 9 demethylation), while in DCs from other donors, flat levels of H3 Lys 4 methylation were observed in spite of a strong transcriptional activation (data not shown), thus arguing against a necessary role for increased Lys 4 methylation in transcriptional induction of this gene. ELC was the only gene where we always detected a reproducible increase in H3 Lys 4 methylation, which correlated with RNApolII recruitment. LPS-induced increase in H3 Lys 4 methylation at ELC was not reversed even at late time points, thus persisting after RNApolII release.
Overall, H3 Lys 4 methylation showed a rather static behavior, consistent with its proposed role as a stable epigenetic mark of active chromatin domains. It is possible, however, that recruitment of H3 Lys 4 HMTases to a few genes (such as ELC) may occur concurrently with activation and contribute to transcriptional induction.
H3 Lys 9 methylation along the ELC/SLC locus
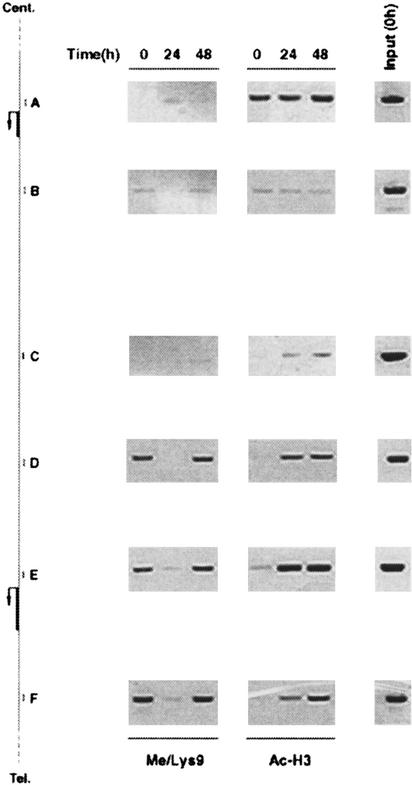
Differentiation-specific patterns of histone modifications are maintained over extended chromatin domains. The average size of the fragments used in our ChIP assays (1 kb) allowed us to analyze how H3 Lys 9 methylation extends to the chromosomal regions surrounding the promoters under investigation. The ELC gene is located on the short arm of chromosome 9 (9p13) in a head to tail orientation with respect to the gene encoding the related chemokine SLC (secondary lymphoid-tissue chemokine; Fig. 3), which has a low level constitutive activity in DCs (data not shown). H3 Lys 9 methylation is detectable over several kilobases, both upstream of and downstream from the ELC coding region (Fig. 3, left panel, regions D,E,F). LPS-induced demethylation and remethylation were also detected in the regions surrounding ELC. The level of H3 Lys 9 methylation dropped significantly at the intergenic regions C and B. Further upstream, the SLC promoter A did not show any methylation at H3 Lys 9, consistent with a low transcriptional activity. Considering that the region E (ELC promoter) lies more than 4 kbp from both regions D and F, that the size of the fragments in the starting extracts is around 1 kbp, and that the density of H3 Lys 9 methylation at the ELC gene is low as compared to heterochromatic genes (Fig. 2d), we can conclude that, presumably, a few nucleosomes scattered along the gene and upstream of it are methylated at Lys 9. This pattern also suggests that H3 Lys 9 methylation may contribute to the tight and independent regulation of a specific gene with respect to adjacent transcription units, thus preventing any kind of potential transcriptional interference.
Figure 3.
Histone H3 methylation at Lys 9 across the ELC/SLC locus. Sequence data were retrieved from the National Center for Biotechnology Information (NCBI; accession no. 17451799). (Left) A schematic representation of the region containing the SLC and ELC genes. Cent., centromere; Tel., telomere. Positions of the sets of primers (A–F) used are shown. (Middle) ChIP assays carried out with an anti-methyl H3 Lys 9 antibody. In the right panels, anti-acetyl H3 ChIP assays show the levels of H3 acetylation over the same region. Input: PCR products obtained with each set of primers using as template a fraction of the ChIP inputs (untreated samples). In DCs from this donor a complete remethylation of the ELC gene and surrounding regions was observed at 48 h after LPS stimulation. The distances between the regions amplified are A > B 3.6 Kbp; B > C 6.5 Kbp; C > D 4.5 Kbp; D > E 4.2 Kbp; E > F 4.6 Kbp.
Conclusions
Our results indicate that H3 Lys 9 methylation at inducible genes accounts for a fraction of euchromatic H3 Lys 9 methylation, is dynamically regulated, and is probably used to generate a time window in which gene activity must be confined. One interesting possibility is that H3 Lys 9 methylation may prevent early induction of a gene, which may occur due to the activation of the required transcription factors at a time before the gene should be transcribed. Thus, H3 Lys 9 methylation and demethylation add an additional regulatory layer to inducible gene expression.
Several mechanistic issues remain to be answered, particularly those regarding the reversal of H3 Lys 9 methylation. Hypothetic mechanisms include demethylation by histone-demethylases, cleavage of methylated tails, and replacement/degradation of methylated histone H3 (Jenuwein and Allis 2001). The possible existence of histone demethylases acting at specific euchromatic regions (possibly by transcription factor-mediated recruitment), without affecting the stable H3 Lys 9 mark at the level of heterochromatin, is particularly intriguing. However, although a histone demethylase activity was described 30 years ago (Paik and Kim 1973), no protein with such an activity has been reported to date. The recent hypothesis that a potential catalytic domain in Elp3-related histone acetyltransferases may have the requisites to carry out histone demethylation reactions is of extreme interest and awaits experimental verification (Chinenov 2002). If the methyl group is removed from H3 Lys 9 through a demethylation reaction, then an obvious possibility is that H3 Lys 9 methylation at the level of strategically positioned nucleosomes interferes with transcription at least in part by preventing acetylation of the same residue. An alternative mechanism for relieving repression mediated by methylated H3 Lys 9 may be represented by the selective transfer or mobilization of the methylated octamers exerted by chromatin remodelling complexes recognizing this modification. In this regard, it should be noticed that the first of the two chromodomains of CHD1 and CHD2, two members of the CHD (Chromodomain/Helicase/DNA-binding) family of SNF2-related helicases, have the essential structural features required for H3 Lys 9 binding (Jacobs and Khorasanizadeh 2002; Nielsen et al. 2002), thus suggesting that they may dock to, and mobilize, nucleosomes methylated at H3 Lys 9.
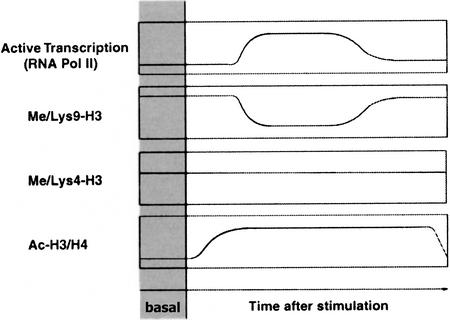
The behavior of H3 Lys 9 methylation in the experimental system we investigated (Fig. 4) suggests that it may contribute to both basal and post-induction repression of tightly regulated inducible genes. However, this is not a universal mechanism regulating inducible gene expression, because a number of inducible genes tested appeared not to be associated with H3 Lys 9 methylation. The genes we found to be methylated at H3 Lys 9 have slower kinetics of activation than non-methylated ones. For instance, RNAPolII recruitment to the IkBα, MIP-1α, and IL-8 genes is already maximal at 1–2 h after stimulation, while it peaks between 4 and 8 h at the IL-12p40 gene, at 24 h at the ELC gene and between 24 and 48 h at the MDC gene. This suggests that the mechanism for H3 Lys 9 removal may be intrinsically inefficient or slow. We therefore envisage the possibility that H3 Lys 9 methylation may be exploited as a repressive mark only at the level of inducible genes with relatively slow kinetics of activation.
Figure 4.
A schematic representation of histone methylation and acetylation changes occurring at a subset of inducible genes. RNApolII recruitment and release kinetics are shown in parallel to histone acetylation and methylation changes occurring at a subset of inducible inflammatory genes.
Materials and methods
Antibodies
Modification-specific antibodies recognizing modified histones were from Upstate Biotechnology Inc, (UBI; Lake Placid, NY). The antibody against RNApolII large subunit rbpI was from Santa Cruz biotechnology (sc-899).
DC preparation
Monocytes were purified by positively sorting peripheral blood mononuclear cells using anti-CD14 conjugated magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). DCs were generated by culturing monocytes for 5 d in RPMI-10% endotoxin-free FCS (Hyclone) supplemented with GM-CSF (50 ng/ml; Leukomax, Novartis, Basel Switzerland) and IL-4 (1000 U/ml).
Chromatin Immunoprecipitation Assay (ChIP)
ChIP was carried out as previously described (Saccani et al. 2002). Sequences of promoter-specific primers and a detailed experimental protocol are available upon request.
Acknowledgments
We are grateful to Antonio Lanzavecchia and Marcus Thelen for critically reading the manuscript. This work was partially supported by the Swiss National Science Foundation (grant no. 31–66720.01 to GN).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gioacchino.natoli@irb.unisi.ch; FAX 41-91-820-0305.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.232502.
References
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Berger SL. An embarrassment of niches: The many covalent modifications of histones in transcriptional regulation. Oncogene. 2001;20:3007–3013. doi: 10.1038/sj.onc.1204324. [DOI] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes & Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y. A second catalytic domain in the Elp3 histone acetyltransferases: A candidate for histone demethylase activity? Trends Biochem Sci. 2002;27:115–117. doi: 10.1016/s0968-0004(02)02058-3. [DOI] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Hwang KK, Eissenberg JC, Worman HJ. Transcriptional repression of euchromatic genes by Drosophila heterochromatin protein 1 and histone modifiers. Proc Natl Acad Sci. 2001;98:11423–11427. doi: 10.1073/pnas.211303598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, Elgin SC. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11:266–273. doi: 10.1016/s0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Karsunky H, Zeng H, Schmidt T, Zevnik B, Kluge R, Schmid KW, Duhrsen U, Moroy T. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet. 2002;30:295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lehming N, Le Saux A, Schuller J, Ptashne M. Chromatin components as part of a putative transcriptional repressing complex. Proc Natl Acad Sci. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol. 2001;11:1981–1985. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- Minc E, Courvalin JC, Buendia B. HP1γ associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet. 2000;90:279–284. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes & Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Paik WK, Kim S. Enzymatic demethylation of calf thymus histones. Biochem Biophys Res Commun. 1973;51:781–788. doi: 10.1016/0006-291x(73)91383-1. [DOI] [PubMed] [Google Scholar]
- Peters AH, Mermoud JE, O'Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ., 3rd KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: A potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher 3rdr, FJ. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Marchio A, Sitterlin D, Transy C, Dejean A. Interaction of SP100 with HP1 proteins: A link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Toney LM, Cattoretti G, Graf JA, Merghoub T, Pandolfi PP, Dalla-Favera R, Ye BH, Dent AL. BCL-6 regulates chemokine gene transcription in macrophages. Nat Immunol. 2000;1:214–220. doi: 10.1038/79749. [DOI] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]