Abstract
Complex disease mapping usually involves a combination of linkage and association techniques. Linkage analysis can scan the entire genome in a few hundred tests. Association tests may involve an even greater number of tests. However, association tests can localize the susceptibility genes more accurately. Using a recently developed combined linkage and association strategy, we analyzed a subset of the Collaborative Study on the Genetics of Alcoholism (COGA) data for the Genetic Analysis Workshop 14 (GAW14). In this analysis, we first employed linkage analysis based on frailty models that take into account age of onset information to establish which regions along the chromosome are likely to harbor disease susceptibility genes for alcohol dependence. Second, we used an association analysis by exploiting linkage disequilibrium to narrow down the peak regions. We also compare the methods with mean identity-by-descent tests and transmission/disequilibrium tests that do not use age of onset information.
Background
The Collaborative Study on the Genetics of Alcoholism (COGA) is a large, multi-site genetic study to identify susceptibility genes for alcohol dependence and related phenotypes [1]. The COGA data have been analyzed using nonparametric sib-pair methods with the two-point linkage program and multipoint linkage program for affected sib pairs [2]. Linkage signals were revealed on chromosomes 1, 2, 4, and 7.
Age of onset data are often collected in studies designed to map a complex disease. If age at onset is genetically mediated, it may carry useful linkage information. Genetic analysis that incorporates variable age of onset may improve the ability to map genes for complex diseases. In this report, we analyzed the COGA data using genetic methods based on additive genetic gamma frailty models to account for age of onset or covariate information [3,4].
Methods
Consider a sibship with n sibs. Let Tj be the random variable of age at disease onset for the jth sib. Let (tj,δj) be the observed data where tj is the observed age at onset if δj = 1, and age at censoring if δj = 0. Consider a marker locus d in the test chromosomal region. We assume that the hazard function of developing disease for the jth individuals at age tj is modelled by the proportional hazards model with random effect Zj,
λj(tj|Zj) = λ0(tj)exp(Xjβ)Zj, for j = 1, 2, ..., n,
where λ0(t) is the unspecified baseline hazard function, and Xj is a vector of observed covariates for the jth sib and β is a vector of regression parameters associated with the covariates. Zj is the unobserved genetic frailty. The genetic frailty is defined as the following
![]()
where Vd = (v1, v2, ..., v2n - 1, v2n) is the inheritance vector [5] of a sibship at locus d, v2j - 1 = 1 or 2, and v2j = 3 or 4 to indicate the origins of the inherited alleles for j = 1, 2, ..., n. Ud1 and Ud2 represent the genetic frailties due to part of the genome on the two chromosomes of the father at locus d. Ud3 and Ud4 represent the same, though for the mother. The random frailty term, Up, takes into account possible genetic contributions to shared familial effects. Gamma distributions were used to model the frailties and retrospective likelihood ratio tests were constructed for linkage analysis [3]. After linkage evidence is established by the linkage analysis [3], we used an association test in the presence of linkage as proposed by Zhong and Li [4] that examines the putative association between the disease and the testing allele at a candidate chromosomal locus in the linked region. Because we use age-at-onset as the outcome, the association test is based on the proportional hazards model.
The dataset includes a total of 143 nuclear and multigenerational families with 1,614 individuals. There are two kinds of diagnostic definitions of alcohol dependence, labelled in the data set as ALDX1 and ALDX2. ALDX1 alcohol-dependent subjects were defined as those individuals who met both the DSM-III-R (Diagnostic and Statistical Manual of the American Psychiatric Association-Revised) criteria for alcohol dependence and the Feighner criteria for alcoholism. ALDX2 alcohol-dependent subjects were defined as those who met the DSM-IV criteria. Both ALDX1 and ALDX2 alcohol dependence phenotypes are coded in four levels: pure unaffected, never drank, unaffected with some symptoms, and affected. We combined the first three codings as unaffected. We extracted genotyped affected sib pairs with their parents from each pedigree to ensure independence between nuclear families. This yielded 142 affected sib pairs with their parents for a total of 568 individuals for the ALDX1 phenotype for alcoholism. For the ALDX2 phenotype, this yielded 117 affected sib pairs with their parents for a total of 468 individuals. We utilized MAPMAKER/SIBS linkage program [6] to estimate the full multipoint probability of each pair of selected sibs sharing 0, 1, or 2 alleles identical by descent (IBD) on each chromosome. We obtained the Kaplan-Meier curves as the approximation to the baseline functions using the available age-of-onset data from all of the founders in the full dataset (287 for ALDX1 and 289 for ALDX2). The founders are individuals who do not have parents in pedigree and are considered as random subjects from the general population. The Kaplan-Meier survival curves for females and males (figures not shown here) indicate strong evidence of sex differences in ages of onset distributions. Alcohol dependence is more common in males than females. The Kaplan-Meier survival curves for smokers and nonsmokers (figures not shown here) also indicate distributional differences between smokers and nonsmokers.
For each of the two disease classifications, ALDX1 and ALDX2, we first performed linkage analysis over the whole genome, excluding the sex chromosome, using the methods of Li and Zhong [3], adjusting sex and smoking status as the covariates as well as the mean IBD test to analyze the microsatellite markers on each chromosome for linkage. The mean IBD test determines whether affected sib pairs share alleles at a specific marker more than the Mendelian expectation under no linkage. After the linkage evidence (p-value < 0.01) to some candidate genes is established, we further applied the association method [4] as well as the transmission/disequilibrium tests [7] to single-nucleotide polymorphism (SNP) markers within the peak regions.
Results
A preliminary multipoint genome scan using mean IBD tests (figures not shown here) indicated some evidence of linkage to ALDX1 in the regions around 145 cM of chromosome 7 (p-value of 0.006), 161 cM of chromosome 6 (p-value of 0.008), and 157 cM of chromosome 12 (p-value of 0.016). Evidence of linkage to ALDX2 was found in the regions around 128 cM of chromosome 7 (p-value of 0.0086), 14 cM of chromosome 8 (p-value of 0.0196), 169 cM of chromosome 6 (p-value of 0.021), 5 cM of chromosome 2 (p-value of 0.041), and 148 cM of chromosome 2 (p-value of 0.049).
Table 1 lists the names and the map positions in centimorgans of the markers near those peaks with p-values less than 0.01 for genome scans using frailty models for linkage incorporating gender as a covariate or gender and smoking status as covariates for both ALDX1 and ALDX2. Frailty models identified the regions with evidence of linkage by the mean IBD tests, but with stronger signals, and also revealed some new regions with significant evidence of linkage. Adjusting for gender and smoking status as covariates, the strongest evidence of linkage to ALDX1 was achieved on a region on chromosome 7 (Table 1). Some evidence of linkage to ALDX2 was also found close to that region. The strongest evidence of linkage to ALDX2 was obtained on a region on chromosome 12. The same region was also found with some evidence of linkage to ALDX1. Figure 1 displays the multipoint linkage scans over chromosome 7 for ALDX1 and chromosome 12 for ALDX2, adjusting gender and smoking status as covariates.
Table 1.
Table of linkage results
| Sex | Sex + Smoking | |||||
| Chr | Markers | Position(cM) | P-value | Markers | Position(cM) | P-value |
| ALDX1 | ||||||
| 2 | D2S1329 | 4.9 | 0.0098 | D2S1319 | 4.9 | 0.008 |
| 2 | D2S1790 | 114.2 | 0.001 | |||
| 2 | D2S2370 | 184.3 | 0.006 | |||
| 2 | D2S1323 | 251.9 | 0.002 | |||
| 3 | D3S2398 | 216.5 | 0.006 | |||
| 4 | D4S1651 | 110.3 | 0.003 | D4S1651 | 110.3 | 0.003 |
| 6 | GATA165G02 | 160.4 | 0.003 | GATA165G02 | 160.4 | 0.003 |
| 7 | D7S490 | 145.5 | 0.004 | D7S490 | 145.5 | 0.0003 |
| 9 | DBH.PCR2.1 | 165.5 | 0.0007 | |||
| 10 | D10S1213 | 134.1 | 0.005 | |||
| 12 | D12S390 | 67.9 | 0.007 | D12S390 | 67.9 | 0.007 |
| 12 | D12S2078 | 156.8 | 0.003 | D12S2078 | 156.8 | 0.003 |
| ALDX2 | ||||||
| 2 | D2S1328 | 147.6 | 0.002 | D2S1328 | 147.6 | 0.002 |
| 3 | D3S2398 | 216.5 | 0.007 | |||
| 4 | GABRB1 | 51.4 | 0.0007 | GABRB1 | 51.4 | 0.0007 |
| 5 | D5S1473 | 34 | 0.003 | |||
| 6 | D6S1007 | 168.6 | 0.009 | |||
| 7 | D7S1799 | 127.7 | 0.003 | D7S1799 | 127.7 | 0.002 |
| 8 | D8S1145 | 13.6 | 0.005 | D8S1145 | 13.6 | 0.005 |
| 9 | DBH.PCR2.1 | 165.5 | 0.001 | |||
| 12 | D12S390 | 67.9 | 0.00004 | |||
| 13 | D13S325 | 34.1 | 0.006 | |||
| 15 | D15S816 | 122.6 | 0.006 | |||
| 16 | D16S423 | 8.2 | 0.004 | |||
| 20 | D20S94 | 80.9 | 0.002 | D20S94 | 80.9 | 0.001 |
Summary of linkage markers (P-value < 0.01) from multipoint genome scan.
Figure 1.
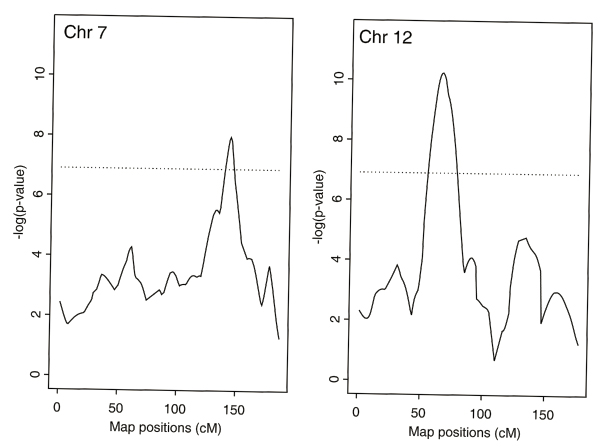
The multipoint linkage scans over chromosome 7 for ALDX1 and chromosome 12 for ALDX2, adjusting gender and smoking status as covariates. The dashed horizontal lines refer to the negative of the natural logarithm of 0.001 (i.e., -log(0.001)) corresponding to the significance level of 0.001.
For the association tests in the presence of linkage, we present here only the SNPs within a 10-cM vicinity of the two linkage peaks with smallest p-values under each of disease criteria. For ALDX2, evidence of association at a 0.001 significant level using frailty models adjusting gender and smoking status as covariates was found for SNPs rs273954 and rs700273 on chromosome 7, and rs710411 on chromosome 9 and ALDX1, and between rs1978161, rs1565933, rs1867299, rs2279400, rs1848125, rs1224438, rs1495042, rs965125, and rs1444588 on chromosome 12, and rs1039559 on chromosome 4. As a comparison, for ALDX1, evidence of association at a 0.05 level using transmission/disequilibrium test on trio (parents and one affected sib) data was found between rs273954 and rs727714 on chromosome 7; for ALDX2, evidence was found for rs12142 and rs951149 on chromosome 4, and rs1705748, rs1705772, rs1843910, rs1846629, and rs1820545 on chromosome 12.
Discussion
Although the markers identified under the two criteria, ALDX1 and ALDX2, are not entirely the same, several common regions were identified. The similarity of the regions supports some common genetic bases for ALDX1 and ALDX2, whereas the differences in the identified regions underscore the importance of using different phenotypes. As studied in [3,4], the methods using frailty models have correct type I error rates. The methods using frailty models incorporate age at onset and covariate factors and can increase the power of detecting linkage evidence over the traditional methods such as mean IBD test, which does not make use of age at onset information (Table 1). It is also interesting to note that the inclusion of smoking as a covariate in the linkage analysis resulted in more candidate genes with significant linkage evidence. We should note that the p-values reported here are not corrected for multiple comparisons.
Abbreviations
COGA: Collaborative Study on the Genetics of Alcoholism
GAW: Genetic Analysis Workshop
IBD: Identical by descent
SNP: Single-nucleotide polymorphism
Authors' contributions
XZ conceived of the study, carried out the genetic studies, performed the statistical analysis and drafted the manuscript. HZ drafted the manuscript, revised it critically for important intellectual content, and gave final approval of the version to be published. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This research is supported in part by grant R01DA12468, DA016750, and DA017713 from the National Institute on Drug Abuse.
Contributor Information
Xiaoyun Zhong, Email: Xiaoyun.Zhong@yale.edu.
Heping Zhang, Email: Heping.Zhang@yale.edu.
References
- Edenberg HJ. The Collaborative Study on the Genetics of Alcoholism: an update. Alcohol Res Health. 2002;26:214–218. [PMC free article] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Eerdewegh PV, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr, Tischfield JA, Crowe RR, Cloninger RC, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. A genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. doi: 10.1002/(SICI)1096-8628(19980508)81:3<207::AID-AJMG1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Li H, Zhong X. Multivariate survival models induced by genetic frailties, with application to linkage analysis. Biostatistics. 2002;3:57–75. doi: 10.1093/biostatistics/3.1.57. [DOI] [PubMed] [Google Scholar]
- Zhong X, Li H. Score tests of genetic association in the presence of linkage based on the additive genetic gamma frailty model. Biostatistics. 2004;5:307–327. doi: 10.1093/biostatistics/5.2.307. [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES. Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet. 1995;57:439–454. [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]


