Abstract
The clonal determination of B-cell lymphoproliferative disorders by immunoglobulin heavy chain (IgH) rearrangement by polymerase chain reaction (PCR) is widely used. However, few attempts have been made to detect immunoglobulin κ light chain (Igκ) gene rearrangement using PCR. We studied 145 cases of B-cell neoplasms, along with 58 atypical and 18 reactive lymphoproliferative disorders, using newly designed degenerate oligoprimers recognizing the framework 3 (FR3κ) and the joint (Jκ) regions of the Igκ gene. PCR products were analyzed on nondenaturing polyacrylamide gel (ndPAGE). Clonal B-cell determination was further investigated using IgH rearrangement and t(11:14) or t(14:18). By combining these methods, we detected either clonality or translocation in 117 of 137 cases (85%) in mature B-cell neoplasms. The additional analysis of Igκ rearrangement improved sensitivity from 66% to 85%. To investigate whether the Ig gene configuration could be characterized using Igκ PCR in B-cell neoplasms showing severe breakdown of genomic DNA, 18 selected cases were analyzed. Successful amplification was detected in 72% of the cases using either FR3/2-JH and/or FR3Jκ oligoprimers. Finally, clonality was detected in 21 of 58 atypical B-cell proliferations, and among them, the atypical marginal cell (54%) and atypical large cell (50%) proliferations showed the highest frequency of clonal immunoglobulin gene products. We concluded that PCR/ndPAGE analysis of Igκ is a sensitive, rapid, and efficient method for assessing clonality in conjunction with IgH and specific translocation analysis. This approach is particularly useful in the characterization of B-cell lymphoproliferative disorders in archival material with poor preservation of the genomic DNA.
The characterization of immunoglobulin (Ig) gene rearrangement has become an important and crucial step in the diagnosis of B-cell lymphoid malignancies. The clonal rearrangements of Ig heavy chains (IgH) and Ig light chains (IgL) in B-cell disorders provide excellent markers for molecular analysis. 1,2 For the past 10 years, Southern blotting has been widely used to detect clonality. 3-5 Recently, however, polymerase chain reaction (PCR) has gradually replaced this approach and has become increasingly accepted as the primary method for detecting Ig clonality. 6,7 As compared to Southern blotting, PCR has the advantage of high sensitivity, low cost, fast turnaround time, and technical simplicity. Furthermore, because small DNA fragments are the targets of PCR, this approach has been successfully applied in the study of formalin-fixed and paraffin-embedded tissue samples. Even in severely damaged archival material, PCR can still detect fragments smaller than 200 bp and identify clonal Ig gene products. 8,9
Several groups have established PCR-based methodologies for the detection of IgH rearrangement. 9-15 Most of the methods were designed using oligonucleotides recognizing FR2 and FR3 fragments of variable regions of the IgH and expanding to the joint regions. However, even using multiple oligoprimers, the overall sensitivity ranges between 50% and 80%, and at least 20% of neoplasms remain undetected. To overcome this difficulty, some investigators have used heavy chain family-specific oligoprimers that recognize either leader or framework 1 regions (FR1) of Ig heavy and light chains to increase the sensitivity. 16,17 However, this method has several disadvantages. It amplifies larger fragments and is often not successful in poorly preserved formalin-fixed and paraffin-embedded material. Furthermore, it requires multiple pairs of oligoprimers and more complex gels. 18,19 Although, because of these technical difficulties, this approach is not practical for routine clinical usage, it has been used to study κ light chain Ig (Igκ) and lambda light chain Ig (Igλ) gene rearrangements. However, most of these studies have been based on a small number of cases, and the overall findings were not sufficient for establishing whether this approach can be routinely applied in clinical diagnostic settings. 20-22
Igκ gene rearrangement analysis is a powerful tool for establishing the clonal nature of B-cell disorders. During B-cell differentiation, the Igκ gene locus undergoes rearrangement after IgH. In the case in which functional κ chain products are not obtained, the Igλ locus subsequently undergoes rearrangement. 23-25 Studies have shown that in all Igκ+ and in a very large majority of Igλ+ B-cell malignancies, either productive or nonproductive Igκ products are formed. Finally, only a small subset of Igλ+ B-cell neoplasms show biallelic deletions of the Igκ variable region (17%) or Igκ in germline configuration (2%). 5,26 Therefore, the rearranged genomic products of Igκ represent an excellent marker for B-cell clonal analysis. We have designed universal FR3κ and Ig κ light chain joint region (Jκ) primers that expand the hypervariable complementary determined region 3 (CDR3) of Igκ chain. This single pair of degenerate oligoprimers should be able to recognize the large majority of all Ig κ light chain variable region (Vκ) members within the six different families of the κ gene. Our primers amplify short products ranging from 126 to 144 bp, and, using a ndPAGE, the polyclonal and monoclonal patterns are well detected. The addition of Igκ analysis to the IgH and specific translocation assays allows the detection of more than 80% of mature B-cell neoplasms.
Materials and Methods
Patients and Cell Lines
A panel of 221 well-characterized specimens consisting of 32 follicle center lymphomas (FCLs) (including small, mixed, and large cells), 35 mantle cell lymphomas (MCLs), 24 B-cell chronic lymphocytic leukemias/small lymphocytic lymphomas (CLLs/SLLs), 24 marginal zone B-cell lymphomas (MZBCLs) (including nodal, extranodal, and splenic marginal zone B-cell lymphomas), 22 diffuse large cell lymphomas (DLCLs), eight B-cell acute lymphoblastic leukemias (B-ALLs), 18 reactive lymphoid proliferations (RLPs), and 58 atypical lymphoid proliferations (ALPs) were included in this study. All of the samples were clinical cases routinely examined in the Hematopathology/Molecular Pathology Laboratory of the New York University Medical Center from 1995 to 1998, in the Department of Pathology of Leuven University, Belgium, or in the Department of Anatomical Pathology of Torino University, Torino, Italy. They were all classified using conventional clinical and histopathological criteria in accordance with the REAL classification. 27 ALPs included cases in which histological and/or immunohistochemical findings did not allow a definite distinction between a malignant and a reactive process. The ALPs were subcategorized according to the predominant cell type (marginal zone, mantle, follicle center, or plasma cells, etc) or to morphology (large, small, or mixed cells) when no predominant cell type was recognized. Only cases in which the amplification of control gene p53 was successful were selected. Reactive tonsil tissues were used as a polyclonal control. Several B- and T-cell lymphoma or leukemia cell lines and Epstein-Barr virus (EBV) immortalized cell lines were used as positive and negative controls. The cell lines included CB33, UH10.1, and RD (EBV immortalized B-lymphoblastoid cell lines 28 ), Karpas 299 (anaplastic large cell lymphoma cell line, a gift from Dr. Lorenzana), CEM, and Jurkat (T-cell lymphoblastic leukemia cell lines; ATCC, Rockville, MD). Patient samples with known t(11:14) and t(14:18) translocations were used as positive controls for Bcl-1 and Bcl-2 gene rearrangement analyses.
DNA Sample Preparation
Genomic DNA was extracted from cryopreserved mononuclear cell suspensions, frozen tissue blocks, or formalin-fixed and paraffin-embedded archival tissue blocks. The frozen tissue samples were digested overnight (55°C) in the digestion buffer (10 mmol/L Tris-HCl, 400 mmol/L NaCl, and 2 mmol/L EDTA) in the presence of 200 μg/μl of proteinase K solution as previously described. 9 The proteinase K was then heat inactivated (95°C for 10 minutes). Samples were centrifuged (15 minutes at 12,000 rpm), supernatants were collected, and DNA concentration was measured. In selected cases, DNA was extracted with phenol/chloroform and precipitated. For DNA obtained from archival formalin-fixed, paraffin-embedded tissue, one to three 5-μm-thick tissue sections were cut and collected in microfuge tubes. Tissues were deparaffinized with xylene (three times) and then extracted with 100% ethyl alcohol. Specimens were then processed as described above for fresh tissue samples.
Polymerase Chain Reaction Analysis
Ig gene rearrangement products were amplified by PCR, using oligonucleotides that were synthesized by the solid-phase triester method (GeneLink, Thornwood, NY). Igκ gene rearrangement was analyzed by PCR using oligonucleotides recognizing the Igκ FR3 region (FR3κ: 5′-TTCAG{C/T}GGCAGCGG{A/G}TCTGGG-3′; codon 62–68) and Igκ joint region (Jκ: 5′-CA{G/C}CTT{G/T}GTCCC{C/T}TGGCCGAA-3′; codon 98–104). Two hundred nanograms of genomic DNA was subjected to PCR amplification in the presence of PCR buffer A (250 μmol/L deoxynucleoside triphosphates (dNTPs), 50 mmol/L KCl, 10 mmol/L Tris-HCl (pH 9.0), 1.2 mmol/L MgCl2, 0.1% Triton X-100, 20 pmol of each oligonucleotide, and 2.5 U of Taq DNA polymerase) using a touchdown PCR (denaturing cycle of 95°C for 3 minutes followed by 4 cycles of 15 seconds at 94°C, 1 minute at 64°C, 1 minute at 72°C, 4 cycles of 15 seconds at 94°C, 1 minute at 62°C, and 1 minute at 72°C, followed by 30 cycles of 15 seconds at 94°C, 1 minute at 60°C, and 1 minute at 72°C, with a final extension at 72°C for 10 minutes) performed on an automated heat block (DNA Thermal-Cycler; Perkin-Elmer Cetus, Norwalk, CT). The PCR products were subsequently mixed with an equal amount of running buffer (98% formamide, 10 mmol/L EDTA, 0.025% xylene cyanol FF, 0.025% bromophenol blue), denatured at 95°C for 3 minutes, incubated on ice for 15 minutes, and loaded onto a 6% nondenaturing polyacrylamide gel (ndPAGE) (MiniProtein II System; Biorad, Hercules, CA) in 1× TBE buffer (90 mmol/L Tris-borate, 2 mmol/L EDTA). Samples were electrophoresed at 90 V at room temperature for 30 minutes. Gels were then stained with ethidium bromide and photographed under ultraviolet light.
ndPAGE is optimal in facilitating the resolution of homoduplex/heteroduplex bandings in detecting Igκ clonality. This technique was originally developed to evaluate the phylogeny in evolution 29 and was subsequently found to be an excellent tool for the study of Ig gene clonality. 30,31 Under a cycle of denaturation and renaturation of the PCR products, the DNA of monoclonal products will perfectly match each other and form a uniform double-stranded population. This population is composed of a single homoduplex population and appears as a distinct single band on ndPAGE. On the other hand, after the denaturation and renaturation cycle, each double-stranded product of polyclonal population will form a heteroduplex with a varying degree of mismatch. The mismatch alters the conformation of every double-stranded DNA, and this heteroduplex population appears as a smear on ndPAGE, which also has slower mobility compared with the homoduplex population.
To investigate the sensitivity of PCR/ndPAGE in detecting clonal Igκ rearrangement, serially diluted genomic DNAs obtained from fresh cells of a patient with B-cell chronic lymphocytic leukemia (B-CLL) (>95% tumor B cells) were mixed with the genomic DNA of a reactive tonsil. A large population of polyclonal B cells (≥50%) exists in the tonsil tissue, and together with remaining T cells, better mimics the subpopulation of the polyclonal reactive B cells within any putative B-cell clonal population.
Heavy chain Ig gene rearrangement products were investigated using genomic DNA (200 ng) in the presence of specific oligonucleotide primers recognizing the human IgH variable region FR 2 (FR2a: 5′-TGG{A/G}TCCG{A/C}CAG{G/C}C{G/A/C}{C/T}C{A/C/G/T}GG-3′; codons 36–42) or FR3 (FR3a: 5′-GGTGGATCGATGAATTCTTACACGGC{C/T}{G/C}TGTATTACTGT-3′; codon 86–92) and universal joint region oligonucleotide primers (JH-770: 5′-ACCTGAGGAGACGGTGACC-3′ or JH-33: 5′-GGATGGTACCAAGCTTTGAGGAGACGGTGACCA-3′; codon 109–115). 9 DNAs were amplified in the presence of buffer A (FR2) or buffer B (FR3) (same as buffer A, except with 1.5 mmol/L MgCl2). The mixture was overlaid with mineral oil and subjected to one cycle of 3 minutes at 94°C, followed by 35 cycles of 15 seconds at 94°C, 1 minute at 57°C (FR2/JH) or 58°C (FR3/JH), and 1 minute at 72°C, followed by a final extension of 7 minutes at 72°C. PCR products were size fractionated on alkaline 2.5% (FR2 products) or 3% (FR3 products) agarose gels by electrophoretic separation, allowing the detection of PCR fragments of 237–288 bp or 86–120 bp for FR2/JH and FR3/JH regions, respectively. The majority of clonal IgH cases showed a pattern with a single, distinct band. This set of oligoprimers was rarely able to successfully amplify the products derived from the rearrangements of both alleles (two bands). A smear was considered as a polyclonal pattern, and nonamplification was interpreted as negative.
The presence of the t(11:14) and t(14:18) translocations was studied by PCR as previously described. 32,33 The expected t(11:14) or t(14:18) products were of variable sizes because of the different translocation breakpoints, ranging from 200–350 bp and 200–250 bp for Bcl-1 and Bcl-2, respectively.
p53 amplification was used as a positive control to determine whether the sample DNA was suitable for PCR analysis. The gene amplification product of p53 spanning exons 8 and 9 (approximately 450 bp) is 150–300 bp larger than IgH or Igκ products; therefore it is ideal to indicate the quality of genomic DNA preservation. The p53 genomic region was amplified using oligonucleotide p53-8–5 (5′-TATCCTGAGTAGTGGTAATC-3′) and p53-9–3 (5′-AAGAAGAAAACGGCATTTTG-3′). 34,35 To evaluate the ability of detecting clonal immunoglobulin gene rearrangements in poorly preserved DNA specimens, an additional 18 B-cell neoplasms of various categories (four DLCLs, four FCLs, one CLL/SLL, one MCL, two MM/PLs, six unclassifiable) with negative p53 control gene amplification were selected and subjected to the evaluation of IgH FR2, IgH FR3, and Igκ FR3 rearrangements.
In all of the experiments, samples of genomic DNA from monoclonal neoplastic B cells, normal or reactive lymph nodes, reactive tonsil tissue, and T-cell leukemia were included as positive and negative controls. In addition, mixtures without a template were included as a negative control to exclude a potential contamination.
Single-Strand Conformation Polymorphism Analysis
Single-strand conformation polymorphism (SSCP) analysis was accomplished according to an adapted version of a previously reported method. 34 Briefly, PCRs were performed with 100 ng of genomic DNA, 10 pmoles of each primer (FR3κ and Jκ), 2.5 μmol/L dNTPs, 1 μCi of [α-32P]dCTP (New England Nuclear) (specific activity, 3000 Ci/mmol), 10 mmol/L Tris (pH 8.8), 50 mmol/L KCl, 1 mmol/L MgCl2, 0.01% gelatin, and 0.5 U Taq polymerase, in a final volume of 10 μl. Thirty cycles of denaturation (94°C), annealing (63°C), and extension (72°C) were performed on an automated heat block. Four microliters of reaction products was mixed with an equal volume of the stop solution (50 μl of 0.1% sodium dodecyl sulfate and 10 mmol/L EDTA), denatured (5 minutes, 95°C), and loaded onto a 6% acrylamide-TBE gel containing 10% glycerol. After electrophoresis, the gels were fixed, air dried, and autoradiographed as previously described. 34
Results
κ Light Chain Ig Gene Rearrangement by PCR
Ig gene rearrangement analysis using PCR has become a common practice in the characterization of human lymphoproliferative disorders. This approach is performed using oligonucleotide primers recognizing a relatively highly conserved region of the IgH. Considering that the amplification of small genomic fragments is successful even in poorly preserved material and the analysis of CDR3 segments is highly informative of Ig gene rearrangement products, we designed a new set of oligonucleotide primers recognizing the FR3 region of all members of the six Ig κ variable (Vκ) families and the κ joint region (Jκ).
Because of the relatively small number of amino acids generating the Igκ CDR3, the absence of diversity regions of these segments, and small number of members of these gene families, the overall length range of κ CDR3 is considerably less heterogeneous (126–144 bp) than that seen in the case of CDR3 of IgH. 20,36,37 Thus, routine agarose gel electrophoresis identifies clonal products relatively poorly and cannot differentiate between small polyclonal and monoclonal products, both of which usually appear as a single band (Figure 1A) ▶ . On the other hand, when the VJκ products are resolved by high-resolution ndPAGE, the polyclonal and clonal products are easily detected. The monoclonal pattern appears as a distinct single band (approximately at 120 bp), running faster than polyclonal products. Polyclonal products are represented by a uniform smear ranging from 120 to 150 bp (Figure 1B) ▶ . To confirm the presence of clonal products identified using ndPAGE, selected cases were also characterized by SSCP analysis (Figure 1C) ▶ . Our results showed 100% concordance between ndPAGE and SSCP (data not shown) .
Figure 1.
Detection of Igκ gene rearrangement. Genomic DNA from human B cell lines (lane 1, CB33; lane 2, RD), reactive tonsil (RT) (lane 3), and patient samples (CLL, MCL, FCL, MZBCL, RLP) was subjected to PCR amplification of FR3Jκ regions and analyzed with 3% agarose gel (A), ndPAGE (B), and SSCP (C). A: Polyclonal and monoclonal patterns were not discriminated on 3% agarose gel and appeared as single bands. Instead, the polyclonal and monoclonal populations were well separated on 6% ndPAGE. B: The polyclonal pattern appears as a clear smear, and the monoclonal pattern shows a distinct single band (approximately at 120 bp). C: The clonality detected using ndPAGE/gel was confirmed by SSCP analyses.
Using this approach in the category of mature B-cell malignancies, Igκ clonal products could be detected in 52% of the cases (Table 1) ▶ . The highest yield was obtained in MCLs and B-cell CLLs/SLLs (66% and 63%, respectively). The lowest frequency of Igκ rearrangement was observed in B-ALLs (13%). This low frequency is largely due to the absence of IgL rearrangements in the early stage of B-cell differentiation. 23 Finally, Igκ monoclonality was also detected in 12% of atypical B-cell proliferation. No clonal bands were observed in reactive cases.
Table 1.
PCR Detection of Immunoglobulin Gene Heavy and Light Chain Rearrangements in Lymphoproliferative Disorders
| Diagnostic categories | IgH FR2 | IgH FR3 | Igκ | Bcl-1/Bcl-2 | Overall* | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| p/t | % | p/t | % | p/t | % | p/t | % | p/t | % | |
| Mature B-cell neoplasms | ||||||||||
| FCL | 7 /32 | 22% | 13 /32 | 41% | 13 /32 | 41% | 7 /32 | 22% | 28 /32 | 88% |
| MCL | 20 /35 | 57% | 21 /35 | 60% | 23 /35 | 66% | 11 /35 | 31% | 33 /35 | 94% |
| CLL/SLL | 11 /24 | 46% | 14 /24 | 58% | 15 /24 | 63% | n /a | n/a | 21 /24 | 88% |
| MZBCL | 7 /24 | 29% | 15 /24 | 63% | 9 /24 | 38% | n /a | n/a | 18 /24 | 75% |
| DLCL | 12 /22 | 55% | 6 /22 | 27% | 11 /22 | 50% | n /a | n/a | 17 /22 | 77% |
| Total | 57 /137 | 42% | 69 /137 | 50% | 71 /137 | 52% | n /a | n/a | 117 /137 | 85% |
| Immature B-cell neoplasm | ||||||||||
| B-ALL | 4 /8 | 50% | 2 /8 | 25% | 1 /8 | 13% | n /a | n/a | 5 /8 | 63% |
| Nonneoplastic | ||||||||||
| ALP | 9 /58 | 16% | 13 /58 | 22% | 7 /58 | 12% | n /a | n/a | 21 /58 | 36% |
| RLP | 0 /18 | 0% | 1 /18 | 5% | 0 /18 | 0% | n /a | n/a | 1 /18 | 5% |
p/t, Positive/total cases; % Pos, frequency of positivity.
*Overall cases that show at least one clonal gene rearrangement product using Ig or Bcl-1/Bcl-2 PCRs.
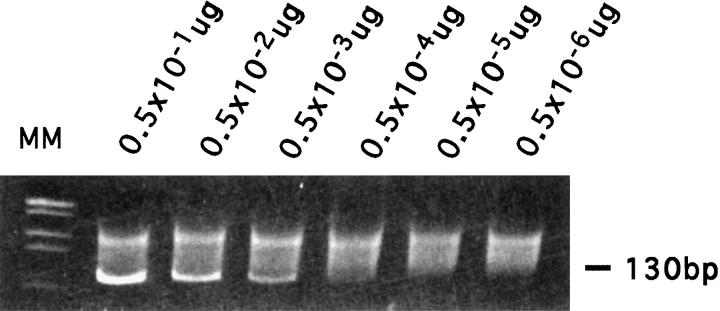
The sensitivity of PCR/ndPAGE in detecting clonal Igκ rearrangement was analyzed by the serial dilution method. After gel electrophoretic separation, a distinct PCR fragment could be identified in dilutions up to 0.5 × 10−3, which represents 0.05% of the total DNA (Figure 2) ▶ .
Figure 2.
Detection of the sensitivity of the Igκ gene rearrangement. DNA from monoclonal B cell (CLL) was serial diluted (0.5, 0.5 × 10−1, 0.5 × 10−2, 0.5 × 10−3, 0.5 × 10−4, 0.5 × 10−5 μg/sample) with DNA from a reactive tonsil and subjected to FR3Jκ PCR amplification. PCR products were separated using 6% ndPAGE. A distinct PCR band could be identified in the dilution up to 0.5 × 10−3, which represented 0.05% of the total DNA.
Detection of B-Cell Clonality Using Multiapproach PCR
All cases were also evaluated by PCR analysis for clonality in the IgH region. Moreover, the t(11:14) and t(14:18) translocation products of MCLs and FCLs, respectively, were also analyzed (Table 1) ▶ . Using two pairs of oligoprimers expanding the IgH CDR2 and CDR3 regions, we detected clonality in 66% of mature B-cell neoplasms overall (data not shown). FR3/JH detected slightly more cases (50%) than did FR2/JH (42%). Among individual categories of tumors, the highest rate of clonality was observed in MCLs and DLCLs for FR2/JH (57% and 55%, respectively) and in MZBCLs and MCLs for FR3/JH (63% and 60%, respectively).
Our study shows that, in mature B-cell neoplasms, the combination of Igκ and IgH rearrangement analysis increased the positive rate to 85% overall as compared to IgH PCR alone (66%). In particular, using this multiple PCR approach, we have detected the highest frequencies of clonality, among mature B-cell neoplasms, in MCLs (94%), FCLs (88%), and B-CLLs/SLLs (88%).
Identification of B-Cell Clonality in Poorly Preserved Tissue Samples
In routine molecular laboratory practice, very often the source of DNA is limited to fixed and paraffin-embedded materials. Frequently, these tissue samples show poor preservation of the genomic DNA and the amplification of large DNA fragments is impaired. To investigate whether shorter PCR products allow the characterization of poorly preserved materials, a total of 18 archival samples of B-cell neoplasms, which showed no amplification of the p53 control gene (450 bp), were analyzed by IgH and Igκ rearrangements. Six cases showed Igκ amplification, and eight cases showed IgH FR3 amplification. Only one case showed amplification of IgH FR2 products. The overall amplification frequency was 72%. Among 13 amplified cases, seven displayed a monoclonal band with either IgH or Igκ rearrangements (54%). Because Igκ and IgH FR3 gene products are substantially smaller than that of the p53 control gene, it is reasonable to conclude that even in poorly preserved tissue, these smaller fragments are still well maintained and allow successful Igκ and IgH FR3 amplifications (Table 2) ▶ .
Table 2.
Effect of Immunoglobulin Heavy and Light Chain Rearrangement in Formalin-Fixed and Paraffin-Embedded Tissue
| Gene segment | No amplification | Monoclonal pattern | Polyclonal pattern | Overall* amplification |
|---|---|---|---|---|
| p53 control gene | 18 | 0 | 0 | 0 |
| IgH FR2 | 17 | 1 | 0 | 1 |
| IgH FR3 | 10 | 3 | 5 | 8 |
| IgL Jκ | 12 | 4 | 2 | 6 |
| Total (18) | 5 (28%) | 7 (39%) | 6 (33%) | 13 (72%) |
*Overall number of cases that could be successfully amplified using FR3, and/or FR2, and/or Jκ.
Identification of B-Cell Clonality in Atypical Lymphoproliferative Disorders and Reactive Nodes
Based on the high frequency of detection of B-cell clonality in lymphomas by our multi-IgH and Igκ approach, we decided to investigate the presence of clonal B-cell populations among those cases in which the neoplastic B-cell nature cannot be established by pure morphology and/or immunohistochemistry. Not only do these atypical lymphoproliferations represent a diagnostic dilemma; their biological features remain unclear. A total of 58 cases of B-cell ALPs were studied. Clonal PCR products were detected in seven and 15 cases using Igκ and IgH, respectively, representing a total of 21/58 (36%) cases (Tables 1 and 3) ▶ ▶ . Among single histological categories, atypical monocytoid and atypical large cell proliferations were most likely to show clonal rearrangements (54% and 50%, respectively). In comparison, we also studied 18 lymph nodes with reactive processes. A single case showed a monoclonal IgH (FR3) rearrangement, but none showed a Igκ rearrangement. Review of the clinical findings demonstrated that the patient had a history of CLL, althought this could not be recognized in the nodal histology.
Table 3.
Detection of Immunoglobulin Gene Rearrangement in Atypical Lymphoproliferative Disorders
| Diagnostic categories | IgH and/or Igκ | |
|---|---|---|
| p/t | % | |
| Atypical marginal cell proliferation | 6 /11 | 54% |
| Atypical follicular proliferation | 3 /13 | 23% |
| Atypical plamacytoid infiltration | 0 /1 | n/a |
| Atypical small cell infiltration | 2 /7 | 29% |
| Atypical large cell infiltration | 3 /6 | 50% |
| Atypical mixed cell infiltration | 7 /20 | 35% |
| Total | 21 /58 | 36% |
To investigate the potential evolution of ALPs, the relationship among clonal PCR products obtained from different tissue samples in the same patients with that histological diagnosis was evaluated. This was done by PCR and SSCP analysis in eight individuals who had sequential biopsies. In two patients identical clones were detected in sequential biopsied samples by IgH or Igκ analysis (data not shown).
Discussion
The CDR3 region of Igκ contributes the greatest variability within the VJκ segment. This variability is largely due to the juxtaposition of VJ regions and the addition of P and N nucleotides. 36,38 The high degree of variability of the CDR3 region of Igκ is an ideal target for separation of polyclonal and monoclonal products. 20,37 Thus we designed a single pair of degenerate oligoprimers spanning the κ CDR3 segment. This oligoprimer pair has the advantage of a single amplification, and hence is a simple and rapid approach in daily clinical practice. As compared to the IgH locus, Igκ does not contain a diversity (D) region resulting in less diversity in κ VJ than the heavy chain VDJ segments. To facilitate the resolution of the banding in detecting Igκ clonality, we used nondenaturing polyacrylamide gel to analyze the FR3Jκ products. This approach offers several advantages: 1) ndPAGE allows the discrimination of a relatively less heterogeneous population of DNA products; 2) when compared with other high resolution gels, ndPAGE gel has the advantage of small size, simplicity of assemblage, short running time (30 minutes), and economy. In our experience, the banding pattern in ndPAGE is similar to the patterns of other studies that used a more complex gel (DGGE, TGGE, etc). 39,40 Offermans et al 41 tested TCR gene rearrangement by both denatured and ndPAGE gels. They found that compared to DGGE, ndPAGE was a relatively simple and rapid procedure with high separation potential. In our experience, ndPAGE is excellent in the separation of FR3Jκ monoclonal and polyclonal products.
Many PCR-based studies have been performed to characterize light chain immunoglobulin gene rearrangements in normal and neoplastic B-cells, using either genomic DNA 21,22 or cDNA 42-45 as templates, and κ leader or FR1 oligoprimers, in conjunction with Jκ- or Cκ-specific primers. When successful, these approaches allow the amplification of the entire VJκ products. However, they require multiple oligoprimers to recognize different members of the six Vκ a families. Thus their routine utilization in the clinical laboratory may not be feasible. Moreover, the identification of predominant clonal light chain products, using family-specific oligoprimers, may be difficult in those cases in which the neoplastic cells represent a minority compared to the normal polyclonal B-cell population. One can envision that, in this scenario, the utilization of fluorescently labeled oligoprimers and automatic DNA sequencing may be applied successfully, as in the case of heavy chain Ig and β-T-cell receptor (β-TCR) gene rearrangement analyses. 46-49 Alternatively, because the CDR3 regions of light chain Ig are sufficiently diverse, oligoprimers spanning the CDR3 region should be quite informative and allow the identification of clonal B-cell populations. The data recently obtained from Shiokawa et al 50 and the findings described in this study clearly demonstrate that this approach can be successful in the analysis of clonal B-cell processes when either Ig light messenger signals 50 or gene rearrangement products are evaluated.
We have studied a panel of 221 well-characterized B-cell lymphoproliferative processes by using Igκ and IgH rearrangements. Among 137 cases of mature B-cell malignancies, using Igκ alone, 52% cases showed clonality, and none of 18 cases of reactive B-cell proliferations did. Although the analysis of the clonal B-cell lymphoproliferative disorders by Igκ identified a percentage of monoclonal cases similar to that identified by IgH FR3, by combining the two methods, the yield in each category of mature B-cell malignancies did significantly increase from 66% to 85%. In the analysis IgH rearrangements, we obtained frequencies similar to those obtained by other investigators, which showed a higher positive rate in IgH FR3 than FR2. 9,51,52 As expected, in the immature/precursor B-cell malignancy category (B-cell ALL), the effect of Igκ is less prominent (13%). This is most likely due to the early lineage development of the B-ALL. 23,25
Toward this end, multiple attempts have been made to improve the overall rate of detection of clonal B-cell lymphoproliferative disorders. In particular, the utilization of DGGE gels, SSCP, and fluorescently labeled oligoprimers has allowed the identification of clonal B-cell products in a greater percentage of cases, even when a single set of oligoprimers has been used. 46,47 On the other hand, the utilization of agarose gels and a single universal JH primer may be responsible for the lower sensitivity seen in our study. One also should consider that our data are derived from the analysis of routinely analyzed clinical cases. These cases were often investigated on a molecular level because a definitive diagnosis could not be easily achieved by routine morphology/immunohistochemistry due to their complexity (ie, small number of tumor cells associated with a large population of benign B-cells, etc). The fact that unselected cases were used in this study may explain the overall lower sensitivity of single pair oligoprimers (FR3-JH). However, we believe that these examples are more representative of a realistic clinical practice.
The sensitivity of the Igκ assay was studied by a DNA serial dilution method. We used tonsil tissue as a polyclonal control, which in our opinion is an appropriate control and is superior to genomic DNA of T-cell lines. 9 The tonsil normally contains polyclonal cell populations composed of large numbers of polyclonal B cells (≥50%) as well as T cells. This better represents the background cell population of clinical specimens. Using this approach, clonal Igκ bands can be detected in up to 0.5 × 10−3 dilution (dilution of 0.05%), a sensitivity equal or close to that of the semi-nested approach. This high sensitivity may be due to the fact that FR3Jκ products were characterized by ndPAGE.
Since the increasing acceptance of the PCR as the primary method in gene rearrangement analysis, numerous studies have been made of formalin-fixed and paraffin-embedded archival tissues. Most of these studies demonstrated a similar frequency of positive rate in fresh and archival materials by using short PCR amplification fragments (less than 200 bp). Relatively good results can be achieved even in decalcified bone marrow tissues by using a semi-nested PCR approach. 9,17,21,53 We also evaluated the effect of Igκ amplification on poorly preserved tissue material. All of the cases were formalin-fixed and paraffin-embedded tissues that showed severe breakdown of genomic DNA. Despite this limitation, in 72% (13/18) of cases amplification of short Ig products was successful, and, combining Igκ and IgH FR3 gene analysis, we were able to detect 54% clonality in seven of 13 cases. This percentage is relatively lower than that demonstrated in ideal samples; however, it allows the detection of a considerable fraction of B-cell neoplasms, even when only poorly preserved tissue is available.
Prior studies have shown that optimal oligoprimer selection for clonality assessment by PCR may be necessary to obtain the highest sensitivity. Interestingly, unique sets of primers may be used as a first choice. In low-grade 13 or intermediate to high-grade 15 lymphoproliferative disorders and in follicular lymphomas 14 oligoprimers recognizing consensus VH-FR3 and major/minor Bcl-2 cluster regions, respectively, should be applied as a first choice. However, with the utilization of a single set of primers, a variable but typically large subpopulation of clonal positive cases remains undetected, and “reserve sets” are necessary. Based on these studies and our present data, we propose a new algorithm. Specifically, samples should first be subjected to the analysis of VH-FR3/JH. Then all negative cases should be characterized using VK-FR3/Jk PCR, followed, if necessary, by VH-FR2/JH and Bcl-1/bcl-2. Finally, the remaining negative cases might be studied using VH-FR1 and/or VL-FR1 oligoprimers. If correctly applied, this new algorithm can be a very powerful screening tool. In this way, the utilization of Southern blot analysis can be limited to the small proportion of PCR-negative cases and to further identification of unique entities among B-cell lymphoproliferative disorders (c-myc in endemic/sporadic Burkitt’s lymphomas, bcl-1 minor cluster region in MCL, etc).
Atypical B-cell lymphoproliferative disorders often represent a diagnostic dilemma, and their malignant potential is usually undetermined. This results in difficult therapeutic choices: often clinicians are obliged to simply follow up these patients to discern those individuals who will evolve into a bona fide B-cell neoplasm. To solve some of these issues, we studied a large panel of atypical disorders (58 cases), using our multiPCR approach. Interestingly, monoclonality was detected in a relatively large number of patients (36%). Among these clonal cases, atypical marginal cell proliferations were the most frequently encountered category, which comprised 54% of total cases, followed by atypical large cell proliferations (50%). The possible neoplastic evolution of ALP was further studied by evaluating IgH and Igκ clonality in eight patients who had undergone sequential biopsies at several-year intervals. Two of these patients showed identical clones, which strongly suggested the neoplastic nature of the original lesions.
The frequent positivity of Ig gene rearrangement in atypical marginal and large cell proliferations reveals the high degree of uncertainty in the histological diagnosis of these processes and therefore the importance of gene rearrangement studies. However, clonal PCR products in atypical proliferation only demonstrate the presence of clonal B-cell populations, but their nature and biological features are uncertain. In fact, these populations may simply represent the oligoclonal and/or clonal expansion of abnormal but not fully transformed cells. In these cases, an immunological deregulation may be operational. 54 On the other hand, we may be facing a different scenario, in which a fully transformed clone may be present. In this second scenario, the neoplastic population may represent only a minority of the cells without major architectural effacement. It is impossible at present to distinguish these two different possibilities, a particularly important issue in view of the different clinical therapeutic approaches that can be visualized. Obviously, in the first case, the pharmacological modulation of the immunological imbalance would be the most appropriate approach, whereas in the second case, a more aggressive therapeutic intervention would be favored.
In conclusion, our study demonstrated that the unique approach of combined IgH and Igκ gene rearrangement analysis, along with a highly sensitive ndPAGE method, is a useful tool in the routine clinical laboratory assessment of B-cell clonality. Using a single pair of primers to detect κ light chain rearrangement on ndPAGE is a sensitive, simple, and cost-effective method as an adjunct to IgH analysis. Owing to the short amplification products in FR3Jκ fragments, it is an ideal tool for analyzing poorly preserved formalin-fixed and paraffin-embedded material, which is most frequently available in routine clinical laboratory practice.
Acknowledgments
We thank Elena Mazzone for her excellent technical assistance. We thank Dr. J. Tiesinga for critical review of the manuscript.
Footnotes
Address reprint requests to Dr. Giorgio Inghirami, New York University, Department of Pathology and Kaplan Comprehensive Cancer Center, 550 First Avenue, New York, NY 10016. E-mail: Inghig01@mcrcr6.med.nyu.edu.
Supported in part by National Institutes of Health grants CA64033, CA66229, and CA14462 to Dr. Inghirami.
J. Gong and G. Frizzera’s present address: Department of Pathology, Cornell University, New York, NY.
References
- 1.Korsmeyer SJ: B-lymphoid neoplasms: immunoglobulin genes as molecular determinants of clonality, lineage, differentiation, and translocation. Adv Intern Med 1988, 33:1-15 [PubMed] [Google Scholar]
- 2.Medeiros LJ, Bagg A, Cossman J: Application of molecular genetics to the diagnosis of hematopoietic neoplasms. Edited by DM Knowles. Baltimore, Williams and Wilkins, 1992, 263–298
- 3.van Dongen JJ, Wolvers-Tettero IL: Analysis of immunoglobulin and T cell receptor genes. Part II. Possibilities and limitations in the diagnosis and management of lymphoproliferative diseases and related disorders. Clin Chim Acta 1991, 198:93-174 [DOI] [PubMed] [Google Scholar]
- 4.Arnold A, Cossman J, Bakhshi A, Jaffe ES, Waldmann TA, Korsmeyer SJ: Immunoglobulin-gene rearrangements as unique clonal markers in human lymphoid neoplasms. N Engl J Med 1983, 309:1593-1599 [DOI] [PubMed] [Google Scholar]
- 5.Beishuizen A, Verhoeven MA, Mol EJ, van Dongen JJ: Detection of immunoglobulin κ light-chain gene rearrangement patterns by Southern blot analysis. Leukemia 1994, 8:2228-2236(discussion 2237–2239) [PubMed] [Google Scholar]
- 6.Sioutos N, Bagg A, Michaud GY, et al: Polymerase chain reaction versus Southern blot hybridization. Detection of immunoglobulin heavy-chain gene rearrangements. Diagn Mol Pathol 1995, 4:8–13 [DOI] [PubMed]
- 7.Lehman CM, Sarago C, Nasim S, Comerford J, Karcher DS, Garrett CT: Comparison of PCR with Southern hybridization for the routine detection of immunoglobulin heavy chain gene rearrangements. Am J Clin Pathol 1995, 103:171-176 [DOI] [PubMed] [Google Scholar]
- 8.Trainor KJ, Brisco MJ, Story CJ, Morley AA: Monoclonality in B-lymphoproliferative disorders detected at the DNA level. Blood 1990, 75:2220-2222 [PubMed] [Google Scholar]
- 9.Inghirami G, Szabolcs MJ, Yee HT, Corradini P, Cesarman E, Knowles DM: Detection of immunoglobulin gene rearrangement of B cell non-Hodgkin’s lymphomas and leukemias in fresh, unfixed and formalin-fixed, paraffin-embedded tissue by polymerase chain reaction. Lab Invest 1993, 68:746-757 [PubMed] [Google Scholar]
- 10.Yamada M, Hudson S, Tournay O, et al: Detection of minimal disease in hematopoietic malignancies of the B-cell lineage by using third-complementarity-determining region (CDR-III)-specific probes. Proc Natl Acad Sci USA 1989, 86:5123-5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deane M, Norton JD: Detection of immunoglobulin gene rearrangement in B lymphoid malignancies by polymerase chain reaction gene amplification. Br J Haematol 1990, 74:251-256 [DOI] [PubMed] [Google Scholar]
- 12.Trainor KJ, Brisco MJ, Wan JH, Neoh S, Grist S, Morley AA: Gene rearrangement in B- and T-lymphoproliferative disease detected by the polymerase chain reaction. Blood 1991, 78:192-196 [PubMed] [Google Scholar]
- 13.Segal GH, Jorgensen T, Masih AS, Braylan RC: Optimal primer selection for clonality assessment by polymerase chain reaction analysis. I. Low grade B-cell lymphoproliferative disorders of nonfollicular center cell type. Hum Pathol 1994, 25:1269–1275 (see Comments) [DOI] [PubMed]
- 14.Segal GH, Jorgensen T, Scott M, Braylan RC: Optimal primer selection for clonality assessment by polymerase chain reaction analysis. II. Follicular lymphomas. Hum Pathol 1994, 25:1276-1282 [DOI] [PubMed] [Google Scholar]
- 15.Lombardo JF, Hwang TS, Maiese RL, Millson A, Segal GH: Optimal primer selection for clonality assessment by polymerase chain reaction analysis. III. Intermediate and high-grade B-cell neoplasms. Hum Pathol 1996, 27:373-380 [DOI] [PubMed] [Google Scholar]
- 16.Deane M, Norton JD: Immunoglobulin heavy chain variable region family usage is independent of tumor cell phenotype in human B lineage leukemias. Eur J Immunol 1990, 20:2209-2217 [DOI] [PubMed] [Google Scholar]
- 17.Kuppers R, Zhao M, Rajewsky K, Hansmann ML: Detection of clonal B cell populations in paraffin-embedded tissues by polymerase chain reaction. Am J Pathol 1993, 143:230-239 [PMC free article] [PubMed] [Google Scholar]
- 18.Davis TH, Yockey CE, Balk SP: Detection of clonal immunoglobulin gene rearrangements by polymerase chain reaction amplification and single-strand conformational polymorphism analysis. Am J Pathol 1993, 142:1841-1847 [PMC free article] [PubMed] [Google Scholar]
- 19.Thunberg U, Rosenquist R, Lindstrom A, et al: Comparative analysis of detection systems for evaluation of PCR amplified immunoglobulin heavy-chain gene rearrangements. Diagn Mol Pathol 1997, 6:140-146 [DOI] [PubMed] [Google Scholar]
- 20.Wagner SD, Martinelli V, Luzzatto L: Similar patterns of V κ gene usage but different degrees of somatic mutation in hairy cell leukemia, prolymphocytic leukemia, Waldenstrom’s macroglobulinemia, and myeloma. Blood 1994, 83:3647-3653 [PubMed] [Google Scholar]
- 21.Weirich G, Funk A, Hoepner I, et al: PCR-based assays for the detection of monoclonality in non-Hodgkin’s lymphoma: application to formalin-fixed, paraffin-embedded tissue and decalcified bone marrow samples. J Mol Med 1995, 73:235-241 [DOI] [PubMed] [Google Scholar]
- 22.Kuppers R, Willenbrock K, Rajewsky K, Hansmann ML: Detection of clonal lambda light chain gene rearrangements in frozen and paraffin-embedded tissues by polymerase chain reaction. Am J Pathol 1995, 147:806-814 [PMC free article] [PubMed] [Google Scholar]
- 23.Korsmeyer SJ, Hieter PA, Ravetch JV, Poplack DG, Waldmann TA, Leder P: Developmental hierarchy of immunoglobulin gene rearrangements in human leukemic pre-B-cells. Proc Natl Acad Sci USA 1981, 78:7096-7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollis GF, Evans RJ, Stafford-Hollis JM, Korsmeyer SJ, McKearn JP: Immunoglobulin lambda light-chain-related genes 14.1 and 16.1 are expressed in pre-B cells and may encode the human immunoglobulin omega light-chain protein. Proc Natl Acad Sci USA 1989, 86:5552-5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felix CA, Reaman GH, Korsmeyer SJ, et al: Immunoglobulin and T cell receptor gene configuration in acute lymphoblastic leukemia of infancy. Blood 1987, 70:536-541 [PubMed] [Google Scholar]
- 26.Feddersen RM, Martin DJ, Van Ness BG: The frequency of multiple recombination events occurring at the human Ig κ L chain locus. J Immunol 1990, 144:1088-1093 [PubMed] [Google Scholar]
- 27.Harris NL, Jaffe ES, Stein H, et al: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 [PubMed] [Google Scholar]
- 28.Inghirami G, Grignani F, Sternas L, Lombardi L, Knowles DM, Dalla-Favera R: Down-regulation of LFA-1 adhesion receptors by C-myc oncogene in human B lymphoblastoid cells. Science 1990, 250:682-686 [DOI] [PubMed] [Google Scholar]
- 29.Delwart EL, Shpaer EG, Mullins JI: Heteroduplex Mobility Assays for Phylogenetic Analysis. 1995:154-160 Academic Press San Diego
- 30.Chhanabhai M, Adomat SA, Gascoyne RD, Horsman DE: Clinical utility of heteroduplex analysis of TCR γ gene rearrangements in the diagnosis of T-cell lymphoproliferative disorders. Am J Clin Pathol 1997, 108:295-301 [DOI] [PubMed] [Google Scholar]
- 31.Langerak AW, Szczepanski T, van der Burg M, Wolvers-Tettero IL, van Dongen JJ: Heteroduplex PCR analysis of rearranged T cell receptor genes for clonality assessment in suspect T cell proliferations. Leukemia 1997, 11:2192-2199 [DOI] [PubMed] [Google Scholar]
- 32.Molot RJ, Meeker TC, Wittwer CT, et al: Antigen expression and polymerase chain reaction amplification of mantle cell lymphomas. Blood 1994, 83:1626-1631 [PubMed] [Google Scholar]
- 33.Crescenzi M, Seto M, Herzig GP, Weiss PD, Griffith RC, Korsmeyer SJ: Thermostable DNA polymerase chain amplification of t(14;18) chromosome breakpoints, and detection of minimal residual disease. Proc Natl Acad Sci USA 1988, 85:4869-4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaidano G, Ballerini P, Gong JZ, et al: p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc Natl Acad Sci USA 1991, 88:5413-5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballerini P, Gaidano G, Gong JZ, et al: Multiple genetic lesions in acquired immunodeficiency syndrome-related non-Hodgkin’s lymphoma. Blood 1993, 81:166-176 [PubMed] [Google Scholar]
- 36.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C: Sequences of proteins of immunological interest, ed 5. Edited by EA Kabat. Bethesda MD, National Institutes of Health, 1991, pp 1229–1239
- 37.Cannell PK, Amlot P, Attard M, Hoffbrand AV, Foroni L: Variable κ gene rearrangement in lymphoproliferative disorders: an analysis of V κ gene usage, VJ joining and somatic mutation. Leukemia 1994, 8:1139-1145 [PubMed] [Google Scholar]
- 38.Foster SJ, Brezinschek HP, Brezinschek RI, Lipsky PE: Molecular mechanisms and selective influences that shape the κ gene repertoire of IgM+ B cells. J Clin Invest 1997, 99:1614-1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greiner TC, Raffeld M, Lutz C, Dick F, Jaffe ES: Analysis of T cell receptor-γ gene rearrangements by denaturing gradient gel electrophoresis of GC-clamped polymerase chain reaction products. Correlation with tumor-specific sequences. Am J Pathol 1995, 146:46-55 [PMC free article] [PubMed] [Google Scholar]
- 40.Kneba M, Bolz I, Linke B, Hiddemann W: Analysis of rearranged T-cell receptor β-chain genes by polymerase chain reaction (PCR) DNA sequencing and automated high resolution PCR fragment analysis. Blood 1995, 86:3930-3937 [PubMed] [Google Scholar]
- 41.Offermans MT, Sonneveld RD, Bakker E, Deutz-Terlouw PP, de Geus B, Rozing J: Denaturing and non-denaturing gel electrophoresis as methods for the detection of junctional diversity in rearranged T cell receptor sequences. J Immunol Methods 1995, 181:101-114 [DOI] [PubMed] [Google Scholar]
- 42.Marks JD, Tristem M, Karpas A, Winter G: Oligonucleotide primers for polymerase chain reaction amplification of human immunoglobulin variable genes and design of family-specific oligonucleotide probes. Eur J Immunol 1991, 21:985-991 [DOI] [PubMed] [Google Scholar]
- 43.Bridges SL, Jr, Lee SK, Johnson ML, et al: Somatic mutation and CDR3 lengths of immunoglobulin κ light chains expressed in patients with rheumatoid arthritis and in normal individuals. J Clin Invest 1995, 96:831-841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welschof M, Terness P, Kolbinger F, et al: Amino acid sequence based PCR primers for amplification of rearranged human heavy and light chain immunoglobulin variable region genes. J Immunol Methods 1995, 179:203-214 [DOI] [PubMed] [Google Scholar]
- 45.Stamatopoulos K, Kosmas C, Papadaki T, et al: Follicular lymphoma immunoglobulin κ light chains are affected by the antigen selection process, but to a lesser degree than their partner heavy chains. Br J Haematol 1997, 96:132-146 [DOI] [PubMed] [Google Scholar]
- 46.Linke B, Bolz I, Pott C, Hiddemann W, Kneba M: Use of UITma DNA polymerase improves the PCR detection of rearranged immunoglobulin heavy chain CDR3 junctions. Leukemia 1995, 9:2133-2137 [PubMed] [Google Scholar]
- 47.Linke B, Bolz I, Fayyazi A, et al: Automated high resolution PCR fragment analysis for identification of clonally rearranged immunoglobulin heavy chain genes. Leukemia 1997, 11:1055-1062 [DOI] [PubMed] [Google Scholar]
- 48.Kneba M, Bolz I, Linke B, Hiddemann W: Analysis of rearranged T-cell receptor β-chain genes by polymerase chain reaction (PCR) DNA sequencing and automated high resolution PCR fragment analysis. Blood 1995, 86:3930-3937 [PubMed] [Google Scholar]
- 49.Foss HD, Anagnostopoulos I, Araujo I, et al: Anaplastic large-cell lymphomas of T-cell and null-cell phenotype express cytotoxic molecules. Blood 1996, 88:4005-4011 [PubMed] [Google Scholar]
- 50.Shiokawa S, Nishimura J, Ohshima K, Uike N, Yamamoto K: Establishment of a novel B cell clonality analysis using single-strand conformation polymorphism of immunoglobulin light chain messenger signals. Am J Pathol 1998, 153:1393-1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segal GH, Wittwer CT, Fishleder AJ, Stoler MH, Tubbs RR, Kjeldsberg CR: Identification of monoclonal B-cell populations by rapid cycle polymerase chain reaction. A practical screening method for the detection of immunoglobulin gene rearrangements. Am J Pathol 1992, 141:1291-1297 [PMC free article] [PubMed] [Google Scholar]
- 52.Achille A, Scarpa A, Montresor M, et al: Routine application of polymerase chain reaction in the diagnosis of monoclonality of B-cell lymphoid proliferations. Diagn Mol Pathol 1995, 4:14-24 [DOI] [PubMed] [Google Scholar]
- 53.Wan JH, Trainor KJ, Brisco MJ, Morley AA: Monoclonality in B cell lymphoma detected in paraffin wax embedded sections using the polymerase chain reaction. J Clin Pathol 1990, 43:888-890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frizzera G: Atypical lymphoproliferative disorders. Edited by DM Knowles. Baltimore, Williams and Wilkins, 1992, 459–496