Abstract
Smooth muscle cell (SMC) proliferation is a key event in renarrowing of blood vessels after balloon angioplasty. Mechanical injury imparted to the arterial wall in experimental models induces the expression of the immediate-early gene, egr-1. Egr-1 binds to and activates expression from the proximal promoters of multiple genes whose products can, in turn, influence the vascular response to injury. Here, we used antisense strategies in vitro to inhibit rat vascular SMC proliferation by directly targeting Egr-1. A series of phosphorothioate antisense oligonucleotides of 15 base length and complementary to various theoretically accessible regions within Egr-1 mRNA were synthesized and assessed for their ability to selectively inhibit SMC proliferation in an Egr-1-dependent manner. Western blot analysis revealed that two oligonucleotides, AS2 and E11, inhibited Egr-1 synthesis in cells exposed to serum without affecting levels of the zinc finger protein Sp1. AS2 and E11 inhibited serum-inducible [3H]thymidine incorporation into DNA, as well as serum stimulation of total cell numbers. Size-matched phosphorothioate oligonucleotides with random, scrambled, sense or mismatch sequences failed to inhibit. Antisense Egr-1 inhibition was nontoxic and reversible. These oligonucleotides also inhibited SMC regrowth after mechanical injury in vitro. Egr-1 thus plays a key regulatory role in SMC proliferation and repair following injury.
Physical disruption of atherosclerotic lesions by balloon angioplasty is a frequently performed procedure that results in increased luminal diameter and immediate symptomatic relief in most cases. In 30 to 40% of patients undergoing the procedure, this initial success is complicated by progressive renarrowing of the artery wall, or restenosis. The use of metallic stents in conjunction with angioplasty has reduced the incidence of restenosis. 1 Restenosis, however, is still a major problem. A dominant cellular event in the renarrowing of the lumen after angioplasty is smooth muscle cell (SMC) proliferation. 2,3 SMCs rich in rough-surfaced endoplasmic reticulum elaborate a large number of growth-regulatory molecules and extracellular matrix components. 2,4 These bioactive mediators are thought to participate in autocrine and paracrine growth loops as SMCs accumulate in the developing neointima.
Early growth response factor-1 is the product of an immediate-early gene and is a prototype member of the zinc finger superfamily of transcription factors. 5-7 The gene appears on human chromosome 5 (5q23–31) and encodes a protein with three tandem Cys2-His2 zinc finger motifs. 5 Egr-1 interacts with the major groove of DNA 8 and associates with the promoter regions of a large number of genes implicated in the pathogenesis of vascular occlusive lesions. These include genes encoding cytokines, adhesion molecules, components of the coagulation cascade, and growth factors such as basic fibroblast growth factor, transforming growth factor-β and platelet-derived growth factor 9-11 (reviewed in Refs. 10 and 12 ). Recent studies have implicated a regulatory role for Egr-1 in the control of cell proliferation. For example, transforming growth factor-α enhancement of cyclin D1 transcription may involve the interaction of Egr-1 with a cis-regulatory element in the D1 promoter. 13 Similarly, a dominant negative form to Egr-1 blocks progression of the cell cycle from late G1 to the S-phase. 14 In the normal rat vessel wall, Egr-1 is expressed at low or undetectable levels. However, it is dramatically induced following balloon dilatation of the carotid artery 15 or on denudation of arterial endothelium. 10,16 Since Egr-1 positively regulates the expression of multiple genes implicated in the proliferative response to injury, targeting this transcription factor in SMCs may ultimately be useful in efforts to reduce the incidence of restenosis after angioplasty.
Gene expression can be suppressed by exploiting specificity conferred by Watson-Crick base pairing. Synthetic antisense oligonucleotides (ODNs) can bind to target mRNA and block gene expression by presenting the mRNA as a heteroduplex such that it is a substrate for endogenous RNase H. Alternatively, the duplex can effect a steric block as the translational machinery moves along the RNA. Synthesis of the target protein is suppressed thereby compromising the ability of the factor to participate in biological processes. In this paper, we used the antisense approach targeting Egr-1 in cultured vascular SMCs.
Materials and Methods
PS-ODN Synthesis, Purification, and Radiolabeling
Phosphorothioate (PS)-ODNs were synthesized commercially and purified by HPLC to greater than 90% purity (Oligos, Etc.). PS-ODNs were 5′-end labeled with [γ32P]ATP (Bresatec) using T4 polynucleotide kinase (New England Biolabs). Unincorporated label was separated by centrifugation using Chromaspin-10 columns (Clontech).
Cell Culture
Rat aortic SMCs (used between passages 3–7) (Cell Applications, Inc.) were cultured in Waymouth’s MB752/1 medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS), 30 μg/ml L-glutamine, 10 units/ml penicillin and 10 μg of streptomycin at 37°C and 5% CO2. Cells were passaged by rinsing the monolayers with phosphate-buffered saline (PBS) and trypsinization.
32P-PS-ODN Uptake
Subconfluent SMCs were arrested overnight in serum-free medium (SFM). The 32P-labeled PS-ODN was incubated with the cells for various times before washing with cold PBS and solubilization in 0.1 mol/L NaCl, 0.01 mol/L Tris-HCl, pH 7.6, 1 mmol/L EDTA, pH 8.0, 1 μg/ml aprotinin, and 100 μg/ml phenylmethylsulfonyl fluoride (PMSF). Cell-associated radioactivity in the lysate was quantitated in a β-scintillation counter (Packard).
Fluorescence Microscopy
SMCs were seeded onto chamber slides (Nunc InterMed) and grown to 80% confluency. Fluoresceinated (FITC) PS-ODN was incubated with the cells 6 hours after the change of medium to serum-free and allowed to incubate for a further 24 hours. The cells were washed with PBS before fixation with 2% paraformaldehyde for 30 minutes at 22°C. The fixed cells were then treated with permeabilizing solution (0.5% Triton X-100, 50 mmol/L NaCl, 3 mmol/L MgCl2, 10 mmol/L PIPES, pH 6.8) for 1 minute at 4°C. The slides were mounted in anti-fade mounting medium (0.1% p-phenylenediamine, 10% PBS, 90% glycerol) before fluorescence imaging using a laser scanning microscope (Olympus).
Assay of DNA Synthesis
DNA synthesis in SMC was assessed using a modification of a protocol described previously. 17 SMCs were seeded into the wells of a 96-well titer plate (3000 cells/well) and incubated for 48 hours. The cells were incubated with SFM for 6 hours before addition of PS-ODN and incubation for a further 18 hours. The cells were incubated for 24 hours with fresh PS-ODN in medium containing a concentration of FBS sufficient to stimulate [3H]thymidine incorporation into DNA submaximally. The cells were pulsed with [3H]thymidine for 6 hours. The cells were washed three times with cold PBS to remove unincorporated label. The cells were then fixed with cold 5% trichloroacetic acid and washed with cold absolute ethanol. Plates were dried at 37°C for 10 minutes and the cells were lysed with 0.1 mol/L NaOH. ACSII scintillant was added to tubes containing the lysate and radioactivity was quantitated using a β-scintillation counter (Packard).
Total Cell Counts
SMCs were seeded into the wells of a 96-well titer plate (3000 cells/well) in complete medium and incubated for 72 hours before incubation in SFM. After 6 hours, PS-ODNs were added to the wells and incubated for a further 18 hours. FBS sufficient to stimulate cell replication submaximally was added together with fresh PS-ODN and the incubation was continued for a further 72 hours. The cells were rinsed with PBS, trypsinized, and the cell suspension quantitated by hemocytometry or Coulter counter.
Western Immunoblot Analysis
Subconfluent SMCs in Petri dishes were incubated with SFM for 6 hours before addition of PS-ODN and a further incubation for 18 hours. Medium containing FBS and fresh PS-ODN was added and incubated for the times indicated. The cells were washed in cold PBS and extracted in 150 mmol/L NaCl, 50 mmol/L Tris-HCl, pH 7.5, 1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 5 mmol/L EDTA, 1% trasylol, 10 μg/ml leupeptin, 1% aprotinin, 2 mmol/L PMSF). Ten-μg protein samples were loaded onto 10% denaturing SDS-polyacrylamide gels and electroblotted onto PVDF nylon membranes (NEN-DuPont). Membranes were air dried before blocking with nonfat skim milk powder in PBS containing 0.1% Tween 20. The membrane was incubated with the antibodies to Egr-1 (Santa Cruz Biotechnology, Inc.) or PDGF-A (Genzyme) then with horseradish peroxidase-linked mouse anti-rabbit Ig secondary antiserum. Bands were visualized by chemiluminescent detection (DuPont, NEN). Where indicated, the membrane was stripped reprobed with antibodies to Sp1 (Santa Cruz).
In Vitro Injury Model
SMCs were grown to confluence in slide chambers and the medium was changed to serum-free. Six hours subsequently, the cells were exposed to 1 μmol/L of PS-ODN and incubated for an additional 18 hours. Two hours before scraping the cells with a sterile toothpick, mitomycin C (20 μmol/L, final concentration) and fresh PS-ODN (1 μmol/L) were added. The cells were incubated for 72 hours before fixation with 4% (v/v) formaldehyde and staining with hematoxylin/eosin.
Results
Selection of PS-ODN Binding Sites in Egr-1 mRNA
Certain regions of any given mRNA are more susceptible to PS-ODN binding and inhibition than others. The basis for this variation in accessibility may reside in regional differences in the conformation of the mRNA. Areas of low theoretical secondary structure were determined for Egr-1 mRNA by application of the MFOLD program to consecutive 50 base-overlapping windows of 250 bases within the mRNA. 18 Regions containing low levels of local secondary structure in all folded structures were selected as potential target sites, and a series of antisense phosphorothioate-modified ODNs of 15 base length were synthesized (Table 1) ▶ .
Table 1.
PS-ODNs Used in This Study
| PS-ODN | Sequence |
|---|---|
| Egr-1 antisense | |
| AS2 | 3′-TAC CGT CGC CGG TTC-5′ |
| E3/4 | 3′-CCG GAA CCG GCG ACG-5′ |
| E6 | 3′-GAT GGC GGT CGG ACC-5′ |
| E7 | 3′-GGT TGT AGT CAA GAG-5′ |
| E8 | 3′-ACT ACA GAG GCG ACG-5′ |
| E9 | 3′-CTC GTG GAC TGG TGT-5′ |
| E10 | 3′-GAG GTG GAT GGG TAG-5′ |
| E11 | 3′-TCG TCT GTT TTC ACA-5′ |
| Control oligonucleotides | |
| E1 | 3′-GTG ATC CAT TAC CGC-5′ |
| E11C | 3′-ACT GTC TAC GTT CTT-5′ |
| AS2C | 3′-CCT GTC GTC TTC ACG-5′ |
| E11S | 3′-TGT GAA AAC AGA CGA-5′ |
| E11M | 3′-TCG TAT TTT TTA ACA-5′ |
Uptake of FITC-PS-ODNs by Cultured Vascular SMCs
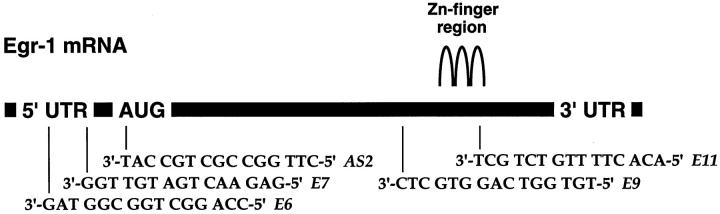
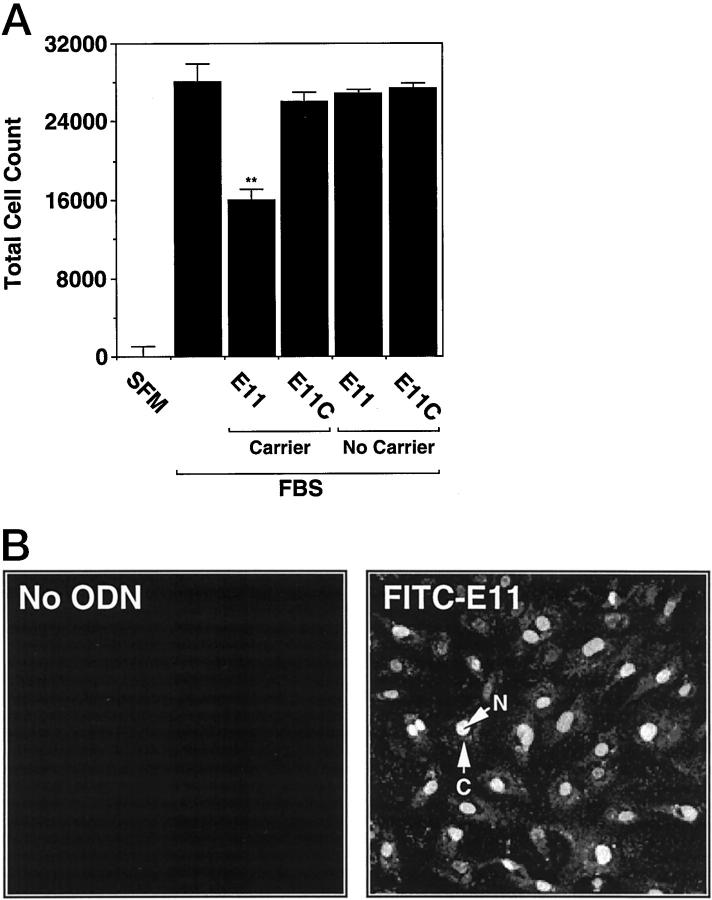
The capacity of an PS-ODN to inhibit gene expression by antisense mechanism(s) is contingent on its ability to enter the cell. We first determined whether a candidate PS-ODN, E11, whose sequence is complementary to a region within Egr-1 mRNA encoding the third zinc finger (Figure 1) ▶ , could localize within SMCs. To facilitate these studies, E11 was fluorescein-labeled at its 5′ end and incubated with cultured SMCs. After 24 hours exposure, the PS-ODN was readily detectable apparently within the cytoplasm and, to a lesser extent, the nucleus (Figure 2 ▶ , bottom). FITC-E1, a size-matched, nonspecific PS-ODN, E1, was taken up with similar efficiency (Figure 2 ▶ , center). Autofluorescence was not observed in cultures in which PS-ODN was omitted (Figure 2 ▶ , center). These results indicate that SMCs incorporate PS-ODN from the culture media and accumulate the nucleic acid intracellularly. PS-ODN accumulation in the extranuclear environment is consistent with PS-ODN localization previously observed in the absence of carrier. 19-22
Figure 1.
Schematic representation of selected antisense PS-ODN target sites within Egr-1 mRNA. Regions of low free energy were determined by the Zuker algorithm. 18
Figure 2.

Uptake of fluorescein-labeled PS-ODNs into cultured vascular SMCs. SMCs in chamber slides were exposed to 1 μmol/L of FITC-labeled PS-ODN 6 hours after the change of medium to serum-free and incubated for 18 hours. The cells were washed, fixed with paraformaldehyde, and fluorescence was assessed by laser scanning microscopy. The figure (FITC-E1 (center), FITC-E11 (bottom), or medium alone (top) shows a representative field in each treatment group (magnification, ×200). The data are representative of two independent experiments. Arrows indicate cytoplasm (C) and nucleus (N).
Time Course Uptake of 32P-labeled E11 by SMCs
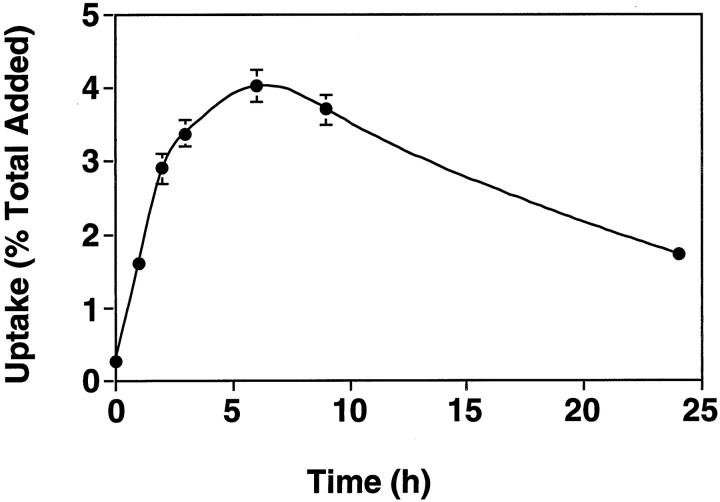
To determine the temporal pattern with which E11 associates with SMCs, the PS-ODN was 5′ end-labeled with 32P by kinase reaction and incubated with the cells for various times. 32P-E11 was taken up most efficiently within the first 4 hours of exposure and optimally after 6 hours (Figure 3) ▶ . Interestingly, 32P-E11 was still associated with the cells even after 24 hours (Figure 3) ▶ . Electrophoretic analysis on denaturing gels revealed that cell-associated 32P-E11 was not fragmented even after 24 hours, since laddering was not observed (data not shown).
Figure 3.
Time course association of 32P-labeled E11 with SMCs. Growth-arrested SMCs were incubated with 32P-E11 (100,000 cpm) for the times indicated. The cells were washed twice with PBS, pH 7.4, and solubilized before assessment of uptake by β-counting. The ordinate is expressed as a percentage of total counts added. The data are representative of two independent experiments performed in duplicate. Standard errors of the mean are indicated in the figure.
Antisense Egr-1 PS-ODNs Inhibit Serum-Inducible DNA Synthesis in SMCs
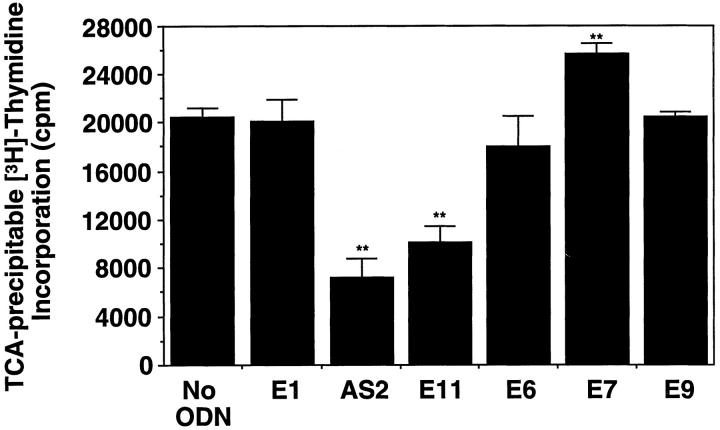
E11 and the entire antisense PS-ODN panel (Table 1) ▶ were assessed for the ability to inhibit SMC proliferation inducible by serum. SMCs were exposed to the PS-ODNs 6 hours after the removal of serum from the medium to induce growth arrest. PS-ODNs were added a second time 18 hours subsequently with a change of medium and the addition of serum. In preliminary titration studies, we determined that the maximal tolerated dose of the nonspecific PS-ODN, E1, was 1 μmol/L (data not shown). Further experiments revealed that PS-ODNs E11, and AS2, whose sequence is complementary to the translation initiation region (Figure 1) ▶ , inhibited [3H]thymidine incorporation into DNA at this concentration in cells exposed to serum by over 50% (Figure 4) ▶ . In contrast, E1 and antisense PS-ODNs E6, E7, and E9 (Figure 4) ▶ failed to inhibit serum-inducible DNA synthesis in SMCs. Antisense PS-ODNs E3/4, E8, and E10 were also unable to influence this mitogenic response (data not shown). Neither AS2, E11, nor any other PS-ODN used in this study possessed any more than three contiguous guanosines in their respective sequences (Table 1) ▶ .
Figure 4.
Antisense Egr-1 PS-ODNs inhibit serum-inducible DNA synthesis in SMCs. SMCs were exposed to 1 μmol/L of PS-ODN 6 hours after the change of medium to serum-free. Medium containing serum (5% FBS) and 1 μmol of PS-ODN was added 18 hours subsequently and incubated for a further 24 hours. TCA-precipitable [3H]thymidine incorporation into DNA was assessed as described under “Materials and Methods.” The data are representative of at least two independent experiments performed in triplicate. Standard errors of the mean are indicated in the figure. **, P < 0.01 (Student’s paired t-test) compared to control (No ODN).
Serum-Inducible SMC Proliferation Is Inhibited by Antisense Egr-1 PS-ODNs
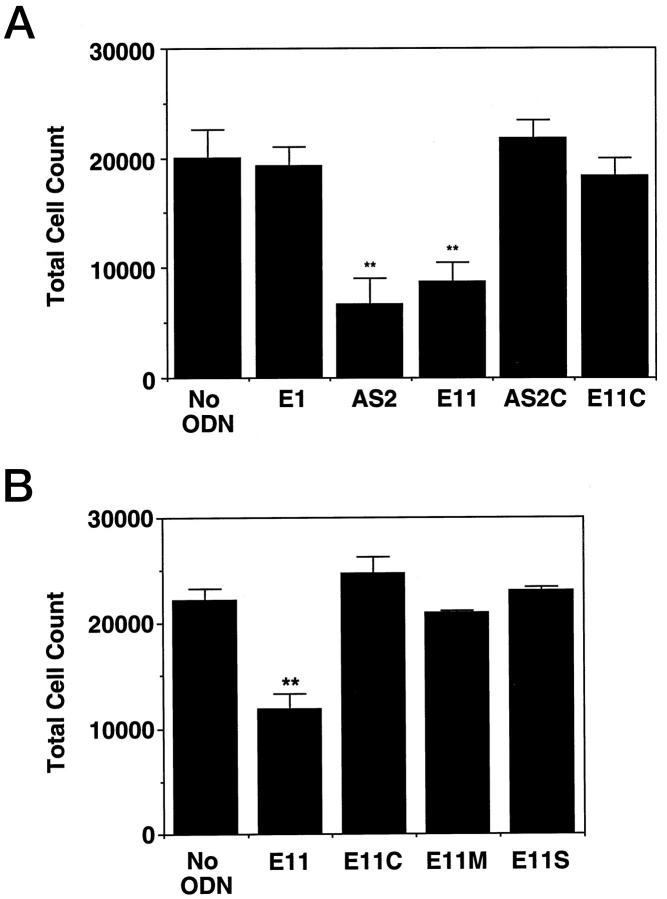
To confirm findings from thymidine incorporation experiments, we measured the total numbers of cells in each treatment. AS2 and E11 each suppressed serum-inducible SMC proliferation by over 50% (Figure 5A) ▶ . In contrast, E1 had no effect (Figure 5A) ▶ . PS-ODNs AS2C and E11C (Table 1) ▶ , with base composition similar to AS2 and E11, respectively, also failed to inhibit proliferation (Figure 5A) ▶ . In addition, neither E11S, the sense counterpart of E11, nor E11M, bearing a 3 nucleotide mismatch (Table 1) ▶ could attenuate SMC replication (Figure 5B) ▶ . These findings indicate that antisense Egr-1 PS-ODNs are able to inhibit SMC replication in a sequence-specific manner and are consistent with the capacity of antisense PS-ODNs to inhibit DNA synthesis and cell proliferation despite localization in the cytoplasm. 23
Figure 5.
Serum-inducible SMC proliferation is inhibited by antisense Egr-1 PS-ODNs. SMCs were exposed to 1 μmol of PS-ODN 6 hours after the change of media to serum-free. Medium containing serum (5% FBS) and 1 μmol/L of PS-ODN E1, AS2, E11, AS2C, E11C (A) or E11, E11C, E11M, E11S (B) was added 18 hours subsequently and incubated for a further 72 hours. Cell numbers were quantitated as described under “Materials and Methods.” The data are representative of at least two independent experiments performed in triplicate. Standard errors of the mean are indicated in the figure. **, P < 0.01 (Student’s paired t-test) compared to control (No ODN).
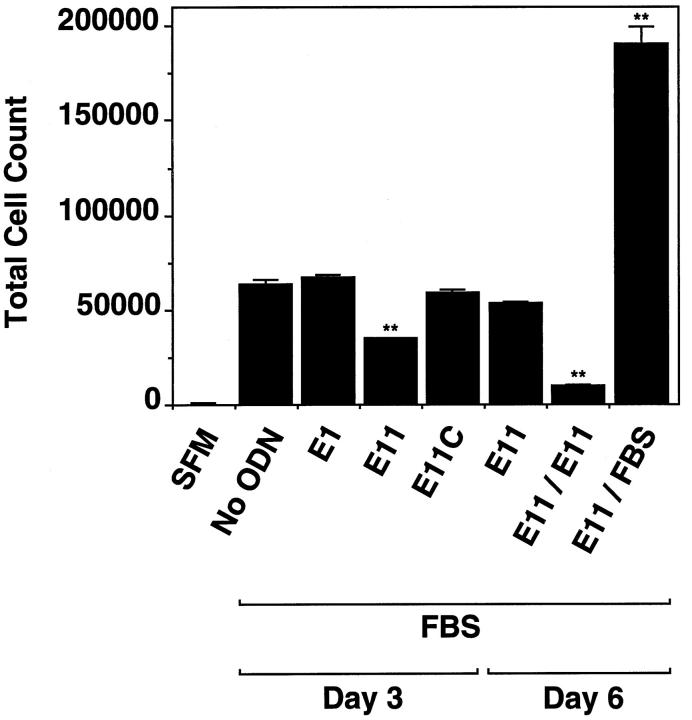
Antisense Egr-1 PS-ODN inhibition of proliferation was not due to cytotoxicity. Trypan blue was still excluded by the cells after 72 hours of exposure to the PS-ODNs (data not shown). Additional experiments revealed that PS-ODN inhibition was reversible. Medium containing serum and E11 was replaced with fresh medium without PS-ODN, and total cell counts were assessed three days subsequently. Removal of the PS-ODN resulted in resumption of cell proliferation (Figure 6) ▶ .
Figure 6.
Antisense Egr-1 PS-ODN inhibition of SMC proliferation is reversible. SMCs were exposed to 1 μmol/L of PS-ODN 6 hours after the change of media to serum-free. Medium containing serum (5% FBS) and 1 μmol of PS-ODN was added 18 hours subsequently and incubated for a further 72 hours before harvest. Alternatively, instead of harvesting, the medium was replaced with fresh 5% FBS without E11 (E11/Serum), 1 μmol of E11 (E11/E11), or was left undisturbed (E11) and trypsinized 72 hours later. Cell numbers were quantitated as described under “Materials and Methods.” SFM, serum-free medium. The data are representative of two independent experiments performed in triplicate. Standard errors of the mean are indicated in the figure. **, P < 0.01 (Student’s paired t-test) compared to control (No ODN).
To determine whether E11 and AS2 had the capacity to inhibit the growth of a related but genotypically modified cell line, we used SV40-transformed SMCs in proliferation assays. The mitogenic responsiveness of this cell line to serum was, like normal SMCs, inhibited by exposure to either PS-ODN (at 1 μmol/L) by over 50%. E1 had no effect in this setting (data not shown).
Antisense Egr-1 PS-ODNs Inhibit Egr-1 Induction in SMCs Exposed to Serum
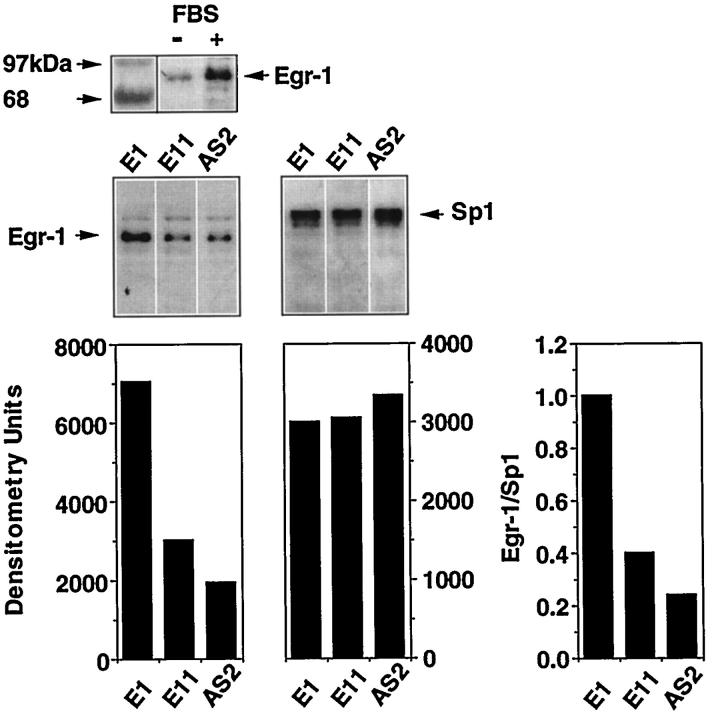
We performed Western blot analysis to determine whether antisense PS-ODN inhibition of SMC proliferation was a consequence of the ability of these molecules to suppress the induction of Egr-1 itself. Initial experiments established that Egr-1 protein was strongly induced in growth-arrested SMCs exposed to serum within 1.5 hours (Figure 7 ▶ , top). Subsequent studies revealed that incubation of E11 or AS2 before the addition of serum inhibited the induction of Egr-1 protein (Figure 7 ▶ , center) by 60% and 75%, respectively (Figure 7 ▶ , bottom). In contrast, identical exposure to E1 had no effect (Figure 7 ▶ , center). When the blot was stripped and reprobed with antibodies directed toward Sp1, we observed that E11 or AS2 did not affect levels of this zinc finger protein (Figure 7 ▶ , center and bottom). PS-ODNs E11 and AS2 can thus inhibit the activation and nuclear accumulation of Egr-1 protein. Additional experiments revealed that E11C, E11S, or E11M could inhibit serum-inducible Egr-1 synthesis (data not shown).
Figure 7.
Antisense Egr-1 PS-ODNs inhibit the induction of Egr-1 in SMCs exposed to serum. Western blot analysis was performed with antibodies to Egr-1 and Sp1, and growth-arrested SMCs exposed to 1 μmol/L of E1, E11, and AS2 for 24 hours before the addition of serum and incubation for 1.5 hours, as described under “Materials and Methods.” The bands were quantitated using a scanning densitometer and Egr-1 values normalized to Sp1. The blot is representative of three independent experiments.
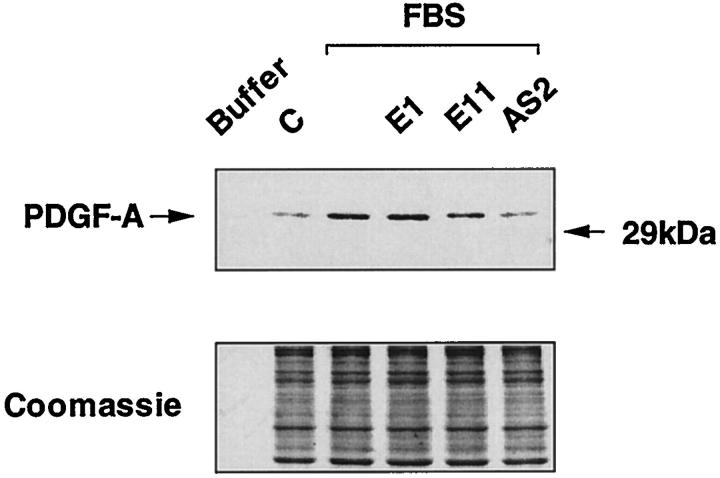
We next determined whether antisense Egr-1 PS-ODNs could affect the expression of an Egr-1-dependent gene, such as platelet-derived growth factor A-chain (PDGF-A). Egr-1 mediates inducible PDGF-A expression in response to multiple extracellular stimuli. 9,12,16,24,25 Moreover, overexpression of Egr-1 can induce PDGF-A promoter-dependent expression. 9,16 PDGF-A levels increased in SMCs exposed to serum within 4 hours (Figure 8) ▶ . Preincubation with PS-ODN E1 did not affect the induction of PDGF-A, whereas AS2 inhibited the increase in PDGF-A (Figure 8) ▶ . Inhibition by E11 was less profound than AS2, consistent with the relative effects of these PS-ODNs on the induction of Egr-1 protein (Figure 7) ▶ . Thus, PS-ODN inhibition of Egr-1 can influence gene expression dependent on this transcription factor.
Figure 8.
Induction of PDGF-A by serum is inhibited by antisense Egr-1 PS-ODN. Western blot analysis was performed with antibodies to PDGF-A and extracts of growth-arrested SMCs exposed to 1 μmol/L of E1, E11, and AS2 for 24 hours before the addition of serum and incubation for 4 hours, as described under “Materials and Methods.” C, unstimulated (control) cells. The Coomassie Blue-stained gel indicates equal protein loading.
SMC Regrowth after Injury Is Inhibited by Antisense PS-ODNs Directed to Egr-1
To determine whether these molecules also had the capacity to modulate regrowth of SMCs after injury, we exposed the PS-ODN panel to SMCs that had been injured by scraping. E1, E11, E11C, or no PS-ODN were added 18 hours before and immediately after the cells were scraped. SMCs not exposed to any PS-ODN migrated from the wound edge into the denuded zone and completely covered this area within 72 hours (Figure 9A) ▶ . Inclusion of E1 in the culture medium had no effect (Figure 9A) ▶ . In contrast, SMC regrowth was attenuated in wells containing E11 (Figure 9, A and B) ▶ . E11C, like E1, failed to affect the reparative response (Figure 9, A and B) ▶ . SMC repair was also inhibited by AS2, whereas AS2C, E11S, and E11M had no effect (data not shown). When fresh medium without PS-ODN was added to the cultures, SMCs previously inhibited by E11 recovered the denuded zone within a further 72 hours (Figure 9A) ▶ . Thus, as well as suppressing the serum-induction of Egr-1 and cell proliferation, PS-ODNs targeting Egr-1 can inhibit the reparative response to injury in vitro in a sequence-specific and reversible manner.
Figure 9.
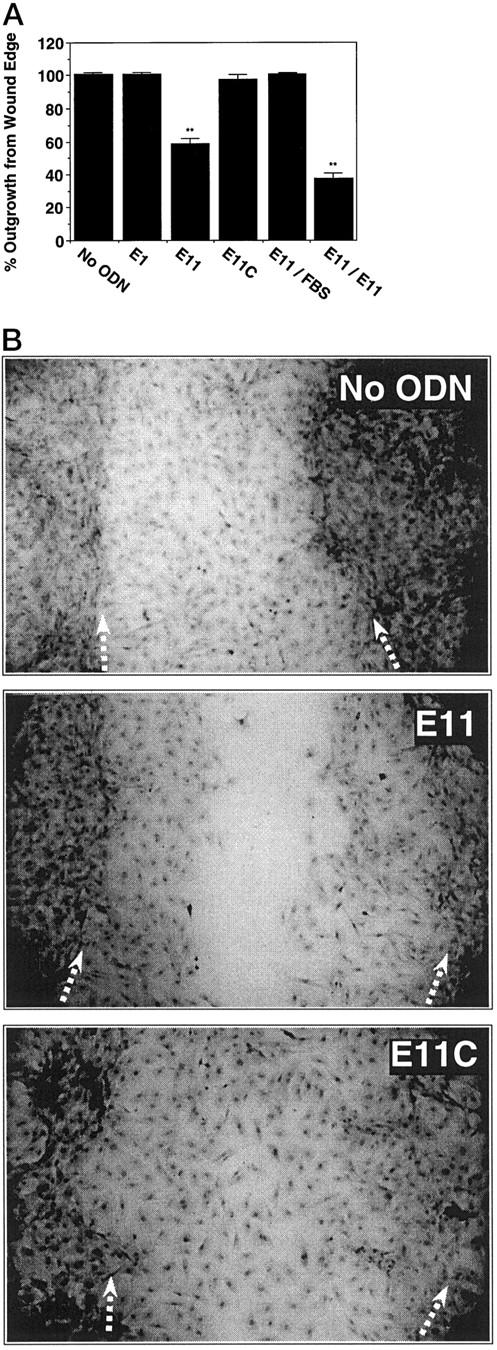
Antisense Egr-1 PS-ODNs inhibit the reparative response of SMCs to injury. SMCs in slide chambers were exposed to 1 μmol/L of PS-ODN 6 hours after the change of medium to serum-free. Eighteen hours subsequently, the cells were injured by scraping with a sterile toothpick. Fresh PS-ODN was added (1 μmol/L, final concentration) and the cells were incubated for a further 72 hours before assessment of the denuded area recovered (A) or staining with hematoxylin/eosin and subsequent photomicrography (B). Magnification, ×40. Arrows denote the wound edge. E11/FBS denotes wells in which fresh medium without E11 was added and incubated for a further 72 hours. E11/E11 denotes wells in which fresh medium containing E11 was added and left for 72 hours. The area occupied by SMCs outgrown from the wound edge in each treatment is expressed as a percentage of the total original denuded area. The data are representative of at least two independent experiments performed in triplicate. Standard errors of the mean are indicated in the figure. **, P < 0.01 (Student’s paired t-test) compared to control (No ODN).
Previous investigators have used the antibiotic mitomycin C to assess whether antisense PS-ODNs or other agents influence SMC migration, as distinct from proliferation, after in vitro injury. 26,27 This compound has also been used to inhibit proliferation in other cell types. 28,29 Mitomycin C pretreatment (3, 10, or 20 μmol/L; 2 hours before injury) had no effect on SMC regrowth after injury (data not shown), consistent with previous observations. 26 Mitomycin C did not influence the inhibitory effect of E11, or any other treatment, on the reparative response to injury. These findings, taken together, indicate that SMC migration, like proliferation (Figures 4 to 6) ▶ ▶ ▶ , is suppressed by antisense PS-ODNs targeting Egr-1.
Finally, we determined whether PS-ODN potency could be augmented by facilitation of nuclear, rather than extranuclear localization under passive conditions (Figure 2) ▶ . Using a dendrimeric carrier, E11 suppressed serum-inducible SMC proliferation at 0.3 μmol/L (Figure 10A) ▶ . At this concentration, E11 was unable to inhibit in the absence of carrier (Figure 10A) ▶ . Fluorescence microscopy revealed that PS-ODN accumulation in SMC nuclei when carrier was used (Figure 10B) ▶ , which contrasts with the spatial pattern observed under passive conditions (Figure 2) ▶ . These results support the notion that PS-ODN potency is influenced by its localization within the cell.
Figure 10.
Effect of transfection agent on Egr-1 PS-ODN inhibition of SMC proliferation. SMCs were exposed to 0.3 μmol/L of PS-ODN 6 hours after the change of media to serum-free either under passive conditions or using Superfect according to manufacturer’s instructions (Qiagen). Medium containing serum (5% FBS) and 0.3 μmol of PS-ODN was added 18 hours subsequently (with or without Superfect) and incubated for a further 24 hours before fluorescence microscopy (A) or 72 hours before assessment of cell counts (B) as described under “Materials and Methods.” Magnification, ×200. The data are representative of at least two independent experiments performed in triplicate. Standard errors of the mean are indicated in the figure. **, P < 0.01 (Student’s paired t-test) compared to control (No PS-ODN). Arrows indicate cytoplasm (C) and nucleus (N).
Discussion
Despite recent advances in the application of balloon angioplasty and stents in the treatment of vascular injury, restenosis remains a significant medical problem largely undertreated by conventional pharmaceuticals. Efforts have thus been directed at identifying novel methods for the treatment of this condition. One approach has been the application of antisense PS-ODN strategies to target and suppress genes that may to be involved in the process of restenosis. Effective gene-specific PS-ODN antagonists potentially have a dual application: first, these agents can themselves be used as therapeutics; and second, they may serve as models in the design of more sophisticated nucleic-based agents, such as ribozymes or catalytic DNA molecules. In this paper, we used antisense strategies to inhibit vascular SMC proliferation in vitro by targeting the zinc finger transcription factor Egr-1. Two antisense Egr-1 PS-ODNs, one directed to the translation initiation site and the other further within the coding region, inhibited the inducible synthesis of Egr-1 in a sequence-specific manner. These PS-ODNs blocked the ability of serum to stimulate DNA synthesis and cell replication in a selective manner. PS-ODN inhibition was reversible and not due to cell death. PS-ODNs also inhibited SMC regrowth after mechanical injury in vitro; experiments with mitomycin C suggest that this was due to PS-ODN inhibition of SMC migration. Finally, PS-ODN inhibition was observed at lower concentrations if localization in the nucleus was facilitated using a carrier. Egr-1, therefore, appears to play a regulatory role in proliferation and the reparative response to injury.
While many groups have applied PS-ODNs to the study and treatment of restenosis in experimental models (c-myb, 26,30 c-myc, 31,32 nuclear factor-κB p65 subunit, 33 cdc2, and PCNA 34 ), there has been considerable speculation as to the precise mechanisms and specificity of the observed effects. A contributory factor to the controversy is the phosphorothioate group in the ODN. Substitution of the phosphodiester backbone of native DNA with phosphorothioate moieties markedly augments resistance to nuclease digestion. 35,36 Phosphorothioates, however, have also been found to bind with high affinity to a large number of proteins 37,38 presumably the result of alterations in the presentation of negative charges. Without appropriate controls, sequence-independent effects caused by phosphorothioate chemistry can be mistakenly attributed to a true antisense effect. Scrambled PS-ODNs serve as useful controls since these have identical net charge as their parent counterparts. Sense and mismatch PS-ODNs help provide further evidence of sequence specificity. The field is confounded by observations that have shown that ODNs bearing four consecutive G nucleotides can inhibit SMC proliferation in a G quartet-specific or sequence-independent 30,39 manner. While such inhibitory ODNs still have clinical potential, these observations raise concerns over the mechanism(s) of action. It was, therefore, essential in this study to determine the levels at which PS-ODNs exerted sequence-independent suppression of SMC growth and, based on such information, to use antisense molecules and appropriate controls at permissible concentrations. We observed that the maximum tolerated dose of an PS-ODN before sequence-independent inhibition of SMC proliferation was 1 μmol/L. The random sequence PS-ODN, E1, as well as size-matched PS-ODNs with scrambled, sense, and mismatch sequences served as important controls. Inhibition was not due to differences in PS-ODN uptake, since an inhibitory and noninhibitory PS-ODN localized intracellularly with similar spatial pattern. Use of various controls in the present study suggests that antisense Egr-1 PS-ODN inhibition of SMC replication is sequence-specific.
The focus for target regions in the antisense literature is typically in the vicinity of the translation initiation codon (such as the AS2 target site; Figure 1 ▶ ) or the 3′ untranslated region. In this paper, regions of accessibility within Egr-1 mRNA were selected based on prediction of areas of low free energy. 18 Clearly, site selection was not perfect as only a subset of the PS-ODN panel could inhibit SMC proliferation at 1 μmol/L. Nevertheless, this method yielded E11, targeting a region within Egr-1 mRNA encoding the third zinc finger (Figure 1) ▶ . A sequence homology search (FastA; EMBL and GenBank) revealed no significant homology between the E11 and AS2 target sites in Egr-1 mRNA and the mRNA sequences of Egr-2, Egr-3, or Egr-4. Improved methods for secondary structure prediction are emerging 40,41 and may be useful in identifying sites that are structurally more accessible. While secondary structure is likely to remain a key determinant in the efficiency with which antisense ODNs bind to their target and inhibit gene expression, the importance of other variables still to be characterized, such as the role of sequence-dependent serum binding proteins and mechanisms of cellular uptake may also emerge as influential factors.
While Egr-1 is expressed at low or undetectable levels in the normal rat artery wall, it is dramatically induced following mechanical injury. 10,15 Interestingly, recent studies indicate that Egr-1 is expressed by SMCs as they migrate from the media toward the intima after injury. 16 Egr-1 binds to the proximal promoters of many genes implicated in the proliferative and chemotactic response to injury. 10,12 Its activation by injury and multiple other stimuli 6,12 combined with its ability to drive the expression of an increasing number of pathophysiologically relevant genes clearly implicates the transcription factor as a potential therapeutic target. That inducible Egr-1 expression in SMCs can be selectively inhibited by the antisense PS-ODNs in this study provides a valuable research tool to delineate further the biological functions of Egr-1. Moreover, the capacity of these PS-ODNs to inhibit SMC proliferation and regrowth after injury in a sequence-specific manner indicates an important regulatory role for Egr-1 in SMC regeneration and suggests that these molecules may be useful in future strategies directed at compromising neointima formation after injury.
Footnotes
Address reprint requests to: Levon M. Khachigian, Ph.D., Centre for Thrombosis and Vascular Research, School of Pathology, The University of New South Wales, Sydney, NSW 2052, Australia, E-mail: l.khachigian@unsw.edu.au.
Supported in part by a Strategic Partnership with Industry (Research and Training) grant from the Australian Research Council (L.M.K.) and a Health Research & Development Infrastructure grant from the N.S.W. Department of Health. L.M.K. is supported by an R. Douglas Wright Fellowship from the National Health and Medical Research Council of Australia.
References
- 1.Fischman DL, Leon MB, Baim D, Schatz RA, Savage MP, Penn I, Detre K, Veltri L, Ricci D, Nobuyoshi M: A randomized comparison of coronary stent placement and balloon angioplasty in the treatment of coronary artery disease. N Engl J Med 1994, 331:496-501 [DOI] [PubMed] [Google Scholar]
- 2.Jackson CL, Schwartz SM: Pharmacology of smooth muscle replication. Hypertension 1992, 20:713-736 [DOI] [PubMed] [Google Scholar]
- 3.O’Brien ER, Schwartz SM: Update on the biology and clinical study of restenosis. Trends Cardiovasc Med 1994, 4:169-178 [DOI] [PubMed] [Google Scholar]
- 4.Ross R: The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature 1993, 362:801-809 [DOI] [PubMed] [Google Scholar]
- 5.Sukhatme VP, Cao X, Chang LL, Tsai-Morris C-H, Stamenkovich D, Ferreira PCP, Cohen DR, Edwards SA, Shows TB, Curran T, Le Beau MM, Adamson ED: A zinc finger encoding gene coregulated with c-Fos during growth and differentiation and after depolarization. Cell 1988, 53:37-43 [DOI] [PubMed] [Google Scholar]
- 6.Gashler A, Sukhatme V: Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acids Res 1995, 50:191-224 [DOI] [PubMed] [Google Scholar]
- 7.Changelian PS, Feng P, King TC, Milbrandt J: Structure of the NGFI-A gene and detection of upstream sequences responsible for its transcriptional induction by nerve growth factor. Proc Natl Acad Sci USA 1989, 86:377-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavletich NP, Pabo CO: Zinc finger-DNA recognition: crystal structure of a zif268-DNA complex at 2.1A. Science 1991, 252:809-817 [DOI] [PubMed] [Google Scholar]
- 9.Khachigian LM, Williams AJ, Collins T: Interplay of Sp1 and Egr-1 in the proximal PDGF-A promoter in cultured vascular endothelial cells. J Biol Chem 1995, 270:27679-27686 [DOI] [PubMed] [Google Scholar]
- 10.Khachigian LM, Lindner V, Williams AJ, Collins T: Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science 1996, 271:1427-1431 [DOI] [PubMed] [Google Scholar]
- 11.Biesiada E, Razandi M, Levin ER: Egr-1 activates basic fibroblast growth factor transcription. J Biol Chem 1996, 271:18576-18581 [DOI] [PubMed] [Google Scholar]
- 12.Khachigian LM, Collins T: Inducible expression of Egr-1-dependent genes: a paradigm of transcriptional activation in vascular endothelium. Circ Res 1997, 81:457-461 [DOI] [PubMed] [Google Scholar]
- 13.Yan YX, Nakagawa H, Lee MH, Rustgi AK: Transforming growth factor-α enhances cyclin D1 transcription through the binding of early growth response protein to a cis-regulatory element in the cyclin D1 promoter. J Biol Chem 1997, 272:33181-33190 [DOI] [PubMed] [Google Scholar]
- 14.Hallahan DE, Dunphy E, Virudachalam S, Sukhatme VP, Kufe DW, Weischselbaum RR: c-jun and Egr-1 participate in DNA synthesis and cell survival in response to ionizing radiation exposure. J Biol Chem 1995, 270:30303-30309 [DOI] [PubMed] [Google Scholar]
- 15.Kim S, Kawamura M, Wanibuchi H, Ohta K, Hamaguchi A, Omura T, Yulimura T, Miura K, Iwao H: Angiotensin II type 1 receptor blockade inhibits the expression of immediate-early genes, and fibronectin in rat injured artery. Circulation 1995, 92:88-95 [DOI] [PubMed] [Google Scholar]
- 16.Silverman ES, Khachigian LM, Lindner V, Williams AJ, Collins T: Inducible PDGF A-chain transcription in vascular smooth muscle cells is mediated by Egr-1 displacement of Sp1 and Sp3. Am J Physiol 1997, 42:H1415-H1426 [DOI] [PubMed] [Google Scholar]
- 17.Khachigian LM, Chesterman CN: Synthetic peptides representing the alternatively spliced exon of the PDGF A-chain modulate mitogenesis stimulated by normal human serum and several growth factors. J Biol Chem 1992, 267:7478-7482 [PubMed] [Google Scholar]
- 18.Zuker M: On finding all suboptimal foldings of an RNA molecule. Science 1989, 244:48-52 [DOI] [PubMed] [Google Scholar]
- 19.Bennett CF, Chiang M-Y, Chan H, Shoemaker JE, Mirabelli CK: Cationic lipids enhance cellular uptake and activity of phosphorothioate antisense oligonucleotides. Mol Pharmacol 1992, 41:1023-1033 [PubMed] [Google Scholar]
- 20.Beltinger C, Saaragovi HU, LeSauteur L, Shah N, DeDionisio L, Christensen L, Raible A, Jarett L, Gewirtz AM: Binding, uptake and intracellular trafficking of phosphorothioate-modified oligodeoxynucleotides. J Clin Invest 1995, 95:1814-1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett MR, Schwartz SM: Antisense therapy for angioplasty restenosis. Circulation 1995, 92:1981-1993 [DOI] [PubMed] [Google Scholar]
- 22.Wyroba E, Pawlowska Z, Kobylanska A, Pluskota E, Maszewska M, Stec WJ, Cierniewski CS: Internalization of oligodeoxynucleotide antisense to type-1 plasminogen activator inhibitor mRNA in endothelial cells: a three dimensional reconstruction by confocal microscopy. Biol Cell 1996, 87:37-43 [PubMed] [Google Scholar]
- 23.Hofer G, Grimmer C, Sukhatme VP, Sterzel RB, Rupprecht HD: Transcription factor Egr-1 regulates glomerular mesangial cell proliferation. J Biol Chem 1996, 271:28306-28310 [DOI] [PubMed] [Google Scholar]
- 24.Khachigian LM, Anderson KA, Halnon NJ, Resnick N, Gimbrone MA, Jr, Collins T: Egr-1 is activated in endothelial cells exposed to fluid shear stress and interacts with a novel shear-stress response element in the PDGF A-chain promoter. Arterioscler Thromb Vasc Biol 1997, 17:2280-2286 [DOI] [PubMed] [Google Scholar]
- 25.Delbridge GJ, Khachigian LM: FGF-1-induced PDGF A-chain gene expression in vascular endothelial cells involves transcriptional activation by Egr-1. Circ Res 1997, 81:282-288 [DOI] [PubMed] [Google Scholar]
- 26.Pitsch RJ, Goodham GR, Minion DJ, Madura JA, Fox PL, Graham LM: Inhibition of smooth muscle cell proliferation and migration in vitro by antisense oligonucleotide to c-myb. J Vasc Surg 1996, 23:783-791 [DOI] [PubMed] [Google Scholar]
- 27.Horodyski J, Powell RJ: Effect of aprotinin on smooth muscle cell proliferation, migration, and extracellular matrix synthesis. J Surg Res 1996, 66:115-118 [DOI] [PubMed] [Google Scholar]
- 28.Chen P, Gupta K, Wells A: Cell movement elicited by epidermal growth factor receptor requires kinase and autophosphorylation but is separable from mitogenesis. J Cell Biol 1994, 124:547-555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho TC, Del Priore LV, Hornbeck R: Effect of mitomycin C on human retinal pigment epithelium. Curr Eye Res 1997, 16:572-576 [DOI] [PubMed] [Google Scholar]
- 30.Villa AE, Guzman LA, Poptic EJ, Labhasetwar V, D’Souza S, Farrell CL, Plow EF, Levy RJ, DiCorleto PE, Topol EJ: Effects of antisense c-myb oligonucleotides on vascular smooth muscle cell proliferation and response to vessel wall injury. Circ Res 1995, 76:505-513 [DOI] [PubMed] [Google Scholar]
- 31.Bennett MR, Lindner V, Deblois D, Reidy MA, Schwartz SM: Effect of phosphorothioated oligonucleotides on neointima formation in the rat carotid artery: dissecting the mechanism of action. Arterioscler Thromb Vasc Biol 1997, 17:2326-2332 [DOI] [PubMed] [Google Scholar]
- 32.Bennett MR, Anglin S, McEwan JR, Jagoe R, Newby AC, Evan GI: Inhibition of vascular smooth muscle cell proliferation in vitro and in vivo by c-myc antisense oligodeoxynucleotides. J Clin Invest 1994, 93:820-828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Autieri MV, Yue T-L, Ferstein GZ, Ohlstein E: Antisense oligonucleotides to the p65 subunit of NF-kB inhibit human vascular smooth muscle cell adherence and proliferation and prevent neointima formation in rat carotid arteries. Biochem Biophys Res Commun 1995, 213:827-836 [DOI] [PubMed] [Google Scholar]
- 34.Morishita R, Gibbons GH, Ellison KE, Nakajima M, Zhang L, Kaneda Y, Ogihra T, Dzau V: Single intraluminal delivery of antisense cdc2 kinase and proliferating-cell nuclear antigen oligonucleotides results in chronic inhibition of neointimal hyperplasia. Proc Natl Acad Sci USA 1993, 90:8474-8478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcus-Sekura C, Woerner A, Shinozuka K, Zon G, Quinnan G: Comparative inhibition of chloramphenicol acetyltransferase gene expression by antisense oligonucleotide analogues having alkyl triester, methyl phosphonate and phosphorothioate linkages. Nucleic Acids Res 1987, 15:5749-5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cazenave C, Stein C, Loreau N, Thuong N, Neckers L, Subasinghe C, Helene C, Cohen J, Toulme J: Comparative inhibition of rabbit globin mRNA translation by modified antisense oligodeoxynucleotides. Nucleic Acids Res 1989, 17:4225-4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krieg AM, Stein CA: Phosphorothioate oligonucleotides: antisense or anti-protein? Antisense Res Dev 1995, 5:241. [DOI] [PubMed] [Google Scholar]
- 38.Guvakova MA, Yakubov LA, Vlodavsky I, Tonkinson JL, Stein CA: Phosphorothioate oligodeoxynucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors, and remove it from low affinity binding sites on extracellular matrix. J Biol Chem 1995, 270:2620-2627 [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Chen HJ, Schwartz A, Cannon PJ, Stein CA, Rabbani LE: Sequence-independent inhibition of in vitro smooth muscle cell proliferation, migration, and in vivo neointimal formation by phosphorothioate oligonucleotides. J Clin Invest 1996, 98:443-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perochondorisse J, Chetouani F, Aurel S, Iscolo N, Michot B: RNA-D2: a computer program for editing and display of RNA secondary structures. Comp Appl Biosci 1995, 11:101-109 [DOI] [PubMed] [Google Scholar]
- 41.Chetouani F, Monestie P, Thebault P, Gaspin C, Michot B: ESSA: an integrated and interactive computer tool for analysing secondary structure. Nucleic Acids Res 1997, 25:3514-3522 [DOI] [PMC free article] [PubMed] [Google Scholar]