Abstract
Primary central nervous system lymphomas (PCNSLs) have recently received considerable clinical attention due to their increasing incidence. To clarify the histogenetic origin of these intriguing neoplasms, PCNSLs from 10 HIV-negative patients were analyzed for immunoglobulin (Ig) gene rearrangements. All tumors exhibited clonal IgH gene rearrangements. Of the 10 cases, 5 used the V4–34 gene segment, and all of these lymphomas shared an amino acid exchange from glycine to aspartate due to a mutation in the first codon of the complementarity-determining region 1. No preferential usage of DH, JH, Vκ, Jκ, Vλ, or Jλ gene segments was observed. All potentially functional rearrangements exhibited somatic mutations. The pattern of somatic mutations indicated selection of the tumor cells (or their precursors) for expression of a functional antibody. Mean mutation frequencies of 13.2% and 8.3% were detected for the heavy and light chains, respectively, thereby exceeding other lymphoma entities. Cloning experiments of three tumors showed ongoing mutation in at least one case. These data suggest that PCNSLs are derived from highly mutated germinal-center B cells. The frequent usage of the V4–34 gene and the presence of a shared replacement mutation may indicate that the tumor precursors recognized a shared (super) antigen.
The central nervous system (CNS) has traditionally been regarded as an immunologically privileged organ. This view was based on the existence of the blood-brain barrier, which separates the brain from the immune system; the lack of a conventional lymphatic system including lymph nodes and draining lymphatic vessels; and the inability of the CNS to reject allografts, as well as the absence of major histocompatibility antigens on brain cells. 1,2 Modern neuroimmunology has modified this concept and demonstrated that the immunologically down-regulated phenotype of the brain is partially mediated by an immunosuppressive activity of brain cells 3,4 and that the CNS is integrated into a complex, tightly regulated neuroimmune network. It is interesting that, although the normal brain is devoid of a significant number of lymphocytes, highly malignant lymphomas may develop in the brain.
Indeed, the incidence of primary CNS lymphomas (PCNSLs) has significantly increased over the last decade. 5 This entity now accounts for up to 6.6% of all primary malignant intracranial tumors. 6 An important risk factor for the development of PCNSL is primary or secondary immunodeficiency. HIV-infected patients show a 1000-fold increased risk as compared with HIV-negative persons. 7-9 However, an increase in the incidence of PCNSL has also been shown in immunocompetent patients, the reason for which is unknown.
Histopathological and immunohistochemical analyses have identified the vast majority of PCNSLs as highly malignant, strongly proliferating B cell non-Hodgkin’s lymphomas. They are categorized as diffuse large-cell lymphomas (DLCL) based on the revised European-American lymphoma classification system. 10 It is interesting that PCNSLs differ in their biological behavior from extracerebral DLCL and appear to be associated with significantly reduced survival. 5 This clinical observation has raised the question whether PCNSLs compose a separate disease entity. In addition, PCNSLs are distinguished from extracerebral lymphomas by the fact that they are strictly confined to the CNS and neither manifest extracerebrally nor metastasize to other organs including the lymphatic system. 11,12
The pathogenesis of PCNSL is still poorly defined. Whereas PCNSLs of HIV-positive and -negative patients share many clinical features including their exclusive localization in the CNS, the underlying pathogenesis may be quite different. The regular association with Epstein-Barr virus (EBV) infection in AIDS-related PCNSL suggests an oncogenic role of EBV-encoded genes. 11,12 In contrast, there is no evidence for an involvement of EBV in the development of AIDS-unrelated PCNSL. 11
To clarify the molecular pathogenesis of PCNSL, the precise characterization of the histogenetic origin and differentiation stage of the tumor cells is a prerequisite. Recently, a series of PCNSLs from both immunocompetent and immunodeficient patients has been shown to express mutations in the 5′-noncoding region of the bcl-6 gene, 12 a marker for transition of B cells through the germinal center (GC) of secondary lymphatic organs. 13-15 These data suggested that PCNSL may in part be related to GC B cells. However, the mere presence of a bcl-6 gene mutation does not distinguish whether the tumor cells correspond to GC or post-GC B cells, ie, memory B cells and plasma cells. To precisely define the cell of origin, a molecular study of immunoglobulin (Ig) gene rearrangements in association with sequence analysis of rearranged V region genes is a suitable tool. Whereas naive IgM+ IgD+ CD27− B cells express rearranged but unmutated V region genes, 16 somatic mutations are introduced into V region genes during the process of somatic hypermutation, 17 which occurs in the microenvironment of the GC in an antigen- and T cell-dependent reaction. 18-20 Thus, GC B cells and their descendants are characterized by somatically mutated V region genes. Cells leaving this anatomical compartment shut down their hypermutation machinery and do not undergo further somatic mutations. Moreover, in somatically mutated V genes, the pattern of somatic mutations can reveal whether the respective B cells were selected for expression of a functional antigen receptor.
Studies on PCNSL have been hampered by the limited amount of tissue available, and, therefore, Ig V region genes have not yet been examined in detail. In extracerebral lymphomas, a preferential rearrangement of VH gene segments was identified for some lymphoid malignancies as compared with normal B cells. For example, an over-representation of the VH1 family has been reported for chronic lymphocytic leukemia 21 and of the VH4 family for other subtypes of B cell lymphoma, 22 and several IgH V gene segments have been associated with autoimmune disorders. 23 A biased usage of gene segments of the VH4 family has been detected in one study of extracerebral DLCL, 24 which, however, was not confirmed in another series of DLCL. 25
To precisely characterize the differentiation stage of PCNSL, we investigated a series of 10 PCNSLs from HIV-negative patients by PCR analysis of their V region genes. Our data point to GC B cells as the histogenetic origin of PCNSL.
Materials and Methods
Tumor Samples
This study included PCNSL samples from 10 HIV-negative patients (Table 1) ▶ . All specimens were from primary tumors, and most were obtained by stereotactic biopsy. Before neurosurgical intervention, all patients were untreated and had received neither corticosteroids nor radiotherapy. Immediately after neurosurgical removal, sections were cut from the tumor, and the tumor samples were stored at −80°C until used for molecular biological analysis. Cryostat sections (10 μm) were cut from the blocks to assure that the frozen tissue contained sufficient numbers of tumor cells.
Table 1.
Clinical Data, as Well as Histopathological, Immunohistochemical, and in Situ Hybridization Findings
| Case number | Age (years) | Sex | Location | EBER† | MIB-1† |
|---|---|---|---|---|---|
| 1 | 51 | f | Temporal | − | +++ |
| 2 | 69 | f | Parieto-occipital | ± | +++ |
| 3 | 74 | f | Occipital | − | +++ |
| 4 | 65 | f | Temporal | − | +++ |
| 5 | 65 | m | Fourth ventricle* | − | +++ |
| 6 | 73 | f | Parietal* | − | ++ |
| 7 | 62 | f | Frontal | − | +++ |
| 8 | 49 | m | Frontal | − | ++ |
| 9 | 67 | f | Occipital | − | +++ |
| 10 | 61 | m | Frontal | − | +++ |
All tumors were CD20 positive and classified as malignant non-Hodgkin’s lymphoma of the diffuse large-cell lymphoma type; only the location of the biopsied lesion is given.
Abbreviations and symbols: f, female; m, male;
* cases with multifocal intracerebral manifestations.
†For the analysis of MIB-1 and Epstein-Barr virus (EBER)- positive cells, a grading system was used: −, negative; +, <20% of the tumor cells stained; ++, 20% to 50% of the tumor cells stained; +++, more than 50% of the tumor cells stained; ±, it could not be decided with certainty whether the EBER-positive cells represented tumor cells or were bystander lymphocytes.
Histopathology
All tumors were histopathologically classified according to the revised European American lymphoma classification. 10 The diagnoses were based on a combination of routine morphology, which included hematoxylin-eosin and Giemsa stains, and immunohistochemistry. Staining was performed on paraffin sections.
The expression of the B cell-specific antigen CD20 (clone L26, mouse IgG2aκ; Dakopatts, Hamburg, Germany), the leukocyte common antigen CD45RB (clone PD7/26, mouse IgG1κ; Dakopatts), CD45RO (clone UCHL-1, mouse IgG2aκ; Dakopatts), the T cell-specific antigen CD3 (clone A0452, rabbit IgG; Dakopatts), the macrophage-specific antigen CD68 (clone KP1, IgG1κ; Dakopatts), and the proliferation-associated nuclear antigen Ki-67 (clone MIB-1, mouse IgG1κ; Dakopatts) were studied by use of an avidin-biotin-peroxidase complex protocol. After incubation with the primary antibody, a biotinylated rabbit anti-mouse or swine anti-rabbit antibody (Dakopatts) was applied to the sections. The sections were then incubated with the avidin-biotin-peroxidase complex (Dakopatts). Peroxidase reaction product was visualized using 3,3′-diaminobenzidine (Sigma Chemical Co., Deisenhofen, Germany) and H2O2 as cosubstrate. Sections were lightly counterstained with hemalum. All incubation steps were carried out at room temperature. Control reactions included omission of the primary antibody or application of an irrelevant antibody of the same IgG class instead of the primary antibody.
EBV-Encoded RNA in Situ Hybridization
Small EBV-encoded RNA (EBER) in situ hybridization was performed with paraffin-embedded tissue sections as described previously. 26 The EBER probes were kindly provided by Dr. G. Niedobitek 27 and were digoxigenin labeled using the DIG RNA labeling kit (Roche Diagnostics GmbH, Mannheim, Germany).
DNA Extraction
DNA was extracted from frozen tissue with the NucleoSpin Tissue Kit (Clontech Laboratories, Heidelberg, Germany). DNA was dissolved in 40 μl of 10 mmol/L Tris–1 mmol/L ethylenediaminetetraacetic acid buffer (pH 7.6). Of this stock DNA solution, 2 to 5 μl, corresponding to 100 ng of DNA, were used in each amplification reaction.
PCR Analysis
Rearrangements of the Ig heavy-chain locus (IgH) were amplified with VH gene segment family-specific primers as published previously. 25,28,29 Primers hybridize either to the framework region (FR) I (FRI) or to the leader peptide region of the VH gene segments. Each of six VH gene segment families (VH1 to -6) was studied in a separate PCR. As downstream primers, a 3′-JH mix (together with VH-leader peptide region primers) 30 or a 5′-JH mix (together with the VH-FRI primers) was used. 29,31 Taq DNA polymerase (Life Technologies, Eggenstein, Germany) was added before PCR. Amplification was carried out for one cycle at 95°C for 3 minutes, 63°C for 30 seconds, and 72°C for 1 minute, followed by 40 cycles at 95°C for 50 seconds, 63°C for 30 seconds, and 72°C for 1 minute. A final extension step at 72°C for 10 minutes was added.
Rearrangements of the light-chain locus (Igκ and Igλ, together designated as IgL) were amplified with Vκ and Vλ gene segment family-specific primers as published previously. 25,28,32 The primers hybridize to the FRI of the Vκ or the Vλ gene segments, respectively. As downstream primers, J gene segment-specific primer mixes (5′-Jκ or -Jλ mix) were used. 20,32 Each V gene segment family was analyzed in a separate PCR. Cycling conditions for rearrangements of the Igκ locus were 95°C for 5 minutes and 80°C for 4 minutes during which Taq DNA polymerase was added, followed by 61°C for 30 seconds and 72°C for 1 minute as the first cycle; and 44 additional cycles at 95°C for 1 minute, 61°C for 30 seconds, and 72°C for 1 minute; plus a final extension step at 72°C for 10 minutes. PCR cycling conditions for rearrangements in the Igλ locus were one cycle at 95°C for 3 minutes, 63°C for 30 seconds, and 72°C for 1 minute; followed by 44 cycles at 95°C for 50 seconds, 63°C for 30 seconds, and 72°C for 1 minute; plus a final extension step at 72°C for 10 minutes. Taq DNA polymerase was added before PCR.
DNA extracted from EBV-transformed B cell lines (kindly provided by Dr. J. Irsch, University of Cologne), which are characterized by rearranged V region genes of the IgH (VH2–VH5 families), as well as Igλ (Vλ1 and Vλ3) or Igκ (Vκ1–Vκ4 families) loci, was used as positive control in each PCR. In addition, the p53 gene was amplified as a cellular control gene (data not shown). As negative control, water instead of tumor DNA was used.
Sequence Analysis of Ig Genes and Cloning of PCR Products
Of each PCR reaction, 10 μl was analyzed on a 2% agarose gel stained with ethidium bromide. PCR products (40 μl) were extracted from a 2% low-melting agarose gel (NuSieve GTG Agarose, Biozym, Hessisch-Oldendorf, Germany) with the QIAEx II Kit (Qiagen, Hilden, Germany) and directly sequenced from both sides with the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer Applied Biosystems, Weiterstadt, Germany) and an automated sequencer (ABI 373 and 377, Perkin Elmer Applied Biosystems).
Sequences were compared with human germline Ig gene sequences with the International ImMunoGeneTics 33 and GenBank 34 databases and DNAPlot 33 and Blast 34 software. Codons were numbered according to Kabat et al. 35 DH gene segments were identified by alignment to germline DH gene segments and were considered homologous when at least 7 consecutive bp or at least 8 bp interrupted by one point mutation matched the germline sequence.
The ratio of replacement (R) to silent (S) mutations (R/S ratio) within the FRs of V genes was determined. This ratio indicates whether the respective B cell has been selected for expression of a functional antigen receptor within a GC. 36 Each single-point mutation in FRI–III of all potentially functional sequences was compared with the respective germline sequence and classified as an R or S mutation. From these data, R/S ratios were calculated.
In addition, PCR products of three PCNSLs (cases 4, 8, and 9) were cloned with the TOPO TA Cloning Kit (Invitrogen, Leek, The Netherlands). From each case, eight clones were sequenced from both sides.
Results
Neuropathological Analysis
The clinical and histopathologic data of the 10 HIV-negative patients analyzed are summarized in Table 1 ▶ . At the time of neurosurgical intervention, the patients’ mean age was 64 years (range 49–74 years). Eight patients had solitary tumors, whereas two patients presented with multifocal intracerebral lesions. All tumors resided in the supratentorial brain parenchyma. One patient (case 5) had an additional tumor focus in the posterior fossa around the fourth ventricle, and in another patient (case 6), the lymphoma also involved the meninges and extended into the overlying subarachnoid space. None of the patients included in this study had lymphoma manifestation outside the CNS.
Histopathologically (Figure 1) ▶ , all tumors were classified as CD20+ B cell non-Hodgkin’s lymphoma of the DLCL type based on the revised European American lymphoma classification. 10 Tumor cells mostly exhibited a centroblast morphology (Figure 1a) ▶ . The mitotic activity was generally high, and the majority of the tumors contained more than 50% MIB-1-positive proliferating cells (Table 1 ▶ , Figure 1b ▶ ). Of 10 PCNSLs, 9 did not harbor EBER; in one tumor (case 2), some small cells appeared to be EBER positive (Table 1) ▶ . However, it could not be decided with certainty whether the EBER-positive cells were tumor cells or bystander lymphocytes. In all tumors, the malignant cells were characteristically intermingled with an infiltrate composed predominantly of leukocyte common antigen-positive, CD45R0+, CD3+ small, mature T lymphocytes, as well as CD68+ macrophages (Figure 1c) ▶ and some CD20+ small, mature, presumably nonmalignant B cells. All lymphomas showed a characteristic angiocentric growth pattern with splitting of the blood vessel walls (Figure 1d) ▶ . In addition, the tumors diffusely infiltrated the adjacent brain parenchyma, which showed edema and reactive astrocytosis. Areas of necrosis were present in some cases.
Figure 1.
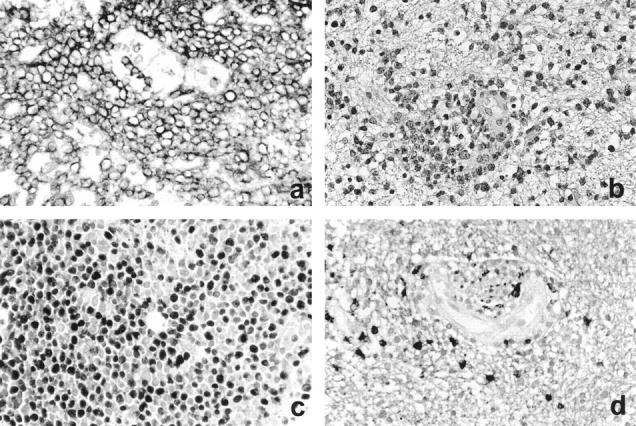
Histopathological characteristics of PCNSL. a: Lymphoma cells strongly express the CD20 antigen (CD20 immunostaining, original magnification, ×280). b: Characteristic growth pattern of a PCNSL with splitting of the blood vessel walls and a diffuse infiltration of the brain parenchyma. Note the marked edema in the affected brain tissue (H&E staining; original magnification, ×220). c: More than 50% of the tumor cells are in a proliferating state as demonstrated by nuclear Ki-67 immunoreactivity (MIB-1 immunostaining; original magnification, ×280). d: The tumor cells are embedded in an infiltrate of small, mature T cells, which reside in a perivascular location and are also scattered throughout the brain (CD3 immunostaining; original magnification, ×280).
IgH and IgL Gene Rearrangements in PCNSL
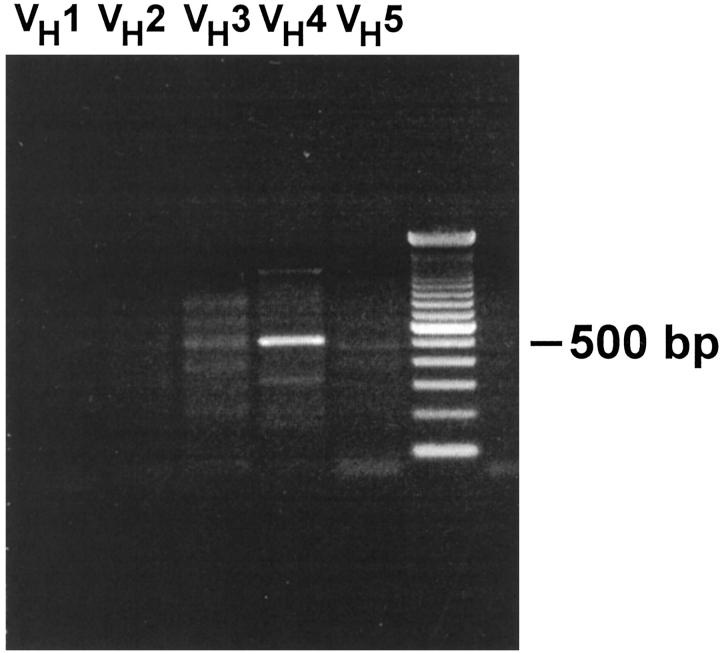
To characterize the differentiation stage of the tumor cells, 10 PCNSL cases were analyzed for rearrangements of the IgH, Igκ, and Igλ chain loci by use of six VH, six Vκ, and eight Vλ family-specific primers in separate reactions together with the respective J gene segment primer mixes. Under these conditions, rearranged Ig genes could be identified in all PCNSLs of this series. For the IgH gene analysis, one or two dominant PCR products were detected in all tumors (Figure 2) ▶ . Occasionally, some additional, diffuse bands for other VH gene families were disclosed in the agarose gels. These faint PCR products were most likely derived from nonmalignant, polyclonal B cells, which are characteristically part of the accompanying infiltrate.
Figure 2.
PCR analysis of rearranged IgH genes. The PCR product of 550 bp indicates the presence of a monoclonal B cell population that has rearranged a gene of the VH4 family. PCR analysis of VH families 3–5 discloses additional faint bands indicating admixed polyclonal, presumably nonmalignant B cells.
In each lymphoma, a monoclonally rearranged IgH gene could be detected. Among 13 dominant PCR products chosen for sequence analysis, 10 turned out to be clonal Ig gene rearrangements, as indicated by clearly readable sequences. Corresponding VH and JH gene segments were identified in all cases. DH gene segments could be identified in five of the tumors. For the light-chain locus, rearranged V genes were observed in seven PCNSLs. Three tumors lacked dominant amplificates, and in these lymphomas, neither monoclonal Igκ nor Igλ gene rearrangements could be detected. This is most probably because highly mutated rearranged gene segments (as they are present in PCNSL; see below) may sometimes not be recognized by the primers.
Sequence Analysis of V Region Genes
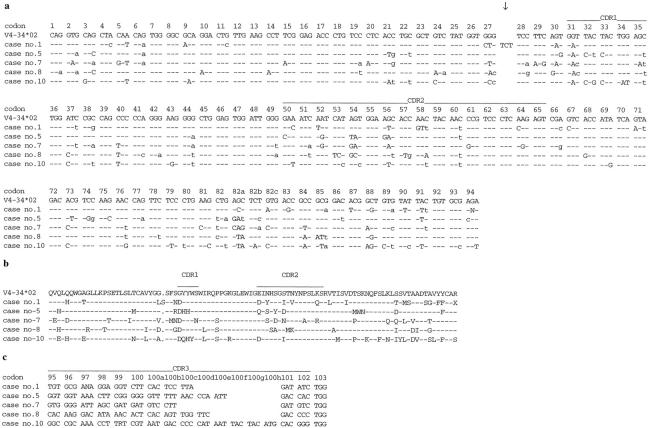
The successful identification of a clonal cell population allowed us to further examine rearranged Ig genes. Results of the sequence analyses are summarized in Table 2 ▶ . It is interesting that examination of the VH gene repertoire revealed a strong bias toward a restricted usage of VH gene families. In this series of PCNSLs, only VH gene segments of the VH3 and VH4 families were found, each family accounting for 5 of the 10 cases. More important, all monoclonal B cell populations from PCNSLs, which used a gene of the VH4 family, had rearranged the V4–34 gene segment (Table 2) ▶ . Sequence analysis of the 10 cases demonstrated that all of these rearranged VH genes were in-frame and did not harbor stop codons, and, thus, they were considered as potentially functional. Remarkably, all PCNSLs using the V4–34 gene segment shared a common mutation in the first codon of complementarity-determining region 1 (Figure 3) ▶ . Translation of the nucleic acid sequence into the amino acid sequence disclosed that this mutation resulted in an exchange from glycine to aspartate.
Table 2.
IgH and IgL Gene Rearrangements in Primary Nervous System Lymphomas
| Case number | VH | DH† | JH | Mutations‡ (n) | Mutation frequency§ | In frame¶ | VL | JL | Mutations‡ (n) | Mutation frequency‡ | In frame¶ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | V4-34*02 | na | J3*02 | 33 | 11.2 | + | − | − | |||
| 2 | V3-23*01 | D2-2*02 | J4*02 | 11 | 3.7 | + | Vκ1-8*01 | Jκ5*01 | 5 | 2.5 | + |
| 3 | V3-23*01 | na | J4*02 | 45 | 22.2 | + | Vκ3-15*01 | Jκ4*01 | 35 | 13.7 | + |
| 4 | V3-7*02 | na | J4*01 | 47 | 16.0 | + | Vκ1-8*01 | Jκ3*01 | 55 | 21.7 | − |
| 5 | V4-34*02 | D3-10*01 | J4*02 | 34 | 11.8 | + | Vκ1-37*01 | Jκ4*01 | 0 | 0 | − |
| Vκ2-40*01 | Jκ5*01 | 4 | 1.5 | + | |||||||
| 6 | V3-30*01 | D3-10*01 | J3*01 | 43 | 14.6 | + | Vκ3-11*01 | Jκ1*01 | 44 | 17.3 | + |
| 7 | V4-34*02 | D3-3*01 | J3*01 | 44 | 15.6 | + | − | − | |||
| 8 | V4-34*02 | na | J5*02 | 42 | 14.4 | + | − | − | |||
| 9 | V3-7*01 | D3-10*01 | J4*02 | 26 | 8.9 | + | Vλ2-8*01 | Jλ3*01 | 13 | 4.9 | + |
| 10 | V4-34*02 | na | J6*03 | 47 | 16.6 | + | Vκ1-5*03 | Jκ1*01 | 24 | 9.1 | + |
†na, the sequence between the VH and the JH gene segment could not be aligned to any known DH gene segment.
‡Values designate the number of point mutations in the V gene segment as compared with the most homologous germline sequence.
§Ratio between the number of point mutations and the base pair length of the respective V gene segment.
¶+, in-frame rearrangements; −, out-of-frame rearrangements. All in-frame rearrangements did not contain a stop codon. Accession numbers of the germline gene segments are as follows: IgH: V3-7*01, M99649; V3-7*02, X92288; V3-23*01, M99660; V3-30*01, M83134; V4-34*02, M99684; D2-2*02, X97051; D3-3*01, X13972; D3-10*01, X13972; J3*01, J00256; J4*01, J00256; J4*02, X86355; J5*02, X86355; J6*03, X86356. Igκ: V1-5*03, X72813; V1-8*01, Z00014; V1-37*01, X59316; V2-40*01, X59316; V3-11*01, X01668; V3-15*01, M23090; J1*01, J00242; J3*01, J00242; J4*01, J00242. Igλ: V2-8*01, X97462; J3*01, X97462. Sequences have been submitted to the GenBank data library under accession numbers AF168797–AF168823.
Figure 3.
Biased usage of the V4–34 gene segment in PCNSL. a: The VH sequences (codons 1–94) of cases 1, 5, 7, 8, and 10 were aligned to the most homologous germline VH gene segment (V4–34*02). Dashes indicate sequence identity. Point mutations are shown either as small letters for silent mutations or as capital letters for replacement mutations. Mutations in codons 93 to 94 of cases 1, 5, and 10 were not taken into account for calculation of the R/S values or the mutation frequency. The arrow points to an insertion of 3 bp between codons 27 and 28 in case 1. b: Translation of the VH sequences from a and alignment of the amino acid sequences. Dashes indicate sequence identity and a point indicates a gap of one amino acid for better alignment. c: Complementarity-determining region 3 and beginning of FRIV of the IgH sequences (codons 95–103) of five PCNSLs, using the V4–34 gene segment. A sequence comparison demonstrates unique sequences for each case.
In addition, five PCNSLs used gene segments of the VH3 family. Among this subgroup of tumors, two lymphomas had rearranged the V3–23 gene segment. Another two tumors shared the same VH3 germline gene segment, but different alleles (V3–7*01 and V3–7*02). Although both alleles differed at codon 48 (V3–7*01, AAG; V3–7*02, AAA), they both encoded the amino acid lysine; thus, there was no difference between these two alleles at the amino acid level. The V3–30 gene segment was used in one case.
In 5 of 10 cases, a DH gene segment could be identified. This was not possible for the other five tumors, because the homology of the sequences to published germline sequences was too low (less than 7 bp). Three tumors (cases 5, 6, and 9) used the D3–10 gene segment; however, sequence analysis revealed that they had rearranged different parts of the gene segment. The remaining two cases (cases 2 and 7) revealed the D2–2 and D3–3 gene segments, respectively. There was no association with a particular VH gene segment. JH gene segments 3 to 6 were found to be rearranged in one (JH5, 6), two (JH3), or five cases (JH4). Again, individual VH gene segments were not preferentially associated with particular JH gene segments.
Mutation frequencies of the rearranged IgH genes were calculated. The mean mutation frequency was 13.2% with a range of 3.7 to 22.2%. This frequency significantly exceeds the average mutation frequency of normal, nonmalignant memory B cells (5–6%). 16
Rearranged Vκ genes were amplified from six of seven PCNSLs, which showed a clonal rearrangement of light-chain genes. Two PCNSLs harbored the same Vκ gene segment, V1–8, whereas the other tumors differed in their Vκ gene segments. Thus, in contrast to the VH gene segments, there was no evidence for a preferential usage of Vκ gene segments in PCNSL. In cases 2, 3, 6 and 10, rearranged Igκ genes were in-frame and did not harbor a stop codon and, therefore, were considered potentially functional. In one tumor (case 4), a rearranged Igκ gene was detected that was out-of-frame. It contained three stop codons in amino acid positions 16, 37, and 97. It is interesting that tumor 5 harbored two rearranged Igκ genes, one of which was in-frame and one of which was out-of-frame. The out-of-frame Igκ gene was unmutated, and the in-frame Igκ gene showed a low mutation frequency of 1.5%. The absence of somatic mutations in the out-of-frame Vκ gene might be due to an inactivation of the nonfunctional Vκ gene by rearrangement of a κ-deleting element, which often abolishes somatic mutations in remaining VκJκ joints. 36 All potentially functional Igκ genes were somatically mutated. The analysis of the Jκ gene segments showed an unbiased usage of the Jκ1, -3, -4, and -5 genes. A clonal, somatically mutated Igλ gene was amplified from one tumor (case 9). This rearrangement was assembled by the gene segments V2–8 and J3, and the rearrangement was potentially functional. The mutation frequency of all potentially functional IgL rearrangements ranged from 1.5% to 17.3% with a mean of 8.3%.
Taken together, sequence analysis of Ig genes of PCNSL demonstrated a biased usage of particular VH, but not DH, JH, Vκ, Jκ, Vλ, and Jλ gene segments. The tumor cells carried a high load of somatic mutations, which indicates that the tumor cells had reached the differentiation stage of GC B cells.
Analysis of the Mutation Pattern
The R/S ratio in FRI–III was calculated for all in-frame IgH and IgL genes. This parameter indicates whether the corresponding B lymphocyte was selected for expression of a functional antibody. 36 The R/S ratios for the FRs were determined as 1.3 (range 0.3–2.2) and 1.3 (range 0–2.5) for the IgH and the IgL genes, respectively. The R/S ratios clearly indicate that the tumor cells (or their precursors) were selected for antibody expression because selected B cells usually show R/S values below 1.5 for the FRs. 36
Intraclonal Diversity in PCNSL
The IgH gene sequences from one tumor (case 4), which were clearly readable, exhibited a few “double peaks.” To investigate whether this observation might be indicative of intraclonal variation, this tumor and two additional tumors (cases 8 and 9) were further analyzed in cloning experiments of the IgH PCR products. Eight clones per tumor were sequenced (Figure 4) ▶ .
Figure 4.
Intraclonal diversity in a subset of PCNSL. PCR products of rearranged IgH genes were cloned in cases 4 (a), 8 (b), and 9 (c). Eight separate clones were analyzed in each case. Only those codons are shown in which sequence heterogeneity was observed. Dashes indicate sequence identity. Point mutations are labeled either as small letters for silent mutations or as capital letters for replacement mutations. gl, germline sequence.
In case 4, all clones had 47 identical point mutations, as compared with their respective germline gene segment (V3–7*02). Among these 8 clones (Figure 4a) ▶ , three sequences did not contain further mutations. The other five clones had introduced additional point mutations into their sequence. Among these, two clones had acquired three identical point mutations, and one of these contained one further sequence change. The remaining three clones harbored one individual mutation each. Surprisingly, the locations of “double peaks” in the sequences derived from direct sequencing were identical in the eight cloned sequences.
In case 8, all clones shared 42 point mutations as compared with the respective germline gene segment (V4–34*02). Six clones had identical sequences, two clones exhibited two additional, identical point mutations, and one of them harbored a third unique point mutation (Figure 4b) ▶ .
All clones derived from tumor 9 had 26 point mutations in common as compared with the respective germline gene segment (V3–7*01). Six clones had an identical sequence, and the other two clones differed from the main clone by a further introduction of another point mutation, which, however, was different in the two clones (Figure 4c) ▶ .
To confirm whether these sequence differences between the cloned PCR products were indeed indicative of intraclonal diversity or simply reflected Taq DNA polymerase errors, we calculated the expected errors for our PCR conditions. Based on a Taq DNA polymerase error rate of 10−5/bp and cycle, 37,38 we expect 1 error/2439 bp. Taking into account the total base pairs obtained from the various sequences (case 4, 2568 bp; case 8, 2640 bp; case 9, 2592 bp), approximately one mutation would be expected to result from Taq DNA polymerase error in each of the cases. Because the number of different mutations in case 4 (seven mutations) is well above this threshold, we cannot exclude with certainty that the three mutations (one unique mutation) in case 8 and the two unique mutations in case 9 may be due to a Taq DNA polymerase error.
Discussion
The aim of the present study was to define the histogenesis and the molecular phenotype of PCNSL, which develop in an environment that normally contains very low numbers of lymphocytes and actively suppresses the proliferation of lymphocytes. 4 To address these issues, we sequenced Ig V region genes of the tumor cells from 10 patients. All PCNSLs of our series carried monoclonally rearranged V region genes and, moreover, had introduced somatic mutations into the rearranged Ig genes. These data indicate that PCNSLs are derived from mature B cells, which have experienced an antigen and have undergone T cell-dependent affinity maturation in the microenvironment of the GC of secondary lymphatic organs. Additional cloning experiments in three tumors demonstrated ongoing mutation in at least one case. This also points to GC B cells and not to memory B cells or plasma cells as the cells of origin in at least a fraction of PCNSLs.
This study extends previous data by Larocca et al, 12 who suggested that PCNSL frequently may be related to GC B cells. Their hypothesis was based on the observation of mutations in the 5′-noncoding region of the bcl-6 gene, which are specifically acquired by B cells at the time of transition through the GC. 14,15 However, bcl-6 gene mutations are present in both GC and post-GC B cells, and, therefore, the analysis of Larocca et al 12 did not distinguish between GC and post-GC B cells. Moreover, in the latter study, mutations in the bcl-6 gene could be identified in only approximately 50% of PCNSLs, which raised the question whether the origin of PCNSL is heterogenous and includes pre-GC B cells. Because all tumors of our series harbored mutated V region genes, we have no evidence for a subset of PCNSL that have not reached the differentiation stage of GC B cells.
Sequence analysis of the rearranged V genes yielded further insights into the characteristics of the tumor cells. Somatic mutations were observed in all potentially functional IgH and IgL genes (Table 2) ▶ . It is interesting that the average mutation frequency was rather high, 13.2% and 8.3% for IgH and IgL genes, respectively. Previously, a similar mutation frequency was reported for V region genes of extracerebral DLCL. 25,36 For IgH, PCNSL exceeded the mutation frequency calculated for other DLCL outside the CNS. The high frequency of somatic mutations in the V genes of PCNSL can be viewed as a further argument that the tumor cells are indeed derived from GC B cells. This high mutation frequency may be acquired during a prolonged interaction of the tumor cell (or its precursor) in the microenvironment of the GC. A transforming event could have occurred before or during proliferation of the tumor cell in the GC. On one hand, affected clones may have become independent of the microenvironment of the GC initiated by the hypermutation machinery itself via translocation of Ig genes 29 or by introducing mutations in non-Ig genes, eg, bcl-6. 14,15 On the other hand, if a memory B cell, which has already inactivated the hypermutation machinery, served as precursor for PCNSL, one would expect to find mutation frequencies typical for memory B cells (5–6%). 16 Thus, the unusually high mutation frequency of Ig genes indicates that the tumor clone is derived from a GC B cell.
To address the issue of ongoing mutation in PCNSL, three PCNSLs (cases 4, 8, and 9) were analyzed in cloning experiments with PCR products of rearranged VH genes. These studies demonstrated intraclonal diversity in at least one case and provide evidence for intracerebrally ongoing mutation of PCNSL. Because ongoing mutation normally requires the presence of both antigen and T cells, it is tempting to speculate that an intracerebral antigen in concert with T cells stimulates further expansion of the tumor clone. The observation of ongoing mutation in the CNS, an extranodal organ devoid of conventional lymphatic structures including GC, was surprising. Although we cannot exclude with certainty that a transforming event took place in the GC of secondary lymphatic organs while the B cell clone still introduced somatic mutations and continued to acquire somatic mutations before leaving the GC, it is unlikely that several different tumor clones leave the GC of lymphatic organs simultaneously and selectively home to the CNS. On the other hand, it is also possible that the transforming event rendered the tumor cells unable to inactivate their hypermutation machinery.
In this study, exclusively genes of the VH3 and VH4 families were rearranged. Usage of different VH3 genes by five of the lymphomas reflects a normal VH gene usage pattern, because approximately 50% of normal human B cells use genes of the VH3 family. 39 However, the five remaining lymphomas showed a rearranged V4–34 gene segment, which is used by 4% to 7% of normal B cells in humans. 39 This finding lends further support to the hypothesis that intracerebral stimuli direct the Ig repertoire and have an important influence on the development and biology of PCNSL. Our data, which were obtained from HIV-unrelated PCNSLs with the morphological characteristics predominantly of centroblasts, are at variance with a recent study of VH genes in the PCNSLs of AIDS patients. 40 In HIV-related PCNSL of the immunoblastic subtype, evidence for biased usage of particular VH families or for intraclonal diversity has not been reported. This difference supports the concept that AIDS-unrelated and AIDS-related PCNSLs represent separate disease entities with distinct pathogenesis.
Surprisingly, all tumors with a rearranged V4–34 gene segment shared a common mutation in the first codon of complementarity-determining region 1 (Figure 3) ▶ , which leads to an amino acid exchange from glycine to aspartate. The constant detection of this exchange in all tumors using the V4–34 gene segment markedly exceeded the expected probability, even considering that the first 4 bp of complementarity-determining region 1 correspond to a mutational hotspot motif. 41 This mutation has also been observed in B cells obtained from 2 of 19 patients with cold agglutinin disease 23,42 and in 1 of 8 mucosa-associated lymphoid tissue lymphomas. 43 However, it appears unlikely that these 4 bp indeed constitute a strong mutational hotspot in V4–34, because mutations at this position have been reported in only 10% of somatically mutated V4–34 gene segments as observed by analysis of cDNA libraries. 44,45 In our study, this shared mutation suggests antigen-selected maturation of the tumor cells. This notion is further supported by the low R/S ratio, which reflects counterselection of R mutations in the FRs and indicates that the tumor cells (or their precursors) have indeed—at least temporarily—been antigen-selected for expression of a functional antibody. In this regard, PCNSLs behave similarly to extracerebral DLCL. 25
Whereas PCNSLs appear to preferentially use the V4–34 gene, it is controversial whether this holds true for extracerebral DLCL. This gene segment was also rearranged in lymphomas of other entities, including high-grade mucosa-associated lymphoid tissue lymphoma, mantle cell lymphoma, and Burkitt’s lymphoma. 22,43,46 A biased usage of VH4 genes has been reported in one analysis of extracerebral DLCL, 24 but could not be confirmed in another study. 25 The apparent discrepancy between the various studies may be explained by the limited number of tumors included and differences in anatomical sites from which the samples were derived. Whether the microenvironment of the CNS favors the observed expansion of lymphomas bearing particular Ig genes remains to be elucidated. The high frequency of usage of the V4–34 gene strongly suggests a functional role of the Ig encoded by this gene in the development of PCNSL. Either of two mechanisms may underlie its biased usage, an antigen-driven or a superantigen-fostered expansion of V4–34-encoded Ig, which may also have autoreactive properties. 47-50 For normal, nonmalignant B lymphocytes, bacterial and viral proteins have been described to function as superantigens, eg, staphylococcal proteins 51,52 and the gp120 protein of the human immunodeficiency virus. 53,54 Interestingly, a survival-fostering effect has been reported for VH3 and VH4—including V4–34-expressing human B cells in response to staphylococcal enterotoxins A and D, which rescued these cells from apoptosis in in vitro experiments. 55,56 In our series, the restricted pattern of a small subset of VH genes, together with a random usage of DH and JH gene segments, would be compatible with a superantigen effect. However, candidates for superantigens involved in the pathogenesis of PCNSL have not yet been identified.
On the other hand, antigens may preferentially stimulate the expansion and intracerebral persistence of B cells, which produce antibodies encoded by the respective V4–34 gene segment. In this regard, a wide variety of viruses are attractive candidates, including polyomaviruses and herpes viruses, which are known to persist in the brain. It is noteworthy that elevated levels of Ig recognized by the 9G4 antibody, which specifically binds to antibodies encoded by the V4–34 gene segment, 23,57 have been reported in association with EBV infections. 50 However, 9 of 10 of our tumors were EBV negative, which is in line with other studies that have found that PCNSLs in HIV-negative patients do not harbor EBV. 11,12 Thus, our data do not provide evidence that an EBV infection is pathogenetically involved in the biased usage of the V4–34 gene segment in these PCNSL.
The identification of GC B cells as the origin of PCNSL in non-AIDS patients and the detailed characterization of their molecular phenotype provide a basis for further detailed studies on the pathogenesis of PCNSL.
Acknowledgments
The authors thank A. Rang, J. Jesdinsky, M. Fahrig, and P. Ruhnau-Declair for expert technical assistance and H.-U. Klatt for photographic help.
Footnotes
Address reprint requests to Dr. Martina Deckert-Schlüter, Department of Neuropathology, Sigmund-Freud-Strasse, 25, D-53105 Bonn, Germany. E-Mail: mds@uni-bonn.de.
Supported by the BONFOR program (grant no. 154/27).
References
- 1.Fontana A, Frei K, Bodmer S, Hofer E: Immune-mediated encephalitis: on the role of antigen-presenting cells in brain tissue. Immunol Rev 1987, 100:185-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lassmann H, Rössler K, Zimprich F, Vass K: Expression of adhesion molecules and histocompatibility antigens at the blood-brain barrier. Brain Pathol 1991, 1:115-123 [DOI] [PubMed] [Google Scholar]
- 3.Tontsch U, Rott O: Cortical neurons selectively inhibit MHC class II induction in astrocytes but not in microglial cells. Int Immunol 1993, 5:249-254 [DOI] [PubMed] [Google Scholar]
- 4.Irani DN, Lin KI, Griffin DE: Regulation of brain-derived T cells during acute central nervous system inflammation. J Immunol 1997, 158:2318-2326 [PubMed] [Google Scholar]
- 5.Lantos PL, VandenBerg SR, Kleihues P: Tumours of the nervous system. Greenfield’s Neuropathology, vol 2, ed 6. Edited by DI Graham, PL Lantos. New York, Oxford University Press, 1997, pp 766–775
- 6.Miller DC, Hochberg FH, Harris NL, Gruber ML, Louis DN, Cohen H: Pathology with clinical correlations of primary central nervous system non-Hodgkin’s lymphoma. The Massachusetts General Hospital experience 1958–1989. Cancer 1994, 74:1383-1397 [DOI] [PubMed] [Google Scholar]
- 7.Goplen AK, Dunlop O, Liestol K, Lingjaerde OC, Bruun JN, Maehlen J: The impact of primary central nervous system lymphoma in AIDS patients: a population-based autopsy study from Oslo. J Acquired Immune Defic Syndr Hum Retrovirol 1997, 14:351-354 [DOI] [PubMed] [Google Scholar]
- 8.Beral V, Peterman T, Berkelman R, Jaffe H: AIDS-associated non-Hodgkin lymphoma. Lancet 1991, 337:805-809 [DOI] [PubMed] [Google Scholar]
- 9.Cote TR, Manns A, Hardy CR, Yellin FJ, Hartge P: Epidemiology of brain lymphoma among people with or without acquired immunodeficiency syndrome. AIDS/Cancer Study Group. J Natl Cancer Inst 1996, 88:675-679 [DOI] [PubMed] [Google Scholar]
- 10.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 [PubMed] [Google Scholar]
- 11.Morgello S: Pathogenesis and classification of primary central nervous system lymphoma: an update. Brain Pathol 1995, 5:383-393 [DOI] [PubMed] [Google Scholar]
- 12.Larocca LM, Capello D, Rinelli A, Nori S, Antinori A, Gloghini A, Cingolani A, Migliazza A, Saglio G, Cammilleri-Broet S, Raphael M, Carbone A, Gaidano G: The molecular and phenotypic profile of primary central nervous system lymphoma identifies distinct categories of the disease and is consistent with histogenetic derivation from germinal center-related B cells. Blood 1998, 92:1011-1019 [PubMed] [Google Scholar]
- 13.Cattoretti G, Chang CC, Cechova K, Zhang J, Ye BH, Falini B, Louie DC, Offit K, Chaganti RS, Dalla-Favera R: BCL-6 protein is expressed in germinal-center B cells. Blood 1995, 86:45-53 [PubMed] [Google Scholar]
- 14.Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti RS, Klein U, Küppers R, Rajewsky K, Dalla-Favera R: BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci USA 1998, 95:11816-11821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen HM, Peters A, Baron B, Zhu X, Storb U: Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science 1998, 280:1750-1752 [DOI] [PubMed] [Google Scholar]
- 16.Klein U, Rajewsky K, Küppers R: Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med 1998, 188:1679-1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajewsky K: Clonal selection and learning in the antibody system. Nature 1996, 381:751-758 [DOI] [PubMed] [Google Scholar]
- 18.Jacob J, Kelsoe G, Rajewsky K, Weiss U: Intraclonal generation of antibody mutants in germinal centres. Nature 1991, 354:389-392 [DOI] [PubMed] [Google Scholar]
- 19.Berek C, Berger A, Apel M: Maturation of the immune response in germinal centers. Cell 1991, 67:1121-1129 [DOI] [PubMed] [Google Scholar]
- 20.Küppers R, Zhao M, Hansmann ML, Rajewsky K: Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J 1993, 12:4955-4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, Schulman P, Vinciguerra VP, Rai K, Rassenti LZ, Kipps TJ, Dighiero G, Schroeder H, Ferrarini M, Chiorazzi N: Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest 1998, 102:1515-1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Funkhouser WK, Warnke RA: Preferential IgH V4–34 gene segment usage in particular subtypes of B-cell lymphoma detected by antibody 9G4. Hum Pathol 1998, 29:1317-1321 [DOI] [PubMed] [Google Scholar]
- 23.Thorpe SJ, Turner CE, Stevenson FK, Spellerberg MB, Thorpe R, Natvig JB, Thompson KM: Human monoclonal antibodies encoded by the V4–34 gene segment show cold agglutinin activity and variable multireactivity which correlates with the predicted charge of the heavy-chain variable region. Immunology 1998, 93:129-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu FJ, Levy R: Preferential use of the VH4 Ig gene family by diffuse large-cell lymphoma. Blood 1995, 86:3072-3082 [PubMed] [Google Scholar]
- 25.Küppers R, Rajewsky K, Hansmann ML: Diffuse large cell lymphomas are derived from mature B cells carrying V region genes with a high load of somatic mutation and evidence of selection for antibody expression. Eur J Immunol 1997, 27:1398-1405 [DOI] [PubMed] [Google Scholar]
- 26.Niedobitek G, Herbst H, Young LS, Brooks L, Masucci MG, Crocker J, Rickinson AB, Stein H: Patterns of Epstein-Barr virus infection in non-neoplastic lymphoid tissue. Blood 1992, 79:2520-2526 [PubMed] [Google Scholar]
- 27.Niedobitek G, Young LS, Lau R, Brooks L, Greenspan D, Greenspan JS, Rickinson AB: Epstein-Barr virus infection in oral hairy leukoplakia: virus replication in the absence of a detectable latent phase. J Gen Virol 1991, 72:3035-3046 [DOI] [PubMed] [Google Scholar]
- 28.Küppers R, Zhao M, Rajewsky K, Hansmann ML: Detection of clonal B cell populations in paraffin-embedded tissues by polymerase chain reaction. Am J Pathol 1993, 143:230-239 [PMC free article] [PubMed] [Google Scholar]
- 29.Goossens T, Klein U, Küppers R: Frequent occurrence of deletions and duplications during somatic hypermutation: implications for oncogene translocations and heavy chain disease. Proc Natl Acad Sci USA 1998, 95:2463-2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braeuninger A, Küppers R, Strickler JG, Wacker HH, Rajewsky K, Hansmann ML: Hodgkin and Reed-Sternberg cells in lymphocyte predominant Hodgkin disease represent clonal populations of germinal center-derived tumor B cells [published erratum appears in Proc Natl Acad Sci USA 1997, 94: 14211]. Proc Natl Acad Sci USA 1997, 94:9337-9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Küppers R, Hansmann M, Rajewsky K: Micromanipulation and PCR analysis of single cells from tissue sections. Weir’s Handbook of Experimental Immunology, vol IV, ed 5. Edited by H La. Blackwell Science, 1996, pp 206.201–206.204
- 32.Küppers R, Willenbrock K, Rajewsky K, Hansmann ML: Detection of clonal lambda light chain gene rearrangements in frozen and paraffin-embedded tissues by polymerase chain reaction. Am J Pathol 1995, 147:806-814 [PMC free article] [PubMed] [Google Scholar]
- 33.Lefranc MP, Giudicelli V, Busin C, Bodmer J, Müller W, Bontrop R, Lemaitre M, Malik A, Chaume D: IMGT, the International ImMunoGeneTics database. Nucleic Acids Res 1998, 26:297-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ: Gapped BLAST, and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997, 25:3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabat EA, Wu TT, Reid-Miller M, Perry HM, Gottesman KS: Sequences of Proteins of Immunological Interest. 1987. U.S. Government Printing Office, Bethesda, MD,
- 36.Klein U, Goossens T, Fischer M, Kanzler H, Braeuninger A, Rajewsky K, Küppers R: Somatic hypermutation in normal and transformed human B cells. Immunol Rev 1998, 162:261-280 [DOI] [PubMed] [Google Scholar]
- 37.Arnheim N, Erlich H: Polymerase chain reaction strategy. Annu Rev Biochem 1992, 61:131-156 [DOI] [PubMed] [Google Scholar]
- 38.Erlich HA, Gelfand D, Sninsky JJ: Recent advances in the polymerase chain reaction. Science 1991, 252:1643-1651 [DOI] [PubMed] [Google Scholar]
- 39.Brezinschek HP, Brezinschek RI, Lipsky PE: Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J Immunol 1995, 155:190-202 [PubMed] [Google Scholar]
- 40.Julien S, Radosavljevic M, Labouret N, Camilleri-Broet S, Davi F, Raphael M, Martin T, Pasquali JL: AIDS primary central nervous system lymphoma: molecular analysis of the expressed VH genes and possible implications for lymphomagenesis. J Immunol 1999, 162:1551-1558 [PubMed] [Google Scholar]
- 41.Neuberger MS, Ehrenstein MR, Klix N, Jolly CJ, Yelamos J, Rada C, Milstein C: Monitoring and interpreting the intrinsic features of somatic hypermutation. Immunol Rev 1998, 162:107-116 [DOI] [PubMed] [Google Scholar]
- 42.Pascual V, Victor K, Spellerberg M, Hamblin TJ, Stevenson FK, Capra JD: VH restriction among human cold agglutinins: the VH4–21 gene segment is required to encode anti-I and anti-i specificities. J Immunol 1992, 149:2337-2344 [PubMed] [Google Scholar]
- 43.Hallas C, Greiner A, Peters K, Müller-Hermelink HK: Immunoglobulin VH genes of high-grade mucosa-associated lymphoid tissue lymphomas show a high load of somatic mutations, and evidence of antigen-dependent affinity maturation. Lab Invest 1998, 78:277-287 [PubMed] [Google Scholar]
- 44.Chapman CJ, Mockridge CI, Hamblin TJ, Stevenson FK: Tracking of the V4–34 (VH4–21) gene in human tonsil reveals clonal isotype switch events and a highly variable degree of somatic hypermutation. Clin Exp Immunol 1996, 105:360-368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kraj P, Friedman DF, Stevenson FK, Silberstein LE: Evidence for the overexpression of the VH4–34 (VH4.21) Ig gene segment in the normal adult human peripheral blood B cell repertoire. J Immunol 1995, 154:6406-6420 [PubMed] [Google Scholar]
- 46.Chapman CJ, Wright D, Stevenson FK: Insight into Burkitt’s lymphoma from immunoglobulin variable region gene analysis. Leuk Lymphoma 1998, 30:257-267 [DOI] [PubMed] [Google Scholar]
- 47.Stevenson FK, Longhurst C, Chapman CJ, Ehrenstein M, Spellerberg MB, Hamblin TJ, Ravirajan CT, Latchman D, Isenberg D: Utilization of the VH4–21 gene segment by anti-DNA antibodies from patients with systemic lupus erythematosus. J Autoimmun 1993, 6:809-825 [DOI] [PubMed] [Google Scholar]
- 48.Stevenson FK, Spellerberg MB, Chapman CJ, Hamblin TJ: Differential usage of an autoantibody-associated VH gene, VH4–21, by human B-cell tumors. Leuk Lymphoma 1995, 16:379-384 [DOI] [PubMed] [Google Scholar]
- 49.Pascual V, Capra JD: VH4–21, a human VH gene segment overrepresented in the autoimmune repertoire. Arthritis Rheum 1992, 35:11-18 [DOI] [PubMed] [Google Scholar]
- 50.Chapman CJ, Spellerberg MB, Smith GA, Carter SJ, Hamblin TJ, Stevenson FK: Autoanti-red cell antibodies synthesized by patients with infectious mononucleosis utilize the VH4–21 gene segment. J Immunol 1993, 151:1051-1061 [PubMed] [Google Scholar]
- 51.Amariglio N, Rechavi G: Do superantigens play a role in lymphoproliferation? Leuk Lymphoma 1996, 22:237-243 [DOI] [PubMed] [Google Scholar]
- 52.Silverman GJ: Human antibody responses to bacterial antigens: studies of a model conventional antigen and a proposed model B cell superantigen. Int Rev Immunol 1992, 9:57-78 [DOI] [PubMed] [Google Scholar]
- 53.Berberian L, Goodglick L, Kipps TJ, Braun J: Immunoglobulin VH3 gene products: natural ligands for HIV gp120. Science 1993, 261:1588-1591 [DOI] [PubMed] [Google Scholar]
- 54.Karray S, Juompan L, Maroun RC, Isenberg D, Silverman GJ, Zouali M: Structural basis of the gp120 superantigen-binding site on human immunoglobulins. J Immunol 1998, 161:6681-6688 [PubMed] [Google Scholar]
- 55.Domiati-Saad R, Lipsky PE: Staphylococcal enterotoxin A induces survival of VH3-expressing human B cells by binding to the VH region with low affinity. J Immunol 1998, 161:1257-1266 [PubMed] [Google Scholar]
- 56.Domiati-Saad R, Attrep JF, Brezinschek HP, Cherrie AH, Karp DR, Lipsky PE: Staphylococcal enterotoxin D functions as a human B cell superantigen by rescuing VH4-expressing B cells from apoptosis. J Immunol 1996, 156:3608-3620 [PubMed] [Google Scholar]
- 57.Potter KN, Li Y, Pascual V, Williams R, Byres LC, Spellerberg M, Stevenson FK, Capra JD: Molecular characterization of a cross reactive idiotope on human immunoglobulins utilizing the VH4–21 gene segment. J Exp Med 1993, 178:1419-1428 [DOI] [PMC free article] [PubMed] [Google Scholar]