Abstract
Twenty-two primary central nervous system lymphomas of immunocompetent adults were studied by comparative genomic hybridization. All were high-grade diffuse large B cell lymphomas. Comparative genomic hybridization revealed an average of 5.5 chromosomal changes per tumor, with gains being more common than losses (3.5 vs. 2.0). The most frequent DNA copy number changes were gains on chromosomes 1, 12, 18 (41% each), 7 (23%), and 11 (18%) and losses involving chromosomes 6 (59%), 18, and 20 (18% each). Commonly involved regions were +12q (41%), +18q (36%), +1q (32%), and +7q (23%), as well as −6q (50%), −6p (18%), −17p, and −18p (14% each). High-level gains were found on 7 chromosomes, mainly involving chromosomes 18q (23%), 12q (18%), and 1q (14%). Minimal common regions of over- and underrepresentation were found on +1q25–31, −6q16–21, +7q11.2, +12p11.2–13, +12q12–14, +12q22–24.1, and +18q12.2–21.3. A significant correlation between loss of DNA copy numbers on chromosome 6q and shorter survival could be established (10.2 vs. 22.3 months; P < 0.05). Our findings suggest that chromosomal imbalances of primary central nervous system lymphomas are similar to those of diffuse large B cell lymphomas at other locations and are probably not related to cerebral presentation; however, they may be prognostically relevant.
Primary central nervous system lymphomas (PCNSL) are defined as extranodal malignant lymphomas presenting in the central nervous system (CNS) in the absence of obvious lymphoma outside the nervous system at the time of diagnosis. Approximately 98% of them are B cell lymphomas with immunohistochemical expression of pan-B markers such as CD20; diffuse large cell lymphomas are the most common subgroup. 1,2 The incidence of PCNSL has been increasing recently in both immunosuppressed and immunocompetent patients from 0.8–1.5% up to 6.6% of primary intracranial neoplasms in some studies. 3-5 The peculiar clinicopathological setting of PCNSL suggests the presence of distinct molecular aberrations underlying their pathogenesis. However, the cytogenetic and molecular genetic profile of PCNSL is still virtually unknown. 6
Comparative genomic hybridization (CGH) is a recently developed technique that identifies imbalances of the entire genome in terms of DNA copy number changes. Its main advantage is that it bypasses the need for cell culture to harvest metaphase spreads. CGH has previously been applied to nodal and other extracerebral lymphomas 7-15 and B-cell leukemias. 14,16,17 However, it has hitherto not been used for the assessment of PCNSL.
To screen PCNSL for DNA copy number changes that may show the location of relevant oncogenes and tumor suppressor genes, to compare our findings with the data gained from extracerebral lymphomas, and to correlate chromosomal gains and losses with clinical features, we applied CGH on primary high-grade non-Hodgkin‘s diffuse large B-cell lymphomas (DLCL) of the brain obtained from 22 immunocompetent patients.
Materials and Methods
Patients and Tumors
Formalin-fixed, paraffin wax-embedded biopsy specimens from 22 patients (6 men, 16 women; mean age 63.5 ± 13.2 years; range, 33–84 years) suffering from previously untreated primary CNS non-Hodgkin’s lymphomas (NHL) were analyzed (Table 1) ▶ . None of the patients suffered from apparent immunodeficiency. The diagnosis of PCNSL was established according to the revised European-American classification of lymphoid neoplasms (REAL classification). 18 All 22 cases were DLCL. Only tumor samples that had been shown histologically to contain more than 50% tumor cells were included. Routine hematoxylin and eosin staining and immunohistochemistry using an avidin-biotin complex (ABC) technique and monoclonal antibodies against CD20 (clone L26) as well as the proliferation antigen Ki-67 (clone MIB-1) were performed. All lymphomas showed a positive immunoreaction for B-cell antigen CD20. Furthermore, all available clinical data were reviewed.
Table 1.
Summary of Findings
| Case No. | Age (yrs.) | Sex | MIB | Gains | Losses | Survival (months) |
|---|---|---|---|---|---|---|
| 1 | 77 | M | 61.3 | 18p+q | 3p,6q,10p | 21 |
| 2 | 72 | F | 62.2 | 6p,7q,9q,12q,18q | 1q,4q,6q,18p | >13 |
| 3 | 33 | F | 51.6 | 1q,11q,18q,20q | 1p+q,6p,8p,11p,13q,17p,20p | >24 |
| 4 | 84 | F | 53.3 | 1q,12q,22q | 5q,6q | nd |
| 5 | 47 | M | 62.2 | 12p+q,21q | 15q | >55 |
| 6 | 51 | F | 77.0 | 18p+q | 6q,9q | 3 |
| 7 | 41 | F | 59.8 | − | − | >23 |
| 8 | 65 | F | 62.5 | 3q,17q,18p+q | 6p+q,17p | nd |
| 9 | 65 | F | 56.5 | 2q,7q,10q | 20p | 22 |
| 10 | 68 | F | 49.4 | 1p,6p,11p+q,12p+q,16p,17p | 6q | 6 |
| 11 | 63 | F | 40.7 | − | 18q,20q | nd |
| 12 | 70 | F | 54.0 | 1q,18q,22q | 6q | nd |
| 13 | 75 | F | 38.5 | 3q,7q,12q,18q | 6p,14q,17p,18p | 6 |
| 14 | 55 | M | 72.6 | 1q,16p+q | 8p | >19 |
| 15 | 68 | M | 66.9 | 1q,7q,13q,18q | 2p,6q,18p | 16 |
| 16 | 51 | F | 50.4 | 11q,12p+q | − | nd |
| 17 | 65 | M | 58.4 | 3p+q,7p+q,11q,12p+q,14q,18p | 6p+q | nd |
| 18 | 80 | F | 49.1 | 1q,4q,6p,7p+q,12p+q | 6q,15q | 2 |
| 19 | 74 | F | 56.2 | 1q,20p+q,21q | 1p,3q | nd |
| 20 | 54 | M | 50.4 | − | − | 12 |
| 21 | 78 | F | 57.5 | 1p,12q | 6q,11q | nd |
| 22 | 62 | F | 43.2 | − | 20q | 17 |
Bold Face type denotes high-level gains. MIB, MIB-1 proliferation index (per cent); >, still alive at time of investigation, nd, no data available.
CGH Analysis
DNA was isolated by phenol-chloroform extraction according to standard protocols. With minor modifications, CGH analysis was performed as described by du Manoir et al. 19 Briefly, tumor DNA was labeled with biotin-16-dUTP (Boehringer Mannheim, Mannheim, Germany) and reference DNA from a healthy male donor with digoxigenin-11-dUTP (Boehringer Mannheim) in a standard nick translation reaction. The DNase concentration in the labeling reaction was adjusted to reveal an average fragment size of 200 to 500 bp. The labeled DNA fragments were purified from remaining nucleotides by column chromatography.
For CGH, 500 ng of tumor DNA, 300 ng of reference DNA, and 30 μg of human Cot1 DNA (Gibco, Karlsruhe, Germany) were coprecipitated and redissolved in 10 μl of hybridization buffer. Denaturation of DNA at 75°C for 5 minutes was followed by a preannealing time of 45 minutes at 37°C. Target metaphase spreads (46,XY), which had been prepared following standard procedures, were denatured separately in 70% formamide/2× SSC for 2 minutes at 72°C. Hybridization was allowed to proceed for 3 to 4 days. Posthybridization washes were carried out to a stringency of 50% formamide/2× SSC at 45°C and 0.1× SSC at 60°C. Biotinylated and digoxiginated sequences were detected simultaneously, using avidin-fluorescein isothiocyanate (FITC; Boehringer Mannheim, 1:200) and anti-digoxigenin-rhodamine (Boehringer Mannheim, 1:40). The slides were counterstained with diauridino-phenylindol (DAPI) and mounted in an antifade solution (Vectashield, Vector Laboratories).
Microscopy and Digital Image Analysis
Separate digitized gray level images of DAPI, FITC, and rhodamine fluorescence were taken with a CCD camera connected to a Leica DMRBE microscope (Leica, Wetzler, Germany). The image processing was carried out by use of Applied Imaging software (Applied Imaging, Sunderland, UK). Average green-red ratios were calculated for each chromosome in at least 10 metaphases.
Statistical Thresholds and Controls
Chromosomal regions with CGH ratio profiles surpassing the 50% CGH thresholds (upper threshold 1.25, lower threshold 0.75) were defined as loci with copy number gains or losses. Based on experiments with normal control DNA, these thresholds have been shown to eliminate false positive results. These values have been used in several studies comparing CGH data with results obtained by other cytogenetic methods and have proven to provide robust criteria for the diagnosis of chromosomal gains and losses. Overrepresentations were diagnosed as high-level gains or amplifications when the fluorescence intensity levels exceeded 1.5 15 or when the FITC fluorescence showed strong focal signals. For the assignment of these high-level amplifications to chromosomal bands, the signal intensities were compared to the DAPI banding on individual chromosomes. As tumor specimens and normal DNA were not sex-matched, X and Y chromosomes were excluded. Also excluded were centromeric and satellite regions of the acrocentric chromosomes and chromosome 19, because of the abundance of highly repetetive DNA sequences and the frequent occurrence of false positive CGH results as shown by interphase fluorescence in situ hybridization using suitable DNA probes. Student’s t-test was used to prove significance.
Results
Our CGH investigation of primary DLCL of the brain revealed DNA copy number changes in 20 of 22 patients (91%; Table 1 ▶ ). In two cases (7 and 20) no chromosomal gains or losses were found by CGH analysis, whereas one case (3) showed a maximum of 12 changes in total. An average of 5.5 chromosomal changes per tumor was found, consisting of more gains (mean, 3.5) than losses (mean, 2.0) of genetic material. The most frequent changes were gains of DNA copies on chromosomes 1, 12, and 18 in 41% each as well as on chromosomes 7 (23%) and 11 (18%; Figure 1 ▶ ). Losses most commonly involved chromosomes 6 (59%), 18, and 20 (18% each). The most frequently affected chromosomal regions were +12q (41%), +18q (36%), +1q (32%), and 7q (23%), as well as −6q (50%), −6p (18%), −17p, and −18p (14% each); a typical CGH image and profile is shown for case 15 in Figure 2 ▶ . High-level gains were found on 7 chromosomes, 1, 3, 7, 11, 12, 17, and 18, mainly involving chromosomes 18q (5 high-level gains), 12q (4), and 1q (3). On each of the frequently affected chromosomes, CGH analysis allowed us to delineate the following minimal common regions of over- and under-representation: +1q25–31, −6q16–21, +7q11.2, +12p11.2–13, +12q12–14, +12q22–24.1, and +18q12.2–21.3.
Figure 1.
Summary of gains and losses of DNA sequences identified by CGH. Gains are shown as black bars on the right side of the chromosome ideogram and losses on the left. High-level amplifications are marked as white inlays within the black bars. Each vertical represents the affected chromosomal region seen in a single tumor specimen in case number order.
Figure 2.
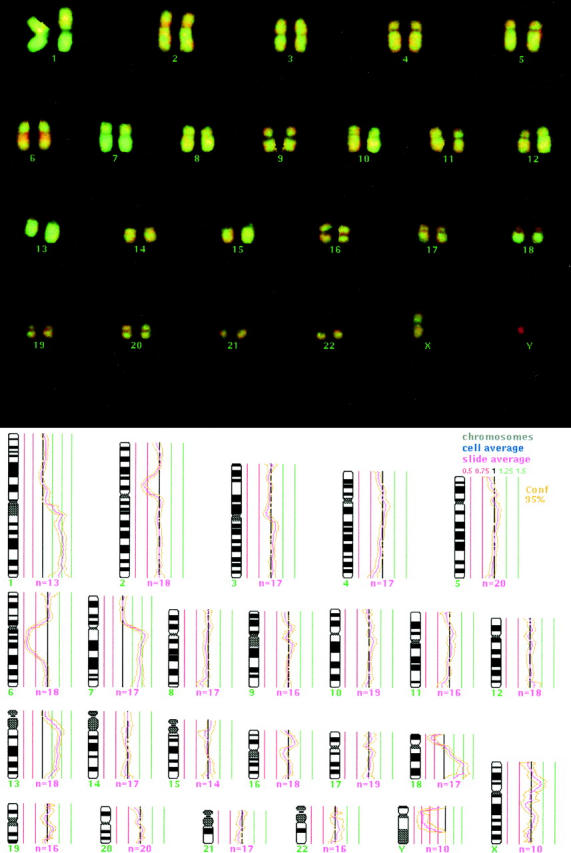
Two-color CGH image of hybridized chromosomes of case 15 with computer-generated CGH ratio superimposed. Green regions represent gains, red regions losses (top). Calculated CGH profile shows gains on 1q, 7q, 13q, and 18q as well as losses on 2p, 6q, und 18p. Average ratio profile of autosomal chromosomes is depicted with a 95% confidence interval. The ratios are plotted alongside the chromosome ideogram. A balanced copy number has a baseline ratio of 1, represented by the central black line; thresholds of copy number gains (1.25) and losses (0.75) are shown (bottom).
The respective MIB-1 proliferation indices were 56.1 ± 9.7%, ranging from 38.5 to 77.0%. The average survival time for patients suffering from primary cerebral DLCL was 17.1 months. At the time of the survey, nine patients had died of the disease after 2 to 22 months, whereas five were still alive after 13 to 55 months (4 free of disease, 1 (case 7) with residual tumor). Eight other patients had been lost to follow-up and no clinical data could be obtained for them. Recurrences occurred in five patients (cases 9, 13, 15, 20, and 22), all of whom died after 6 to 22 months.
The only statistically significant correlation between shorter survival and loss of DNA could be found for chromome −6q (P = 0.045). Loss on chromosome 6p and gains on chromosomes 1, 7, 12, and 18 or their respective long arms as well as combinations of the above imbalances did not significantly correlate with survival, nor did the sum of chromosomal imbalances (gains plus losses), the number of gains alone, or the number of losses alone. Furthermore, no correlations could be found between any of the chromosomal changes and tumor proliferation, gender, or age of the patient, or between Ki-67/MIB-1 proliferation index and survival. Moreover, neither clusters of specific gains or losses nor specific combinations of chromosomal changes could be identified.
Discussion
The rapidly growing number of studies applying CGH to different entities impressively demonstrates the potential of this approach to detect chromosomal gains and losses in tumor genomes. No CGH study on PCNSL has been reported in the literature, and conventional and interphase cytogenetic as well as molecular data on PCNSL have also been scarce. Cytogenetic analyses of single cases of PCNSL in non-AIDS patients revealed gains of 6q, 7p, and chromosome 12 and losses of chromosomes 6, 7, 8, 14q, and 19 as well as translocations (1;14), (6;14), (13;18), and (14;21), findings similar to those observed in nodal B cell lymphoma. 6,20 In comparison with systemic DLCL where amplifications for CDK4, BCL2, MDM2, MYC and REL have been demonstrated, 8,9,13,14,21,22 the mutational spectrum of oncogenes and tumor suppressor genes in PCNSL is still largely unknown. 6 CDKN2A and CDKN2B mutations were found in 4 of 5 PCNSL 23 whereas TP53 mutations were found in 2 of 5 sporadic PCNSL. 24 Cobbers et al, 25 in their analysis of 20 PCNSL of immunocompetent patients, found CDKN2A deletions in 50% but detected no amplifications for CDK4, CCND1, BCL2, MDM2, MYC or REL, and only one case showed a TP53 mutation. Larocca et al 26 found a frequent association of PCNSL with BCL6 mutations but no alterations for C-MYC or BCL2. Immunohistochemical studies on PCNSL of non-AIDS cases reported varying overexpression of p53 (50%, 25 30%, 27 and none 28 ), bcl-2, 25-29 and bcl-6 26,27 whereas c-myc, 27 mdm2, 25,27 p16, 25 and cyclin D1 25 were not expressed.
One of the mechanisms for activating proto-oncogenes is gene amplification resulting in an enhanced expression of the corresponding gene product. In non-Hodgkin’s lymphomas such gene amplifications have rarely been identified. Using CGH, a technique that has not only proven to be very sensitive for the detection of high-level DNA amplifications of units as small as 90 kb 30 but also points to the chromosomal localization of the amplified sequences, we analyzed 22 primary CNS non-Hodgkin’s lymphomas of DLCL histological subtype. As shown in previous studies on systemic lymphomas, chromosomal gains were more frequent than losses. 9,10,12,15 The most common imbalances involved gains on chromosomes 1, 12, 18, and 7 (high-level gains mainly affected 18q, 12q, and 1q) as well as losses on chromosomes 6, 18, and 20. The most frequently involved chromosomal regions were +12q, +18q, +1q, and +7q, as well as −6q, −6p, −17p, and −18p. Similar changes were observed in a series of 32 DLCL, albeit extracerebral, in which CGH revealed DNA copy number gains on chromosomes 1q, 3, 6p, 7, 11, 12, and 18, as well as losses on 6q, 1p, and 8p. 10
Amplifications of 18q and BCL2 (18q21) are commonly found in nodular DLCL and suggest that, in addition to 14/18 translocation, BCL2 amplification might be another mechanism for BCL2 protein overexpression. BCL2 is a proto-oncogene that is known to inhibit apoptosis and deregulation plays an important role in many cases of DLCL 10 as well as most cases of nodular follicle center lymphoma 7,11 and marginal zone B cell lymphoma. 12,31 However, two recent studies on PCNSL in immunocompetent patients found no amplifications, mutations, or other alterations of BCL2, 25,26 whereas overexpression of bcl-2 protein was shown to be consistently present by several authors. 25-29
Findings similar to ours were also reported for DLCL of the gastrointestinal tract, where CGH and fluorescence in situ hybridization revealed gains on 1q and 12 and losses on 6q and 17p 15,21 ; gains on chromosome 12 were found in 10/10 15 and 9/31 21 cases, respectively. Our identification of amplified genes on chromosome 12, in particular 12q, is in accordance with previous studies, as amplification on chromosome 12 was found to be a common cytogenetic finding in nodal B cell neoplasms; these changes have also been documented by CGH investigations performed on B cell chronic lymphatic leukemia, 16,17 follicular lymphomas, 7 and mediastinal thymic B cell lymphomas. 9 Several candidate genes are located on chromosome 12 and have been proposed to play a role in tumorigenesis: CCND2 (12p13), FGF6 (12p13), KRAS2 (12p12.1), CDK4 (12q13), MDM2 (12q13–14), and GLI (12q13–14) 14,16,22 ; however, no CDK4 amplification was found in PCNSL by Cobbers and coworkers. 25 Thus, the relevant genes on 12q involved in PCNSL pathogenesis remain to be determined.
Gains on 1q were among the most common changes and have also been found by CGH to be present in nodular DLCL 10 and DLCL of the gastrointestinal tract 15,21 as well as marginal zone B cell lymphomas 12,31 and follicular lymphomas. 32 A candidate gene on 1q23–31, a frequently highly amplified region, has not been put forward; however, it corresponds to the location of the proto-oncogene TRK/TRKC. The next most frequent gains after chromosomes 1, 12, and 18 affected amplification on 7q, which has also been found on a number of B cell neoplasms in general 14 as well as extranodal systemic DLCL, 8,10 follicular lymphomas, 11 high-grade MALT, 15 and mediastinal thymic B cell lymphomas. 9 Here, too, candidate proto-oncogenes and tumor suppressor genes have not yet been located.
Gains of 3q in our study have been found in only 3 of 22 patients; all of them, however, showed copy number changes in the region of BCL6 (3q27), 2 as high-level amplifications. Similar findings have been reported for PCNSL 26 as well as for non-Hodgkin’s lymphomas in general, 33,34 systemic and gastrointestinal DLCL, 10,21 and marginal zone B cell lymphomas. 12,31
Deletion of 6q was found to be the single most common chromosomal change among our patients and has so far also frequently been discovered in other hematological malignancies and solid tumors. 10 It is a recurrent cytogenetic event in many B cell neoplasms and three regions have been isolated, possibly containing different genes involved in lymphoma development 35 ; 27% of NHL had structural abnormalities of chromosome 6, which are among the most common recurring karyotypic abnormalities in NHL, most of them 6q deletions the frequency of which ranged from 14 to 31% in six large series of NHL. 36 Deletions of 6q have been found almost consistently in nodal and extranodal DLCL, 8,10,15,21 follicular lymphomas, 7,11 and chronic B cell leukemias. 16 These structural aberrations have occasionally been correlated with clinical features of non-Hodgkin’s lymphoma like tumor progression, transformation,and survival. 32,35 In our study, loss of chromosomal material on 6q was significantly correlated with shorter survival compared to patients without loss of 6q (10.2 vs.22.3 months; P < 0.05).
Frequent chromosomal gains of chromosome 2p have been reported in several studies on nodal and extranodal DLCL, 8 primary mediastinal thymic B-cell lymphomas, 9 and follicular lymphomas 11 and were identified to correspond to amplifications of REL (2p12–13) 8,21,22 and N-MYC (2p24.1). 14,21,22 However, no case of DNA copy number gains on 2p were found in our series, so that amplifications of REL and N-MYC do not seem to play a role in PCNSL, a finding that corroborates data recently published by Cobbers et al 25 and Larocca et al. 26 Similarly, gains on chromosome 8 identified in several studies on follicular lymphomas, 7,11 systemic DLCL, 22 and chronic B cell lymphomas 16 could not be found among our patients.
Current therapy regimens consisting of radiotherapy and/or chemotherapy in non-AIDS-associated PCNSL show response rates of 85% with a median survival of 17 to 44 months 1 and 2- and 5-year survival rates of 40 to 70% and 25 to 45%, respectively. 37,38 Clinically, gene amplifications often have been associated with a more aggressive tumor phenotype and shorter overall survival in several tumor types. 39 Three cytogenetic studies on systemic nodal and extranodal non-Hodgkin’s lymphomas found that in follicular lymphomas, six or more chromosomal breaks and structural abnormalities of chromosomal regions 1p, 6q, or 17p, 32 as well as gains on chromosomes 5, 6, 17, and 18, 33 were associated with a poorer prognosis, whereas a significantly shorter survival in high-grade large B cell lymphomas of the gastrointestinal tract was reported to be associated with two or more chromosomal aberrations. 21 We found that among our patients with PCNSL losses of chromosome 6q were significantly correlated with a shorter median survival of 10.2 months, compared to 22.3 months in patients with unaffected chromosome 6q. However, prognosis did not depend on proliferation index, age, or gender. Furthermore, we could not identify clusters of specific gains or losses, specific combinations of chromosomal changes, or a correlation between chromosomal changes and proliferation. Clearly, possible correlations among these parameters should be reassessed in a larger series of PCNSL.
In conclusion, our results suggest that most of the chromosomal regions affected in PCNSL are similar to those found in systemic extracerebral DLCL, whereas other regions implicated in several investigations on nodal non-Hodgkin lymphoma, eg, gains on chromosomes 2 and 8, do not seem to play a major role in PCNSL. Chromosomal imbalances of PCNSL do not seem to account for cerebral location; however, they may be prognostically relevant.
Acknowledgments
The invaluable help and skillful assistance of Ms. Katja Porthuis and Ms. Ulrike Neubert is appreciated. We also thank all of the heads of neurosurgical departments, in particular Prof. Wassmann (University Hospital, Münster), Prof. Brandt (Bathildis-Hospital, Bad Pyrmont), Prof. Busch (St. Barbara Hospital, Hamm), and Prof. Rama (Paracelsus Hospital, Osnabrück) and the many general practitioners who provided us with surgical specimens and clinical data.
Footnotes
Address reprint requests to Christian H. Rickert, M.D., University of Münster, Institute of Neuropathology, Domagkstrasse 19, D-48129 Münster, Germany. E-mail: rickchr@uni-muenster.de.
Supported by a grant from the Marohn-Stiftung.
References
- 1.Jellinger K, Paulus W: Primary central nervous system lymphoma: an update. J Cancer Res Clin Oncol 1992, 119:7-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgello S: Pathogenesis and classification of primary central nervous system lymphoma: an update. Brain Pathol 1995, 5:383-393 [DOI] [PubMed] [Google Scholar]
- 3.Grant JW, Isaacson PG: Primary central nervous system lymphoma. Brain Pathol 1992, 2:97-109 [DOI] [PubMed] [Google Scholar]
- 4.Miller DC, Hochberg FH, Harris NL, Gruber ML, Louis DN, Cohen H: Pathology with clinical correlations of primary central nervous system non-Hodgkin’s lymphoma: the Massachusetts General Hospital experience 1958–1989. Cancer 1994, 74:1383-1397 [DOI] [PubMed] [Google Scholar]
- 5.Cote TR, Manns A, Hardy CR, Yellin FJ, Hartge P: Epidemiology of brain lymphoma among people with or without acquired immunodeficiency syndrome. AIDS/Cancer Study Group. J Natl Cancer Inst 1996, 88:675-679 [DOI] [PubMed] [Google Scholar]
- 6.Paulus W, Jellinger K, Morgello S: Malignant lymphomas. Pathology and Genetics of Tumours of the Nervous System. Edited by Kleihues P, Cavenee WK. Lyon, France, International Agency for Research on Cancer, 1997, pp 154–159
- 7.Bentz M, Werner CA, Döhner H, Joos S, Barth TFE, Siebert R, Schröder M, Stilgenbauer S, Fischer K, Möller P, Lichter P: High incidence of chromosomal imbalances and gene amplifications in the classical follicular variant of follicle center lymphoma. Blood 1996, 88:1437-1444 [PubMed] [Google Scholar]
- 8.Houldsworth J, Mathew S, Rao PH, Dyomina K, Louie DC, Parsa N, Offit K, Chaganti RSK: Rel proto-oncogene is frequently amplified in extranodal diffuse large cell lymphoma. Blood 1996, 87:25-29 [PubMed] [Google Scholar]
- 9.Joos S, Otano-Joos MI, Ziegler S, Brüderlein S, du Manoir S, Bentz M, Möller P, Lichter P: Primary mediastinal (thymic) B-cell lymphoma is characterized by gains of chromosomal material including 9p and the REL gene. Blood 1996, 87:1571-1578 [PubMed] [Google Scholar]
- 10.Monni O, Joensuu H, Franssila K, Knuutila S: DNA copy number changes in diffuse large B-cell lymphoma: comparative genomic hybridization study. Blood 1996, 87:5269-5278 [PubMed] [Google Scholar]
- 11.Avet-Loiseau H, Vigier M, Moreau A, Mellerin MP, Gaillard F, Harousseau JL, Bataille R, Milpied N: Comparative genomic hybridization detects genomic abnormalities in 80% of follicular lymphomas. Br J Haematol 1997, 97:119-122 [DOI] [PubMed] [Google Scholar]
- 12.Dierlamm J, Rosenberg C, Stul M, Pittaluga S, Wlodarska I, Michaux L, Dehaen M, Verhoef G, Thomas J, de Kelver W, Bakker-Schut T, Cassiman JJ, Raap AK, De Wolff-Peeters C: Van den Berghe H, Hagemeijer A: Characteristic pattern of chromosomal gains and losses in marginal zone B cell lymphoma detected by comparative genomic hybridization. Leukemia 1997, 11:747-758 [DOI] [PubMed] [Google Scholar]
- 13.Monni O, Joensuu H, Franssila K, Klefstrom J, Alitalo K, Knuutila S: BCL2 overexpression associated with chromosomal amplification in diffuse large B-cell lymphoma. Blood 1997, 90:1168-1174 [PubMed] [Google Scholar]
- 14.Werner CA, Döhner H, Joos S, Trümper LH, Baudis M, Barth TFE, Ott G, Möller P, Lichter P, Bentz M: High-level DNA amplifications are common genetic aberrations in B-cell neoplasms. Am J Pathol 1997, 151:335-342 [PMC free article] [PubMed] [Google Scholar]
- 15.Chan WY, Wong N, Chan ABW, Chow JHS, Lee JCW: Consistent copy number gain in chromosome 12 in primary diffuse large cell lymphomas of the stomach. Am J Pathol 1998, 152:11-16 [PMC free article] [PubMed] [Google Scholar]
- 16.Bentz M, Huck K, du Manoir S, Joos S, Werner CA, Fischer K, Döhner H, Lichter P: Comparative genomic hybridization in chronic B-cell leukemias shows a high incidence of chromosomal gains and losses. Blood 1995, 85:3610-3618 [PubMed] [Google Scholar]
- 17.Brito-Babapulle V, Garcia-Marco J, Maljaie SH, Hiorns L, Coignet L, Conchon M, Catovsky D: The impact of molecular cytogenetics on chronic lymphoid leukaemia. Acta Haematol 1997, 98:175-186 [DOI] [PubMed] [Google Scholar]
- 18.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, De Wolf Peeters C, Falini B, Gatter KC, Grogan TM, Isaacson PG, Knowles DM, Mason DY, Müller-Hermelink HK, Pileri SA, Piris MA, Ralfkiaer E, Warnke RA: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 [PubMed] [Google Scholar]
- 19.du Manoir S, Speicher MR, Joos S, Schrock E, Popp S, Dohner H, Kovacs G, Robert Nicoud M, Lichter P, Cremer T: Detection of complete and partial chromosome gains and losses by comparative genomic in situ hybridization. Hum Genet 1993, 90:590-610 [DOI] [PubMed] [Google Scholar]
- 20.Itoyama T, Sadamori N, Tsutsumi K, Tokunaga Y, Soda H, Tomonaga M, Yamamori S, Masuda Y, Oshima K, Kikuchi M: Primary central nervous system lymphomas: immunophenotypic, virologic, and cytogenetic findings of three patients without immune defects. Cancer 1994, 73:455-463 [DOI] [PubMed] [Google Scholar]
- 21.Barth TF, Döhner H, Werner CA, Stilgenbauer S, Schlotter M, Pawlita M, Lichter P, Möller P, Bentz M: Characteristic pattern of chromosomal gains and losses in primary large B-cell lymphomas of the gastrointestinal tract. Blood 1998, 91:4321-4330 [PubMed] [Google Scholar]
- 22.Rao PH, Houldsworth J, Dyomina K, Parsa NZ, Cigudosa JC, Louie DC, Popplewell L, Offit K, Jhanwar SC, Chaganti RS: Chromosomal and gene amplification in diffuse large B-cell lymphoma. Blood 1998, 92:234-240 [PubMed] [Google Scholar]
- 23.Kumanishi T, Zhang S, Ichikawa T, Endo S, Washiyama K: Primary malignant lymphoma of the brain: demonstration of frequent p16 and p15 gene deletions. Jpn J Cancer Res 1996, 87:691-695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koga H, Zhang S, Ichikawa T, Washiyama K, Kuroiwa T, Tanaka R, Kumanishi T: Primary malignant lymphoma of the brain: demonstration of the p53 gene mutations by PCR-SSCP analysis and immunohistochemistry. Noshuyo Byori 1994, 11:151-155 [PubMed] [Google Scholar]
- 25.Cobbers JMJL, Wolter M, Reifenberger J, Ring GU, Jessen F, An HX, Niederacher D, Schmidt EE, Ichimura K, Floeth F, Kirsch L, Borchard F, Louis DN, Collins P, Reifenberger G: Frequent inactivation of CDKN2A and rare mutation of TP53 in PCNSL. Brain Pathol 1998, 8:263-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larocca LM, Cacello D, Rinelli A, Nori S, Antinori A, Gloghini A, Cingolani A, Migliazza A, Saglio G, Cammilieri-Broet S, Raphael M, Carbone A, Gaidano G: The molecular and phenotypic profile of primary central nervous system lymphoma identifies distinct categories of the disease and is consistent with histogenetic derivation from germinal center-related B cells. Blood 1998, 92:1011-1019 [PubMed] [Google Scholar]
- 27.Nozaki M, Tada M, Mizugaki Y, Takada K, Nagashima K, Sawamura Y, Abe H: Expression of oncogenic molecules in primary central nervous system lymphomas in immunocompetent patients. Acta Neuropathol 1998, 505–510 [DOI] [PubMed]
- 28.Deckert-Schlüter M, Rang A, Wiestler OD: Apoptosis and apoptosis-related gene products in primary non-Hodgkin’s lymphoma of the central nervous system. Acta Neuropathol 1998, 96:157-162 [DOI] [PubMed] [Google Scholar]
- 29.Krogh-Jensen M, Johansen P, D’Amore F: Primary central nervous system lymphomas in immunocompetent individuals: histology, Epstein-Barr virus genome, Ki-67 proliferation index, p53 and bcl-2 gene expression. Leuk Lymphoma 1998, 30:131-142 [DOI] [PubMed] [Google Scholar]
- 30.Joos S, Scherthan H, Speicher MR, Schlegel J, Cremer T, Lichter P: Detection of amplified genomic sequences by reverse chromosome painting using genomic tumor DNA as probe. Hum Genet 1993, 90:584-589 [DOI] [PubMed] [Google Scholar]
- 31.Dierlamm J, Pittaluga S, Stul M, Wlodarska I, Michaux L, Thomas J, Verhoef G, Verhest A, Depardieu C, Cassiman JJ, Hagemeijer A, De Wolff-Peeters C: Van den Berghe H: BCL6 gene rearrangements also occur in marginal zone B-cell lymphoma. Br J Haematol 1997, 98:719-725 [DOI] [PubMed] [Google Scholar]
- 32.Tilly H, Rossi A, Stamatoullas A, Lenormand B, Bigorgne C, Kunlin A, Monconduit M, Bastard C: Prognostic value of chromosomal abnormalities in follicular lymphoma. Blood 1994, 84:1043-1049 [PubMed] [Google Scholar]
- 33.Schouten HC, Sanger WG, Weisenburger DD, Anderson J, Armitage JO: Chromosomal abnormalities in untreated patients with non-Hodgkin’s lymphoma: associations with histology, clinical characteristics, and treatment outcome. Blood 1990, 75:1841-1847 [PubMed] [Google Scholar]
- 34.Bastard C, Deweindt C, Kerckaert JP, Lenormand B, Rossi A, Pezzella F, Fruchart C, Duval C, Monconduit M, Tilly H: LAZ3 rearrangements in non-Hodgkin’s lymphoma: correlation with histology, immunophenotype, karyotype, and clinical outcome in 217 patients. Blood 1994, 83:2423-2427 [PubMed] [Google Scholar]
- 35.Offit K, Parsa NZ, Gaidano G, Filippa DA, Louie D, Pan D, Jhanwar SC, Dalla-Favera R, Chaganti RSK: 6q deletions define distinct clinico-pathologic subsets of non-Hodgkin’s lymphoma. Blood 1993, 82:2157-2162 [PubMed] [Google Scholar]
- 36.Offit K, Chaganti RSK: Chromosomal aberrations in non-Hodgkin’s lymphoma: biologic and clinical correlations. Hematol Oncol Clin North Am 1991, 5:853-869 [PubMed] [Google Scholar]
- 37.Bessell EM, Graus F, Punt JA, Firth JL, Hope DT, Moloney AJ, Lopez Guillermo A, Villa S: Primary non-Hodgkin’s lymphoma of the CNS treated with BVAM or CHOD/BVAM chemotherapy before radiotherapy. J Clin Oncol 1996, 14:945-954 [DOI] [PubMed] [Google Scholar]
- 38.O’Brien PC, Roos DE, Liew KH, Trotter GE, Barton MB, Walker QJ, Poulsen MG, Olver IN: Preliminary results of combined chemotherapy and radiotherapy for non-AIDS primary central nervous system lymphoma. trans-Tasman Radiation Oncology Group (TROG). Med J Aust 1996, 165:424-427 [PubMed] [Google Scholar]
- 39.Schwab M, Amler L: Amplification of cellular oncogenes: a predictor of clinical outcome in human cancer. Genes Chromosomes Cancer 1990, 1:181-193 [DOI] [PubMed] [Google Scholar]



