Abstract
High-mobility group I (HMGI) proteins are architectural transcription factors expressed predominantly during embryonic development. Their genetic loci are the most frequent targets of chromosomal rearrangements in uterine leiomyomas and other benign tumors. It was therefore suggested that both HMGI genes are involved in the neoplastic transformation of benign tumors. By Western analysis we found that 16 of 33 uterine leiomyomas expressed high levels of HMGI-C or HMGI(Y) proteins, whereas they were not detected in the corresponding myometrium. Immunohistochemistry demonstrated that the expression of HMGI-C is restricted to leiomyoma smooth muscle cells but is not expressed in vascular smooth muscle cells or the connective tissue of the tumor. Northern blotting confirmed the protein expression data for HMGI-C, whereas HMGI(Y) mRNA and protein levels did not correlate, suggesting that posttranscriptional mechanisms are involved in the regulation of HMGI(Y) expression. Three of the uterine leiomyomas analyzed expressed HMGI-C gene products with altered molecular weight. Two of them were proved to consist of the entire DNA-binding domain but lacked sequences of the C-terminal acidic tail. Conversely, other tumors expressed HMGI-C or HMGI(Y) genes that were not affected by mutations of the coding region. Thus we identified uterine leiomyomas that expressed mutated HMGI-C, whereas other uterine leiomyomas expressed wild-type HMGI-C or HMGI(Y). On the basis of our data we assume that the enhanced expression of functionally active HMGI proteins, whether they are wild-type or not, is important for the pathogenesis of uterine leiomyomas.
Uterine leiomyomas are benign smooth muscle cell tumors of the myometrium. They are the most common solid tumors in women, occurring with an incidence of up to 77%. 1 Although they are benign, they can lead to abnormal uterine bleeding, pelvic pain, spontaneous abortion, and possibly cause infertility. Because effective medical treatment is not available, the ultimate treatment is hysterectomy or, in some cases, myomenucleation. Up to now, the molecular pathogenesis of these tumors has been unknown.
The neoplastic transformation of normal myometrium to leiomyoma tissue probably involves somatic mutations and deregulation of genes crucial for cell growth and differentiation. Approximately 36% of uterine leiomyomas contain chromosomal abnormalities. 2 Translocations involving chromosome 12q13–15 are the most frequent chromosomal alterations in uterine leiomyomas and are frequently observed in lipomas, hamartomas of the lung and breast, fibroadenomas of the breast, and endometrial polyps. 2-4 These tumors have the common property of being of mesenchymal origin and benign. Recently it was found that the gene coding for HMGI-C is disrupted by 12q13–15 rearrangements. 2-7 A closely related gene, HMGI(Y), coding for the two proteins HMGI and HMGY, which are generated by an alternative splicing mechanism, is located on chromosome 6p21 and is also a common target of chromosomal rearrangements in uterine leiomyomas and other benign tumors. 8-12 It was therefore suggested that both genes, HMGI-C and HMGI(Y), may play a causative role in the development of benign tumors.
HMGI-C and HMGI(Y) proteins consist of a DNA-binding domain within the N-terminus, which is called an AT-hook, and an acidic tail in the C-terminus. The chromosomal breakpoints of HMGI-C and HMGI(Y) rearrangements in uterine leiomyomas have been mapped either within the coding region or in the 3′-UTR, but can also be found in the 5′ region of the gene more than 100 kb upstream of the transcription start site. 2,4,6,10,13 Translocations within the coding region generate truncated or chimeric genes in which the N-terminal DNA-binding domain is separated from the C-terminal acidic tail of the protein and fused to ectopic sequences. Transcripts derived from chimeric or truncated HMGI-C and HMGI(Y) genes have been detected in uterine leiomyomas as well as other benign tumors by 3′-rapid amplification of cDNA ends (RACE). 3-5,9,11,13
HMGI proteins bind to the minor groove of AT-rich DNA sequences, thereby inducing a bend within the DNA. 14 Neither of the proteins stimulates initiation of transcription on its own, but both enhance promoter binding of transcriptional activators like ATF2, Elf-1, NF-κB, SRF, and Tst-1/Oct-6. 15-22 These properties classify HMGI proteins as architectural transcription factors that contribute to the transcriptional activation of specific genes by their ability to organize the framework of transcriptional initiation complexes. 23,24 On other promoters HMGI proteins can function as transcriptional repressors. 25-27 These promoters are thought to be induced by a release of HMGI-C or HMGI(Y) and the subsequent binding of sequence-specific transcriptional activators.
Several lines of evidence point to an important role for HMGI proteins in cell differentiation. Both proteins are expressed predominantly during embryonic stages. In adult stages only a very low ubiquitous expression has been reported for HMGI(Y). 28 HMGI-C is not detectable in differentiated adult tissues. 29-31 In contrast, HMGI(Y) and/or HMGI-C is expressed at high levels in human prostate carcinomas, 32 human thyroid carcinomas, 33 human colorectal carcinomas, 34 human breast cancer, 35 and human uterine cervix carcinomas. 36 In addition, a marked decrease in HMGI(Y) expression was observed during the differentiation of mouse F9 teratocarcinoma cells. 37 Finally, inactivation of the HMGI-C gene in mice induces a pygmy phenotype that is caused by growth retardation of mesenchymal tissues. 38
In the present study we analyzed HMGI-C and HMGI(Y) expression in myometrial and uterine leiomyoma tissues derived from 33 patients undergoing hysterectomy and obtained evidence that not only mutated but also wild-type HMGI proteins may play an important role in the transformation process of uterine leiomyomas.
Materials and Methods
Tissue Samples
Tumor samples and their corresponding normal myometrial tissues were obtained from women undergoing hysterectomy for uterine leiomyomas. Immediately after receipt, part of each sample was frozen in liquid nitrogen and stored at −80°C for mRNA or protein extraction. For immunohistochemistry a second part was fixed in 4% formaldehyde or embedded in TissueTek (Miles Diagnostic), frozen in dry ice, and stored at −80°C. All samples were collected with the approval of the local ethical committee.
Western Blot Analysis
Liquid nitrogen frozen tissue samples were pulverized and homogenized in sodium dodecyl sulfate (SDS) loading buffer (0.1 mol/L Tris, pH 6.8, 4% SDS, 0.1% Bromphenol-blue, 20% glycerol, 0.2% dithiothreitol). HepG2 cells (American Type Culture Collection) were lysed in SDS loading buffer in a concentration of 2000 cells/μl. Samples were boiled for 10 minutes at 95°C, cleared by centrifugation (10 minutes, 12°C, 100,000 × g), and stored at −80°C. The extracted proteins were separated on a 15% polyacrylamide/0.1% SDS gel. To check for equal protein loading the separated proteins were visualized by Coomassie staining. For HMGI protein detection the proteins were electrotransferred to a nitrocellulose membrane (Amersham-Buchler). Nonspecific interactions were blocked by preincubating the membranes with 0.75% blocking reagent (Boehringer Mannheim) in phosphate-buffered saline (PBS) for 1 hour at room temperature. The membranes were then incubated with primary HMGI-C antibodies (crude rabbit anti-peptide serum, 1:5000 in 0.75% blocking reagent) or HMGI(Y) N-19 antibodies (Santa Cruz; 1:4000 in 0.75% blocking reagent) overnight, followed by an additional incubation with horseradish peroxidase-conjugated secondary antibodies (Amersham-Buchler; 1:3000 in 0.75% blocking reagent) for 4 hours. After each antibody incubation the membranes were washed three times with PBS. Bound antibodies were visualized using the ECL detection system (Amersham-Buchler). The anti-peptide antibodies specific for HMGI-C were obtained by immunization of rabbits with the synthetic peptide GAGQPSTSAQGQ (Eurogentec).
Preparation of HMGI-C and HMGI(Y) Northern Blot Probes
The coding region of human HMGI-C and HMGI(Y) was amplified from total RNA of the cell line HepG2 (American Type Culture Collection) by reverse transcriptase polymerase chain reaction (RT-PCR), using the gene-specific primers ICTr.up (5′-CAGGATGAGCGCACGCGGTGAG) and Ex5.dw for HMGI-C or Y.up and Y.dw for HMGI(Y). The PCR products were cloned into plasmid pCR3.1-Uni (Invitrogen), and the correct sequence was confirmed by DNA sequencing. To generate Northern blot probes the cDNA was removed by HindIII/EcoRI digestion, purified by agarose gel electrophoresis, and labeled with 32P, using the Prime it-kit (Stratagene).
Northern Blot Analysis
For the isolation of total RNA, liquid nitrogen-frozen tissue samples were pulverized and dissolved in TRIzol reagent (Life Technologies). RNA isolation was performed according to the manufacturer’s instructions. Twenty micrograms of total RNA per lane was separated under denaturing conditions on a 1.2% agarose/6.3% formaldehyde gel and transferred onto GeneScreen Plus membrane (NEN). The RNA was immobilized by a UV cross-linker (Stratagene). The blots were prehybridized with QuikHyb solution (Stratagene) for 20 minutes at 68°C and hybridized for 2 hours at 68°C with 32P-labeled cDNA probes in QuickHyb solution in the presence of 0.1 mg/ml denatured salmon sperm DNA. Hybridized filters were washed twice for 15 minutes at room temperature in 2× standard saline citrate (SSC) (0.3 mol/L NaCl, 30 mmol/L sodium citrate, pH 7.2)/0.1% SDS and once for 15 minutes at 60°C in 0.1× SSC/0.1% SDS and exposed to Biomax films (Kodak). Quantification of bound probes was performed with a phosphorimager (Molecular Dynamics). Before reprobing the filters were stripped with 0.1× SSC/0.1% SDS for 30 minutes at 95°C.
RT-PCR
RT-PCR was performed as previously described. 39 Amplification of the HMGI-C DNA-binding motive sequence was performed using primers Ex1.up (5′-CGAAAGGTGCTGGGCAGCTCCGG) and Ex3.dw (5′-CCATTTCCTAGGTCTGCCTCTTG). Amplification of the whole HMGI-C coding sequence was performed using primers Ex1.up and Ex5.dw (5′-CTAGTCCTCTTCGGCAGACTC). Amplification of the whole coding sequence of HMGI(Y) was performed using primers Y.up (5′-GGAAGATGAGTGAGTCGAGCTCG) and Y.dw (5′-GCAGGCGGCACGCATG GGTCA). The quality of the cDNA was proved by amplification of the housekeeping gene cytochrome c oxidase 1A, using primers 1A.up (5′-CGTCACAGCCCATG CATTTG) and 1A.dw (5′-GGTTAGGTCATACGGAGGTCT). The PCR products were separated by 1.0% agarose gel electrophoresis and visualized by ethidium bromide staining. PCR products were sequenced with an ABI 377 sequencer (Perkin Elmer).
Immunohistochemistry
Tissue was fixed for 24 hours in 4% formaldehyde/PBS and embedded in paraffin. For staining of HMGI-C, 5-μm sections were spread on slides (Menzel), dried for 5 days, deparaffinized, partially digested with 0.1% pepsin in 0.1 N HCl for 15 minutes, and rinsed with PBS. Slides were then incubated with normal goat serum for 30 minutes to reduce nonspecific binding of secondary antibodies and with primary HMGI-C antibodies 1:200 in PBS for 1 hour. Slides were then washed two times in PBS/0.2% TWEEN20, incubated with biotinylated goat anti-rabbit secondary antibodies (Zymed, 1:1000 in PBS) for 1 hour, and again washed two times in PBS/0.2% TWEEN20 after incubation with avidin-biotin-peroxidase complexes (Vecstain Elite ABC Kit) for 1 hour. Staining was performed with diaminobenzidin (Zymed Substrate Kit). All steps were performed at room temperature.
Results
Characterization of HMGI-C Antibodies
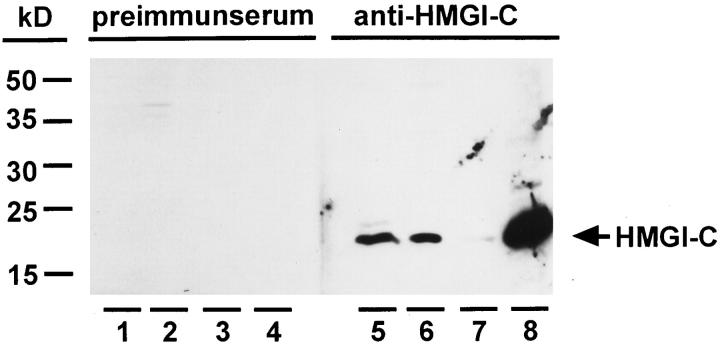
To obtain antibodies raised against HMGI-C, rabbits were immunized with the synthetic peptide GAGQPSTSAQGQ derived from the N-terminus of human HMGI-C. In contrast to preimmunserum, these antibodies detected a single protein that migrated at 18 kd in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Figure 1) ▶ . In accordance with the expression pattern of HMGI-C, 31 this protein is detectable in mouse embryonic tissues as well as various mouse and human cell lines, for example HepG2, 40 but not in adult mouse tissues (Figure 1 ▶ , lanes 5 and 6 and data not shown). Furthermore, we demonstrated that the detected protein is soluble in mouse embryonic whole cell extracts supplemented with 5% trichloroacetic acid (Figure 1 ▶ , lanes 7 and 8), a feature characteristic of high-mobility group proteins. 40,41 Altogether, the migration in SDS-PAGE, the tissue specificity, and the solubility in 5% trichloroacetic acid indicate that the detected protein is HMGI-C.
Figure 1.
Characterization of HMGI-C antibodies. Rabbit preimmunserum (lanes 1–4) was compared with an antiserum raised against a synthetic peptide derived from the N-terminus of HMGI-C (lanes 5–8). Western blot analysis was carried out with 20 μg of whole-cell extract from mouse embryo day 13 p.c. (lanes 1 and 5); whole-cell extract of 4 × 10 4 cells from the human hepatoma cell line HepG2 (lanes 2 and6); 20 μg precipitate of mouse embryo whole-cell extract supplemented with 5% trichloroacetic acid (lanes 3 and7); and 0.5 μg of supernatant of mouse embryo whole-cell extract supplemented with 5% trichloroacetic acid (lanes 4 and8).
HMGI-C Expression in Uterine Leiomyomas
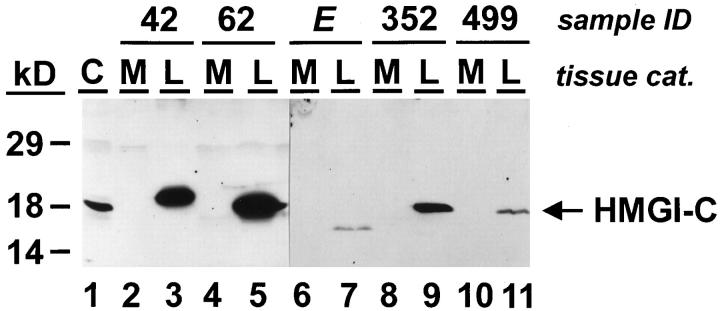
To determine the number of uterine leiomyomas that express HMGI-C we analyzed 33 uterine leiomyoma samples and their normal corresponding myometrial tissue by Western analysis. This study revealed that none of the 33 myometrium samples expressed HMGI-C, whereas high amounts of HMGI-C proteins were detected in nine of 33 uterine leiomyoma samples (Figure 2 ▶ and Table 1 ▶ ). Interestingly, two of these HMGI-C-positive uterine leiomyomas expressed HMGI-C proteins with abnormal molecular masses of 15 kd and 19.5 kd, respectively (Figure 2 ▶ , lanes 3 and 7), whereas the other tumors expressed HMGI-C proteins of the predicted molecular mass of 18 kd (Figure 2 ▶ , lanes 5, 9, and 11). It is noteworthy that these two uterine leiomyomas exclusively expressed the aberrant form of HMGI-C and not the wild-type form of HMGI-C.
Figure 2.
HMGI-C protein expression in uterine leiomyomas (L) and their corresponding myometrium (M). Western analysis was carried out using a polyconal rabbit antibody specific for HMGI-C. Fifty micrograms of whole-cell extracts from human myometrium (lanes 2, 4, 6, 8, and10) and uterine leiomyomas (lanes 3, 5, 7, 9, and11) were analyzed. Ten micrograms of whole-cell extract from the human hepatoma cell line HepG2 (C) was used as a control for HMGI-C expression (lane 1).
Table 1.
HMGI-C and HMGI(Y) Expression in Uterine Leiomyomas and Corresponding Normal Myometrium
| No. | Sample ID | I-C and I(Y) Western | I-C Northern | I(Y) Northern | I-C Immunohisto | ||||
|---|---|---|---|---|---|---|---|---|---|
| M | L | M | L | M | L | M | L | ||
| 1 | 40 | − | I-C | − | + | + | + | − | + |
| 2 | 42 | − | I-C* | − | +* | + | + | − | + |
| 3 | 80 | − | I-C | − | + | + | + | − | + |
| 4 | 246 | − | I-C | − | + | + | + | ||
| 5 | 352 | − | I-C | − | +* | + | + | − | + |
| 6 | 418 | − | I-C | − | + | + | + | − | + |
| 7 | 499 | − | I-C | − | + | + | + | − | − |
| 8 | 62 | − | I-C+I(Y) | − | + | + | + | − | − |
| 9 | 48 | − | I(Y) | − | − | + | + | − | − |
| 10 | 631 | − | I(Y) | − | − | + | + | ||
| 11 | 2273 | − | I(Y) | − | − | + | + | ||
| 12 | 2288 | − | I(Y) | − | − | + | + | ||
| 13 | 1 | − | − | − | − | + | + | − | − |
| 14 | 28 | − | − | − | − | + | + | ||
| 15 | 29 | − | − | − | − | + | + | − | − |
| 16 | E | − | I-C* | − | + | ||||
| 17 | 41 | − | I(Y) | − | − | ||||
| 18 | 55 | − | I(Y) | ||||||
| 19 | 57 | − | I(Y) | ||||||
| 20 | 50 | − | − | − | − | ||||
| 21 | 56 | − | − | − | − | ||||
| 22 | 92 | − | − | ||||||
| 23 | 138 | − | − | − | − | ||||
| 24 | 196 | − | − | ||||||
| 25 | 265 | − | − | − | − | ||||
| 26 | 268 | − | − | ||||||
| 27 | 488 | − | − | − | − | ||||
| 28 | 547 | − | − | ||||||
| 29 | 710 | − | − | ||||||
| 30 | 763 | − | − | ||||||
| 31 | 789 | − | − | ||||||
| 32 | D | − | − | − | − | ||||
| 33 | H | − | − | − | − |
*HMGI-C isoforms with altered molecular mass.
HMGI(Y) Expression in Uterine Leiomyomas
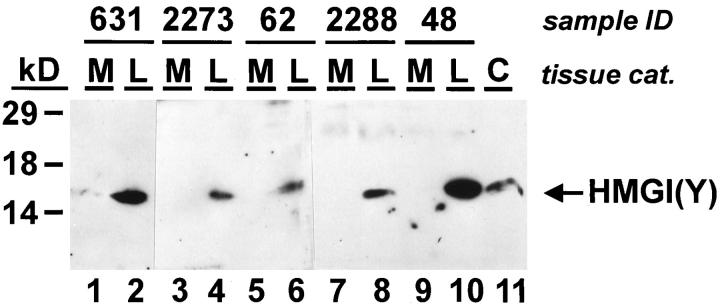
To determine the expression of HMGI(Y) proteins, a commercially available polyclonal antibody raised against the N-terminus of human HMGI(Y) was used. Because of the low sensitivity of this antibody the HMGI(Y) proteins were not detected in normal myometrium (Figure 3 ▶ , lanes 1, 3, 5, 7, and 9), although HMGI(Y) is expressed at low levels in differentiated tissues. 28 In contrast, high levels of HMGI(Y) were found in eight of 33 uterine leiomyomas (Figure 3 ▶ , lanes 2, 4, 6, 8, and 10, and Table 1 ▶ ). As SDS-PAGE does not separate the two protein isoforms of the HMGI(Y) gene (ie, HMGI and HMGY), the band recognized by Western analysis refers to both proteins. None of the tissues analyzed displayed HMGI(Y) proteins with an abnormal molecular mass. Interestingly, only one uterine leiomyoma, sample L62, displayed increased expression of both proteins, HMGI(Y) and HMGI-C (Figure 2 ▶ , lane 5, and Figure 3 ▶ , lane 6), whereas other uterine leiomyomas expressed either HMGI-C or HMGI(Y). Taken together, we found that HMGI-C or HMGI(Y) protein expression was strongly induced in 16 of 33 uterine leiomyomas (Table 1) ▶ .
Figure 3.
HMGI(Y) protein expression in uterine leiomyomas (L) and their corresponding myometrium (M). Western analysis was carried out using a polyconal goat antibody specific for HMGI(Y). Fifty micrograms of whole-cell extracts from human myometrium (lanes 1, 3, 5, 7, and9) and uterine leiomyomas (lanes 2, 4, 6, 8, and10) were analyzed. Whole-cell extract of 2 × 10 4 cells from the human hepatoma cell line HepG2 was used as a control for HMGI-C expression (C, lane 11).
Investigation of HMGI-C and HMGI(Y) mRNA in Uterine Leiomyomas
To confirm these data obtained by Western analysis we selected 15 of the 33 myometrial and uterine leiomyoma samples, of which 12 were positive and three negative for HMGI protein expression to perform Northern analysis (Table 1) ▶ . Examples of this analysis are shown in Figure 4 ▶ . The integrity of the RNA preparations was proved by ethidium bromide staining (not shown) and by hybridization with a cDNA probe for the housekeeping gene cyclophilin 21 (Figure 4, A and B ▶ , lower panel). HMGI-C mRNA expression was analyzed with a probe derived from the coding region of human HMGI-C. As expected, HMGI-C transcripts were expressed in mouse embryonic tissues of day 13 p.c. (Figure 4A ▶ , lane 1), but not in human myometrium (Figure 4A ▶ , lanes 2, 4, 6, 8, and Table 1 ▶ ). High levels of HMGI-C mRNA were found in all of the analyzed tumor samples that expressed HMGI-C proteins in Western analysis (Figure 4A ▶ , lanes 3, 5, 7, 9, and Table 1 ▶ ), whereas no expression was detected in uterine leiomyomas negative for HMGI protein expression and in those tumors exclusively expressing HMGI(Y) (Table 1) ▶ . Interestingly, we found that uterine leiomyoma sample L42, which displayed an altered form of HMGI-C in Western analysis (Figure 2 ▶ , lane 3), expressed a truncated mRNA transcript (Figure 4A ▶ , lane 9). In addition, a truncated HMGI-C mRNA was also observed in uterine leiomyoma L352 (Figure 4A ▶ , lane 5), although this sample expressed HMGI-C proteins of the predicted molecular mass (Figure 2 ▶ , lane 9).
Figure 4.
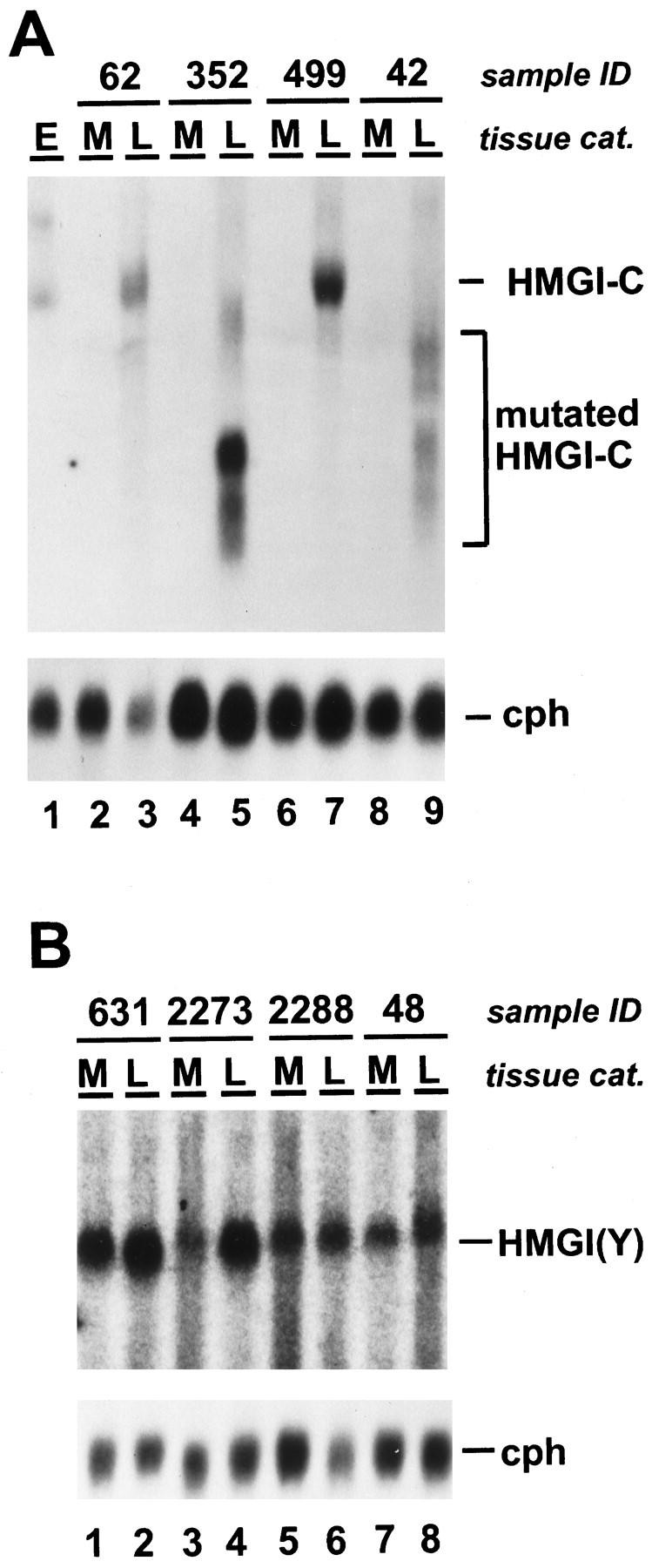
A: HMGI-C mRNA expression in uterine leiomyomas (L) and their corresponding myometrium (M). Northern blot analysis was carried out using probes specific for HMGI-C (upper panel) and the housekeeping gene cyclophilin 23 (lower panel). Total RNA from mouse embryo day 13 p.c. (E) was used as a control for HMGI-C expression (lane 1). Total RNA from myometrium (lanes 2, 4, 6, and8) and uterine leiomyomas (lanes 3, 5, 7, and9) was analyzed. HMGI-C mRNA of the expected size (HMGI-C), truncated HMGI-C mRNA isoforms (mutated HMGI-C), and cyclophilin 23 mRNA (cph) are indicated. B: HMGI(Y) mRNA expression in uterine leiomyomas (L) and their corresponding myometrium (M). Northern blot analysis used probes specific for HMGI(Y) (upper panel) and cyclophilin 23 (lower panel). Total RNA from myometrium (lanes 1, 3, 5, and7) and uterine leiomyomas (lanes 2, 4, 6, and8) was analyzed. HMGI(Y) mRNA and cyclophilin 23 mRNA (cph) are indicated. Cyclophilin 23 was hybridized to control mRNA loading and integrity.
To determine the HMGI(Y) mRNA expression by Northern analysis we used a probe derived from the coding region of human HMGI(Y). All 15 myometrial tissues analyzed displayed a low expression of HMGI(Y) mRNA (Figure 4B ▶ and Table 1 ▶ ), reflecting that the sensitivity of our HMGI(Y) Northern analysis is higher than that of our Western analysis. Surprisingly, we found that HMGI(Y) mRNA expression was not significantly enhanced in any of the five HMGI(Y) protein-positive uterine leiomyomas, as was observed for all of the HMGI(Y) protein-negative uterine leiomyomas (Figure 4B ▶ and Table 1 ▶ ). HMGI(Y) transcripts of altered molecular mass were not detected in any of the samples analyzed.
Sequence-Specific RT-PCR and cDNA Sequencing of HMGI-C and HMGI(Y)
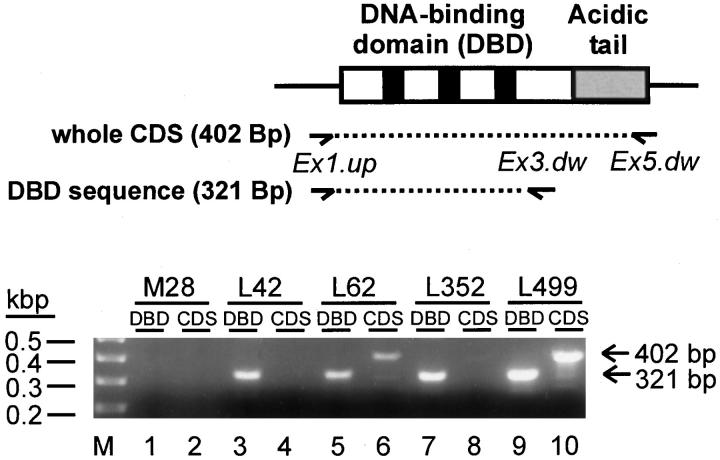
To analyze the truncated HMGI-C transcripts (Figure 4A ▶ , lanes 5 and 9) in more detail and to prove whether the other uterine leiomyomas expressed HMGI-C or HMGI(Y) transcripts that are affected by small mutations that are not visible in Northern and Western analysis, we performed sequence-specific RT-PCR and subsequently sequenced the amplified RT-PCR products. For the amplification of HMGI-C cDNA we used two different primer sets (Figure 5) ▶ . The use of primers Ex1.up and Ex3.dw generates an amplification product of 321 bp that consists of sequences of the HMGI-C DNA-binding domain (DBD). Primers Ex1.up and Ex5.dw generate an amplification product of 402 bp that consists of the whole coding sequence (CDS) of HMGI-C. As expected, neither PCR product was obtained from myometrial tissue M28 (Figure 5 ▶ , lanes 1 and 2). In contrast, the 321-bp and the 402-bp PCR product were amplified from the uterine leiomyomas L62 and L499 (Figure 5 ▶ , lanes 5 and 6, lanes 9 and 10), which are characterized by the expression of HMGI-C mRNA and protein of the predicted molecular mass. Sequencing of the amplified cDNA revealed that both tumors expressed HMGI-C that is not affected by mutations within the coding region. Interestingly, two other uterine leiomyomas, L42 and L352, which are characterized by the expression of truncated HMGI-C mRNAs (Figure 4A ▶ , lanes 5 and 9), showed an amplification of only the shorter 321-bp PCR product that consists of the DNA-binding domain sequence, whereas the larger 402-bp PCR product, which consists of the entire HMGI-C coding sequence, was not amplified (Figure 5 ▶ , lanes 3 and 4, lanes 7 and 8). This result proved that uterine leiomyomas L42 and L352 expressed HMGI-C from a gene that consists of HMGI-C 5′-sequences coding for the DNA-binding domain but did not contain 3′-sequences coding for the HMGI-C acidic tail.
Figure 5.
RT-PCR amplification of the whole coding sequence (CDS) and the DNA-binding domain sequence (DBD) of HMGI-C. Total RNA from human myometrium M28 (lanes 1 and2), uterine leiomyomas L42 (lanes 3 and4), L62 (lanes 5 and 6), L352 (lanes 7 and8), and L499 (lanes 9 and10) was analyzed. The figure illustrates the HMGI-C cDNA binding sites for PCR primers (arrows) and the two PCR-products (dotted lines). Primers Ex1.up bind to exon 1 sequences 5′-ward to the translation start codon, primers Ex3.dw bind to exon 3 sequences in the 3′-region of the DBD sequences, and primers Ex5.dw bind to exon 5 sequences 5′-ward to the HMGI-C CDS. Primer pair Ex1.up and Ex3.dw produces a 321-bp DNA that consists of the entire DBD sequence. Primer pair Ex1.up and Ex5.dw produces a 402-bp DNA that consists of the whole CDS.
To analyze the nucleotide sequence of the coding region of the HMGI(Y) cDNA in uterine leiomyomas, we made a RT-PCR analysis of four uterine leiomyomas that expressed high levels of HMGI(Y) proteins (data not shown). Sequencing of the entire coding region of HMGI(Y) revealed that these uterine leiomyomas exclusively expressed wild-type HMGI(Y) that is not affected by mutations. Taken together, we identified uterine leiomyomas expressing either mutated HMGI-C proteins that consist of the DNA-binding domain but did not contain sequences of the acidic tail or uterine leiomyomas that expressed wild-type HMGI-C or HMGI(Y) not affected by mutations.
HMGI-C Immunohistochemistry on Myometrium and Uterine Leiomyomas
To analyze the cellular distribution of HMGI-C in uterine leiomyomas we performed immunohistochemistry on paraffin sections of 20 uterine leiomyomas and their corresponding myometrium (Table 1) ▶ . The immunostaining confirmed the data obtained by Western and Northern analysis. HMGI-C was not expressed in normal myometrium (Figure 6A ▶ , Table 1 ▶ ) but was clearly detected in uterine leiomyomas that were positive for HMGI-C expression in Western and Northern analysis (Figure 6B ▶ , Table 1 ▶ ). As expected, the expression of HMGI-C was predominantly nuclear, even in tumors that expressed aberrant HMGI-C (Figure 6B) ▶ . Furthermore, the immunostaining revealed that HMGI-C expression is restricted to leiomyoma smooth muscle cells and is not found in vascular smooth muscle cells (Figure 6B, V) ▶ or the connective tissue of the tumor (Figure 6B) ▶ .
Figure 6.
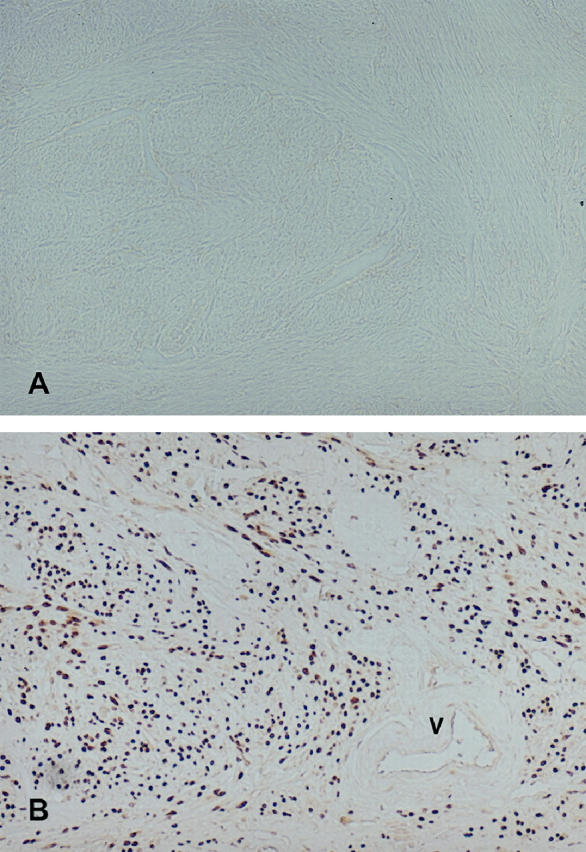
HMGI-C immunohistochemistry (×200) on paraffin sections. A: Human myometrium M42. B: Uterine leiomyoma L42. Brown staining revealed HMGI-C expression. An atherial blood vessle (V) of uterine leiomyoma L42 is indicated.
Discussion
In this report we analyzed the expression of the high-mobility group I proteins HMGI-C and HMGI(Y) in myometrial and uterine leiomyoma tissues derived from 33 patients undergoing hysterectomy. By using HMGI-C- and HMGI(Y)-specific antibodies we could directly demonstrate for the first time that HMGI proteins are expressed in a large subgroup of uterine leiomyomas. Altogether, 48.5% of the 33 tumors analyzed expressed high levels of HMGI-C, HMGI(Y), or both HMGI genes. In contrast, we could not detect HMGI protein expression in any of the corresponding normal myometrial samples. These data prove that not only embryonic tissues 28,31 and some malignant carcinomas 32-36 but also a large subgroup of uterine leiomyomas display increased expression of HMGI genes. It is also in accordance with the report by Williams et al, 10 who found DNA-binding activity for the PRDII motive of the β-interferon promoter in eight of 16 uterine leiomyoma protein extracts but not in protein extracts of the adjacent myometrium. As the PRDII motive cooperatively binds NFκB with either HMGI(Y) 15 or HMGI-C, 22 it is most likely that Williams and co-workers detected HMGI-C and HMGI(Y) proteins in the uterine leiomyomas they analyzed.
The results of various cytogenetic studies have revealed that uterine leiomyomas are affected by chromosomal alterations that target the HMGI-C locus on chromosome 12q14–15 and the HMGI(Y) locus on chromosome 6p21. Some of these chromosomal rearrangements affect the coding region of HMGI-C and HMGI(Y), which leads to the generation of chimeric genes in which an ectopic sequence is fused to the DNA-binding domain of HMGI-C or HMGI(Y). 3,4 Karyotype analysis of metaphase chromosomes from a primary cell line derived from uterine leiomyoma sample L62 revealed a clonal translocation t(12;14)(q15;24) that might affect the HMGI-C locus on chromosome 12q14–15 (U. Fuhrmann, M. Klotzbücher, and J. Bullerdiek, unpublished results). Indeed, we found that the primary uterine leiomyoma L62 expressed high levels of HMGI-C. Interestingly, despite this chromosomal rearrangement, no molecular mass alteration of the HMGI-C protein or mRNA was observed in this uterine leiomyoma, suggesting that the chromosomal breakpoint maps outside of the transcribed region of the HMGI-C gene. It is tempting to speculate that the chromosomal translocation in this tumor accounts for the transcriptional activation of the HMGI-C gene. The expression of HMGI-C in this tumor might be induced by the deletion of a silencer promoter element or by translocation of an active enhancer motive into the HMGI-C locus.
Three of 33 uterine leiomyomas we analyzed expressed HMGI-C gene products that are mutated within the coding region of HMGI-C. One is sample L42, which is characterized by the expression of a 19.5-kd protein instead of the predicted 18 kd of wild-type HMGI-C, and the appearance of a truncated HMGI-C mRNA. Another is sample L352, which expressed a HMGI-C protein of the predicted molecular mass but a truncated HMGI-C mRNA and finally sample LE, which is characterized by the expression of a 15-kd truncated variant of HMGI-C. Unfortunately, we did not obtain enough tissue to isolate and analyze the HMGI-C mRNA from this uterine leiomyoma. But sequence-specific RT-PCR and cDNA sequencing of the HMGI-C mRNA derived from uterine leiomyomas L42 and L352 clearly demonstrated that both tumors expressed HMGI-C transcripts that preserved the entire DNA-binding domain sequence but failed to contain sequences of the acidic tail. As uterine leiomyomas are frequently affected by chromosomal rearrangements that lead to the generation of chimeric or truncated isoforms of HMGI-C, it is tempting to speculate that the three uterine leiomyoma samples LE, L42, and L352 expressed HMGI-C from an allele that was the target of chromosomal rearrangements. Furthermore, we clearly demonstrated by Northern and/or Western analysis that uterine leiomyomas LE, L42, and L352 exclusively expressed the altered form of HMGI-C. The second allele, which is most probably unaffected by mutations, is transcriptionally silent—as it is in normal myometrial cells. This revealed that the transcriptional induction of the mutated allele is caused by the chromosomal rearrangement itself, and not by proteins that are involved in the regulation of HMGI-C gene expression. However, defects in proteins that regulate HMGI gene expression might be an alternative mechanism that can cause up-regulation of HMGI proteins in uterine leiomyomas not affected by HMGI gene rearrangements. In addition, we identified five uterine leiomyomas characterized by an enhanced HMGI(Y) protein expression in which the HMGI(Y) mRNA content was not increased compared to the corresponding myometrium. Defects in the posttranscriptional regulation of HMGI(Y) might account for the HMGI(Y) overexpression in these uterine leiomyomas.
By cDNA sequencing we proved that some uterine leiomyomas expressed HMGI-C or HMGI(Y) that was not affected by mutations within the coding region. Thus our expression studies revealed uterine leiomyomas that expressed mutated HMGI-C as well as uterine leiomyomas that expressed wild-type HMGI-C and/or HMGI(Y). This opens the discussion about whether mutated HMGI proteins as well as wild-type HMGI proteins can both be involved in the pathogenesis of uterine leiomyomas. All chimeric and truncated HMGI genes identified so far have in common that they preserved the AT-hook DNA-binding domain. 3-7,9,11,13 This is also confirmed by our RT-PCR analysis. Ectopic sequences fused to the HMGI-C or HMGI(Y) genes by chromosomal rearrangements did not display any common properties. Thus we would assume that the N-terminal DNA-binding domain is the most important functional domain of HMGI proteins. Indeed, Chin and co-workers 21 generated a HMGI(Y) deletion mutant that consists of the N-terminal DNA-binding domain only but is still functional with regard to serum response factor (SRF) binding, cooperative binding to serum response elements, and stimulation of SRF-mediated transactivation. 21 The C-terminal acidic tail of HMGI proteins might be important for the fine-tuning of HMGI proteins by phosphorylation or acetylation, etc, or might even be involved in properties other than transcriptional regulation of target genes, but it is obviously not essential for the transcriptional regulation of target genes. Therefore we suggest that the elevated expression of HMGI proteins that are functionally active with regard to transcriptional regulation, whether they are wild-type, C-terminal-deleted, or chimeric proteins, is an important event in the pathogenesis of uterine leiomyomas.
Acknowledgments
We thank Jenny List, Kathrin Ratajzcak, Franziska Köpp, and Gisela Repenthin for excellent technical assistance. We also gratefully thank Dr. Huber-Schumacher, Dr. Kleine-Tebbe, and Prof. Dr. Lichtenegger (Charite-Virchow Klinikum, Berlin) for their help in the collection of myometrium and uterine leiomyoma samples; Prof. Bullerdiek (University of Bremen) for karyotype analysis; and Dr. Hinzmann (Metagen, Berlin) for DNA sequencing. We also thank James Johnston for critical reading of the manuscript as well as Prof. Dr. Lübbert (Benjamin Franklin Klinikum, Berlin) and Dr. Lessl for helpful discussions.
Footnotes
Address reprint requests to Dr. Ulrike Fuhrmann, Schering AG, Female Health Care Research, D-13342 Berlin, Germany.
References
- 1.Cramer SF, Patel A: The frequency of uterine leiomyomas. Am J Clin Pathol 1990, 94:435-438 [DOI] [PubMed] [Google Scholar]
- 2.Hennig Y, Wanschura S, Deichert U, Bartnitzke S, Bullerdiek J: Rearrangements of the high mobility group protein family genes and the molecular genetic origin of uterine leiomyomas and endometrial polyps. Mol Hum Reprod 1996, 277–283 [DOI] [PubMed]
- 3.Ashar HR, Fejzo MS, Tkachenko A, Zhou X, Fletcher JA, Weremowicz S, Morton CC, Chada K: Disruption of the architectural factor HMGI-C: DNA-binding AT hook motifs fused in lipomas to distinct transcriptional regulatory domains. Cell 1995, 82:57-65 [DOI] [PubMed] [Google Scholar]
- 4.Schoenmakers EF, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJ: Recurrent rearrangements in the high mobility group protein gene HMGI-C, in benign mesenchymal tumours. Nature Genet 1995, 10:436-444 [DOI] [PubMed] [Google Scholar]
- 5.Kazmierczak B, Rosigkeit J, Wanschura S, Meyer-Bolte K, Van de Ven WJ, Kayser K, Krieghoff B, Kastendiek H, Bartnitzke S, Bullerdiek J: HMGI-C rearrangements as the molecular basis for the majority of pulmonary chondroid hamartomas: a survey of 30 tumors. Oncogene 1996, 12:515-521 [PubMed] [Google Scholar]
- 6.Schoenberg Fejzo M, Ashar HR, Krauter KS, Powell WL, Rein MS, Weremowicz S, Yoon SJ, Kucherlapati RS, Chada K, Morton CC: Translocation breakpoints upstream of the HMGIC gene in uterine leiomyomata suggest dysregulation of this gene by a mechanism different from that in lipomas. Genes Chromosom Cancer 1996, 17:1-6 [DOI] [PubMed] [Google Scholar]
- 7.Geurts JM, Schoenmakers EF, Van de Ven WJ: Molecular characterization of a complex chromosomal rearrangement in a pleomorphic salivary gland adenoma involving the 3′-UTR of HMGIC. Cancer Genet Cytogenet 1997, 95:198-205 [DOI] [PubMed] [Google Scholar]
- 8.Kazmierczak B, Bol S, Wanschura S, Bartnitzke S, Bullerdiek J: PAC clone containing the HMGI(Y) gene spans the breakpoint of a 6p21 translocation in a uterine leiomyoma cell line. Genes Chromosom Cancer 1996, 17:191-193 [DOI] [PubMed] [Google Scholar]
- 9.Xiao S, Lux ML, Reeves R, Hudson TJ, Fletcher JA: HMGI(Y) activation by chromosome 6p21 rearrangements in multilineage mesenchymal cells from pulmonary hamartoma. Am J Pathol 1997, 150:901-910 [PMC free article] [PubMed] [Google Scholar]
- 10.Williams AJ, Powell WE, Collins T, Morton CC: HMGI(Y) expression in human uterine leiomyomata Involvement of another high-mobility group architectural factor in a benign neoplasm. Am J Pathol 1997, 150:911-918 [PMC free article] [PubMed] [Google Scholar]
- 11.Tkachenko A, Ashar HR, Meloni AM, Sandberg AA, Chada KK: Misexpression of disrupted HMGI architectural factors activates alternative pathways of tumorigenesis. Cancer Res 1997, 57:2276-2280 [PubMed] [Google Scholar]
- 12.Dal Cin P, Wanschura S, Christiaens MR, Van den Berghe I, Moerman P, Polito P, Kazmierczak B, Bullerdiek J, Van den Berghe H: Hamartoma of the breast with involvement of 6p21 and rearrangement of HMGIY. Genes Chromosom Cancer 1997, 20:90-92 [DOI] [PubMed] [Google Scholar]
- 13.Kazmierczak B, Hennig Y, Wanschura S, Rogalla P, Bartnitzke S, Van de Ven W, Bullerdiek J: Description of a novel fusion transcript between HMGI-C, a gene encoding for a member of the high mobility group proteins and the mitochondrial aldehyde dehydrogenase gene. Cancer Res 1995, 55:6038-6039 [PubMed] [Google Scholar]
- 14.Nissen MS, Reeves R: Changes in superhelicity are introduced into closed circular DNA by binding of high mobility group protein I/Y. J Biol Chem 1995, 270:4355-4360 [DOI] [PubMed] [Google Scholar]
- 15.Thanos D, Maniatis T: The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-β gene. Cell 1992, 71:777-789 [DOI] [PubMed] [Google Scholar]
- 16.Du W, Thanos D, Maniatis T: Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell 1993, 74:887-898 [DOI] [PubMed] [Google Scholar]
- 17.Lewis H, Kaszubska W, De Lamarter JF, Whelan J: Cooperativity between two NF-kappa B complexes, mediated by high-mobility-group protein I(Y), is essential for cytokine-induced expression of the E-selectin promoter. Mol Cell Biol 1994, 14:5701-5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.John S, Reeves RB, Lin JX, Child R, Leiden JM, Thompson CB, Leonard WJ: Regulation of cell-type-specific interleukin-2 receptor α-chain gene expression: potential role of physical interactions between Elf-1, HMG-I(Y), and NF-kappa B family proteins. Mol Cell Biol 1995, 15:1786-1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leger H, Sock E, Renner K, Grummt F, Wegner M: Functional interaction between the POU domain protein Tst-1/Oct-6 and the high-mobility-group protein HMG-I/Y. Mol Cell Biol 1995, 15:3738-3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Himes SR, Coles LS, Reeves R, Shannon MF: High mobility group protein I(Y) is required for function and for c-Rel binding to CD28 response elements within the GM-CSF and IL-2 promoters. Immunity 1996, 5:479-489 [DOI] [PubMed] [Google Scholar]
- 21.Chin MT, Pellacani A, Wang H, Lin SS, Jain MK, Perrella MA, Lee ME: Enhancement of serum-response factor-dependent transcription and DNA binding by the architectural transcription factor HMG-I(Y). J Biol Chem 1998, 273:9755-9760 [DOI] [PubMed] [Google Scholar]
- 22.Mantovani F, Covaceuszach S, Rustighi A, Sgarra R, Heath C, Goodwin GH, Manfioletti G: NF-kappa B mediated transcriptional activation is enhanced by the architectural factor HMGI-C. Nucleic Acids Res 1998, 26:1433-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grosschedl R, Giese K, Pagel J: HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet 1994, 10:94-100 [DOI] [PubMed] [Google Scholar]
- 24.Wolffe AP: Architectural transcription factors. Science 1994, 264:1100-1101 [DOI] [PubMed] [Google Scholar]
- 25.Kim J, Reeves R, Rothman P, Boothby M: The non-histone chromosomal protein HMG-I(Y) contributes to repression of the immunoglobulin heavy chain germ-line epsilon RNA promoter. Eur J Immunol 1995, 25:798-808 [DOI] [PubMed] [Google Scholar]
- 26.Klein-Hessling S, Schneider G, Heinfling A, Chuvpilo S, Serfling E: HMG I(Y) interferes with the DNA binding of NF-AT factors and the induction of the interleukin 4 promoter in T cells. Proc Natl Acad Sci USA 1996, 93:15311-15316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arlotta P, Rustighi A, Mantovani F, Manfioletti G, Giancotti V, Tell G, Damante G: High mobility group I proteins interfere with the homeodomains binding to DNA. J Biol Chem 1997, 272:29904-29910 [DOI] [PubMed] [Google Scholar]
- 28.Chiappetta G, Avantaggiato V, Visconti R, Fedele M, Battista S, Trapasso F, Merciai BM, Fidanza V, Giancotti V, Santoro M, Simeone A, Fusco A: High level expression of the HMGI (Y) gene during embryonic development. Oncogene 1996, 13:2439-2446 [PubMed] [Google Scholar]
- 29.Manfioletti G, Giancotti V, Bandiera A, Buratti E, Sautiere P, Cary P, Crane-Robinson C, Coles B, Goodwin GH: cDNA cloning of the HMGI-C phosphoprotein, a nuclear protein associated with neoplastic and undifferentiated phenotypes. Nucleic Acids Res 1991, 19:6793-6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogalla P, Drechsler K, Frey G, Hennig Y, Helmke B, Bonk U, Bullerdiek J: HMGI-C expression patterns in human tissues Implications for the genesis of frequent mesenchymal tumors. Am J Pathol 1996, 149:775-779 [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, Benson KF, Przybysz K, Liu J, Hou Y, Cherath L, Chada K: Genomic structure and expression of the murine Hmgi-c gene. Nucleic Acids Res 1996, 24:4071-4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamimi Y, van der Poel HG, Karthaus HF, Debruyne FM, Schalken JAA: A retrospective study of high mobility group protein I(Y) as progression marker for prostate cancer determined by in situ hybridization. Br J Cancer 1996, 74:573-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiappetta G, Bandiera A, Berlingieri MT, Visconti R, Manfioletti G, Battista S, Martinez-Tello FJ, Santoro M, Giancotti V, Fusco A: The expression of the high mobility group HMGI (Y) proteins correlates with the malignant phenotype of human thyroid neoplasias. Oncogene 1995, 10:1307-1314 [PubMed] [Google Scholar]
- 34.Fedele M, Bandiera A, Chiappetta G, Battista S, Viglietto G, Manfioletti G, Casamassimi A, Santoro M, Giancotti V, Fusco A: Human colorectal carcinomas express high levels of high mobility group HMGI(Y) proteins. Cancer Res 1996, 56:1896-1901 [PubMed] [Google Scholar]
- 35.Rogalla P, Drechsler K, Kazmierczak B, Rippe V, Bonk U, Bullerdiek J: Expression of HMGI-C, a member of the high mobility group protein family, in a subset of breast cancers: relationship to histologic grade. Mol Carcinog 1997, 19:153-156 [DOI] [PubMed] [Google Scholar]
- 36.Bandiera A, Bonifacio D, Manfioletti G, Mantovani F, Rustighi A, Zanconati F, Fusco A, Di Bonito L, Giancotti V: Expression of HMGI(Y) proteins in squamous intraepithelial and invasive lesions of the uterine cervix. Cancer Res 1998, 58:426-431 [PubMed] [Google Scholar]
- 37.Vartiainen E, Palvimo J, Mahonen A, Linnala-Kankkunen A, Maenpaa PH: Selective decrease in low-Mr HMG proteins HMG I and HMG Y during differentiation of mouse teratocarcinoma cells. FEBS Lett 1988, 228:45-48 [DOI] [PubMed] [Google Scholar]
- 38.Zhou X, Benson KF, Ashar HR, Chada K: Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995, 376:771-774 [DOI] [PubMed] [Google Scholar]
- 39.Lessl M, Klotzbuecher M, Schoen S, Reles A, Stockemann K, Fuhrmann U: Comparative messenger ribonucleic acid analysis of immediate early genes and sex steroid receptors in human leiomyoma and healthy myometrium. J Clin Endocrinol Metab 1997, 82:2596-2600 [DOI] [PubMed] [Google Scholar]
- 40.Patel UA, Bandiera A, Manfioletti G, Giancotti V, Chau KY, Crane-Robinson C: Expression and cDNA cloning of human HMGI-C phosphoprotein. Biochem Biophys Res Commun 1994, 201:63-70 [DOI] [PubMed] [Google Scholar]
- 41.Goodwin GH, Johns EW: Isolation and characterisation of two calf-thymus chromatin non-histone proteins with high contents of acidic and basic amino acids. Eur J Biochem 1973, 40:215-219 [DOI] [PubMed] [Google Scholar]