Abstract
The glycosaminoglycan hyaluronan (HA) and HA-binding proteins (HABPs) serve important structural and regulatory functions during development and in maintaining adult tissue homeostasis. Here we have identified and partially characterized the sequence and expression pattern of three putative novel HABPs. DNA sequence analysis revealed that two of the novel HABPs, WF-HABP and BM-HABP, form a unique HA-binding subfamily, whereas the third protein, OE-HABP, is more closely related to the LINK subfamily of HABPs. Northern blotting experiments revealed that the expression of BM-HABP was highly restricted, with substantial expression detected only in human fetal liver. In contrast, WF-HABP and OE-HABP mRNAs were detected in a number of tissues, with particularly prominent expression in highly vascularized tissues such as the heart, placenta, and lung. Additional studies showed that OE-HABP was expressed by cultured human endothelial cells, smooth muscle cells, and differentiated monocytes. However, only endothelial cells expressed WF-HABP mRNA, and its expression was regulated by growth state, being most prominent in quiescent endothelial cells. We further characterized the expression of WF-HABP in vivo and found that its expression colocalized with CD31-positive cells and was prominently expressed in microvessels in the human aorta and in atherectomy samples. Our data suggest that WF-HABP is an endothelial cell-specific HA receptor and that it may serve a unique function in these cells. The WF-HABP gene was localized to chromosome 3p21.31 and the OE-HABP gene to 15q25.2–25.3.
Hyaluronan 1,2 (HA) is a high-molecular-weight linear polysaccharide consisting of alternating N-acetyl-d-glucosamine and d-glucuronic acid residues linked by β(1–4) and β(1–3) bonds. 3,4 It is found in the extracellular matrix (ECM) of most animal tissues, being most abundant in the soft connective tissues, and is found at high levels in the blood vessel, dermis, brain, and muscle. 2,5-8 HA is more abundant in the rapidly growing fetal tissues, 5 and besides its structural role in cartilage, HA is thought to modulate cell proliferation and migration during tissue development, regeneration, and remodeling. 6 A particularly intriguing element is the finding that HA fragments are potent angiogenic molecules. 9,10 However, the mechanism by which HA regulates cellular activity such as angiogenesis remains poorly understood.
A number of proteins have been identified that bind HA with high specificity and affinity and are believed to mediate the biological activity of HA. These HA-binding proteins (HABPs) or hyaladherins have been classified into two broad groups: matrix HABPs and cell-surface HA receptors. 4 They include aggrecan, link protein, 11 versican, hyaluronectin, neurocan, 12 the CD44 family of receptors, 13,14 receptor for hyaluronan-mediated motility (RHAMM), 14,15 and tumor necrosis factor-stimulated gene 6 (TSG-6). 16 Although there are exceptions (eg, RHAMM), the majority of the hyaladherins contain the structural motif (C-X(15)-A-X(3,4)-G-X(3)-C-X(2)-G-X(8,9)-P-X(7)-C), as it is characterized in the GCG program, 17 that was first identified in the cartilage link protein. 18 TSG-6 is a particularly interesting member of this family, given its postulated anti-inflammatory role in arthritis. 19,20 We previously determined that TSG-6 was expressed in SMCs in response to growth factors and cytokines and is expressed transiently in vivo after vascular injury. 21 In the current study, we used the cDNA sequence of TSG-6 to search an expressed sequence tag (EST) database and identified three novel cDNAs with sequence similarity to TSG-6. These cDNAs were characterized by DNA sequencing, and the expression pattern of these novel genes was determined by Northern blotting and by in situ hybridization. The chromosomal localization of two genes was determined by fluorescence in situ hybridization.
Materials and Methods
Clones
The original cDNA clones for WF-HABP, BM-HABP, and OE-HABP were isolated from human white fat, bone marrow, and osteoblast cDNA libraries, respectively. The 6.8-kb cDNA (KIAA0246 22 genebank accession number D87433) coding for the entire WF-HABP cDNA was kindly provided by Dr. Takahiro Nagaze (Kazuza DNA Research Institute, Kisarazu, Chiba, Japan).
DNA Sequencing and Analysis
(pBluescript) Plasmids containing cDNAs of WF-HABP, BM-HABP, and OE-HABP were isolated from bacteria with a Qiagen Plasmid Midi Kit (Qiagen, Valencia, CA) or a Wizard Plus SV Minipreps DNA Purification System (Promega, Madison, WI). Samples were prepared for sequencing with an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer Corp., Foster City, CA) and sequence-specific 18- to 22-mer oligos (CyberSyn, Lenni, PA; Integrated DNA Technologies, Coralville, IA). Fluorescence-based sequence analysis was subsequently performed on the ABI model 373A DNA sequencer (PE Applied Biosystems, Foster City, CA).
The cDNA encoding DNA fragments of WF-HABP, BM-HABP, and OE-HABP were sequenced. The resulting sequences were used to search the NCBI BLAST 23 EST database for additional homologous cDNAs. The National Center for Biotechnology Information BLAST SWISSPROT database was also searched with the putative protein sequence (derived from the cDNA) for similarities with previously described proteins to determine any possible relationships among them. Sequence alignments and searches were done using MOTIF program (http://www.motif.genome.ad.jp) and GCG (Wisconsin Package Version 9.0, Genetics Computer Group, Madison, WI). 17 DNA and protein sequences of the novel HABPs were manipulated and analyzed using GCG sequence analysis programs.
Cell Culture
Human peripheral blood promyelocytic leukemia cells (HL60, ATCC CCL 240) and human histocytic lymphoma cells (U937, ATCC CRL 1593) were obtained from the American Type Culture Collection (Rockville, MD). Cells were grown in RPMI 1640 supplemented with 10% fetal bovine serum (FBS). Human diploid fetal lung fibroblasts (HFL1, ATCC CCL 153) were obtained from the American Type Culture Collection, and human saphenous vein smooth muscle cells (SMCs) 21 were kindly provided by Dr. Peter Libby (Tufts University School of Medicine, Boston, MA). Cells were grown in Dulbecco’s minimum essential medium and M-199 complemented with 10% FBS. Human umbilical vein endothelial cells (HUVECs), strain H101, were a generous gift from Dr. Susan Garfinkel. Cells were grown in M-199 containing 10% FBS and 10 ng/ml of fibroblast growth factor-1 (FGF-1)/heparin. 24,25 HUVECs were growth arrested for 48 hours in complete media with 10% serum without growth factor 25 and for some experiments were treated with FGF-1, recombinant interleukin-1 (IL-1), or 12-O-tetradecanoylphorbol-13-acetate (TPA). HL-60 and U937 cells were grown under normal conditions for 2 days and then induced with 0.15 mg/ml of TPA for 72 hours to elicit a differentiation response.
RNA Preparation and Northern Blot Analysis
Total RNA was isolated from cultured cells by selective retention on a silica gel-based membrane with an RNeasy Mini Kit (Qiagen). Briefly, cells were pelleted, lysed, and homogenized under denaturing conditions in the presence of guanidinium isothiocyanate. Total RNA was separated from contaminating proteins and DNA by centrifugation and subsequently eluted from the column with water. Five micrograms of purified total RNA per lane was size-fractionated on 1% agarose gel containing 0.5 mol/L formaldehyde, transferred to Zetabind nylon membrane (AMF/Cuno, Meriden, CT) by electroblotting, and UV cross-linked. 26,27 Immobilized RNA was hybridized at 55°C overnight with 1–5 × 10 6 cpm/ml of [α-32P]dCTP-labeled cDNA probes prepared by random primed DNA labeling (Boehringer-Mannheim, Indianapolis, IN). After hybridization, membranes were washed with increasing stringency at 55°C, for 2 × 15 minutes, in each of the following buffers: wash buffer A: 0.5% bovine serum albumin, 5% sodium dodecyl sulfate, 40 mmol/L NaH2PO4, 1 mmol/L EDTA; wash buffer B: 1% sodium dodecyl sulfate, 40 mmol/L NaH2PO4, 1 mmol/L EDTA. 28,29 Blots were air dried and exposed to Kodak X-Omat AR film (Eastman Kodak Company, Rochester, NY) at −80°C. An 800-bp human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA was used as a control probe. 28 Blots containing mRNA obtained from human heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas were acquired from Clontech (Palo Alto, CA). These blots were probed with WF-HABP, BM-HABP, and OE-HABP cDNAs and hybridized with the GAPDH control probe.
Immunohistochemistry
Paraffin-embedded serial 5-μm-thick sections of the human tissues analyzed by Northern blotting and several atherectomy specimens were used. Endogenous peroxidase activity was quenched in methanol with 0.3% H2O2. The sections were reacted with a monoclonal mouse anti-human CD31 antibody specific for endothelial cells (Dako, Glostrup, Denmark) at room temperature for 1 hour. Antibodies were diluted 1:50 in phosphate-buffered saline (PBS) containing 10% normal horse serum and 1% bovine serum albumin. Primary antibody was detected by the indirect avidin-biotin-horseradish peroxidase method (ABC Elite Kit; Vector Laboratories, Burlingame, CA). Normal mouse IgG (1:1000) were used as negative controls. Myer’s hematoxylin was used to counterstain the sections.
In Situ Hybridization
In situ hybridization was performed on paraffin-embedded human tissues and atherectomy specimens. WF-HABP mRNA probes (sense and antisense) were labeled with digoxigenin-11-uridine-5′-triphosphate via in vitro transcription (Dig RNA labeling kit; Boehringer Mannheim). The tissues were cut into serial 5-μm-thick sections onto silanized double-positive glass slides (Fisher Scientific, Pittsburgh, PA). Tissue sections were deparaffinized at 60°C for 60 minutes, washed extensively in xylene, and rehydrated in decreasing ethanol series. Endogenous peroxidase activity was quenched in PBS containing 3% H2O2 for 20 minutes. To facilitate probe penetration, tissue sections were deproteinized in 2 mg/ml pepsin solution in 0.2 N HCl. 30 Sections were equilibrated, prehybridized, and hybridized according to manufacturer’s specifications (Novagen, Madison, WI). Hybridization was carried out in a humid chamber at 50°C for 18 hours with a probe concentration of 1 ng/μl. After hybridization, sections were subjected to successive stringent washes as follows: 2× standard saline citrate (SSC) 50°C for 30 minutes; 2× SSC containing 0.02 mg/ml RNase A, 37°C, for 30 minutes; 2× SSC containing 50% formamide, 50°C, for 30 minutes; two washes with 1× SSC containing 0.067% sodium para-phosphate, 50°C, for 30 minutes each.
Signal amplification was carried out according to Tyramide Signal Amplification for chromogenic in situ hybridization protocol (NEN Life Science Products, Boston, MA). Anti-digoxigenin antibody (sheep Fab fragments conjugated with horseradish peroxidase (POD); Boehringer Mannheim) was diluted 1:50 in PBS containing blocking buffer. Signal was visualized with a diaminobenzidine substrate kit (Vector Laboratories). Myer’s hematoxylin was used to counterstain the sections. To confirm the reliability of the method, protamine antisense probe was hybridized to sections of mouse testes as a positive control in each experiment. Furthermore, hybridization with the sense WF-HABP probe and no probe were used as negative controls.
Fluorescence in Situ Hybridization
Genomic DNA WF-HABP and OE-HABP clones were isolated by screening a human P1 library and labeled with digoxigenin dUTP by nick translation for hybridization. Labeled probes were combined with sheared human DNA and hybridized to normal metaphase chromosomes derived from TPA-stimulated peripheral blood lymphocytes in a solution containing 50% formamide, 10% dextran sulfate, and 2× SSC. First, clone-specific hybridization signals were detected in one-color experiments by incubating the hybridized slides in fluoresceinated antidigoxigenin antibodies followed by counterstaining with diamidino phenylindole (DAPI). In the second experiment, a biotin-labeled probe that is specific for the centromere of the chromosome labeled in the first experiment was cohybridized with either of the digoxigenin-labeled WF-HABP or OE-HABP clones correspondingly. Probe detection for two color experiments was accomplished by incubating the slides in fluoresceinated antidigoxigenin antibodies and Texas red avidin followed by counterstaining with DAPI.
Results
Identification and Characterization of Novel HABPs
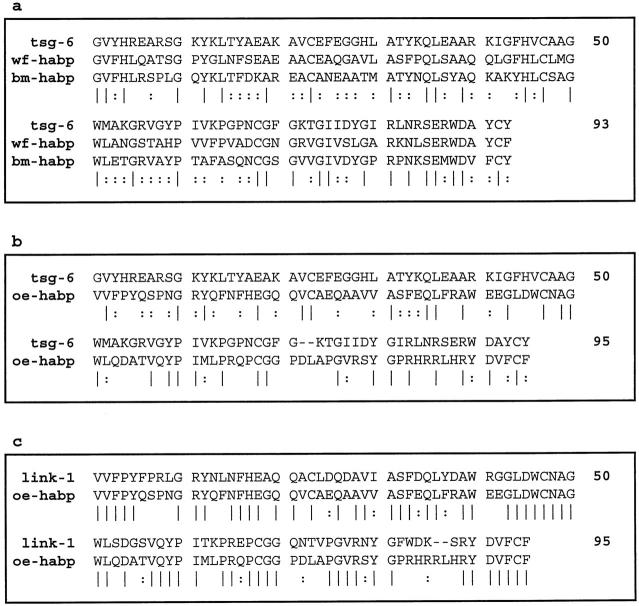
We used the DNA sequence of TSG-6 to search an EST database and identified three cDNA clones that encode novel proteins containing putative HA-binding domains. Based on the HA-binding motifs and the original tissues from which these cDNAs were derived, we have designated these white fat (WF)-HABP, bone marrow (BM)-HABP, and osteoblast (OE)-HABP. WF-HABP and BM-HABP each contain a single HA-binding domain showing the highest sequence identity with the HA-binding domain of TSG-6, with a sequence identity of 51% between WF-HABP and TSG-6 and a sequence identity of 48% between BM-HABP and TSG-6 (Figure 1a) ▶ . Furthermore, comparison of the available sequence between WF-HABP and BM-HABP revealed a total sequence identity of 45% over a 359-amino-acid stretch, indicating that they are closely related. In contrast, the putative HA-binding motif of OE-HABP was found to be more homologous to HA-binding domains of LINK and aggrecan, with a 65% sequence identity to the first HA-binding domain of LINK and 45% identity to the first HA-binding domain of aggrecan (Figure 1b) ▶ .
Figure 1.
Sequence comparison of the novel HA-binding proteins with TSG-6 and LINK-1. a: Protein sequence of the HA-binding domains of human TSG-6, WF-HABP, and BM-HABP were compared using the Multiple Sequence Alignment program of the GCG package. b: Sequence comparison of the HA-binding domains of TSG-6 and OE-HABP, using the Bestfit Sequence Alignment program of the GCG package. c: Sequence comparison of the HA-binding domains of LINK-1 and OE-HABP, using the Bestfit program. A vertical line ‖ indicates a position in the alignment that is perfectly conserved; a : position in the alignment is well conserved. Numbers indicate amino acid position in the HA binding domain and thus are the same for all of the aligned sequences sharing similarities in these domains.
HABP Expression in Human Tissues and Cultured Cells
As a first step toward characterizing the expression pattern of these three novel HABP genes, we examined their mRNA levels in various human tissues and cells by Northern blotting analysis. As shown in Figure 2 ▶ , WF-HABP mRNA was detectable in all of the human tissues we examined, with only trace levels detected in the brain and kidney. The highest level of expression was observed in the heart, placenta, and lung, with lower levels found in the liver, pancreas, and skeletal muscle. Four major transcripts of 9.5, 4.5, 3.0, and 2.4 kb were detected. The 9.5-kb band appeared to be the predominant transcript and was especially prominent in the placenta and the heart. In contrast, BM-HABP mRNA was apparent only in the liver and appeared as a weak, diffuse band between 2 and 5 kb. To further explore the expression pattern of BM-HABP, we next examined whether this gene was expressed at an elevated level in human fetal brain, lung, liver, and kidney. We found that a distinct 9.5-kb mRNA was expressed at an elevated level in fetal liver, with weak expression also detected in the lung (results not shown). For OE-HABP, a 2.2-kb transcript was detected in lung, placenta, and heart, with the highest expression observed in the lung.
Figure 2.
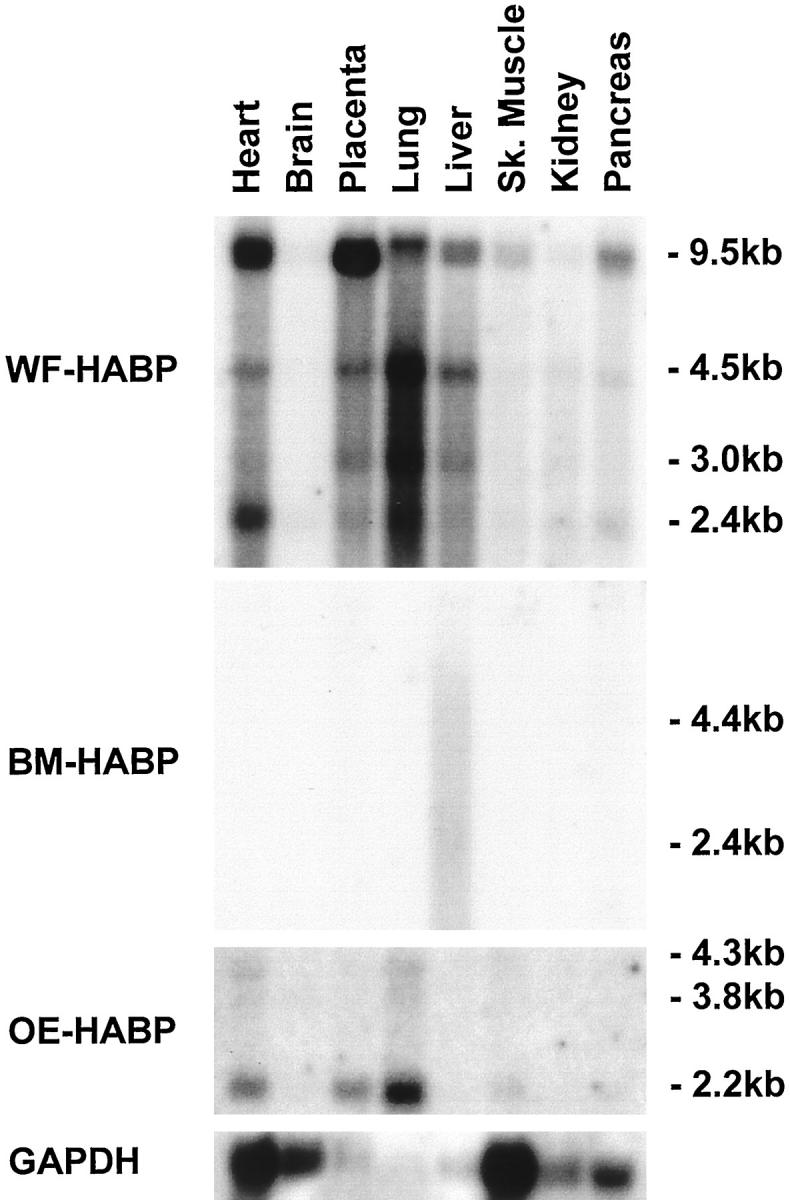
Analysis of the novel HA-binding protein mRNA levels in various human tissues. Total mRNAs isolated from a panel of human tissues were analyzed by Northern blotting for expression levels of White Fat HABP (top panel), Bone Marrow HABP (second panel from top), and Osteoblast HABP (third panel from top). The bottom panel shows control GAPDH mRNA levels. Sk. Muscle = Skeletal muscle.
The expression pattern of WF-, BM-, and OE-HABPs was also examined in human smooth muscle cells (SMCs), human fetal lung fibroblasts (HFLs), human umbilical vein endothelial cells (HUVECs), as well as in the HL-60 and U937 cell lines (Figure 3) ▶ . There was no detectable expression of BM-HABP in any of the cell lines examined (data not shown). Both HUVECs and SMCs but not HFLs, HL-60, or U937s expressed the 2.2-kb OE-HABP transcript. However, U937 cells stimulated with TPA expressed a major new 4.3-kb transcript and minor mRNAs of 3.8 and 3.0 kb. Thus it appears that three of the major cell types relevant to vascular physiology and pathology all express OE-HABP. The most striking mRNA expression pattern was found with WF-HABP, with HUVECs exclusively expressing transcripts of 9.5, 4.5, and 3.0 kb, with a minor 2.4-kb band also detectable in U937, HFLs, and SMCs.
Figure 3.
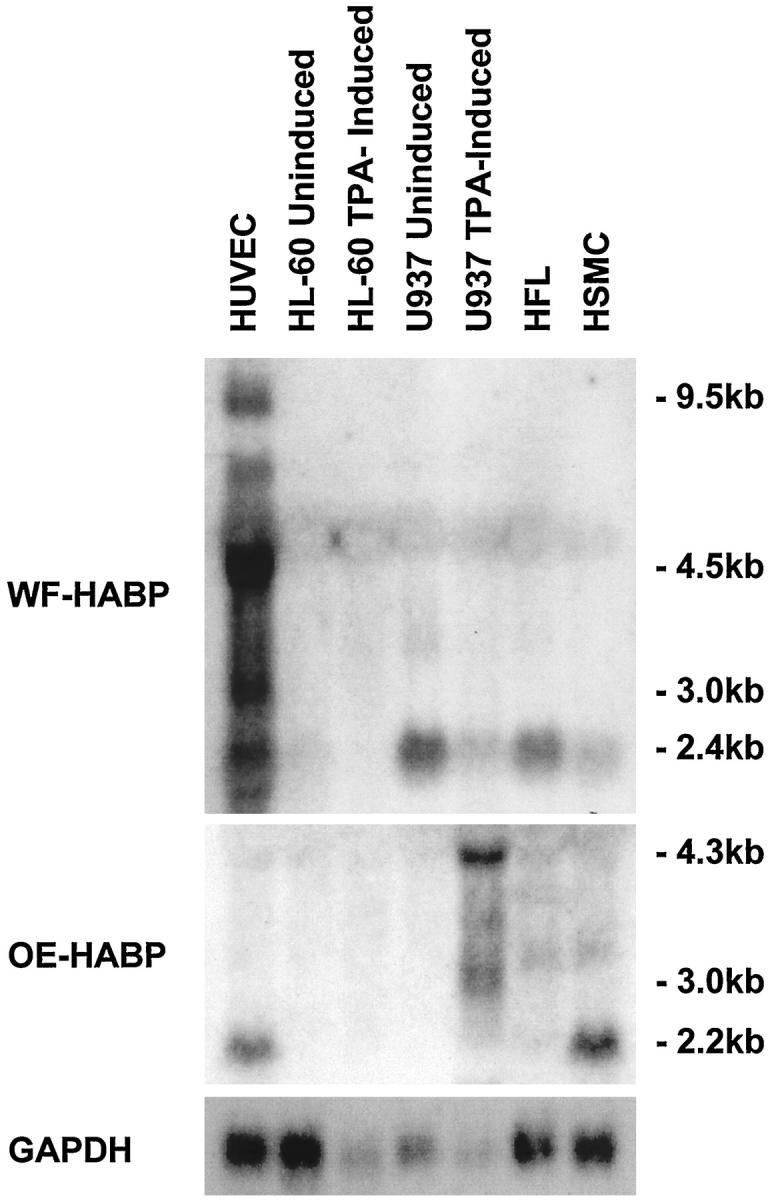
Analysis of the novel HA-binding protein mRNA levels in various human cells. Human umbilical vein endothelial cells (HUVEC), human promyelocytic leukemia cells (HL-60) without and after TPA induction, human histocytic lymphoma cells (U937) without and after TPA induction, human fetal lung firoblasts (HFL), and human smooth muscle cells (HSMC) were grown to confluence and harvested for RNA isolation. mRNA expression of White Fat HABP (top panel) and Osteoblast HABP (middle panel) were analyzed by Northern blotting. The bottom panel shows control GAPDH mRNA levels. Note that higher-molecular-weight mRNA transcripts of WF-HABP are observed exclusively in HUVECs.
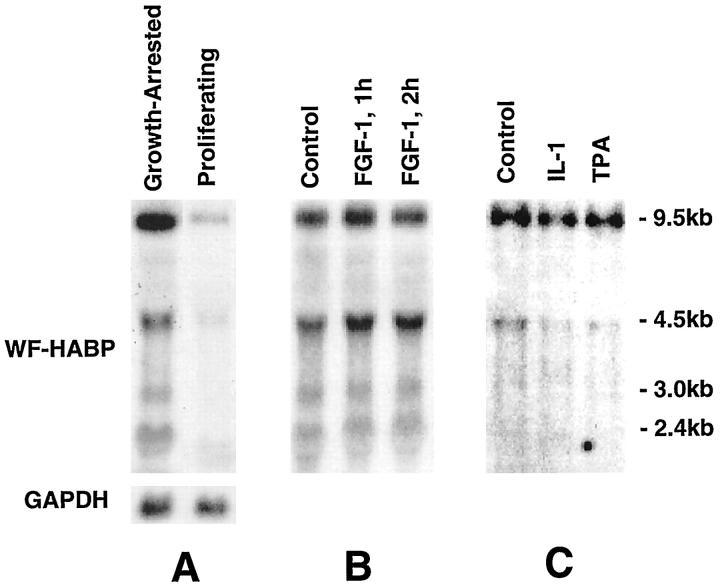
Given that endothelial cells express a unique set of WF-HABP mRNA transcripts, we analyzed the regulation of this gene in HUVECs in greater detail. We observed that growth-arrested HUVECs expressed a high level of WF-HABP mRNA that decreased dramatically in proliferating cells grown in the presence of FGF-1 (Figure 4A) ▶ . However, the expression of WF-HABP did not appear to be directly regulated by FGF-1, inasmuch as stimulation of growth-arrested cells with this factor for 1–2 hours did not decrease WF-HABP levels (Figure 4B) ▶ . Consistent with a correlation with cellular growth state, expression of WF-HABP began to decrease by 24 hours after FGF-1 treatment (results not shown). In addition, we treated growth-arrested cells with IL-1 or TPA for 4 hours and found that these factors also were unable to alter WF-HABP mRNA expression (Figure 4C) ▶ .
Figure 4.
The expression of WF-HABP mRNA in cultured human umbilical vein endothelial cells in response to cellular growth state and to treatment with FGF-1, IL-1, and TPA. A: Growth-arrested (minus FGF-1) and proliferating (plus FGF-1) HUVECs. B: Growth-arrested cells were treated with 10 ng/ml of FGF-1 for the indicated times. C: Growth-arrested cells were treated with 10 ng/ml rIL-1α or 20 ng/ml TPA for 4 hours. Endothelial cells were growth arrested by culturing without FGF-1/heparin for 48 hours and treated with the indicated factors. The cells were then harvested for RNA isolation, and WF-HABP mRNA expression was analyzed by Northern blotting. The bottom of A shows the corresponding control GAPDH mRNA level.
Determination of Cell Types That Express WF-HABP in Vivo
Our in vitro data are consistent with the hypothesis that WF-HABP mRNA expression is restricted to endothelial cells. To determine whether endothelial-specific expression is also observed in vivo, we used in situ hybridization to characterize the expression pattern of WF-HABP in various tissues. We first examined the placenta, which contains a high level of WF-HABP mRNA and is blood vessel enriched. A strong hybridization signal was detected in cells lining fetal blood vessels and in capillaries found inside the terminal villi (results not shown). Syncytial trophoblasts, composing the external layer of the terminal villi, were always negative. This pattern was virtually identical to the immunostained pattern for the endothelial specific antigen CD31, suggesting that the major cell type in the placenta expressing WF-HABP is endothelial cells. WF-HABP-positive signals in the brain were considerably weaker but were also restricted to small blood vessels and capillaries. CD31 staining revealed a similar but more robust expression pattern in the brain, suggesting that brain endothelium also expresses the WF-HABP message but at much lower levels (data not shown).
We next examined the expression pattern of WF-HABP in other vascular tissues, including human aorta and atherectomy specimens. A positive RNA signal was observed only in endothelial cells lining microvessels in the adventitia (Figure 5a) ▶ and the medial layer (Figure 5b) ▶ of the aorta. No signal was detected in smooth muscle cells of the medial layer. Specificity was confirmed by the absence of the signal when hybridization was performed without probe (Figure 5, c and d) ▶ or with a corresponding sense probe (results not shown). We observed nonspecific staining of collagen fibers in the adventitia, even in the presence of no probes, perhaps because of the binding of anti-digoxigenin antibody to collagen (Figure 5c) ▶ . To confirm that cells expressing WF-HABP mRNA were indeed endothelial cells, we also performed immunohistochemical analysis on adjacent sections of the same specimen, using an antibody that specifically recognizes CD31. We found that the signal for CD31 antigen was distributed in a manner analogous to that for WF-HABP (Figure 5, e and f) ▶ . We next examined the expression of WF-HABP mRNA in human atherosclerotic lesions. We found a strong WF-HABP-specific signal in vessel-abundant regions of the specimens (Figure 6a ▶ ; compare with negative control in Figure 6b ▶ ). Again, this signal colocalized with the distribution of endothelial-specific CD31 antigen (Figure 6c) ▶ . No signal was detected in the SMC or in macrophage-rich regions (data not shown). However, within the intimal hyperplasia regions of moderate cellularity (myxomatous tissue), 31,32 occasional stellate cells that contain α-smooth muscle actin were also found to be positive for WF-HABP signal (data not shown). We conclude from these studies that WF-HABP mRNA is expressed predominantly by endothelial cells in various tissues, and its expression is especially prominent in diseased blood vessels.
Figure 5.
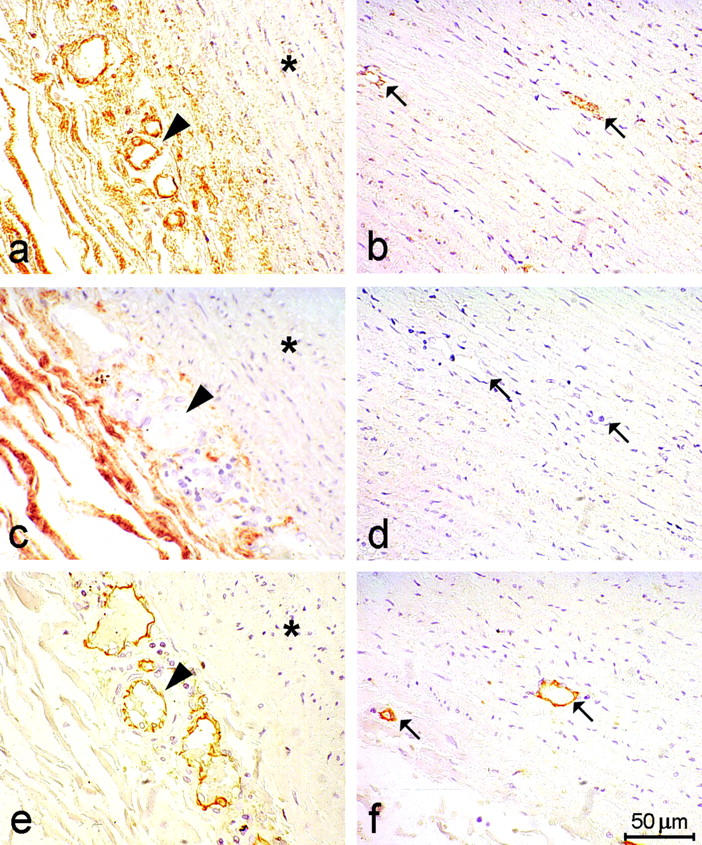
Analysis of WF-HABP mRNA expression in the human aorta. Nonradioactive in situ hybridization with digoxigenin-labeled antisense WF-HABP mRNA probe was performed on 5-μm paraffin-embedded sections. a, c, and e: Similar adventitial regions of the aorta. b, d, and f: Similar medial regions of the aorta. a and b show sections that were hybridized to the WF-HABP riboprobe. c and d show sections treated as in a and b but without the riboprobe. e and f show sections immunostained with an antibody to CD31. Arrowheads mark microvessels in the adventia. Arrows mark microvessels in the media. Asterisks in a, c, and e denote the medial region proximal to the adventia. Positively reacting cells are stained brown. The antibody to CD31 is endothelial cell-specific.
Figure 6.
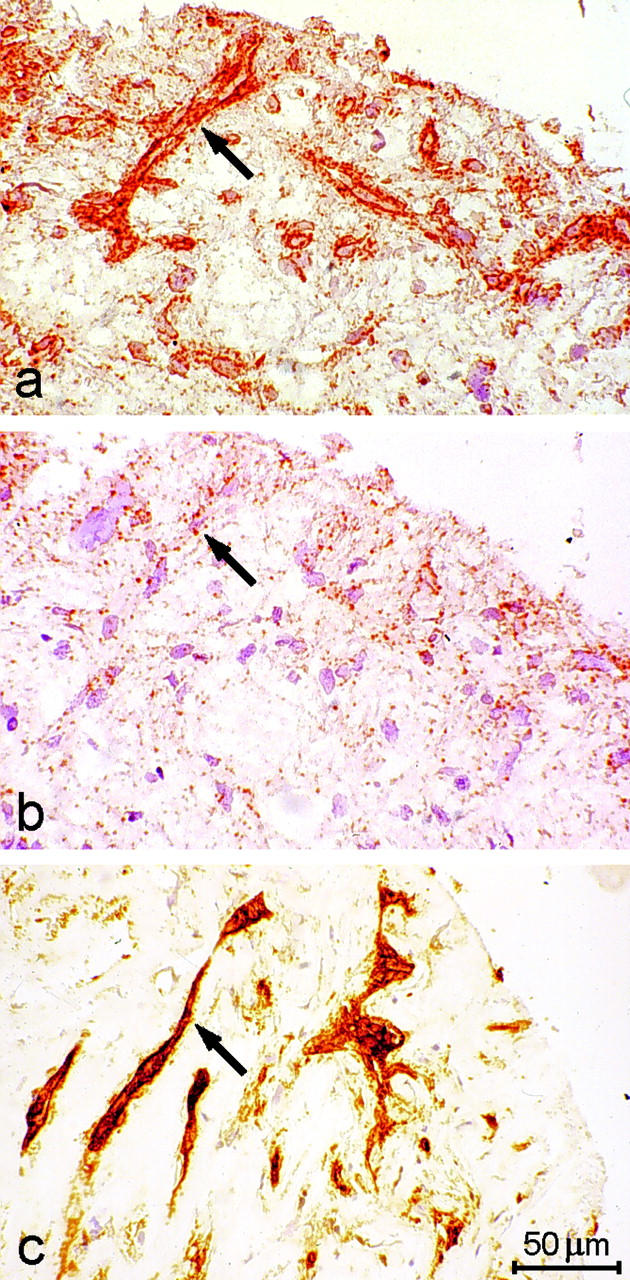
Analysis of WF-HABP mRNA expression in a human atherectomy specimen. Nonradioactive in situ hybridization was carried out as described in Materials and Methods. a: An atherectomy section hybridized to the WF-HABP riboprobe. b: As in a but without the riboprobe. c: A section immunostained with an antibody to CD31. Immunohistochemistry for CD31 antigen identifies blood vessels in the same atherectomy specimen. Arrows indicate blood vessels, and positive cells are stained brown.
Domain Structure of WF-HABP
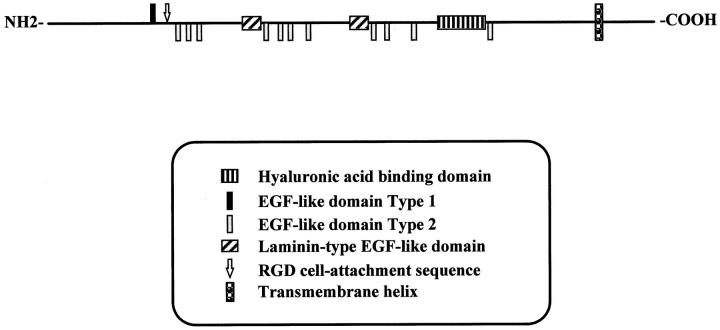
The endothelial cell-specific expression of WF-HABP encouraged us to characterize the full-length cDNA of this gene. We completed sequencing of the entire 1522-bp cDNA and compared the consensus sequence with various databases. This cDNA sequence completely matched a DNA sequence (KIAA0246) deposited in the GenBank database that contains an open reading frame of 6777 bp. 22 In addition to the HA-binding domain, the deduced protein sequence of the presumed full-length WF-HABP (KIAA0246) contained a number of additional motifs, including one type 1 and 11 type 2 EGF-like domains, as well as two laminin-type EGF-like domains and an RGD cell-binding motif. Furthermore, a transmembrane domain was found 72 amino acids away from the C-terminus. The domain structure of WF-HABP (KIAA0246) is summarized in a graphical form in Figure 7 ▶ . We predict that WF-HABP is a novel HA-binding protein that resides predominantly on the endothelial cell surface.
Figure 7.
The domain structure of human WF-HABP. The motif scheme was constructed based on comparison of the deduced WF-HABP protein sequence to known protein domains. The type 1 EGF-like repeat starts at amino acid 431. The RGD cell-attachment site is at position 765. There are 11 type 2 EGF-like repeats that localize between amino acids 521-1964. The two laminin-type EGF-like domains start at amino acids 999 and 1638 respectively. The HA-binding domain spans amino acids 1873 and 1918. The transmembrane motif is between 2125 and 2141aa.
Chromosomal Localization of WF-HABP and OE-HABP
We determined the chromosomal localization of the WF-HABP and OE-HABP genes. The initial experiment for WF-HABP localization resulted in specific labeling of the short arm of a group A chromosome, which was believed to be chromosome 3 on the basis of size, morphology, and banding pattern. In the second experiment, a probe specific for the centromere of chromosome 3 was cohybridized with the WF-HABP probe. The experiment resulted in the specific labeling of the centromere in red and the short arm in green on chromosome 3 (Figure 8A) ▶ . Measurements of 10 specifically labeled chromosomes 3 demonstrated that WF-HABP is located at a position that is 46% of the distance from the centromere to the telomere of the chromosome arm, an area corresponding to 3p21.31. Using the same technique, we localized OE-HABP to a position that is 83% of the distance from the centromere to the telomere of chromosome arm 15q, an area that corresponds to band 15q25.2–25.3 (Figure 8B) ▶ . A total of 80 metaphase cells were analyzed for each gene, with more than 95% of the cells exhibiting specific labeling.
Figure 8.
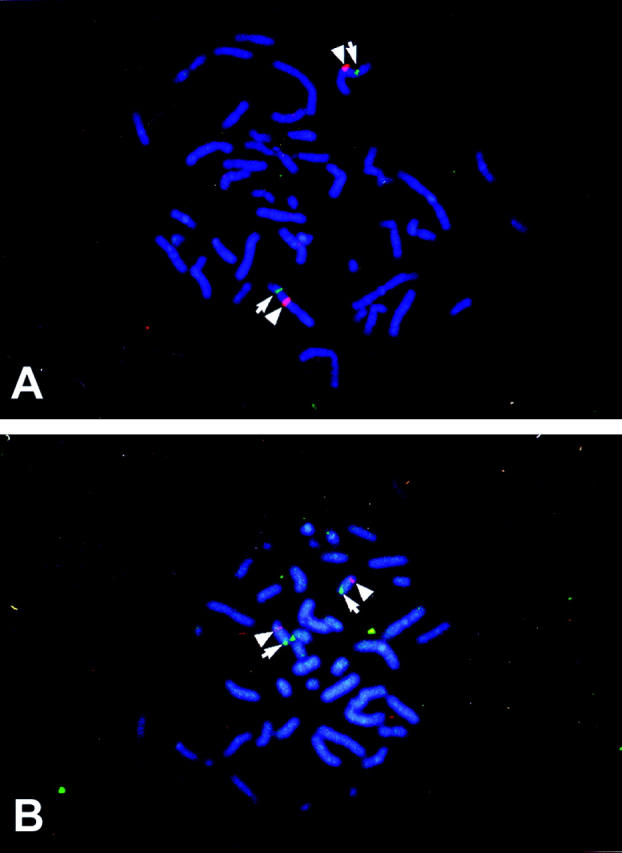
Chromosomal localization of WF-HABP and OE-HABP. A: WF-HABP was localized to chromosome 3 by fluorescence in situ hybridization to a position that is 46% of the distance from the centromere to the telomere of chromosome arm 3p, an area corresponding to 3p21.31. B: OE-HABP was localized to chromosome 15, at a position located 83% of the distance from the centromere to the telomere of chromosome arm 15q, an area that corresponds to 15q25.2–25.3. Red fluorescent staining (pointed to by a triangle) marks the centromere of chromosomes 3 and 15, and green fluorescence (marked by an arrow) specifies the location of the WF-HABP and OE-HABP genes, respectively.
Discussion
We have identified three novel cDNAs with sequence similarity to the HA-binding domain of TSG-6. Comparison of their sequence with all known HABPs revealed that the putative HA-binding domains of WF-HABP and BM-HABP share the greatest sequence homology with TSG-6. Furthermore, WF-HABP and BM-HABP have a high degree of sequence homology outside of the HA-binding domain, with an overall identity of 45% over the available amino acid sequences. Our data indicate that these genes are closely related and define a distinct subfamily of HABPs. WF-HABP was found to be identical to KIAA0246, a gene of unknown function characterized via high-throughput sequencing and encoding a polypeptide of 2212 amino acids. 22 We determined that this putative HA-binding protein contains a total of 16 EGF-like repeats that are similar to the EGF-like motif found in fibrillin, delta, jagged, and notch. 33,34 In addition, we identified a classical transmembrane domain that spans amino acids 2125–2141 of the protein. Our analysis suggests that WF-HABP is a transmembrane protein with a 72-amino-acid intracellular C-terminus segment, whereas the majority of the protein, including all of the EGF-like repeats and the HA-binding domain, is extracellular. The C-terminus has no homology with CD44 or any known protein motifs. However, the overall structure of WF-HABP is somewhat reminiscent of the protein structure of the jagged and notch proteins, and it will be interesting to determine the possible role of WF-HABP in cell-cell and cell-matrix interactions.
The tissue expression pattern of the three novel HABPs is quite distinct, with BM-HABP mRNA being expressed predominantly in the fetal liver and possibly weak expression in the adult liver and the fetal lung. Given that this cDNA was obtained from a bone marrow library and that fetal liver is a major hemopoietic organ, 35 it is possible that BM-HABP expression may be restricted to a hemopoietic stem cell population. In contrast, we found that the closely related WF-HABP was expressed in all of the tissues examined and was particularly prominent in the placenta, lung, and heart, with only the brain expressing a very low level of WF-HABP mRNA. The ubiquitous expression of this gene in various tissues is consistent with our finding that WF-HABP is localized to the endothelium, because blood vessels are found in virtually all tissues. It is possible that the difference in expression level observed in the various tissues is due to the degree of vascularization of these tissues. An alternative explanation is that the expression of WF-HABP may be differentially regulated in various tissues or in different-sized blood vessels. Our data generally agree with the results reported by Nagase et al, 22 who also observed a broad distribution of expression of this gene in various human tissues. However, they were unable to detect the expression of WF-HABP in the heart, whereas we observed prominent expression of this gene in the same organ. One possible explanation for this difference is that we may have examined tissues with a larger amount of intact blood vessels. In addition, we found that WF-HABP expression is considerably more prominent in quiescent endothelial cells versus proliferating cells, and it is possible that the heart tissues examined by Nagase et al 22 may contain a more activated endothelium. However, we also observed a very high level of expression of WF-HABP in fetal vessels of the placenta, but the growth or activation state of these cells is unknown. The expression pattern of WF-HABP by endothelial cells as a function of activation state, vessel size, or location warrants further investigation.
HA degradation fragments have been shown to enhance endothelial cell growth, migration, and tube formation in vitro 10,36 and angiogenesis in vivo. 37 In contrast, native high-molecular-weight HA is believed to inhibit angiogenesis. 9 There is some evidence to support a role for CD44 in mediating the activity of angiogenic HA. 38 Here we report the first HABP with an expression pattern that is mostly restricted to blood vessel endothelium and to cultured endothelial cells. Furthermore, our finding that expression of WF-HABP is regulated by the growth state of the endothelial cell suggests that this protein may play a role in endothelial cell growth and differentiation. Vascular endothelial cell growth factor receptor is an example of a protein with an endothelial cell restricted expression pattern that has a crucial role in angiogenesis. Given the potent angiogenic activity of HA degradation fragments, we speculate that WF-HABP may also have a critical role in modulating the angiogenic process and endothelial cell behavior.
Acknowledgments
We express our gratitude to Ms. Elizabeth Smith for her outstanding histological assistance and Dr. Christian Haudenschild for his invaluable histopathology expertise.
Footnotes
Address reprint requests to Dr. Gene Liau, Genetic Therapy, 938 Clopper Road, Gaithersburg, MD 20878. E-mail: gene.liau@pharma.novartis.com.
Supported by an American Heart Association grant-in-aid and National Institutes of Health grant HL37510.
References
- 1.Meyer K, Palmer JW: The polysaccharide of the vitreous humor. J Biol Chem 1934, 107:629-634 [Google Scholar]
- 2.Laurent TC, Fraser JRE: Hyaluronan. FASEB J, 1992, 6(7):2397–2404 [PubMed]
- 3.Brimacombe JS, Weber JM: Mucopolysaccharides. 1964. Elsevier, Amsterdam
- 4.Toole BP: Hyaluronan and its binding proteins, the hyaladherins. Curr Opin Cell Biol 1990, 2:839-844 [DOI] [PubMed] [Google Scholar]
- 5.Delpech B: Hyaluronan: fundamental principles and applications in cancer. J Intern Med 1997, 242:41-48 [DOI] [PubMed] [Google Scholar]
- 6.Toole BP: Proteoglycans and hyaluronan in morphogenesis and differentiation. Hay ED eds. Cell Biology of Extracellular Matrix. 1991, :pp 305-339 Plenum Press, New York [Google Scholar]
- 7.Laurent T, Laurent U, Fraser R: The structure and function of hyaluronan: an overview. Immunol Cell Biol 1996, 74:A1-A7 [DOI] [PubMed] [Google Scholar]
- 8.Fraser JRE, Laurent TC, Laurent UBG: Hyaluronan: its nature, distribution, function and turnover. J Intern Med 1997, 242:27-33 [DOI] [PubMed] [Google Scholar]
- 9.Deed R, Rooney P, Kumar P, Norton JD, Smith J, Freemont AJ, Kumar S: Early-response gene signaling is induced by angiogenic oligosaccharides of hyaluronan in endothelial cells. Inhibition by non-angiogenic, high-molecular-weight hyaluronan. Int J Cancer 1997, 71:251-256 [DOI] [PubMed] [Google Scholar]
- 10.West DC, Kumar S: Hyaluronan and Angiogenesis. Ciba Foundation Symposium 143. 1989, :pp 187-207 Wiley, Chichester [DOI] [PubMed] [Google Scholar]
- 11.Manuskiatti W, Maibach H: Hyaluronic acid and skin: wound healing and aging. Int J Dermatol 1996, 35:539-544 [DOI] [PubMed] [Google Scholar]
- 12.Knudson CB, Knudson W: Hyaluronan-binding proteins in development, tissue homeostasis, and disease. FASEB J 1993, 7:1233-1241 [PubMed] [Google Scholar]
- 13.Underhill, CB: CD44: the hyaluronan receptor. J Cell Sci 1992, 103:293–298 [DOI] [PubMed]
- 14.Sherman L, Sleeman J, Herrlich P, Ponta H: Hyaluronate receptors: key players in growth, differentiation, migration and tumor progression. Curr Opin Cell Biol 1994, 6:726-733 [DOI] [PubMed] [Google Scholar]
- 15.Yang B, Zhang L, Turley EA: Identification of two hyaluronan-binding domains in the hyaluronan receptor RHAMM. J Biol Chem 1993, 268:8617-8623 [PubMed] [Google Scholar]
- 16.Lee TH, Wisniewski HG, Vilcek J: A novel secretory tumor necrosis factor-inducible protein (TSG-6) is a member of the family of hyaluronate binding proteins, closely related to the adhesion receptor CD44. J Cell Biol 1992, 116:545-557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devereux J, Haeberli P, Smithies O: A comprehensive set of sequence analysis programs for VAX. Nucleic Acids Res 1984, 12:387-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goetinck PF, Stirpe NS, Tsonis PA, Carlone D: The tandemly repeated sequences of cartilage link protein contain the sites for interaction with hyaluronic acid. J Cell Biol 1987, 105:2403-2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wisniewski HG, Maier R, Lotz M, Lee S, Klampfer L, Lee TH, Vilcek J: TSG-6: a TNF-, IL-1-, and LPS-inducible secreted glycoprotein associated with arthritis. J Immunol 1993, 151:6593-6601 [PubMed] [Google Scholar]
- 20.Wisniewski HG, Hua JC, Poppers DM, Naime D, Vilcek J, Cronstein BN: TNF/IL-1-inducible protein TSG-6 potentiates plasmin inhibition by inter-a-inhibitor, and exerts a strong anti-inflammatory effect in vivo. J Immunol 1996, 156:1609-1615 [PubMed] [Google Scholar]
- 21.Ye L, Mora R, Akhayani N, Haudenschild C, Liau G: Growth factors and cytokine-regulated hyaluronan-binding protein TSG-6 is localized to the injury-induced rat neointima and confers enhanced growth in vascular smooth muscle cells. Circ Res 1997, 81:289-296 [DOI] [PubMed] [Google Scholar]
- 22.Nagase T, Seki N, Ishikawa K, Ohira M, Kawarabayasi Y, Ohara O, Tanaka A, Kotani H, Miyajima N, Nomura N: Prediction of the coding sequences of unidentified human genes. VI. The coding sequences of 80 new genes (KIAA0201-KIAA0280) deduced by analysis of cDNA clones from cell line KG-1 and brain. DNA Res 1996, 3:321-329 [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol 1990, 215:403-410 [DOI] [PubMed] [Google Scholar]
- 24.Garfinkel S, Hu X, Prudovsky IA, McMahon GA, Kapnik EM, McDowell SD, Maciag T: FGF-1-dependent proliferative and migratory responses are impaired in senescent human umbilical vein endothelial cells and correlate with the inability to signal tyrosine phosphorylation of fibroblast growth factor receptor-1 substrates. J Cell Biol 1996, 134:783-791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garfinkel S, Wessendorf JH, Hu X, Maciag T: The human diploid fibroblast senescence pathway is independent of interleukin-1 alpha mRNA levels and tyrosine phosphorylation of FGFR-1 substrates. Biochim Biophys Acta 1996, 1314:109-119 [DOI] [PubMed] [Google Scholar]
- 26.Goldberg DA: Isolation and partial characterization of the Drosophila alcohol dehydrogenase gene. Proc Natl Acad Sci 1980, 77:5794-5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsang SS, Yin X, Guzzo-Arkuran C, Jones VS, Davison AJ: Loss of resolution in gel electrophoresis of RNA: a problem associated with the presence of formaldehyde gradients. Biotechniques 1993, 14:380-381 [PubMed] [Google Scholar]
- 28.Winkles JA, Friesel R, Alberts GF, Janat MF, Liau G: Elevated expression of basic fibroblast growth factor in an immortalized rabbit smooth muscle cell line. Am J Pathol 1993, 143:518-527 [PMC free article] [PubMed] [Google Scholar]
- 29.Liau G, Yamada Y, deCrombrugghe B: Coordinate regulation of the levels of type IV and type I collagen mRNA in most but not all mouse fibroblasts. J Biol Chem 1985, 260:532-536 [PubMed] [Google Scholar]
- 30.Nuovo GJ: PCR In Situ Hybridization Protocols and Applications, ed 3. Philadelphia and New York, Lippincott-Raven Publishers, 1997, pp 123–193
- 31.Wight TN, Lara S, Riessen R, Le Baron R, Isner J: Selective deposits of versican in the extracellular matrix of restenotic lesions from human peripheral arteries. Am J Pathol 1997, 151:963-973 [PMC free article] [PubMed] [Google Scholar]
- 32.Tjurmin AV, Ananyeva NM, Smith EP, Gao Y, Hong MK, Leon MB, Haudenschild CC: Studies on the histogenesis of myxomatous tissue of human coronary lesions. Arterioscler Thromb Vasc Biol 1999, 19:83-97 [DOI] [PubMed] [Google Scholar]
- 33.Artavanis-Tsakonas S, Matsuno K, Fortini ME: Notch signaling. Science 1995, 268:225-232 [DOI] [PubMed] [Google Scholar]
- 34.Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J: Notch4/int-3, a mammary proto-oncogene, is an endothelial-specific mammalian Notch gene. Development 1996, 122:2251-2259 [DOI] [PubMed] [Google Scholar]
- 35.Timens W, Kamps WA: Hematopoiesis in human fetal and embryonic liver. Microsc Res Tech 1997, 39:387-397 [DOI] [PubMed] [Google Scholar]
- 36.Montesano R, Kumar S, Orci L, Pepper MS: Synergistic effect of hyaluronan oligosaccharides and vascular endothelial growth factor on angiogenesis in vitro. Lab Invest 1996, 75:249-262 [PubMed] [Google Scholar]
- 37.West DC, Hampson IN, Arnold F, Kumar S: Angiogenesis induced by degradation products of hyaluronic acid. Science 1985, 228:1324-1326 [DOI] [PubMed] [Google Scholar]
- 38.Slevin M, Krupinski J, Kumar S, Gaffney J: Angiogenic oligosaccharides of hyaluronan induce protein tyrosine kinase activity in endothelial cells and activate a cytoplasmic signal transduction pathway resulting in proliferation. Lab Invest 1998, 78:987-1003 [PubMed] [Google Scholar]