Abstract
Suppression subtractive hybridization was used to clone genes associated with proliferation of oval cells in rat liver regenerating after a 70% partial hepatectomy combined with the feeding of 2-acetylaminofluorene. A subset of the identified genes comprised interferon-γ receptor α subunit (IFN-γRα), gp91phox, interleukin-1β (IL-1β), lymphocyte function-associated molecule-1α (LFA-1), eukaryotic initiation factor-2-associated 67-kd protein (eIF-2-associated 67-kd protein), and α-fetoprotein, which constitute part of the cellular program modulated by IFN-γ. Therefore, expression analysis performed by Northern blotting and immunohistochemistry were extended to include IFN-γ, the IFN-γ receptor β subunit (IFN-γRβ), three secondary response genes induced by interaction of IFN-γ with IFN-γ receptor complexes, ie, IL-1β-converting enzyme (ICE), intercellular adhesion molecule-1 (ICAM-1), and urokinase-type plasminogen activator receptor (uPAR), and a cytokine inducing IFN-γ expression, ie, interleukin-18 (IL-18). The Northern blot analysis showed that all examined genes were modulated when progenitor-like oval cells were activated and recruited for liver regeneration. Immunohistochemistry localized the subunits of the IFN-γ receptor complex, IFN-γRα and IFN-γRβ, the secondary response genes uPAR and ICAM-1, the IFN-γ-inducing factor IL-18, and ICE to the ductular structures of oval cells. In contrast, during liver regeneration after a 70% partial hepatectomy, only modulation of IL-1β and ICE was observed. Our results, therefore, indicate that IFN-γ-mediated events may be particularly important when cells in the bile ductules must respond to liver damage by production of ductular oval cells.
The extent to which the adult human and rodent livers are capable of responding to loss of its cellular mass is remarkable. In a normal adult liver, this response can be provoked by partial resection, also called partial hepatectomy (PHx), and is usually accomplished by replication of existing mature epithelial cells, provided the remaining cells are left functionally intact. 1 However, it is now widely acknowledged that impairment of the regenerative capacity of hepatocytes may result in proliferation and migration of cells forming ductular structures and initially continuous with the biliary tree. In rodents, the ductular structures are composed of the so-called oval cells, and their capacity to proliferate and differentiate into hepatocytes in association with impaired regeneration has led to the conclusion that they may represent progeny of facultative liver stem cells. 2,3 Oval cells, named because of their morphological appearance as small, proliferating epithelial cells with an ovoid nucleus and scant basophilic cytoplasm, were first observed in livers of rats exposed to chemical carcinogens. 4 In recent years, considerable interest has evolved in the hepatic oval cells, as they may give rise to new hepatocytes and bile duct cells, but may also represent a target population for hepatic carcinogens. To better understand the biology of oval cells and their role in liver pathogenesis, elucidation of the mechanisms involved in their activation, migration, and differentiation has become an important issue.
Several experimental rodent models have been used to study oval cell proliferation and differentiation in vivo. 5 We have focused our efforts on studying the mechanisms involved in oval cell activation, proliferation, and differentiation in the rat liver using the modified Solt-Farber protocol, in which feeding of 2-acetylaminofluorene (2-AAF) is combined with a 70% partial hepatectomy (AAF/PHx). Feeding with 2-AAF activates cells in the bile ductules to undergo replication and acquire an oval cell phenotype with expression of hepatocyte specific genes, and the partial hepatectomy provides the necessary stimulus to induce massive proliferation and migration of the activated oval cells into the liver parenchyma, where they differentiate to give rise to mature hepatocytes. 2,6,7 Our studies have indicated the involvement of a number of growth modulators, including stem cell factor (SCF), transforming growth factor α (TGF-α), epidermal growth factor (EGF), hepatocyte growth factor (HGF), urokinase-type plasminogen activator (uPA), leukemia inhibitory factor (LIF), and their receptors in oval cell proliferation, and the cellular localization of the different components indicates that an intricate network of paracrine and autocrine mechanisms of growth modulation is involved. 8-13
Despite the research efforts, our knowledge of the mechanisms involved in oval cell activation and differentiation is still limited. To acquire a better understanding of the mechanisms involved, we have begun identifying and characterizing genes that are highly expressed when oval cells are activated and recruited for liver regeneration. In the present study, we have used suppression subtractive hybridization to clone genes with increased expression during oval cell proliferation.
We report here the identification of several genes that we believe may play a role in generating the oval cell response in vivo. A common feature of the identified genes is their connection to the cellular programs orchestrated by interferon-γ (IFN-γ). Using Northern blot and immunohistochemical studies, we further investigated the expression of a number of genes connected to the modulating network of IFN-γ. We show that these genes, which include IFN-γ itself, the IFN-γ receptor subunits α (IFN-γRα) and β (IFN-γRβ), primary and secondary response genes induced by interaction of IFN-γ with IFN-γ receptor complexes (ie, gp91phox, interleukin-1β-converting enzyme (ICE), intercellular adhesion molecule-1 (ICAM-1), and urokinase-type plasminogen activator receptor (uPAR)), cytokines inducing IFN-γ expression (ie, interleukin-18 (IL-18) and interleukin-1β (IL-1β)), and cell adhesion molecules orchestrating lymphocyte-epithelial cell interactions (ie, lymphocyte function-associated molecule-1α (LFA-1)) are all modulated when progenitor-like oval cells are activated and recruited for liver regeneration in the AAF/PHx protocol. However, during liver regeneration from existing mature liver cells, modulation of the majority of these genes cannot be observed. Therefore, our results indicate that IFN-γ-mediated events may be particularly important when cells in the bile ductules are activated in response to liver damage by production of ductular oval cells.
Materials and Methods
Animal Models
In experiments combining the feeding of 2-AAF with a two-thirds partial hepatectomy (AAF/PHx), 8- to 9-week-old male Fischer 344 rats (120–150 g body weight) were given a daily dose of 20 mg/kg 2-AAF by gavage for 4 consecutive days. On day 5 after initiation of the 2-AAF treatment, resection of approximately 70% of the liver (left lateral and median lobe PHx) was performed, followed by administration of 2-AAF for an additional 4 days. All animals were kept under standardized conditions with access to food and water ad libitum and maintained according to the guidelines established at the National Institutes of Health.
For Northern blot analysis, animals were sacrificed 96 hours after initiation of 2-AAF treatment and 7, 8, 10, and 11 days after PHx was performed. After excision of the liver, sections of all liver lobes were fixed in formalin, and the remaining liver tissue snap-frozen in liquid nitrogen. Liver tissue for Northern blot analysis was also obtained from animals sacrificed 1, 6, 12, 18, 24, 30, 36, 42, 48, 54, or 60 hours after a 70% hepatectomy (PHx) without administration of 2-AAF. For isolation of nonparenchymal cell populations after exposure to 2-AAF, animals were treated according to the AAF/PHx protocol or received a daily dose of 20 mg/kg 2-AAF by gavage for up to 96 hours and used for cell isolation as described below.
Isolation of Nonparenchymal Liver Cells and Construction of the Suppression Subtractive Hybridization Library
Although a number of cell types comprised of parenchymal hepatocytes as well as nonparenchymal bile duct, stellate, endothelial, and Kupffer cells may be involved in generating the oval cell response, we were particularly interested in identifying genes that were differentially expressed in the nonparenchymal cell compartments in the liver regenerating from oval cells. Because our previous studies in the AAF/PHx protocol have shown the presence of numerous ductular structures of proliferating oval cells and no overt differentiation of oval cells into foci of basophilic hepatocytes and intestinal-type structures at day 7 after PHx, 2,7 nonparenchymal cell populations containing oval cells were separated from hepatocytes by perfusion of rat livers at this time point (AAF/PHx 7d) and, together with nonparenchymal cell populations isolated from normal liver (control), used for generation of the suppression subtractive hybridization library. The isolation procedure has been described in detail by Bisgaard et al. 11 In brief, liver cells were released by a three-step perfusion procedure in situ, employing first a perfusion with Hanks’ balanced salt solution without calcium and magnesium, second a perfusion with 0.2% pronase, and third a perfusion with 0.1% pronase, 0.1% collagenase type I, and 0.007% DNase I. Viable nonparenchymal cell populations were purified by centrifugation through a two-step Percoll gradient. To ensure that the isolated nonparenchymal cells were comprised of the expected cell populations represented by oval, bile duct, stellate, Kupffer, and endothelial cells, we analyzed the expression of a number of factors expected to be differentially expressed between AAF/PHx cells and cells isolated from control liver. The parameters included γ-glutamyltranspeptidase (γ-GT) as a marker for epithelial cells of ductal origin, α-fetoprotein 14 as a marker for oval cells, and HGF 10 and uPA 11 as markers for cells of nonepithelial origin. In the nonparenchymal cell populations from control liver approximately 10 to 20% of the cells stained positive for γ-GT, indicating their ductal origin. In accordance with the increased numbers of γ-GT-positive cells observed in livers regenerating by recruitment of oval cells, 13 30 to 50% of the cells isolated from AAF/PHx 7d livers were positive for γ-GT. Other genes expressed in nonparenchymal cell populations during liver regeneration from oval cells, including α-fetoprotein, HGF, and uPA, were all increased in nonparenchymal cell populations from AAF/PHx-treated animals relative to cell populations obtained from control liver when assessed by Northern blot analysis (data not shown). Thus, the isolated nonparenchymal populations were comprised of the expected cell populations and were subsequently used in the suppression subtractive hybridization analysis. The final nonparenchymal cell preparations contained less than 1% hepatocytes.
Total RNA from isolated nonparenchymal cell preparations was obtained by lysis of cell pellets in RNAstat Reagent (Tel-Test Inc., Friendswood, TX) and polyadenylated RNA (poly(A)+) selected by passage over an oligo(dT) column (Invitrogen Corp., Carlsbad, CA). The suppression subtractive hybridization was performed with the Clontech PCR-Select cDNA Subtraction Kit (Clontech Laboratories Inc., Palo Alto, CA) according to the manufacturer’s instructions. Samples of 2-μg poly(A)+ RNA were reverse-transcribed to cDNA. In the present study, we were interested in identifying genes that were modulated in nonparenchymal cells from rat liver regenerating after the combined feeding of 2-AAF and a PHx (AAF/PHx 7d mRNA) as compared to nonparenchymal cells isolated from normal rat liver (control mRNA). The cDNA synthesized from AAF/PHx 7d mRNA was therefore used as tester cDNA and cDNA generated from control mRNA as driver cDNA. In brief, the tester and driver cDNA were digested with RsaI and the tester cDNA ligated to the adapter DNA. After two repeated hybridizations of tester and driver cDNA, remaining unhybridized sequences (representing cDNAs expressed highly in the tester sample) were amplified by polymerase chain reaction (PCR) using flanking and nested primers that anneal the adaptor cDNA and the subtracted PCR products ligated into the pCR 2.1 plasmid vector (TA Cloning kit, Invitrogen). The identity of the cDNA inserts was revealed by sequence analysis of purified plasmids using the Thermo Sequenase Cycle Sequencing Kit (Amersham Life Science, Cleveland, OH), and comparison to known DNA sequences using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST/).
Isolation of Nonparenchymal Cells for Northern Blot Analysis
Cell populations enriched in bile epithelial and/or oval cells were isolated according to the protocol described by Bisgaard et al. 11 Animals treated with 2-AAF were sacrificed 0 (normal), 3, 6, 24, 48, and 96 hours after initiation of the 2-AAF treatment regimen. Nonparenchymal cell populations were isolated by perfusion and Percoll purification as described above. Kupffer cells were removed from the nonparenchymal cell populations by selective adherence to plastic tissue culture dishes. Removal of macrophages, endothelial cells, and red blood cells was achieved by selective panning using the mouse monoclonal antibody OX43 (MCA276; Serotec, Accurate Chemical and Scientific Corp., Westbury, NY). Cell preparations were snap-frozen in liquid nitrogen and stored at −70°C until processed for total RNA isolation and Northern blot analysis.
Northern Blot Analysis
Polyadenylated RNA was prepared from snap-frozen liver tissue by ultracentrifugation through a cesium chloride cushion and enrichment by oligo(dT)-cellulose chromatography. Samples of 5 μg poly(A)+ RNA or 20 μg total RNA were electrophoresed in 1% agarose/0.2 formaldehyde gels, transferred onto nylon membranes, and hybridized to cDNA probes labeled with [32P]dCTP (rediprime, Amersham Life Science) using the QuickHyb solution from Stratagene (La Jolla, CA). The following rat cDNAs were obtained from the subtraction analysis: IFN-γRα (homology to mouse IFN-γRα, accession number M28995, 85% identity); gp91phox (homology to mouse gp91phox, accession number U43384, 96% identity); IL-1β (homology to rat IL-1β, accession number M98820, 98% identity); eIF-2-associated 67-kd protein (homology to rat eIF-2-associated 67-kd protein, accession number L10652, 100% identity); LFA-1 (homology to mouse LFA-1, accession number M60778, 91% idenitity); α-fetoprotein (homology to rat α-fetoprotein, accession number X02361, 98% identity); ebnerin (homology to rat ebnerin, accession number U32681, 100% identity). A cDNA fragment of mouse uPA (kindly provided by Dr. J. L. Degen, Children’s Hospital Medical Center, Cincinnati, OH) and rat uPAR (kindly provided by Dr. D. Dichek, J. David Gladstone Institutes, San Fransisco, CA) were also used as probes.
The cDNA probes for IFN-γ, IL-18, ICE, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were generated by reverse transcription and subsequent amplification by polymerase chain reaction (RT-PCR). Poly(A)+ RNA (0.5 μg) isolated from whole liver homogenate at day 10 in the AAF/PHx protocol (AAF/PHx 10d) was reverse transcribed using MMLV-Reverse transcriptase and oligo(dT)18 primers (Advantage RT-for-PCR kit, Clontech). The cDNAs provided templates for PCR using the following forward and reverse primers: rat IFN-γ (404 bp), 5′-tactgccacggcacagtcattgaa-3′ and 5′-gcagcgactccttttccgcttcct-3′; rat IL-18 (446 bp), 5′-actgtacaaccgcagtaatacgg-3′ and 5′-agtgaacattacagatttatccc-3′; rat ICE (1189 bp), 5′-atggccgacaaggtcctgagggc-3′ and 5′-agaaacgttttgtcagggtca-3′; and rat GAPDH (651 bp), 5′-accacagtccatgccatcac-3′ and 5′-tccaccaccctgttgctgta-3′. Oligonucleotide primers were derived from published sequences in the GenBank/EMBL/DDBJ databank. All RT-PCR products were ligated into the pCRII plasmid vector (TA Cloning kit Dual Promoter, Invitrogen) and sequenced to confirm their identity.
Immunohistochemistry
Livers were fixed in 10% neutral formalin and processed for routine histology. Immunohistochemistry was performed on 5-μm tissue sections as previously descibed. 11 Specific antibody binding was revealed by the appropriate Vectastain ABC Elite kits with diaminobenzidine as substrate in combination with an enhancing solution (Vector Laboratories, Burlingame, CA) followed by light counterstaining with Gill’s hematoxylin.
The primary antibodies were as follows: a goat polyclonal antibody against the carboxy terminus of the IL-18 precursor of mouse origin (diluted 1:100); a rabbit polyclonal antibody against the carboxy terminus of the p20 subunit of mouse ICE (diluted 1:500); a rabbit polyclonal antibody raised against the carboxy terminus of the mouse interleukin-1 receptor I (IL-1RI) precursor (1:25); a rabbit polyclonal antibody directed against the carboxy terminus of IFN-γRα (diluted 1:500); a rabbit polyclonal antibody against the carboxy terminus of the IFN-γRβ precursor of mouse origin (diluted 1:250); and a goat polyclonal antibody raised against the carboxy terminus of mouse ICAM-1 (1:1000). These antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The rabbit polyclonal antibody against rat uPAR (diluted 1:10) was obtained from American Diagnostica (Greenwich, CT).
Results
Identification of Genes by Suppression Subtractive Hybridization
In the present study, we used the suppression subtractive hybridization technique to clone the genes expressed in nonparenchymal cell populations during activation and proliferation of oval cells in the AAF/PHx protocol. A subset of the identified genes encoded components of the gene network connected to the proinflammatory cytokine IFN-γ, ie, IFN-γRα, gp91phox, IL-1β, LFA-1, eIF-2-associated 67-kd protein, and α-fetoprotein (Table 1) ▶ .
Table 1.
Summary of Genes and Gene Products
| Gene products identified by suppression subtractive hybridization and Northern blot analysis | ||
|---|---|---|
| Gene name/alternative name | Remarks | References |
| Interferon-γ receptor α (IFN-γRα) | Exhibits ligand binding properties towards IFN-γ. | 30 |
| gp91phox | Heavy chain of cytochrome b558. Gp91 phox is induced as a primary response gene by IFN-γ. | 31 |
| Interleukin-1β (IL-1β) | IL-1β binds to IL-1RI and induces transcription and translation of IFN-γ in hepatocytes. | 17 |
| Lymphocyte function-associated molecule 1-α (LFA-1) β2αL integrin; CD18, αL subunit; CD11a | LFA-1 is regulated in response to IFN-γ. Binds ICAM-1 and ICAM-2, which are induced as secondary response genes by IFN-γ. | 32 |
| Eukaryotic initiation factor-2-associated 67-kd protein | Protects eIF-2 from inhibitory phosphorylation by IFN-γ induced eIF-2 kinases. | 33, 34 |
| α-fetoprotein | Suppresses IFN-γ production. | 35 |
| Genes studied by Northern blot analysis and/or immunohistochemistry | ||
|---|---|---|
| Gene name/alternative name | Remarks | References |
| Interferon-γ (IFN-γ) | High-affinity ligand of IFN-γRα. | 20 |
| Interleukin-18 (IL-18); Interferon gamma-inducing factor (IGIF) | Induces IFN-γ production. | 16 |
| Interleukin-1β converting enzyme (ICE) caspase-1 | ICE is induced as a secondary response gene by IFN-γ. ICE is involved in the activation of IL-1β and IL-18. | 19, 36 |
| IFN-γ receptor β (IFN-γRβ) | β-chain of the IFN-γ receptor complex, involved primarily in signaling. | 20 |
| Urokinase-type plasminogen activator receptor (uPAR) | uPAR is induced by IFN-γ as a secondary response gene. | 37 |
| Urokinase-type plasminogen activator (uPA) | uPA is induced by IFN-γ. | 37 |
| Intercellular adhesion molecule-1 (ICAM-1); CD54 | ICAM-1 is induced as a secondary response gene by IFN-γ, binds to LFA-1. | 20 |
In the AAF/PHx protocol of liver regeneration, treatment with 2-AAF induces cells in the bile ductules to enter DNA synthesis and acquire an oval cell phenotype characterized by expression of α-fetoprotein, albumin, and hepatic transcription factors within 96 hours after exposure to the chemical. 6 The subsequent partial hepatectomy then provides the necessary growth stimulus for the massive proliferation, expansion, and finally differentiation of the AAF-induced oval cells in the liver parenchyma. 2,7 To investigate if the genes cloned by suppression subtractive hybridization were modulated when oval cells were activated and recruited for liver regeneration, we performed a Northern blot analysis with whole liver mRNA extracts derived from a time course experiment with the AAF/PHx protocol. The time points of the experiment were chosen to investigate the modulation of the identified genes during i) the induction of the oval cell phenotype in response to 2-AAF, which occurs within 96 hours after initiation of the 2-AAF exposure, 6 ii) the expansion of the oval cell populations into the liver parenchyma which can be observed at PHx 7d and 8d, 2,7 and iii) differentiation of oval cells resulting in development of foci containing basophilic hepatocytes, which can be observed at PHx 10d and 11d. 2,7 The Northern blot analysis revealed that the steady-state mRNA levels of the cloned genes increased in a manner consistent with activation of oval cells by 2-AAF (Figure 1) ▶ . However, whereas transcripts for IFN-γRα, gp91phox, IL-1β, LFA-1, and eIF-2-associated 67-kd protein were detected at low levels in normal liver and increased in response to treatment with 2-AAF, α-fetoprotein and ebnerin mRNA were detected in livers only after treatment with 2-AAF. In the present study, high steady-state levels of transcripts for all genes were detected 10 and 11 days after partial hepatectomy, a time when the number of oval cells peaked, as revealed by morphological studies (data not shown). Although the analysis of GAPDH expression was intended to provide an internal control for the amount of mRNA loaded, the expression of the gene was increased during liver regeneration in the AAF/PHx protocol (Figure 1) ▶ . Similar results were obtained for β-actin, which excluded a densitometric analysis as an appropriate numerical basis for comparison of the Northern blots. However, the GAPDH expression did confirm the integrity of the isolated mRNA.
Figure 1.
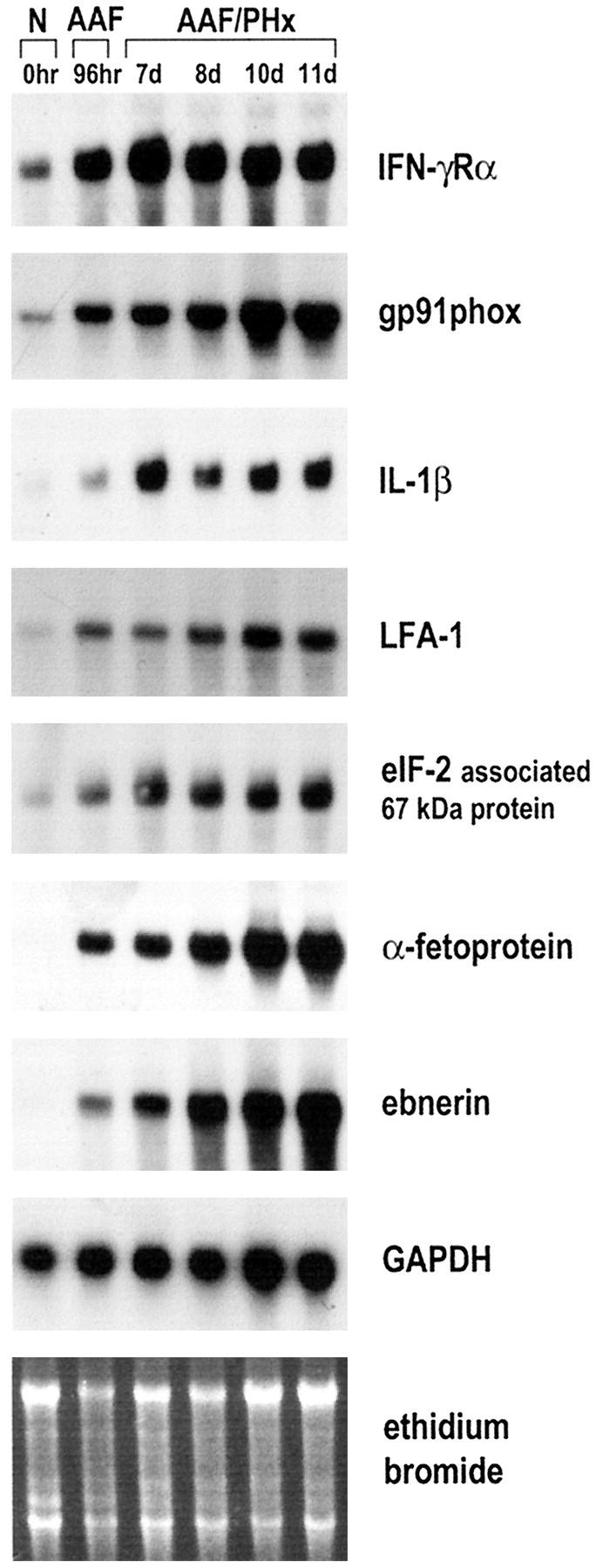
Northern blot analysis of genes identified by suppression subtractive hybridization analysis in the AAF/PHx protocol of liver regeneration. Poly(A)+-selected RNA was isolated from snap-frozen normal rat liver (N) and livers from AAF-treated animals 96 hours after initiation of the protocol (AAF) and 7, 8, 10, and 11 days after the partial hepatectomy (AAF/PHx). Five micrograms of mRNA were loaded in each lane. 32P-labeled cDNA probes cloned by suppression subtractive hybridization were used for hybridization. Hybridization with GAPDH and staining with ethidium bromide were used to assess integrity and equal loading of RNA samples.
Genes Identified by Suppression Subtractive Hybridization Are Selectively Expressed in Nonparenchymal Cells during Oval Cell Proliferation
As demonstrated in Figure 1 ▶ , the steady-state mRNA levels of the studied genes were increased in the liver within 96 hours after treatment with 2-AAF had begun. However, the increased expression could be due to an increased number of cells expressing a certain gene or, alternatively, to an induction of gene expression in certain cell populations. To evaluate the modulation of the cloned genes in response to 2-AAF with special emphasis on the modulation in various cell populations in the liver, we next separated the cells from rat livers exposed to 2-AAF for up to 96 hours by selective perfusion techniques and extracted total RNA. The separated cell populations consisted of either purified hepatocytes or nonparenchymal cells devoid of Kupffer cells, macrophages, endothelial cells, and red blood cells. Northern blot analysis showed that although none of the genes except GAPDH was expressed at significant levels in hepatocytes (data not shown), they were expressed in cell populations enriched in bile epithelial and/or oval cells (Figure 2) ▶ . gp91phox, IL-1β, α-fetoprotein, and ebnerin showed low expression in samples from normal rats but were induced within 96 hours after exposure to 2-AAF. In contrast, nonparenchymal cell populations isolated from normal rats showed high steady-state levels of IFN-γRα and eIF-2-associated 67-kd protein, the expression of which did not change significantly in response to 2-AAF. Therefore, we have identified at least two categories of genes by suppression subtractive hybridization. One category represents genes with induced expression in response to 2-AAF. The other category of genes is constitutively expressed in a specific cell type, but the relative abundance of the cells in the nonparenchymal cell populations isolated at day 7 in the AAF/PHx protocol may be increased as compared to nonparenchymal populations isolated from control liver.
Figure 2.
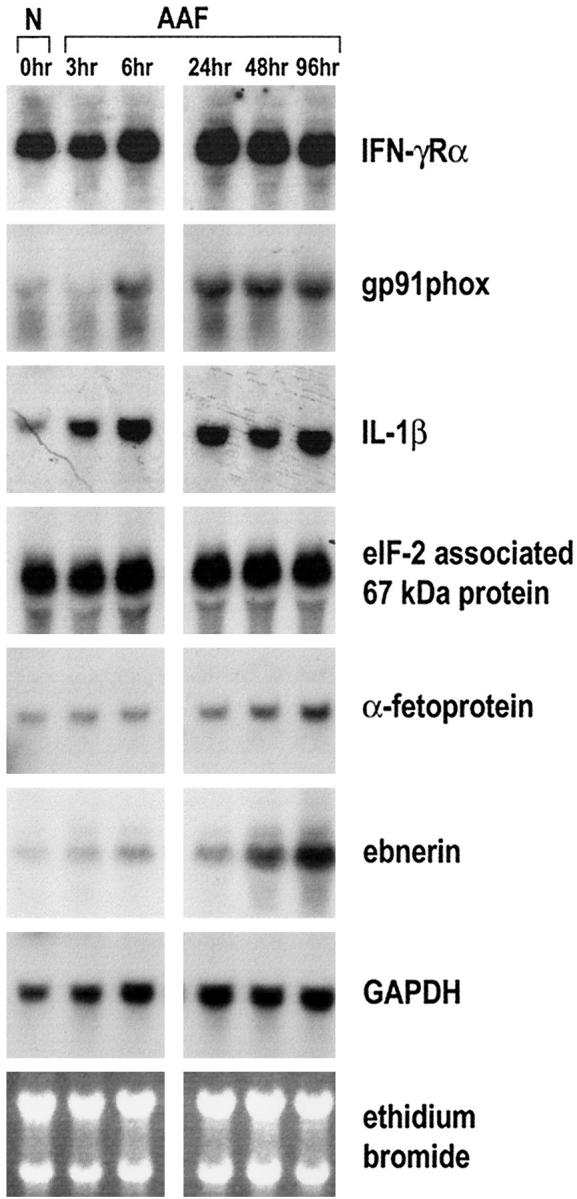
Northern blot analysis of genes identified by suppression subtractive hybridization analysis in selected nonparenchymal cell populations isolated from rat liver by pronase perfusion after administration of AAF. Animals were administered 2-AAF at 0 hours and killed for cell isolation and subsequent RNA extraction at the time points indicated. Twenty micrograms of total RNA were loaded in each lane. 32P-labeled cDNA probes cloned by suppression subtractive hybridization analysis were used for hybridization. Hybridization with GAPDH and staining with ethidium bromide were used to assess integrity and equal loading of RNA samples.
Of the identified genes, only expression of α-fetoprotein has previously been shown to be a feature of activated bile ductular cells and expanding oval cells in the AAF/PHx protocol. 6,14 Ebnerin expression has not been reported in oval cells, but a homologous gene has been shown to be expressed in the bile ducts of the adult mouse liver. 15 Therefore, we conclude that the suppression subtractive hybridization technique has provided us with a number of genes that are modulated when cells in the ductular structures are activated to acquire an oval cell phenotype in response to 2-AAF and stimulated to give rise to expanding populations of ductular oval cells in the AAF/PHx protocol of liver regeneration.
Of known oval cell markers, only α-fetoprotein was identified in our initial screen of the suppression subtractive hybridization library. However, other genes expressed in oval cells, such as the embryonic form of glutathione-S-transferase (Yp form) and γ-GT, would also be expected to be represented in the library. At the present time we have sequenced 50 clones containing cDNA fragments but are currently expanding our analysis of the library. We expect that the increased number of clones analyzed will result in detection of other known oval cell markers and identify genes which, as shown for ebnerin, may be induced in nonparenchymal liver cells in response to 2-AAF.
IFN-γ, IGIF/IL-18 and ICE Are Expressed during Liver Regeneration from Oval Cells
Since the subtraction suppression hybridization had identified several genes expressed as components of the cellular programs orchestrated by IFN-γ (Table 1) ▶ , we investigated the expression of IFN-γ in the AAF/PHx protocol of liver regeneration. As shown by the Northern blot analysis in Figure 3 ▶ , IFN-γ steady-state mRNA levels were not detectable in normal liver, but increased to low but detectable levels during oval cell activation and proliferation. Having established that IFN-γ expression is induced in the AAF/PHx protocol, we then wanted to study the expression of genes involved in IFN-γ production (ie, IL-18 and ICE). As demonstrated in Figure 3 ▶ , IL-18 transcripts were detected at very low levels in normal rat liver. Treatment with 2-AAF resulted in increased steady-state levels of IL-18 transcripts, and, as oval cells expanded into the parenchyma in response to the PHx, the levels increased further and remained elevated throughout the time frame of the experiment. Immunohistochemistry on tissue sections at day 7 after PHx showed that IL-18 protein was highly expressed in sinusoidal lining cells as well as in the ductular structures of oval cells (Figure 4B) ▶ . Hepatocytes did not show significant staining.
Figure 3.
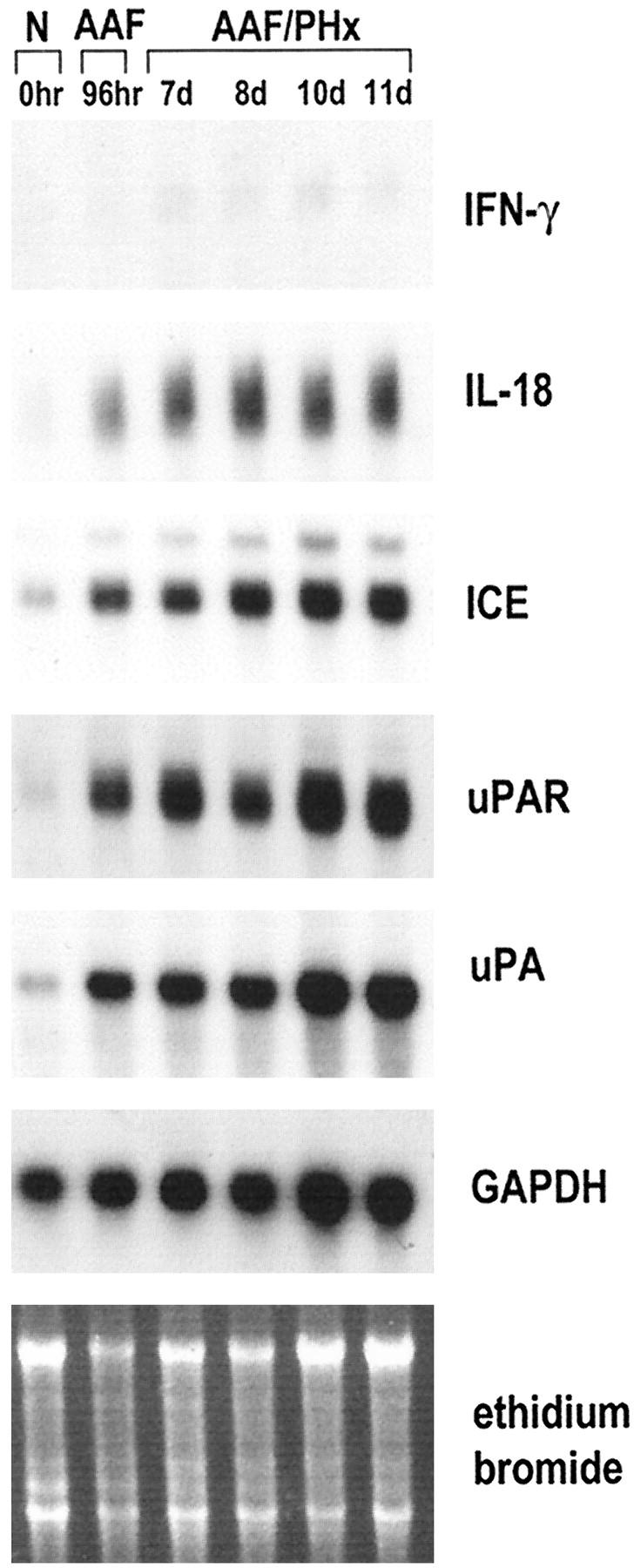
Northern blot analysis of IFN-γ, IL-18, ICE, uPAR, and uPA expression during oval cell activation and proliferation in the AAF/PHx protocol of liver regeneration. The experiment was as described in Figure 1 ▶ . Hybridization with GAPDH and staining with ethidium bromide were used to assess integrity and equal loading of RNA samples.
Figure 4.
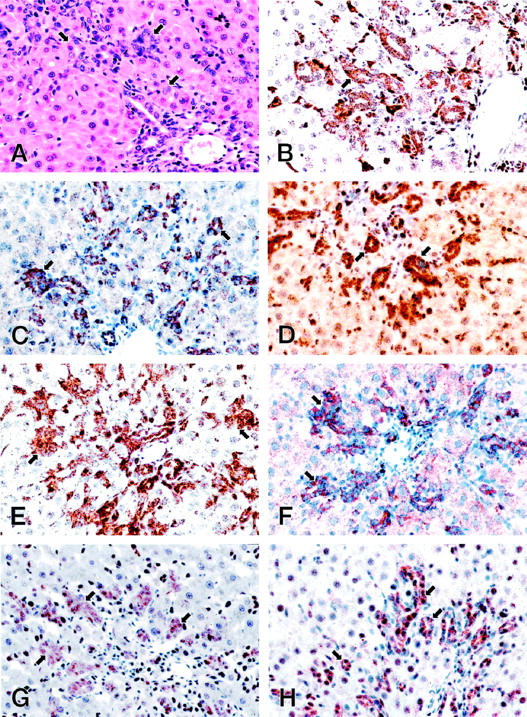
Cellular localization of IL-18, ICE, IL-1RI, IFN-γRα, IFN-γRβ, uPAR, and ICAM-1 in the rat liver regenerating by recruitment of oval cells. Paraffin sections from rat liver at day 7 after partial hepatectomy in the AAF/PHx protocol were (A) stained with H&E to demonstrate the histological features; or immunostained using the peroxidase method with diaminobenzidine as detection substrate and antibodies against IL-18 (B); ICE (C); IL-1RI (D); IFN-γRα (E); IFN-γRβ (F); uPAR (G); and ICAM-1 (H). Original magnification for all sections, ×400. Portal areas with strongly stained ductular structures of oval cells extending into the liver parenchyma are shown.
IL-18 is a strong inducer of IFN-γ production. 16 However, in hepatocytes IL-1β has also been shown to induce IFN-γ production. 17 Having demonstrated that expression of both IL-1β and IL-18 is induced in populations containing oval cells, we next wanted to examine the expression of ICE, the protease that converts the precursors of the two cytokines to their active forms. 18,19 Low steady-state mRNA levels of ICE could be detected in normal liver, but the levels were highly elevated after treatment with 2-AAF and increased further as oval cells proliferated and expanded into the liver parenchyma (Figure 3) ▶ . Immunohistochemistry for ICE protein showed strong staining over the ductular structures of oval cells (Figure 4C) ▶ . Therefore, proliferating oval cells appeared able to produce the precursors of IL-β and IL-18 and to process the proteins to their active forms. Strong immunohistochemical staining for IL-1RI, the high-affinity receptor for IL-1β, was detected in oval cells as well as in the hepatocytes (Figure 4D) ▶ , suggesting that IL-1β may be acting by both autocrine and paracrine mechanisms during liver regeneration in the AAF/PHx protocol.
The Receptors for IFN-γ Are Highly Expressed in Ductular Structures of Oval Cells
IFN-γ interacts with a specific cell surface receptor consisting of two subunits: the α-chain, exhibiting high-affinity ligand binding properties, and the β-chain, required primarily for signaling. 20 In the AAF/PHx protocol, IFN-γRα protein was detected in ductular structures of oval cells as well as in cells located in the sinusoids (Figure 4E) ▶ . The protein was not detected in mature hepatocytes.
Ductular structures of oval cells also showed a strong positive reaction for IFN-γRβ protein, whereas mature hepatocytes stained weakly positive (Figure 4F) ▶ . Unlike IFN-γRα, IFN-γRβ was not detected in other nonparenchymal cell populations in the liver. Therefore, in the AAF/PHx protocol, the ductular structures of oval cells express the highest levels of the IFN-γ receptor subunits, which are both required for proper cellular signaling by IFN-γ.
Genes Regulated in Response to IFN-γ Signaling Are Induced in Oval Cells
A number of genes are regulated in response to ligation of IFN-γ to its receptor. 20 gp91phox, which is induced as a primary response gene, was cloned in the present study by suppression subtractive hybridization (Figure 1) ▶ and shown to be induced in nonparenchymal cell populations containing activated bile duct and/or oval cells (Figure 2) ▶ . Furthermore, other genes induced by IFN-γ, including the protease uPA and its receptor uPAR, were also induced in these cell populations as revealed by Northern blot analysis and immunohistochemistry (Figures 3 and 4G) ▶ ▶ , as was ICAM-1, which is characterized as a secondary response gene to IFN-γ (Figure 4H) ▶ . These observations support the hypothesis that IFN-γ is an important player in the establishment of the oval cell phenotype in the AAF/PHx protocol.
Expression of Genes in the IFN-γ Modulating Network during Liver Regeneration from Existing Mature Epithelial Cells (PHx)
In recent years it has become apparent that many of the numerous growth factors and cytokines, notably HGF, TGF-α, EGF, tumor necrosis factor α (TNF-α), and IL-6, that initiate or facilitate the normal regenerative response from existing liver cells (ie, hepatocytes and bile duct cells) after PHx are also implicated in regeneration from oval cells. 1,3 An increase in HGF mRNA is observed in hepatic stellate cells 3 to 6 hours after a 70% PHx and gradually returns to undetectable levels within 72 to 96 hours after PHx. 21 Similarly, TGF-α mRNA is induced in hepatocytes within 2 to 3 hours after PHx, rises to a peak between 12 and 24 hours, and remains elevated for at least 48 hours after PHx. 22 In the AAF/PHx protocol of liver regeneration, HGF and TGF-α mRNA are induced 96 hours after exposure to 2-AAF, are further increased in response the PHx, and remain expressed at high levels throughout the period of oval cell expansion and differentiation, to return to levels seen in normal liver at the end of the regeneration process. 23 These observations prompted us to investigate if the IFN-γ modulating network was also involved in regeneration after a 70% PHx. A Northern blot analysis spanning the first 60 hours after PHx showed that only IL-1β mRNA reached steady-state levels comparable to those observed in the AAF/PHx protocol of liver regeneration (Figure 5) ▶ . Interleukin-1β transcripts were detected in two major peaks 12 and 36 hours after PHx. Low steady-state levels of transcripts for IFN-γRα, IL-18, and ICE were detectable in all samples when probe hybridizations were performed in parallel with the AAF/PHx experiments and exposure times of the autoradiographs were identical. A small elevation in transcript levels over control for IL-18 and ICE was observed 42 hours after PHx, when DNA synthesis in the bile duct epithelium is at its maximum. 1 Transcripts for IFN-γ, LFA-1, α-fetoprotein, and ebnerin were undetectable (data not shown). Therefore, the IFN-γ-modulating network appears to be a major player in the regenerative process only when oval cells are activated and recruited for tissue repair as exemplified by the AAF/PHx model.
Figure 5.
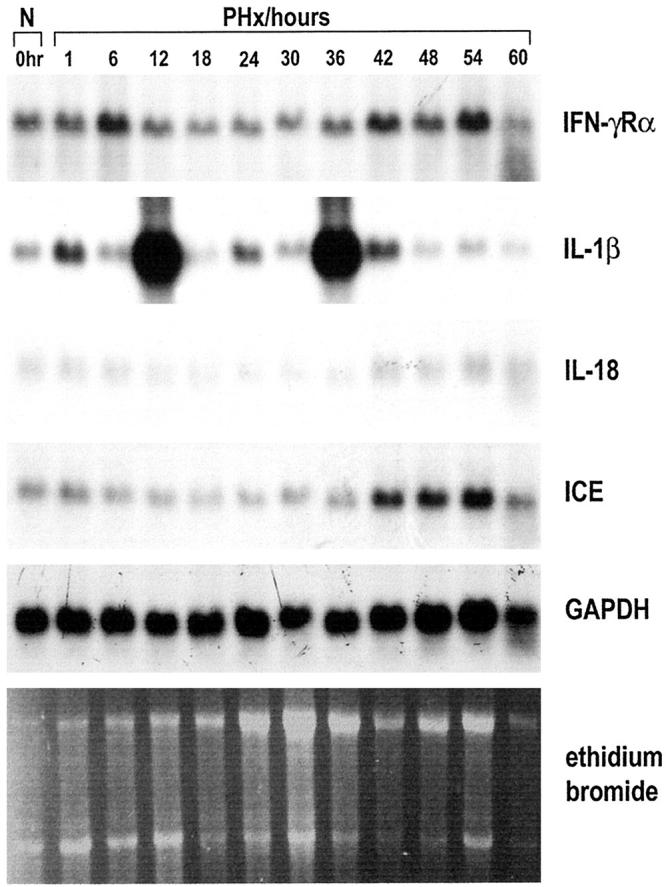
Northern blot analysis of 5 μg poly(A)+-selected RNA hybridized to 32P-labeled cDNA probes for IL-1β, IFN-γRα, IL-18, and ICE. A 70% PHx was performed and animals sacrificed 0 (normal), 1, 6, 12, 24, 30, 36, 42, 48, 54, and 60 hours after the operation. Hybridization with GAPDH and staining with ethidium bromide were used to assess integrity and equal loading of RNA samples.
Discussion
IFN-γ is a pleiotropic cytokine orchestrating a remarkable range of distinct cellular programs. 20 The effects are implemented by complex patterns of cell-specific gene regulation mediated by binding of the cytokine to IFN-γ receptors on the cell surface, followed by activation of intracellular signaling cascades and activation of both primary and secondary response genes. The IFN-γ response is itself regulated by other cytokines including IL-18 and IL-1β. 16,17 The present study has for the first time demonstrated that genes that are integral parts of the cellular programs activated by IFN-γ are modulated when oval cells are activated and recruited for tissue repair in the AAF/PHx protocol. The identified genes include IFN-γ itself, the receptors for IFN-γ (IFN-γRα and IFN-γRβ), primary (gp91phox) and secondary response genes (ICE, ICAM-1 and uPAR), cytokines inducing IFN-γ expression (IL-18 and IL-1β), and, finally, cell adhesion molecules orchestrating lymphocyte-epithelial cell interactions (LFA-1 and ICAM-1; Table 1 ▶ ).
We have shown that ductular structures of oval cells in the regenerating liver express the α and β subunits of the IFN-γ receptor complex. Because both subunits are required for proper signaling on ligation of IFN-γ, these cell populations may be primary targets of IFN-γ when cells in the bile epithelial structures are activated to generate proliferating ductular structures of oval cells. In the AAF/PHx model of liver regeneration, the oval cells later differentiate to give rise not only to new hepatocytes but also to intestinal-like structures. 2,3 Evidence supporting a role for IFN-γ as a major player in ductal cell proliferation and tissue regeneration has been obtained from studies in transgenic mice engineered to constitutively express the mouse IFN-γ gene in either hepatocytes, under control of the human serum amyloid P component gene promoter, or in pancreatic islet cells driven by the human insulin gene promoter. 24,25 Hepatocyte-specific production of IFN-γ was sufficient to induce chronic inflammatory disease with persistent liver cell damage and proliferation of cells in ductular structures. 24 Similarly, in the pancreas, expression of the cytokine in islet cells elicited destruction of islets and, to some extent, of acinar tissue. 25 However, IFN-γ expression in islet cells also initiated an islet regeneration process evidenced by a flurry of mitotic activity in pancreatic duct cells and the appearance of three major endocrine cells in the duct walls and in the islet-like cell aggregates. Additionally, some pancreatic duct cells expressed albumin and α-fetoprotein and occasionally differentiated into hepatocytes and intestinal-like cell types. In summary, the studies performed with transgenic mouse models expressing IFN-γ in both liver and pancreas underscore the importance of this cytokine in the mitogenic activation of cells in the hepatic and the pancreatic ducts and the cytokine’s ability to modulate cellular differentiation pathways, at least in the pancreas.
We have established that IFN-γ and several of the genes it regulates through ligation to the IFN-γ receptor complex are modulated in cell populations responding on exposure to 2-AAF. However, the mechanisms by which IFN-γ may activate cells in bile ductules to enter DNA synthesis and divide, and whether the same mechanisms are involved when progeny of oval cells migrates and expand into the liver parenchyma, are not known. It is well recognized that cytokines modulating the immune system such as IFN-γ act in synergy with growth factors in complex networks to regulate biological processes. Recently, in vitro studies have shown that IFN-γ by itself has very little effect on cellular DNA synthesis. In contrast, the cytokine dramatically enhances DNA synthesis in response to mitogenic growth factors such as EGF. 26 The interaction between IFN-γ and EGF appears to be mediated by signal transducer and activator of transcription 1α (STAT1α), a member of the STAT family of transcription factors. Because a number of mitogenic growth factors including TGF-α, EGF, and HGF has been shown to be involved when oval cells proliferate in the AAF/PHx protocol, 9,10,13 it is possible that IFN-γ may prime ductal cells in vivo through activation of signal transducers that may augment the mitogenic response of the cells to other cytokines and growth factors.
We have previously reported that cells in the bile ductules enter DNA synthesis after exposure to 2-AAF. 6 In the present study we have shown that expression of uPAR, which is induced as a secondary response gene to IFN-γ, is already up-regulated 96 hours after exposure to 2-AAF and is expressed in the infiltrating ductular structures of oval cells. As an essential component in the plasminogen activator/plasmin system uPAR acts as the cellular receptor for uPA, and formation of the receptor-ligand complex results in proteolytic activation of plasminogen to plasmin on the surface of activated ductular and/or oval cells. 11 The plasminogen activator/plasmin cascade is involved in fibronolysis and degradation of extracellular matrix (ECM) components, but has also been implicated in activation of latent growth factors. 27 One of these factors, the heparin-binding glycoprotein HGF, a powerful mitogen for liver epithelial cells in vitro and considered to play a major role in liver regeneration in the AAF/PHx protocol, 10 is converted to its biologically active form by the uPA/uPAR complex. 28 Therefore, we find it reasonable to hypothesize that another priming effect of IFN-γ is to induce expression of uPAR on the surface of bile ductular cells. After binding of uPA, the uPAR/uPA complex will mediate generation of plasmin in the immediate vicinity of the ductular structures, degradation of the surrounding extracellular matrix, and localized liberation and activation of HGF. Mitosis may then be initiated by a paracrine interaction of activated HGF with its receptor on the ductular cells. Supporting this hypothesis is our recent demonstration that both uPA and HGF enhance the mitogenic response in the bile duct epithelium on infusion in 2-AAF treated animals. 13
As discussed above, a number of the cellular responses of hepatic ductular cells to 2-AAF may be orchestrated by ligation of IFN-γ to its cell surface receptor complex. However, IFN-γ production is itself subject to complex regulatory pathways. One of the factors involved in IFN-γ production is IL-18. 29 The cytokine is synthesized as a leaderless precursor requiring ICE for cleavage into the active molecule. In the present study, we have demonstrated that the steady-state levels of IL-18 and ICE transcripts increase when oval cells are recruited for liver regeneration in the AAF/PHx protocol. Furthermore, we have demonstrated the presence of both IL-18 and ICE protein in the ductular structures of oval cells. Taken together, these observations support an important function of IL-18 in activation and expansion of oval cell populations, possibly as a primary mediator of IFN-γ production.
In summary, using suppression subtractive hybridization, Northern blot, and immunohistochemical analyses in the AAF/PHx model of liver regeneration, we have identified several genes that are modulated when oval cells are activated in response to the liver damage inflicted. A common feature of the identified genes is their connection to the cellular programs orchestrated by IFN-γ, indicating an important role for IFN-γ as a growth-modulating molecule when epithelial cells in the bile ductules are activated and induced to produce a large population of oval cells.
Acknowledgments
We thank Gerda Demant Olesen for excellent technical assistance, Dr. Eric Santoni-Rugiu for valuable comments on the manuscript, and Dr. Kim Rasmussen for preparation of the figures.
Footnotes
Address reprint requests to Dr. Hanne Cathrine Bisgaard, Department of Life Sciences and Chemistry, Roskilde University, Building 16.1, Universitetsvej 1, DK-4000, Denmark. E-mail: hcb@virgil.ruc.dk.
Supported by grants from the Danish Cancer Society (to H. C. B. and L. J. R.), the Novo Nordisk Foundation (to H. C. B.), the Danish Natural Science Research Council (to H. C. B.) and the Danish Medical Research Council (to L. J. R.). P. N. was supported by a NATO linkage grant.
References
- 1.Michalopoulos GK, De Frances MC: Liver regeneration. Science 1997, 276:60-66 [DOI] [PubMed] [Google Scholar]
- 2.Evarts RP, Hu Z, Omori N, Omori M, Marsden ER, Thorgeirsson SS: Precursor-product relationship between oval cells and hepatocytes: comparison between tritiated thymidine and bromodeoxyuridine as tracers. Carcinogenesis 1996, 17:2143-2151 [DOI] [PubMed] [Google Scholar]
- 3.Alison M: Liver stem cells: a two compartment system. Curr Opin Cell Biol 1998, 10:710-715 [DOI] [PubMed] [Google Scholar]
- 4.Farber E: Similarities in the sequence of early histological changes induced in the liver by ethionine, 2-acetylaminofluorene, and 3′-methyl-4-dimethyl aminoazobenzene. Cancer Res 1956, 16:142-148 [PubMed] [Google Scholar]
- 5.Alison M, Golding M, Lalani E, Sarraf C: Wound healing in the liver with particular reference to stem cells. Phil Trans R Soc Lond B 1998, 353:877-894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisgaard HC, Nagy P, Santoni-Rugiu E, Thorgeirsson SS: Proliferation, apoptosis, and induction of hepatic transcription factors are characteristics of the early response of biliary epithelial (oval) cells to chemical carcinogens. Hepatology 1996, 23:62-70 [DOI] [PubMed] [Google Scholar]
- 7.Nagy P, Bisgaard HC, Thorgeirsson SS: Expression of hepatic transcription factors during liver development and oval cell proliferation. J Cell Biol 1994, 126:223-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujio K, Evarts RP, Hu Z, Marsden ER, Thorgeirsson SS: Expression of stem cell factor and its receptor, c-kit, during liver regeneration from putative stem cells in the adult rat. Lab Invest 1994, 70:511-516 [PubMed] [Google Scholar]
- 9.Evarts RP, Nakatsukasa H, Marsden ER, Hu Z, Thorgeirsson SS: Expression of transforming growth factor-α in regenerating liver and during hepatic differentiation. Mol Carcinog 1992, 5:25-31 [DOI] [PubMed] [Google Scholar]
- 10.Hu Z, Evarts RP, Fujio K, Marsden ER, Thorgeirsson SS: Expression of hepatocyte growth factor during hepatic differentiation and liver development. Am J Pathol 1993, 142:1823-1830 [PMC free article] [PubMed] [Google Scholar]
- 11.Bisgaard HC, Santoni-Rugiu E, Nagy P, Thorgeirsson SS: Modulation of the plasminogen activator/plasmin system in the rat liver regenerating by recruitment of oval cells. Lab Invest 1998, 78:237-246 [PubMed] [Google Scholar]
- 12.Omori N, Evarts RP, Omori M, Hu Z, Marsden ER, Thorgeirsson SS: Expression of leukemia inhibitory factor and its receptor during liver regeneration in the adult rat. Lab Invest 1996, 75:15-24 [PubMed] [Google Scholar]
- 13.Nagy P, Bisgaard HC, Santoni-Rugiu E, Thorgeirsson SS: In vivo infusion of growth factors enhances the mitogenic response of rat hepatic ductal (oval) cells after administration of 2-acetylaminofluorene. Hepatology 1996, 23:71-79 [DOI] [PubMed] [Google Scholar]
- 14.Bisgaard HC, Nagy P, Ton PT, Hu Z, Thorgeirsson SS: Modulation of keratin 14 and α-fetoprotein expression during hepatic oval cell proliferation and liver regeneration. J Cell Physiol 1994, 159:475-484 [DOI] [PubMed] [Google Scholar]
- 15.Cheng H, Bjerknes M, Chen H: CRP-ductin: a gene expressed in intestinal crypts and pancreatic and hepatic ducts. Anat Rec 1996, 244:327-343 [DOI] [PubMed] [Google Scholar]
- 16.Okamura H, Tsutsui H, Komatsu T, Yutsudo M, Hakura A, Tanimoto T, Torigoe K, Okura T, Nukada Y, Hattori K, Akita K, Namba M, Tanabe F, Konishi K, Fukada S, Kurimoto M: Cloning of a new cytokine that induces IFN-γ production in T cells. Nature 1995, 378:88-91 [DOI] [PubMed] [Google Scholar]
- 17.Schroeder RA, Gu JS, Kuo PC: Interleukin 1β-stimulated production of nitric oxide in rat hepatocytes is mediated through endogenous synthesis of interferon γ. Hepatology 1998, 27:711-719 [DOI] [PubMed] [Google Scholar]
- 18.Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, Alliston KO, Ayala JM, Casano FJ, Chin J, Ding GJ, Egger LA, Gaffney EP, Limjuco G, Palyha OC, Raju SM, Rolando AM, Salley JP, Yamin T, Lee TD, Shively JE, MacCross M, Mumford RA, Schmidt JA, Tocci MJ: A novel heterodimeric cysteine protease is required for interleukin-1β processing in monocytes. Nature 1992, 356:768-774 [DOI] [PubMed] [Google Scholar]
- 19.Gu Y, Kuida K, Tsutsui H, Ku G, Hsiao K, Fleming MA, Hayashi N, Higashino K, Okamura H, Nakanishi K, Kurimoto M, Tanimoto T, Flavell RA, Sato V, Harding MW, Livingston DJ, Su SS: Activation of interferon-γ inducing factor mediated by interleukin-1β converting enzyme. Science 1997, 275:206-209 [DOI] [PubMed] [Google Scholar]
- 20.Boehm U, Klamp T, Groot M, Howard JC: Cellular responses to interferon-γ. Annu Rev Immunol 1997, 15:749-795 [DOI] [PubMed] [Google Scholar]
- 21.Zarnegar R, DeFrances MC, Kost DP, Lindroos P, Michalopoulos GK: Expression of hepatocyte growth factor mRNA in regenerating liver after partial hepatectomy. Biochem Biophys Res Commun 1991, 177:559-565 [DOI] [PubMed] [Google Scholar]
- 22.Mead JE, Fausto N: Transforming growth factor α may be a physiological regulator of liver regeneration by means of an autocrine mechanism. Proc Natl Acad Sci USA 1989, 86:1558-1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evarts RP, Hu Z, Fujio K, Marsden ER, Thorgeirsson SS: Activation of hepatic stem cell compartment in the rat: role of transforming growth factor α, hepatocyte growth factor, and acidic fibroblast growth factor in early proliferation. Cell Growth Differ 1993, 4:555-561 [PubMed] [Google Scholar]
- 24.Toyonaga T, Hino O, Sugai S, Wakasugi S, Abe K, Shichiri M, Yamamura K: Chronic active hepatitis in transgenic mice expressing interferon-γ in the liver. Proc Natl Acad Sci USA 1994, 91:614-618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu D, Sarvetnick N: Epithelial cell proliferation and islet neogenesis in IFN-γ transgenic mice. Development 1993, 18:33-46 [DOI] [PubMed] [Google Scholar]
- 26.Marra F, Choudhury GG, Abboud HE: Interferon-γ-mediated activation of STAT1α regulates growth factor-induced mitogenesis. J Clin Invest 1996, 98:1218-1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vassilli J-D, Sappino AP, Belin D: The plasminogen activator/plasmin system. J Clin Invest 1991, 88:1067-1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naldini L, Vigna E, Bardelli A, Follenzi A, Galimi F, Comoglio PM: Biological activation of pro-HGF (hepatocyte growth factor) by urokinase is controlled by a stoichiometric reaction. J Biol Chem 1995, 270:603-611 [DOI] [PubMed] [Google Scholar]
- 29.Hunter CA, Timans J, Piscane P, Menon S, Cai G, Walker W, Aste-Amezega M, Chizzonite R, Bazan JF, Kastelein R: Comparison of the effects of interleukin-1α, interleukin-1β, and interferon-γ-inducing factor on the production of interferon-γ by natural killer. Eur J Immunol 1997, 27:2787-2792 [DOI] [PubMed] [Google Scholar]
- 30.Arany I, Tyring SK, Brysk H, Brysk MM: Induction by interferon-γ of its receptor varies with epithelial differentiation and cell type. Arch Dermatol Res 1998, 290:331-334 [DOI] [PubMed] [Google Scholar]
- 31.Eklund EA, Skalnik DG: Characterization of a gp91-phox promoter element that is required for interferon γ-induced transcription. J Biol Chem 1995, 270:8267-8273 [DOI] [PubMed] [Google Scholar]
- 32.Barker JN, Allen MH, MacDonald DM: The effect of interferon-γ on the distribution of LFA-1 and ICAM-1 in normal human skin. J Invest Dermatol 1989, 93:439-442 [DOI] [PubMed] [Google Scholar]
- 33.Wu S, Gupta S, Chatterjee N, Hileman RE, Kinzy TG, Denslow ND, Merrick WC, Chakrabarti D, Osterman JC, Gupta NK: Cloning and characterization of complementary DNA encoding the eukaryotic initiation factor 2-associated 67-kd protein (p67). J Biol Chem 1993, 268:10776-10781 [PubMed] [Google Scholar]
- 34.Kumar A, Yang YL, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams BR: Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J 1997, 16:406-416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamashita T, Nakane A, Watanabe T, Miyoshi I, Kasai N: Evidence that α-fetoprotein suppresses the immunological function in transgenic mice. Biochem Biophys Res Commun 1994, 30:1154-1159 [DOI] [PubMed] [Google Scholar]
- 36.Tamura T, Ishihara M, Lamphier MS, Tanaka N, Oishi I, Aizawa S, Matsuyama T, Mak TW, Taki S, Taniguchi T: An IRF-1-dependent pathway of DNA damage-induced apoptosis in mitogen-activated T lymphocytes. Nature 1995, 376:596-599 [DOI] [PubMed] [Google Scholar]
- 37.Sitrin RG, Todd RF, III, Mizukami IF, Gross TJ, Shollenberger SB, Gyetko MR: Cytokine-specific regulation of urokinase receptor (CD87) expression by U937 mononuclear phagocytes. Blood 1994, 15:1268-1275 [PubMed] [Google Scholar]


