Abstract
We have previously reported that the cyclin D1 (CCND1) GG870 genotype was associated with poorly differentiated tumors and reduced disease-free interval in patients with squamous cell carcinoma of the head and neck (SCCHN). We have now examined the association of this and a second CCND1 polymorphism with gene expression and outcome in SCCHN patients. Analysis of a CCND1 G/C1722 polymorphism revealed that CCND1 CC1722 genotype was associated with poorly differentiated tumors [P = 0.005; odds ratio (OR), 5.7; 95% CI, 1.7 to 19.2), and reduced disease-free interval (P = 0.003; Hazard Ratio (HR), 7.3; 95% CI, 1.1 to 27.2.) independently from the influence of CCND1 GG870 genotype. Patients whose tumors were negative for cyclin D1 were associated with reduced disease-free interval (P = 0.028; HR, 4.1; 95% CI, 1.4 to 14.2). Although G/C1722 genotypes were not associated with expression, we found a significant trend between reduced expression of cyclin D1 in patients with the CCND1 GG870 genotype (P = 0.04). Splicing of CCND1 mRNA in head and neck tissues was modulated by CCND1 A/G870 alleles, thus CCND1 transcript a was spliced equally from CCND1 A870 and G870 alleles, whereas CCND1 transcript b was spliced mainly from the CCND1 A870 allele. Our analysis has also identified differences in cyclin D1 genotype and protein expression and the pathogenesis of SCCHN in males and females. Thus, CCND1 CC1722 genotype was more common in female patients (P = 0.019; OR, 3.3; 95% CI, 1.3 to 10) and cyclin D1 expression was more frequent (chi-square1, 3.96; P = 0.046) and at higher levels (P = 0.004) in tumors from female patients. In summary, our data show that the two CCND1 polymorphic sites are independently associated with tumor biology and clinical outcome. CCND1 A/G870 alleles affect gene expression in head and neck tissues. We also provide preliminary evidence that the molecular genetics of SCCHN development may be influenced by patient gender.
Squamous cell carcinoma of the head and neck (SCCHN) comprise ∼5% of newly diagnosed malignancies in Northern Europe and the United States. Annually, more than 500,000 new cases are registered worldwide and the incidence of the disease is increasing. 1 Survival rates for the disease are poor, clinical outcome can vary among patients with tumors from the same site, with comparable tumor stage, nodal status, and histological grade. 2-4 Chronic consumption of tobacco and alcohol are recognized risk factors although it is unclear which traits determine tumor behavior and therefore prognosis. Studies have demonstrated elevated levels of cell proliferation in a high proportion of SCCHN tumors, and proliferation rates have been related to patient survival, and used in patient treatment strategies. 5-7 Thus, genes that encode regulators of cell proliferation may prove useful in establishing patient prognosis or as targets in therapy regimens.
The cyclin D1 gene (CCND1) encodes cyclin D1 protein, which is expressed in response to mitogenic signals promoting transition through the restriction point in the G1 phase of the cell cycle. 8 Increased expression of cyclin D1 has been associated with increased cell proliferation. 9,10 CCND1 amplification leading to deregulated CCND1 expression is common in tumors from patients with SCCHN. 11,12 Cyclin D1 protein overexpression has been shown to correlate with reduced 5-year and overall survival in SCCHN patients. 13 Other studies have shown that cyclin D1 protein overexpression is associated with poor prognosis in primary hypopharyngeal, laryngeal, esophageal, and oral squamous cell carcinomas. 14-16 Furthermore cyclin D1 antisense experiments have demonstrated that the gene may be a potential target for therapeutic intervention in SCCHN. 17
CCND1 is polymorphic with a common A/G substitution at nucleotide 870 in the conserved splice donor region of exon 4 of the gene. 18 CCND1 alleles have been shown to be associated with splicing of CCND1 mRNA in both normal and tumorous lung tissue. In heterozygotes, transcript a is spliced equally from CCND1 G870 and A870 alleles, whereas transcript b is spliced mainly from the CCND1 A870 allele. 18 In non-small cell lung cancer patients, CCND1 AA870 genotype is associated with poor prognosis. 18 In contrast we have recently demonstrated in SCCHN that the CCND1 GG870 genotype was associated with poorly differentiated tumors and independently from tumor differentiation, with reduced patient disease-free interval. 19,20 A second common G/C polymorphism at nucleotide 1722 within CCND1 3′UTR has also been described (G/C1722 sequence information is available at the NCBI SNP database at http://www.ncbi.nlm.nih.gov/SNP), although the influence of this polymorphism on tumorigenesis has not been examined. 21
In this study we have further investigated the role of CCND1 allelism in SCCHN. We have examined the relationship of the CCND1 G/C1722 polymorphism with A/G870 alleles and clinical outcome. In addition we have further studied the influence of CCND1 alleles on mRNA splicing and protein expression in head and neck tissue.
Materials and Methods
Patients
SCCHN patients (n = 294) were studied from our original cohort of 384 patients previously described. 19,20 These patients were selected because of the availability of DNA and do not represent a clinical subgroup. The clinical pathological characteristics are similar to those of the total cohort. 19 Briefly, the case group comprised German Caucasians suffering a single histologically confirmed oral cavity pharyngeal or laryngeal squamous cell carcinoma. Patients were recruited at first presentation or during follow-up between 1994 and 1996. Malignancies were staged using the TMN classification system. 22 All tumors were squamous cell carcinoma and were histologically graded as: well (G0–1), moderately (G2), and poorly (G3) differentiated. Margins of the resected specimen were examined by a histopathologist and judged for tumor-free margins using the international R0-R2 system; R0, microscopically proven free tumor margins; R1, microscopic infiltration and macroscopically free margin; R2, macroscopic tumor infiltration of the margin. Where details were available we examined the association of cyclin D1 genotype and protein expression with factors known to influence clinical outcome: tumor site (n = 268), tumor size (T1 to T4) (n = 258), histological differentiation (G0/1 to G3) (n = 168) and the presence of nodes at time of surgery (n = 166). We also examined the association of genotypes and protein expression with tumor recurrence (defined as disease-free interval) in 151 patients. The study to identify associations with disease-free interval was performed only in patients in which a R0 resection could be achieved during initial treatment. Patients suffering a tumor re-growth during the first 6 months after initial treatment and those suffering extra-capsular tumor spread in any of the resected lymph nodes were excluded to avoid misjudged recurrences because of residual tumor growth. Lymph node involvement was determined before surgery using ultrasound, computed tomography, and nuclear magnetic resonance imaging and later histologically proven in the neck dissection specimen.
DNA and RNA Extraction
Peripheral blood samples were collected in ethylenediaminetetraacetic acid. Tumor and histologically normal head and neck tissues (salivary gland or muscle) were collected at time of surgery and snap-frozen in liquid nitrogen and stored at −70°C before use. DNA was extracted from frozen tissue and blood using a phenol-chloroform method. 23 mRNA was isolated from frozen normal and tumor tissues using the MicrofastTrack RNA isolation kit (Invitrogen, Groningen, The Netherlands).
Restriction Fragment Length Polymorphism-Polymerase Chain Reaction (PCR) Genotyping
CCND1 G/C1722 genotypes were identified in DNA isolated from peripheral blood using a restriction fragment length polymorphism-PCR based assay. 24 Briefly reactions were performed in 25 μl containing 1× Taq polymerase buffer (Promega, Southampton, UK), 100 μmol/L dNTPs, 0.25 μg of each primer, 2% dimethyl sulfoxide, 0.25 U of Taq polymerase (Promega, UK) and 0.1 μg of DNA. Reactions were performed on an automated thermal cycler with an initial denaturation of 94°C (2 minutes) and cycled 34 times with annealing temperature of 57°C (1 minute), an extension at 72°C (1 minute), and denaturation of 94°C (1 minute). PCR products were digested with HaeIII following the manufacturer’s guidelines (New England Biolabs, Hitchin, Hertfordshire, UK). Alleles were resolved on 3% agarose gels stained with ethidium bromide.
Reverse Transcriptase-PCR Analysis of CCND1 mRNA
mRNA extracted from snap-frozen tissues was reverse-transcribed to cDNA using a superscript preamplification system (Gibco-BRL Life Technologies, Paisley, UK). Analysis of CCND1 transcripts a and b was performed using a nested PCR strategy. 18 Reactions containing 2 μl of cDNA were performed in 100-μl volumes containing 1× PCR buffer (Promega), 100 μmol/L dNTPs, 0.5 μg each primer, 2 U of Taq polymerase (Promega). Primary reactions were cycled 32 times, following which 1 μl of PCR product was nested for 22 cycles. PCR conditions were initial denaturation temperature 94°C (2 minutes), annealing 55°C (1 minute), elongation 74°C (1 minute), and denaturation 94°C (1 minute). PCR products were digested with the restriction enzyme ScrFI (New England Biolabs) and alleles visualized on 8% acrylamide gels and silver stained.
Immunohistochemistry
Paraffin-embedded tumor material was available in 135 cases from the cohort of 294 patients. Immunohistochemistry was performed using a Shandon Sequenza (Shandon Scientific Limited, Cheshire, UK) as described. 25 A mouse monoclonal cyclin D1 antibody (DCS-6; Novocastra, Newcastle-upon-Tyne, UK) was used. A cyclin D1-positive breast tumor provided positive control material. Negative controls had no primary antibody applied and included a section of normal head and neck tissue. Slides were graded as negative (0 to <10% cells stained), low (>10 to 50% cells stained), moderate (>50 to 75% cells stained), and strong (>75% cells stained).
Statistics
Statistical analysis was performed using Stata, version 5 (Stata Corporation, College Station, TX). Pearson chi-square tests were used to identify linkage between CCND1 genotypes and associations of genotypes with gender. Association of genotypes and protein expression with clinicopathological parameters were analyzed and corrected for imbalances in age and gender using logistic regression. Associations between cyclin D1 protein and genotype and cyclin D1 protein expression with gender were analyzed using the Armitage trend test. Cox’s proportional hazard regression model was used in the analysis of the effects of cyclin D1 staining and genotypes on disease-free interval. Kaplan-Meier curves were generated for graphical representation of associations with disease-free interval. A probability level of 5% was considered statistically significant.
Results
The frequencies of CCND1 G/C1722 genotypes in 294 patients with SCCHN are shown (Table 1) ▶ . Allele frequencies conformed to Hardy Weinberg equilibrium. Furthermore significant linkage disequilibrium was demonstrated between G/C1722 and A/G870 alleles, thus 32 of 34 (94%) individuals with the CCND1 CC1722 genotype were also GG870 (Table 2) ▶ . The distribution of CCND1 G/C1722 genotypes was significantly different between female and male patients with the CCND1 CC1722 genotype being more common in female patients (Table 1) ▶ . This association remained significant after correction for smoking and alcohol consumption (P = 0.036; OR, 3.7). CCND1 A/G870 genotypes were not associated with patient gender. 19,20
Table 1.
Distribution of CCND1 G/C1722 Genotypes and Tumor Protein Expression in SSCHN Patients
| CCND1 G/C1722 genotype | Cyclin D1 protein expression | ||||
|---|---|---|---|---|---|
| GG (%) | GC (%) | CC (%) | Positive* (%) | Negative (%) | |
| Total cases | 117 (39.8) | 141 (48.0) | 36 (12.2) | 112 (83.0) | 23 (17.0) |
| Male | 106 (41.7) | 121 (47.6) | 27 (10.6) | 87 (79.8) | 22 (20.2) |
| Female† | 11 (27.5) | 20 (50.0) | 9 (22.5) | 25 (96.1) | 1 (3.9) |
| Tumor site | |||||
| Laryngeal SCC | 81 (41.5) | 90 (46.2) | 24 (12.3) | 56 (82.4) | 12 (17.6) |
| Pharyngeal SCC | 16 (32.7) | 26 (53.1) | 7 (14.3) | 27 (81.8) | 6 (18.2) |
| Oral Cavity SCC | 9 (37.5) | 12 (50.0) | 3 (12.5) | 13 (81.2) | 3 (18.8) |
| Multiple sites | 11 (42.3) | 13 (50.0) | 2 (7.7) | 16 (88.9) | 2 (11.1) |
| T-factor | |||||
| T1 | 36 (38.7) | 51 (54.8) | 6 (6.5) | 22 (73.3) | 8 (26.7) |
| T2 | 27 (42.2) | 29 (45.3) | 8 (12.5) | 25 (92.6) | 2 (7.4) |
| T3 | 21 (42.0) | 21 (42.0) | 8 (16.0) | 14 (100) | 0 (0.0) |
| T4 | 19 (37.3) | 25 (49.0) | 7 (13.7) | 31 (75.6) | 10 (24.4) |
| Histological differentiation‡ | |||||
| Well | 5 (55.6) | 4 (44.4) | 0 (0.00) | 5 (71.4) | 2 (28.6) |
| Moderate | 47 (40.2) | 58 (49.6) | 12 (10.3) | 45 (76.3) | 14 (23.7) |
| Poor | 11 (26.2) | 21 (50.0) | 10 (23.8) | 24 (80.0) | 6 (20.0) |
| Lymph nodes | |||||
| Negative | 38 (40.0) | 49 (51.6) | 8 (8.4) | 43 (79.6) | 11 (20.4) |
| Positive | 23 (32.4) | 34 (47.9) | 14 (19.7) | 30 (76.9) | 9 (23.1) |
*Positive protein expression was defined as >10% tumor cells expressing cyclin D1 and was more frequent in tumors from female patients (chi-square21 = 3.96; P = 0.046).
†Using logistic regression analysis corrected for imbalances in age, with CCND1 GG1722 as a reference, the CCND1 CC1722 genotype was more common in female patients P = 0.025; OR, 3.3; 95% CI, 1.2 to 9.51.
‡Using logistic regression correcting for age and gender in the model CCND1 CC1722 was associated with poor tumor differentiation (G0/1, G2 versus G3), P = 0.005; OR, 5.7; 95% CI, 1.7 to 19.2.
Table 2.
Association of CCND1 A/G870 and G/C1722 Genotypes in SCCHN Patients
| CCND1 G/C1722 genotypes | CCND1 A/G870 genotypes | ||
|---|---|---|---|
| AA (%) | AG (%) | GG (%) | |
| GG (%) | 54 (47.4) | 49 (43.0) | 11 (9.6) |
| G/C (%) | 11 (8.1) | 87 (64.0) | 38 (27.9) |
| CC (%) | 0 (0.0) | 2 (5.9) | 32 (94.1) |
Significant linkage disequilibrium was demonstrated between CCND1 G/C1722 and A/G870 alleles (P < 0.001; chi-square4 = 136.3).
We next examined the data for associations between the CCND1 G/C1722 genotypes and factors that influence outcome in the total group. There was no association between patient genotype and age at tumor presentation (data not shown) and no association was found between CCND1 genotypes and tumor stage and tumor site (Table 1) ▶ . However, we found using logistic regression with correction for age and gender, CCND1 CC1722 genotypes were associated with poorly compared to well and moderately differentiated tumors (Table 1) ▶ . As before, we found that CCND1 GG870 genotype was associated with poorly differentiated tumors 19 (P = 0.016; OR, 2.2; 95% CI, 1.15 to 4.21), however on including both CCND1 genotypes in a model with age and gender we found CCND1 CC1722 remained significantly associated with tumor differentiation (P = 0.036; OR, 4.91; 95% CI, 1.1 to 21.8), whereas CCND1 GG870 was no longer significantly associated (P = 0.76; OR, 1.17; 95% CI, 0.43 to 3.23). CCND1 CC1722 genotype was more common in patients with tumor nodes, although this did not achieve statistical significance (P = 0.075; OR, 2.8; 95% CI, 0.9 to 8.9). We further examined the association of CCND1 G/C1722 genotypes with differentiation and tumor nodes in the cases with laryngeal tumors. CCND1 CC1722 genotypes were associated with poorly differentiated tumors (P = 0.006; OR, 9.2; 95% CI, 1.9 to 44.9) and the presence of tumor nodes in laryngeal tumors (P = 0.046; OR, 4.3; 95% CI, 1.0 to 18.1).
Cyclin D1 expression was detected in 112 of 135 (83%) of tumors examined (Table 1) ▶ . The proportion of tumor cells expressing cyclin D1 was variable between different tumors. Thus expression was low in 35 of 135 (25.9%) cases, moderate in 56 of 135 (41.4%) cases, and high in 21 of 135 (15.5%) cases. Cyclin D1 protein expression was not associated with patient age at presentation neither did we find an association with tumor stage, differentiation, or the presence of tumor nodes (Table 1) ▶ . Analysis of cyclin D1 expression with patient gender revealed that tumor expression of cyclin D1 was high in both sexes (Table 1) ▶ . However, significantly more tumors from females expressed cyclin D1 compared with those from males (Table 1) ▶ . Furthermore, a significant trend was observed between an increase in the proportion of cells expressing cyclin D1 in the tumors from female patients, compared with those from males (Table 3) ▶ .
Table 3.
Association of Cyclin D1 Expression in Tumors with CCND1 A/G870 Genotypes and Patient Gender in SCCHN
| Cyclin D1 protein* | Patient gender | CCND1 genotype | ||
|---|---|---|---|---|
| Female† | Male | CCND1 AG870/AA870 | CCND1 GG870‡ | |
| Negative | 1 (4.3) | 22 (95.7) | 14 (63.6) | 8 (36.4) |
| Low | 5 (14.3) | 30 (85.7) | 22 (66.7) | 11 (33.3) |
| Moderate | 12 (21.4) | 44 (78.6) | 42 (79.2) | 11 (20.8) |
| High | 8 (38.1) | 13 (61.9) | 17 (85.0) | 3 (15.0) |
Figures in parenthesis are percent.
*Levels of cyclin D1 expression are defined as the proportion of tumor cells expressing protein and are, negative (<10%), low (10 to 50%) moderate (50 to 75%) and high (>75%).
†Female patients are associated with high levels of cyclin D1 expression (Armitage test for trend P = 0.004).
‡CCND1 GG870 genotype is associated with low expression of cyclin D1 protein (Armitage test for trend, P = 0.049).
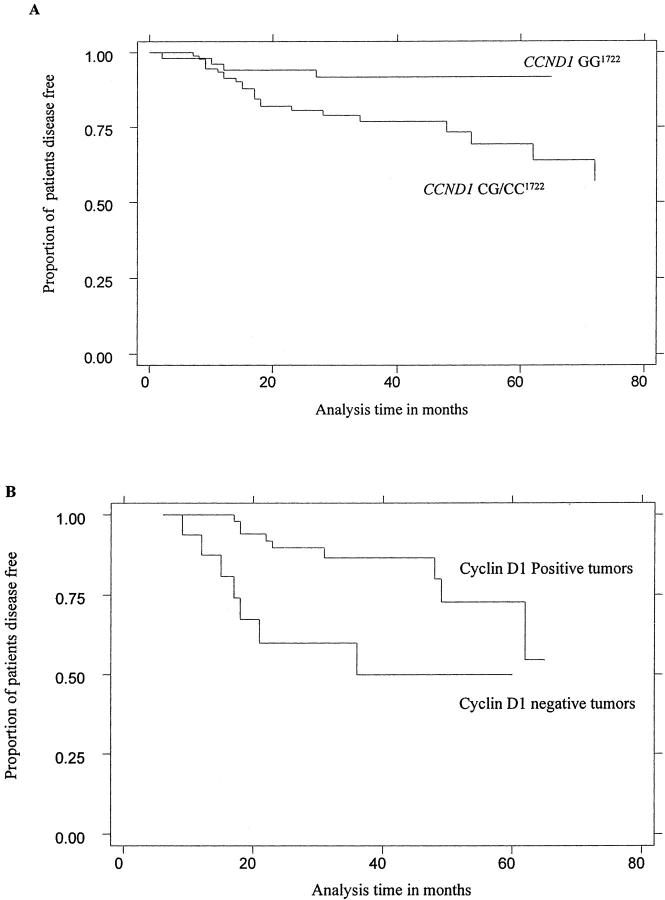
We next examined CCND1 genotype and protein expression data for associations with patient disease-free interval. The Kaplan-Meier plot demonstrated that CCND1 CC1722 genotype was associated with reduced time to tumor recurrence (Figure 1A) ▶ . Using Cox’s proportional hazards model to correct for imbalances in patient age and gender, with CCND1 GG1722 genotype as reference, CCND1 CC1722 was significantly associated with an increased proportion of patients having tumor recurrence after 2 years (Table 4) ▶ . On including tumor differentiation in the model this association was reduced (P = 0.042; HR, 3.76; 95% CI, 1.05 to 13.47). As previously reported CCND1 GG870 genotype was associated with reduced disease-free interval 19 (Table 4) ▶ . Including both genotypes in the model, although not significant CCND1 GG870 (P = 0.12; HR, 2.0; 95% CI, 0.83 to 4.85) and CCND1 CC1722 (P = 0.102; HR, 3.2; 95% CI, 0.8 to 12.9) were both associated with an increase in the proportion of patients suffering a tumor recurrence after 2 years. Patients whose tumors did not express cyclin D1 were associated with reduced disease-free interval (Figure 1B) ▶ . Accordingly, the tumor recurrence after 2 years was significantly higher in patients in this group (Table 4) ▶ . Furthermore absence of tumor cyclin D1 protein expression seemed to be associated with disease-free interval in patients with laryngeal tumors (P = 0.047; HR, 3.7; 95% CI, 1.01 to 5.89).
Figure 1.
Kaplan-Meier survival plots showing the association between CCND1 G/C1722 genotypes and disease-free interval (A) and the association between cyclin-D1 protein and disease-free interval (B) in SCCHN patients.
Table 4.
Association of CCND1 Genotypes, Tumor Cyclin D1 Protein Expression, and Prognostic Indicators with Tumor Recurrence in SCCHN
| Prognostic indicator | P | HR | 95% CI | Proportion of patients with recurrence |
|---|---|---|---|---|
| CCND1 G/C1722* | ||||
| GG | ref | 5.7 | ||
| CG | 0.362 | 1.6 | 0.6–4.8 | 17.6 |
| CC | 0.003 | 7.3 | 11.0–27.2 | 26.9 |
| CCND1 A/G870* | ||||
| AA | ref | — | — | 5.0 |
| AG | 0.456 | 1.5 | 0.5–4.2 | 10.2 |
| GG | 0.010 | 3.9 | 1.4–11.0 | 28.0 |
| Cyclin D1 protein* | ||||
| Positive | ref | — | — | 10.3 |
| Negative | 0.028 | 4.1 | 1.4–14.2 | 40.2 |
| Differentiation† | ||||
| G1/2 | ref | 9.6 | ||
| G3 | <0.001 | 3.1 | 1.7–5.7 | 28.2 |
| Stage† | ||||
| T1, 1.1,2 | ref | — | — | 10.3 |
| T3,4 | 0.134 | 1.6 | 0.9–3.1 | 18.0 |
*P values were obtained using Cox’s regression corrected for age and gender tumor site and T-stage.
†P values were obtained using Cox’s regression corrected for age and gender.
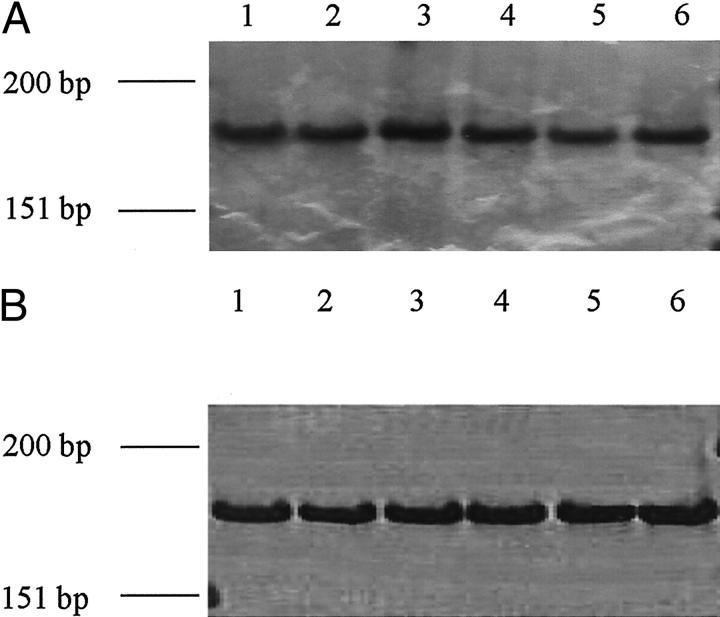
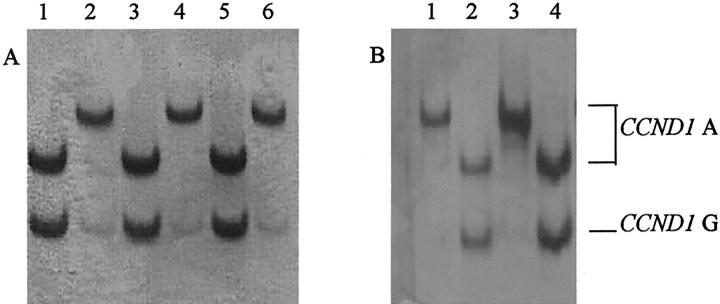
We further examined the influence of CCND1 alleles on CCND1 expression. Cyclin D1 protein was not significantly associated with CCND1 G/C1722 genotypes (data not shown). However, a higher proportion of tumors displayed low expression of cyclin D1 from patients with the CCND1 GG870 genotype compared with tumors from patients with the CCND1 AA870 and A/G870 genotypes (Table 3) ▶ . The influence of CCND1 A/G870 alleles on CCND1 mRNA splicing was examined. We examined the expression of CCND1 mRNA from 13 matched normal and tumor tissue pairs of which 9 were CCND1 AG 870, 2 were AA870, and 2 were GG870 genotypes. Expression of both CCND1 transcripts was detected in tissues from patients of each genotype (Figure 2, A and B) ▶ . Restriction fragment length polymorphism analysis of cDNA from heterozygotes demonstrated that CCND1 A/G870 alleles modulate CCND1 mRNA splicing in head and neck tissues. Thus in nine of nine normal tissues and five of nine corresponding tumors, CCND1 transcript a was spliced equally from the CCND1 A870 and CCND1 G870 alleles, however CCND1 transcript b was spliced mainly from CCND1 A870. A representative example of CCND1 mRNA splicing from three different individuals with the CCND1 AG870 genotype is shown (Figure 3) ▶ .
Figure 2.
CCND1 mRNA is alternately spliced in SCCHN patients with different CCND1 A/G870 genotypes. The expression of CCND1 transcript a (A) and CCND1 transcript b (B) was detected by PCR on cDNA synthesized from tumors with CCND1 AA870 (lanes 1 and 2) CCND1 AG870 (lanes 3 and 4) and CCND1 GG870 (lanes 5 and 6) genotypes. PCR products were run on polyacrylamide gels and silver stained. φX174 HinfI digest was used as a size marker.
Figure 3.
CCND1 alleles modulate transcript splicing in head and neck tissues. Restriction fragment length polymorphism-PCR analysis was performed on cDNA synthesized from tumor tissues from three patients with the heterozygote CCND1 AG870 genotype (patient 1, lanes 1 and 2; patient 2, lanes 3 and 4; and patient 3, lanes 5 and 6) (A), and normal tissues from two of these patients (patient 1, lanes 1 and 2; patient 2, lanes 3 and 4) (B). CCND1 alleles were resolved through digestion of PCR products with ScrFI and separated on polyacrylamide gels and silver stained. CCND1 alleles are marked with arrows. ScrFI digests of CCND1 transcript a-specific PCR product show transcript a is spliced equally from CCND1 allele A870 and G870 alleles in both tumor (A: lanes 1, 3, and 5) and normal (B: lanes 2 and 4) tissues; whereas digests of CCND1 transcript b-specific PCR products show that transcript b is spliced mainly from CCND1 A870 allele in both tumor (A: lanes 2, 4, and 6) and normal (B: lanes 1 and 3) tissues.
Discussion
We have demonstrated for the first time, that polymorphism in the 3′-UTR region of CCND1 is associated with tumorigenesis and clinical outcome in SCCHN patients. CCND1 CC1722 was associated with poorly differentiated tumors and reduced disease-free interval. We have previously reported that CCND1 GG870 was associated with poor tumor differentiation and reduced disease-free interval independent from tumor differentiation. 19 However, our preliminary data suggest that although CCND1 G870 and C1722 alleles are in linkage disequilibrium, their influence on tumor pathology and patient outcome seems to be separate. Thus CCND1 GG870 is an independent marker for tumor recurrence in SCCHN, whereas CCND1 CC1722 is associated with tumor recurrence by a mechanism associated with tumor biology and mainly through differentiation. Similar associations with tumor differentiation, tumor nodes, and clinical outcome were found with CCND1 CC1722 in laryngeal tumors. However case numbers were not large enough to test for significance in pharyngeal and oral tumors although similar trends were observed.
We have also examined the expression of cyclin D1 protein in these tumors. Cyclin D1 expression was frequently deregulated, with the proportion of cells expressing the protein varying between different tumors. Our data are consistent with findings in other studies. 26,27 However, surprisingly, in our cohort of patients, absence of expression of cyclin D1 correlated with reduced disease-free interval. In contrast previous work has associated up-regulated expression of cyclin D1 with poor outcome in SCCHN. 13-16 Although it is assumed, that deregulated expression leads to poor outcome directly through increased cellular proliferation of tumor cells, in a recent study cyclin D1 expression in SCCHN tumors has been associated with an increase in apoptosis and cyclin D1-negative tumors were more proliferatively active. 27 Cyclin D1 expression has been associated with good prognosis in non-small cell lung cancer and bladder cancer 24,28 and both good and bad prognosis in studies on breast and cancer. 29-32 It is possible that these contradictory results in part reflect the many different mechanisms through which deregulated expression of cyclin D1 can occur in cancer. 11,12,24,29,33 It would be interesting in the future, to examine the association between CCND1 alleles with the mechanism of protein overexpression such as CCND1 amplification in addition to biological markers of cellular proliferation and to relate these findings to clinical parameters including tumor differentiation and patient disease-free interval. Further understanding of these mechanisms and their effect on tumor biology in specific tissues may increase understanding of cyclin D1 as a prognostic marker.
To explore mechanisms underlying the link between CCND1 genotypes and clinical outcome in SCCHN, we investigated the data for associations with gene expression. Firstly, we found a significant trend between a reduction in the proportion of cyclin D1-expressing cells within the tumors of patients with CCND1 GG870. Furthermore, CCND1 GG870 was associated with reduced disease-free interval in our patients thus providing a link between CCND1 A/G870 alleles with cyclin D1 expression and clinical outcome in SCCHN. Secondly, we found that CCND1 A/G870 alleles modulate the splicing of CCND1 mRNA in head and neck tissues. CCND1 transcripts a and b were detected from each CCND1 A/G870 allele. However, transcript b was spliced mainly from the CCND1 A870, whereas transcript a was spliced equally from CCND1 G870 and CCND1 A870 alleles. It would be expected therefore, that CCND1 AA870 homozygotes express more transcript b than CCND1 GG870 homozygotes. Although we did not measure transcript levels, it is possible that physiological differences in the ratio of CCND1 transcripts a to b because of interindividual genotypic variation may influence the development of SCCHN. Interestingly, we found that in four of nine of the tumors allele-specific splicing of the transcripts was deregulated (S. L. Holley and P. R. Hoban, unpublished data). It is not clear from this preliminary data whether this is a specific mechanism for tumor development or indicates a general loss of splicing fidelity in these tumors.
CCND1 transcript a is identical to the reported cyclin D1 cDNA, 34 however, transcript b fails to splice at the exon 4/intron 4 boundary, terminates downstream of exon 4 and does not contain exon 5. 18 The difference between the proteins predicted by the nucleotide sequences of the two transcripts is in the carboxy terminal PEST rich region (destruction box) encoded by exon 5, which facilitates turnover of the START cyclins. 35 The terminal region of transcript b has no PEST-rich sequence. Functional differences in the expression of the alternate CCND1 transcripts have also been demonstrated. 36 Further, the monoclonal antibody, DCS-6, used in our immunohistochemical analysis, cross-reacts with recombinant cyclin D1 proteins from both CCND1 transcript a and b and protein from transcript b has been detected with this antibody in lymphoma cells and solid tissues. 36-38 Therefore it is not clear in our immunohistochemical analysis which specific protein is being detected in the SCCHN tumors. We did not find any associations between CCND1 G/C1722 alleles and protein expression. Because of its location in exon 5, we assume that CCND1 G/C1722 is a marker reflecting the activity of CCND1 transcript a. Alternatively CCND1 G/C1722 alleles may be in linkage with other alleles that influence cyclin D1 function or that of a gene close by.
Our study has highlighted potential differences in the genetics and biology of tumors from SCCHN patients of different gender. The distribution of G/C1722 genotypes was different between male and female patients. We also found that significantly more tumors from female patients and a higher proportion of cells in those tumors expressed cyclin D1, than those from male patients. Our cohort was too small to significantly examine the data for differences in outcome based on CCND1 and gender. However it was noted that tumor site significantly differed with patient gender. Thus oral (21% female versus 7% male) and pharyngeal (38% female versus 21% male) tumors were more common in female patients whereas laryngeal tumors were more common in male patients (40% females versus 71% males). SCCHN is documented as a predominantly elderly male disease associated with heavy smoking and high alcohol consumption. 1-3 In our cohort, female patients were associated with less alcohol (30% females compared with 73% of males) and tobacco (62% females compared with 88% males) consumption. This data would indicate that SCCHN etiology in males and females may be different. The role of cyclin D1 in tumor development in patients of differing gender is unclear and the gene may merely be a marker for these differences. However, CCND1 amplification and protein expression has been related to tobacco exposure in SCCHN. 39 In breast cancer patients cyclin D1 expression and estrogen receptor status are significantly correlated and associated with clinical outcome. 40 Furthermore, previous functional studies have suggested that cyclin D1 interacts in a ligand-specific manner with the estrogen receptor in breast cells and the androgen receptor in prostate cells. 41,42
In conclusion, accumulating data demonstrate that CCND1 is important in the development of SCCHN and that the gene has potential both as a prognostic marker, and as a target in the treatment of the disease. Our data suggest that CCND1 A/G870 alleles influence expression of CCND1, and that specific antibodies to transcript b are required to validate expression studies and to identify the specific cyclin D1 protein in SCCHN tumors. We realize, in some cases, numbers were small, and that our findings require independent confirmation in a separate and larger cohort, however, for the first time, we demonstrate the potential of CCND1 G/C1722 alleles as an independent prognostic marker in this disease. Our data also highlights differences in the molecular genetics of SCCHN tumors associated with patient gender. Collectively these findings have implications in rationalizing the protein as a target for treatment of patients with SCCHN.
Footnotes
Address reprint requests to Paul R Hoban, Centre for Cell and Molecular Medicine, University of Keele School of Postgraduate Medicine, North Staffordshire Hospital, Hartshill Road, Hartshill, Stoke-on-Trent ST47NY, UK. E-mail: p.r.hoban@keele.ac.uk.
Supported by the North Staffordshire Medical Institute (grant no. 51272958).
References
- 1.Johnson NW, Ranasinghe AW, Warnakulasuriya KAAS: Potentially malignant lesions and conditions of the mouth and oropharynx: natural history—cellular and molecular markers of risk. Eur J Cancer Prev 1993, 2:31-51 [DOI] [PubMed] [Google Scholar]
- 2.Muir C, Weiland L: Upper aerodigestive tract cancers. Cancer 1995, 75:147-153 [DOI] [PubMed] [Google Scholar]
- 3.Janot F, Klijanienko J, Russo A, Marnet JP, de Braud F, El-Naggar AK, Pignon JP, Luboinski B, Cvitkovic E: Prognostic value of clinicopathological parameters in head and neck squamous cell carcinoma: a prospective analysis. Br J Cancer 1996, 73:531-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magnano M, Bussi M, de Stefani A, Milan F, Lerda W, Ferrero V, Gervasio F, Ragona R, Gabriele P, Valenta G, Cortesina G: Prognostic factors for head and neck tumour recurrence. Acta Otolaryngol 1995, 115:833-838 [DOI] [PubMed] [Google Scholar]
- 5.Kotelnikov VM, Coon J, Haleem A, Taylor S, Hutchinson J, Panje W, Caldarelli D, Preisler HD: Cell kinetics of head and neck cancers. Clin Cancer Res 1995, 1:527-537 [PubMed] [Google Scholar]
- 6.Corvo R, Giaretti W, Sanguineti G, Geido E, Orecchia R, Guenzi M, Margarino G, Bacigalupo A, Garaventa G, Barbieri M, Vitale V: In vivo cell kinetics in head and neck squamous cell carcinomas predicts local control and helps guide radiotherapy regimen. J Clin Oncol 1995, 48:1843-1850 [DOI] [PubMed] [Google Scholar]
- 7.Tomasino RM, Daniele E, Bazan V, Morello V, Tralongo V, Nuara R, Nagar C, Salvato M, Ingria F, Restivo S, Dardanoni G, Vecchione A, Russo A: Prognostic significance of cell kinetics in laryngeal squamous cell carcinomas: clinical pathological associations. Cancer Res 1995, 55:6103-6108 [PubMed] [Google Scholar]
- 8.Sherr CJ: Cancer cell cycles. Science 1996, 274:1672-1677 [DOI] [PubMed] [Google Scholar]
- 9.Quelle DE, Ashmun RA, Shurtleff SA, Kato JY, Bar-Sagi D, Roussel MF, Sherr CJ: Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev 1993, 7:1559-1571 [DOI] [PubMed] [Google Scholar]
- 10.Musgrove EA, Lee CSL, Buckley MF, Sutherland RL: Cyclin D1 induction in breast cancer cells shorten G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc Natl Acad Sci USA 1994, 91:8022-8026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callender T, El Naggar AK, Lee MS, Frankenthaler R, Luna MA, Batsakis JG: PRAD-1 (CCND1)/cyclin D1 oncogene amplification in head and neck squamous cell carcinoma. Cancer 1994, 74:152-158 [DOI] [PubMed] [Google Scholar]
- 12.Jares P, Fernadez PL, Campo E, Nadal A, Bosch F, Aiza G, Nayach I, Trassera J, Cardesa A: PRAD-1/cyclin D1 gene amplification correlates with messenger mRNA overexpression and tumour progression in human laryngeal carcinomas. Cancer Res 1994, 54:4813-4817 [PubMed] [Google Scholar]
- 13.Michalides R, van Veelen N, Hart A, Loftus B, Wientjens E, Balm A: Overexpression of cyclin D1 correlates with recurrence in a group of forty seven operable squamous cell carcinomas of the head and neck. Cancer Res 1995, 55:975-978 [PubMed] [Google Scholar]
- 14.Masuda M, Hirakawa N, Nakashima T, Kuratomi Y, Komiyama S: Cyclin D1 overexpression in primary hypolaryngeal carcinomas. Cancer 1995, 78:390-395 [DOI] [PubMed] [Google Scholar]
- 15.Pignatora L, Pruneri G, Carboni N, Capaccio P, Cesana BM, Neri A, Buffa R: Clinical relevance of cyclin D1 protein overexpression in laryngeal squamous cell carcinoma. J Clin Oncol 1998, 16:3069-3077 [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa T, Furihata M, Ohtsuki Y, Murakami H, Inoue A, Ogoshi S: Cyclin D1 overexpression related to retinoblastoma protein expression as a prognostic marker in human oesophageal squamous cell carcinoma. Br J Cancer 1998, 77:92-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakashima T, Clayman GL: Antisense inhibition of cyclin D1 in human head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surgery 2000, 126:957-961 [DOI] [PubMed] [Google Scholar]
- 18.Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WDJ, Heighway J: Alternate splicing produces a novel cyclin D1 transcript. Oncogene 1995, 11:1005-1011 [PubMed] [Google Scholar]
- 19.Matthias C, Branigan K, Jahnke V, Leder K, Haas J, Heighway J, Jones PW, Strange RC, Fryer AA, Hoban P: Polymorphism within the cyclin D1 gene is associated with prognosis in patients with squamous cell carcinoma of the head and neck. Clin Cancer Res 1998, 4:2411-2418 [PubMed] [Google Scholar]
- 20.Matthias C, Jahnke V, Jones PW, Hoban PR, Alldersea J, Worrall S, Anthony A, Fryer AA, Strange RC: Cyclin D1, glutathione S-transferase and cytochrome P450 genotypes and outcome in patients with upper aerodigestive tract cancers: assessment of the importance of individual genes using multivariate analysis. Cancer Epidemiol Biomarkers 1999, 8:815-823 [PubMed] [Google Scholar]
- 21.Heighway J: Hae III polymorphism with 3′ untranslated region of PRAD 1. Nucleic Acids Res 1991, 19:5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phermanek P, Sobin LH (Eds): Union Internationale Contre le Cancer. TNM Classification of Malignant Tumours, ed 4. Berlin, UICC, Springer-Verlag, 1987
- 23.Sambrook J, Fritsch EF, Maniatis T (Eds): Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, Cold Spring Harbor Laboratory Press, 1989
- 24.Betticher D, Heighway J, Haselton PS, Altermatt MJ, Ryder WDJ, Cerny T, Thatcher N: Prognostic significance of CCND1 (cyclin D1) overexpression in primary resected non-small cell lung cancer. Br J Cancer 1996, 73:294-300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhar KK, Branigan K, Parkes J, Howells REJ, Hand P, Musgrove C, Strange RC, Fryer AA, Redman CWE, Hoban PR: Expression and subcellular localisation of cyclin D1 protein in ovarian epithelial tumour cells. Br J Cancer 1999, 81:1174-1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartkova J, Lukas J, Muller H, Strauss M, Gusterson B, Bartek J: Abnormal patterns of D-type cyclin expression and G1 regulation in human head and neck cancers. Cancer Res 1995, 55:949-956 [PubMed] [Google Scholar]
- 27.Kotelnikov VM, Coon JS, IV, Mundle S, Kelanic S, LaFolette S, Taylor SIV, Hutchinson J, Panje W, Caldarelli DD, Preister HD: Cyclin D1 expression in squamous cell carcinomas of the head and neck and in oral mucosa in relation to proliferation and apoptosis. Clin Cancer Res 1997, 1:95-101 [PubMed] [Google Scholar]
- 28.Bringuier PP, Tamimi E, Schuuring E, Schalken J: Expression of cyclin D1 and EMS in bladder tumours: relationship with chromosome 11q13 amplification. Oncogene 1996, 12:1747-1783 [PubMed] [Google Scholar]
- 29.Gillet C, Fontl V, Smith R, Fisher C, Bartek J, Dickson C, Barnes D, Peters G: Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res 1994, 54:1812-1817 [PubMed] [Google Scholar]
- 30.Pelosio P, Barbareschi M, Bonoldi E, Marchetti A, Verderio P, Caffo O, Bevilacqua P, Boracchi P, Buttitta F, Barbazza R, Dalla Palma P, Gasparini G: Clinical significance of cyclin D1 expression in patients with node-positive breast carcinoma treated with adjuvant therapy. Anal Oncol 1996, 7:695-703 [DOI] [PubMed] [Google Scholar]
- 31.McIntosh GG, Anderson JJ, Milton I, Steward M, Parr AH, Thomas MD, Henry JA, Angus B, Lennard TWJ, Horne CHW: Determination of the prognostic value of cyclin D1 overexpression in breast cancer. Oncogene 1995, 11:885-891 [PubMed] [Google Scholar]
- 32.Seshardri R, Lee CSL, Hui R, McCaul K, Horsfall DJ, Sutherland RL: Cyclin D1 amplification is not associated with reduced overall survival in primary breast cancer but may predict early relapse in patients with features of good prognosis. Clin Cancer Res 1996, 2:1177-1184 [PubMed] [Google Scholar]
- 33.Palmero I, Peters G: Perturbation of cell cycle regulators in human cancer. Cancer Surv 1996, 27:351-367 [PubMed] [Google Scholar]
- 34.Motokura T, Bloom T, Kim HG, Juppner H, Ruderman JV, Kronenberg HM, Arnold A: A novel cyclin encoded by a bcl-1 linked candidate oncogene. Nature 1991, 350:512-515 [DOI] [PubMed] [Google Scholar]
- 35.Pines J: Cyclins and their associated cyclin-dependent kinases in the human cell cycle. Biochem Soc Trans 1993, 21:921-925 [DOI] [PubMed] [Google Scholar]
- 36.Sawa H, Oshima TA, Ukita H, Murakami H, Chiba Y, Kamada H, Hora M, Saito I: Alternatively spliced forms of cyclin D1 modulate entry into cell cycle in an inverse manner. Oncogene 1998, 16:1701-1712 [DOI] [PubMed] [Google Scholar]
- 37.Hosokawa Y, Joh T, Maeda Y, Arnold A, Seto M: Cyclin D1/PRAD1/BCL-1 alternative transcript [B] protein product in B-lymphoid malignancies with t (11;14)(q13;32) translocation. Int J Cancer 1999, 81:616-619 [DOI] [PubMed] [Google Scholar]
- 38.Ramljak D, Calvert RJ, Wiesenfeld PW, Diwan BA, Catipovic B, Marasas WF, Victor TC, Anderson LM, Gelderblom WE: A potential mechanism for fumonisin B (1)-mediated hepatocarcinogenesis: cyclin D1 stabilization associated with activation of Akt and inhibition of GSK-3β activity. Carcinogenesis 2000, 21:1537-1546 [PubMed] [Google Scholar]
- 39.Davidson BJ, Lydiatt WM, Abate MP, Schantz SP, Chaganti RSK: Cyclin D1 abnormalities and tobacco exposure in head and neck squamous cell carcinoma. Head and Neck Journal for the Sciences and Specialities of the Head and Neck 1996, 18:512-521 [DOI] [PubMed] [Google Scholar]
- 40.Kenny FS, Hui R, Musgrove EA, Gee JM, Blamey RW, Nicholson RI, Sutherland RL, Robertson JF: Overexpression of cyclin D1 messenger RNA predicts for poor prognosis in estrogen receptor-positive breast cancer. Clin Cancer Res 1999, 5:2069-2076 [PubMed] [Google Scholar]
- 41.Zwijsen RM, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides RJ: CDK-independent activation of estrogen receptor by cyclin D1. Cell 1997, 88:405-415 [DOI] [PubMed] [Google Scholar]
- 42.Knudsen KE, Cavenee WK, Arden KC: D-Type cyclins complex with the androgen receptor and inhibit its transcriptional transactivation ability. Cancer Res 1999, 59:2297-2301 [PubMed] [Google Scholar]