Abstract
The cellular lineage of sinonasal T/NK (natural killer) cell lymphoma remains controversial. Lineage assignment is difficult because T cells and NK cells have a similar morphology and surface markers. Consequently, the assignment must depend heavily on the status of T-cell receptor (TCR) rearrangement. A monoclonal TCR rearrangement supports a T lineage; however, a corresponding monoclonality test for NK cells has not yet been established. Each NK cell bears a distinct set of killer cell immunoglobulin (Ig)-like receptors (KIRs) that are randomly distributed over three groups. In principle, restriction of the KIR repertoire signifies a monoclonal or possibly oligoclonal NK-cell proliferation, just as Ig light-chain restriction usually indicates a monoclonal B-cell neoplasm. Using a novel group-specific reverse transcriptase-polymerase chain reaction, we found a restricted KIR repertoire in most sinonasal lymphomas (9 of 10), but only rarely in T-cell lymphomas (2 of 10) or reactive conditions involving T/NK cells (1 of 10). KIR+ sinonasal lymphomas usually lacked a monoclonal TCR-γ rearrangement pattern, expressed another NK cell receptor, NKG2a, and were usually CD56-positve, cyclin-dependent kinase-6 (CDK6)-positive, CD44-negative, a phenotype already reported to indicate a true NK cell lineage. We conclude that, although sinonasal lymphomas have heterogeneous genotypes and phenotypes, a restricted KIR repertoire without TCR-γ rearrangement provides preliminary support for the monoclonality hypothesis and can be used for defining a true NK-cell lineage in a subset of sinonasal lymphomas.
Nasal T/NK (natural killer) cell lymphoma, a rare disease in Western populations, is more common among Orientals and South Americans. 1,2 The lymphoma typically grows in an angiocentric pattern, causing ulceration and necrosis of the nasal cavity and of the surrounding facial tissue; hence the old name “angiocentric lymphoma,” which reflects its unique histopathology.
Despite the well-characterized angiocentric histopathology, the origin of this lymphoma remains controversial. The lymphoma often expresses a NK-cell marker, CD56, and some T-cell-related antigens such as CD2, CD43, and/or CD45RO. However, it lacks other T-lineage antigens, such as CD4, CD5, CD8, and surface CD3, and rarely has a T-cell-receptor gene rearrangement (TCR-GR). 3,4 The ambiguous phenotype partially reflects the fact that NK cells are developmentally close to T cells, both possibly arising from a bipotent T/NK progenitor. 5-8 Consequently, the lymphoma was simply designated as angiocentric lymphoma in the REAL (revised European American lymphoma) classification, without further specification of its cellular origin, T- or NK-cell.
After the REAL classification, attempts have been made to separate nasal lymphoma of true NK-cell lineage from that of T-cell lineage. For example, it was concluded that lymphomas with expression of CD56 and CD3e, lack of CD5, and without TCR-GR were of NK lineage. 3,9 Recently, we demonstrated that nuclear expression of cyclin-dependent kinase-6 and surface loss of CD44 without TCR-GR favor a true NK-cell nasal lymphoma. 10
NK cells use both killer cell immunoglobulin-like receptors (KIRs) and C-type lectin receptors (NKG2) to recognize major histocompatibility complex class I molecules on autologous cells. 11 The receptor-major histocompatibility complex binding delivers an inhibitory signal and protects autologous cells from NK-mediated cytotoxicity. Whenever NK cells interact with a target cell without major histocompatibility complex class I molecules as a result of viral infection or tumor transformation, a cytotoxic effect is initiated, leading to destruction of the target cells. 12,13 Despite the very important immunological functions of these receptors, very limited data are available on the status of their expression in lymphomas.
It is known that expression of NKG2 is susceptible to interleukin (IL)-15 regulation. 14 A recent report also demonstrated NKG2A expression in each of four cases of sinonasal lymphoma, but rarely in lymphomas of T- or B-cell origin. 15 NKG2A thus seems to be a marker for sinonasal lymphoma. However, its use as a marker for lineage or monoclonality assignment has yet to be determined.
It is already known that the KIR receptors belong to 12 families, that each NK cell displays, on average, 3 to 4 families of KIRs, and that most people have a KIR repertoire of 6 to 9 KIR families. 16,17 Therefore, a monoclonal population derived from a single NK cell would have a KIR repertoire restricted to three to four families. In contrast, a polyclonal NK-cell population would display an unrestricted distribution of the KIR repertoire covering six to nine families. Because the rationale of NK-cell repertoire restriction in a monoclonal or an oligoclonal NK-cell population is similar to the phenomenon of light-chain restriction in B-cell lymphoma, we proposed to use a restricted NK-cell repertoire as a criterion for assigning a sinonasal lymphoma to the NK lineage.
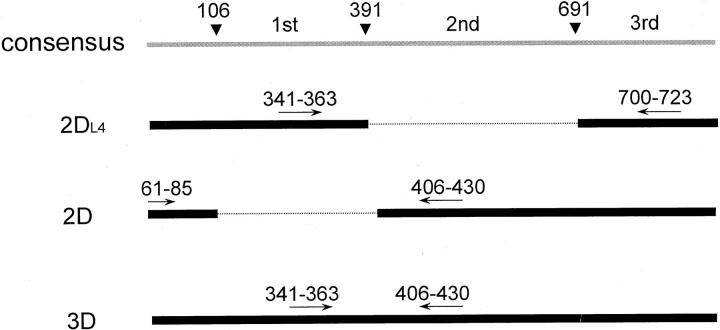
To obtain a simple experimental design, we noticed that the 12 families share a consensus sequence composed of three immunoglobulin (Ig)-like domains. 18 By sequence analysis, the 12 families can be broadly divided into three groups, 2DL4, 2D, and 3D, that differ in the constituent domains. Group 2DL4 has the first and the third domains, group 2D has the second and the third domains, and group 3D has all three domains. 18 If the KIR repertoire from a sinonasal lymphoma can be shown to be composed of only one or two of the three groups, then a restricted pattern is demonstrated, and a monoclonal NK-cell nature can be confirmed.
Based on these observations, we believe that a restricted KIR repertoire is a marker for NK cell differentiation. To exclude the less likely event, in which KIR might be expressed by a minor subset of T cells, we propose that a restricted KIR repertoire without T-cell receptor rearrangement could be used for assigning a subset of sinonasal lymphoma to a true NK cell lineage.
Materials and Methods
Tissue Samples
Ten cases of lymphoma of the nasal cavity and paranasal sinuses were diagnosed in the Pathology Department of the National Taiwan University Hospital between 1993 and 1999. The diagnosis was made initially by a combination of morphology and immunohistochemistry. All 10 cases were CD3-positive or CD45RO-positive by immunohistochemistry. At the time of diagnosis none of the patients had lymphoma involvement outside the nasal cavity or the adjacent paranasal sinuses. In addition, 10 cases of T-cell lymphoma and 10 cases of reactive conditions involving T/NK cells were used as controls. The reactive controls included hyperplastic tonsils, lymph nodes with T-zone expansions, B-cell lymphomas, mononuclear cells from normal peripheral blood, and gestational endometrium. The last type of tissue was chosen because of its known unrestricted KIR repertoire. 19,20
RNA Extraction
RNA was extracted from formalin-fixed, paraffin-embedded tissue by the Trizol method. Briefly, a 20- to 60-μm section of the tumor was mixed with 1 ml of xylene for 2 minutes for removal of paraffin. After centrifugation, residual xylene was dissolved in ethanol and removed by evaporation. The deparaffinized tissue was mixed with 180 μl of lysis buffer and 20 μl of proteinase K at 55°C overnight. Then 1 ml of Trizol (phenol/guanidine isothiocyanate) and 0.2 ml of chloroform were added. After vortexing and centrifugation, the aqueous phase containing RNA was saved. The RNA was precipitated by isopropanol and redissolved in diethyl pyrocarbonate-treated water for future use.
Group-Specific Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) for KIR
The nucleotide sequences for human KIR have been published. 18 The KIR transcripts have a consensus structure containing a leader sequence followed by three Ig-like domains. The first domain extends from nucleotide 106 to nucleotide 391, the second domain from nucleotide 392 to nucleotide 691, and the third domain starts from nucleotide 692 (Figure 1) ▶ . Probably because of alternative splicing, group 2DL4 skips the second domain, group 2D skips the first domain, and group 3D retains both domains. Hence, group 2DL4 has a unique junction between the first and the third domains, group 2D has a unique junction between the leader sequence and the second domain, and group 3D has a unique junction between the first and second domains.
Figure 1.
Group-specific RT-PCR across the junctions of Ig-like domains of KIR. The KIR transcripts have a consensus structure containing a leader sequence followed by multiple Ig-like domains. Because of alternative splicing, group 2DL4 skips the second domain, group 2D skips the first domain, and group 3D retains both domains. Three pairs of primers were designed, each specific for a KIR subgroup.
The unique junction can be used for development of a novel group-specific RT-PCR. We have designed three pairs of group-specific primers, each pair for one specific junction (Table 1 ▶ and Figure 1 ▶ ). For example, the pairs 341 to 363 and 700 to 723 can amplify group 2DL4 to yield a specific PCR product with 83 nucleotides. Although a second product of 383 nucleotides because of nonspecific hybridization to group 3D is theoretically possible, this product can easily be separated by electrophoresis, and is not detected in our system, presumably because the much larger size makes its RT-PCR inefficient. Similarly, the pairs 61 to 85 and 406 to 430 are group 2D-specific, and they yield a PCR product of 85 nucleotides. Finally, the pairs 341 to 363 and 406 to 430 placed at the first and the second domains can amplify group 3D specifically, because it is the only transcript containing both domains. The PCR product has 90 nucleotides.
Table 1.
Primers for Group-Specific RT-PCR Across the Junctional Region of KIR
| KIR | Sequence | Position* | R or F† |
|---|---|---|---|
| 2D | 5′-TGGGTGGGCCAGGAGGAAGGTTT-3′ | 430-406 | R |
| 5′-CATGGCGTGTGTTGGGTTCTTCTTG-3′ | 61-85 | F | |
| 2DL4 | 5′-GCTGAGAGAGAAGGTTCTCATAT-3′ | 723-700 | R |
| 5′-CACTCCCCCACTGGGTGGTCGGC-3′ | 341-363 | F | |
| 3D | 5′-TGGGTGGGCCAGGAGGAAGGTTT-3′ | 430-406 | R |
| 5′-CACTCCCCCACTGGGTGGTCGGC-3′ | 341-363 | F | |
| β-actin | 5′-CTGGAAGGTGGACAGCGAGGC-3′ | 1839-1819 | R |
| 5′-CCCAGCACAATGAAGATCAAG-3′ | 1630-1650 | F |
*The numbers for KIR refer to the nucleotide position in cDNA. The numbers for β-actin refer to the nucleotide position in genomic DNA.
†Reverse or forward primer.
Reverse transcription was done by use of purified RNA, 500 μmol/L each dNTP 0.5 μmol/L antisense primer, 200 U reverse transcriptase (Superscript II; Life Science, Grand Island, NY), and 4 μl 5× buffer (1× equals 50 mmol/L Tris-HCl at pH 8.3, 75 mmol/L KCl, 3 mmol/L MgCl2, 10 mmol/L dithiothreitol) in a total reaction volume of 20 μl at 42°C for 50 minutes. For each case of sinonasal lymphoma, four reverse-transcription reactions in four separate tubes were started with equal amounts of purified RNA and performed in parallel, three for each of the KIR groups and one for β-actin 21 as the positive control. In addition, four corresponding negative controls were run simultaneously under exactly the same conditions, except that the reverse transcriptase was omitted.
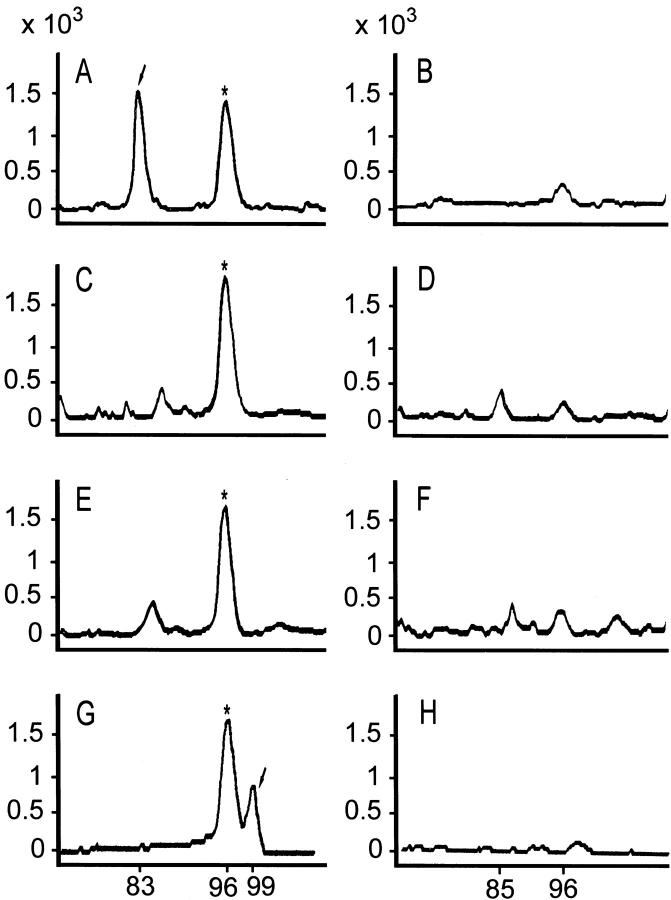
One-tenth of the reaction mixture was used for PCR. The 20-μl PCR reaction mixture included 50 mmol/L Tris-HCl, pH 9.1, 3.5 mmol/L MgCl2, 16 mmol/L ammonium sulfate, 150 μg/ml bovine serum albumin, 200 μmol/L of each dNTP, 2 μmol/L [TAMRA]-dCTP, 0.3 μmol/L of each primer set, and 1 U of Taq polymerase. Each cycle consisted of denaturation at 94°C for 45 seconds, annealing at 40°C for 45 seconds, and extension at 72°C for 45 seconds. The PCR reaction was performed for 35 cycles. At the end of the 35 cycles, a portion of the PCR products was loaded and separated by a DNA analyzer (ABI377 with GeneScan software; Perkin-Elmer, Foster City, CA). A typical result is shown in Figure 2 ▶ .
Figure 2.
Separation of RT-PCR products for KIR and NKG2A by DNA analyzer. RT-PCR products for KIR and NKG2A were mixed with RT-PCR β-actin product in equal portions and separated by electrophoresis on the DNA analyzer. Four pairs of GeneScan tracings for a case of sinonasal lymphoma are shown, with the size of the PCR product on the x axis and the size of the peak on the y axis. These tracings are KIR 2D4L (A) and its negative control (B), KIR 2D (C) and its negative control (D), KIR 3D (E) and its negative control (F), and NKG2A (G) and its negative control (H). The expected sizes of the PCR products for KIR 2D4L, KIR 2D, KIR 3D, β-actin, and NKG2A are 83, 85, 90, 96, and 99, respectively. This case is KIR 2D4L+ (arrow in A), KIR 2D−, and KIR 3D−, and NKG2A+ (arrow in G). Note also a small peak of size 85, corresponding to KIR 2D, can be seen in D, and small peaks of size 96, corresponding to β-actin, can be seen throughout all of the negative controls (B, D, F, and H).
The absolute size of the peak was quantified by measurement of the intensity of the incorporated [TAMRA]-dCTP. The background intensity of the negative control was subtracted, leaving the true intensity. Typically, for each case of lymphoma, three values were obtained, one for each of the KIR groups. The relative sizes of the three groups are listed in Table 2 ▶ . If the peak was indistinguishable from background, a minus sign was entered in Table 2 ▶ .
Table 2.
Killer Immunoglobulin-Like Receptor Repertoire and T Cell Receptor γ Rearrangement in Sinonasal Lymphoma
| Age (yrs) | Sex | Clonal TCR-γ* | KIR2DL4 | KIR 2D | KIR3D | NKG2A | CD3 | CD56 | CDK6 | CD 44 | Response to chemotherapy | Diagnosis/outcome | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sinonasal lymphoma | ||||||||||||||||||||||||||
| 1 | 58 | m | − | − | − | − | + | + | + | +++ | − | Refractory | Died in 3 months, multiple visceral metastases | |||||||||||||
| 2 | 60 | m | − | 100% | 0% | 0% | + | + | + | +++ | + | Responsive | No progression in 1 year | |||||||||||||
| 3 | 56 | m | − | 100% | 0% | 0% | + | + | + | ++ | + | Responsive | No progression in 1 year | |||||||||||||
| 4 | 45 | m | − | 31% | 0% | 69% | + | + | + | +++ | +/− | Partial | Died in 10 months, lung metastasis, multiple LAP | |||||||||||||
| 5 | 62 | m | − | 100% | 0% | 0% | + | + | + | ++ | − | Refractory | Died in 2 months, hepatic metastasis | |||||||||||||
| 6 | 24 | m | − | 81% | 19% | 0% | + | + | + | ++ | − | Responsive | No progression in 1 year | |||||||||||||
| 7 | 67 | m | − | 0% | 100% | 0% | + | + | + | ++ | − | Responsive | No progression in 1 year | |||||||||||||
| 8 | 54 | m | M | 0% | 100% | 0% | + | + | − | ++ | − | Partial | Died in 11 months, lung metastasis | |||||||||||||
| 9 | 42 | m | M | 52% | 48% | 0% | + | + | +/− | +++ | − | Partial | Died in 2 years, extensive mid-facial destruction | |||||||||||||
| 10 | 28 | m | M | 100% | 0% | 0% | − | + | + | ++ | − | Refractory | Died in 2 months, testes and abdominal LAP | |||||||||||||
| T-cell lymphoma | ||||||||||||||||||||||||||
| 1 | 26 | f | M | − | − | − | − | + | − | +++ | + | Lymphoblastic lymphoma, still in CR for 2 years | ||||||||||||||
| 2 | 18 | f | M | − | − | − | + | + | − | +++ | + | Lymphoblastic lymphoma, still in CR for 3 years | ||||||||||||||
| 3 | 53 | m | −† | − | − | − | − | + | − | ++ | + | PTCL, died in 4 months, multiple LAP | ||||||||||||||
| 4 | 68 | m | M | − | − | − | − | + | − | + | + | PTCL, still in CR for 5 years | ||||||||||||||
| 5 | 49 | m | M | − | − | − | + | + | − | − | + | PTCL, died in 3 months, multiple LAP and BM metastasis | ||||||||||||||
| 6 | 68 | f | M‡ | − | − | − | − | + | − | − | + | PTCL, died in 6 months, skin and BM metastasis | ||||||||||||||
| 7 | 77 | f | M | − | − | − | − | + | − | − | + | PTCL, alive but with slow progression of LAP over 5 years, under symptomatic treatment only | ||||||||||||||
| 8 | 38 | f | M | − | − | − | − | + | − | − | + | PTCL, died in 9 months, multiple LAP | ||||||||||||||
| 9 | 51 | f | M‡ | 0% | 100% | 0% | − | + | − | − | − | PTCL, died in 2 years, multiple LAP | ||||||||||||||
| 10 | 84 | m | M | 0% | 100% | 0% | − | + | − | − | − | PTCL, CR for 2 years, recurred and refractory to salvage chemotherapy | ||||||||||||||
| Controls | ||||||||||||||||||||||||||
| 1 | 23 | f | − | − | − | − | − | Hypertrophic tonsil | ||||||||||||||||||
| 2 | 63 | m | − | − | − | − | − | Hypertrophic tonsil | ||||||||||||||||||
| 3 | 66 | f | − | − | − | − | − | T-zone hyperplasia | ||||||||||||||||||
| 4 | 72 | m | − | − | − | − | − | T-zone hyperplasia | ||||||||||||||||||
| 5 | 73 | m | − | − | − | − | − | B-cell lymphoma | ||||||||||||||||||
| 6 | 65 | f | − | 80% | 0% | 20% | − | B-cell lymphoma | ||||||||||||||||||
| 7 | 41 | f | nd | 26% | 14% | 60% | + | Gestational endometrium | ||||||||||||||||||
| 8 | 34 | f | nd | 18% | 19% | 63% | + | Gestational endometrium | ||||||||||||||||||
| 9 | 31 | f | nd | 2% | 12% | 86% | + | Normal peripheral blood | ||||||||||||||||||
| 10 | 29 | m | nd | 6% | 38% | 56% | + | Normal peripheral blood | ||||||||||||||||||
Semiquantitative evaluation of killer inhibitory receptor (KIR) transcripts. −, <1%; +, >1% of β-actin.
*For TCR-γ, M means a monoclonal rearrangement was found; a negative sign means a monoclonal rearrangement was not detected.
†Cytogenetic data show a monoclonal population.
‡This case had a polyclonal TCR-γ rearrangement, but further analysis showed a monoclonal TCR-β rearrangement.
Abbreviations: TCR-γ, T-cell-receptor γ gene; LAP, lymphadenopathy; PTCL, peripheral T-cell lymphoma; CR, complete remission; BM, bone marrow; ND, not determined.
RT-PCR for NKG2A
RT-PCR for NKG2A was done with the primers 5′-ATAGATAATGAAGAAGAAAT-3′ (633 to 652) and 5′-CATTGTCACCCATGGATGATG-3′ (731 to 711). The numbers in parentheses are the nucleotide positions of the RNA transcript. The RT-PCR conditions used and the analysis of the data were the same as those for the RT-PCR for KIR.
DNA Extraction
Genomic DNA was extracted from paraffin-embedded tissue blocks by use of a QIAamp kit (Qiagen, Valencia, CA). Briefly, a 20- to 60-μm section was mixed with 1 ml of xylene at room temperature for 2 minutes for removal of paraffin. After centrifugation, residual xylene was dissolved in ethanol and removed by evaporation. The deparaffinized tissue was mixed with 180 μl of lysis buffer and 20 μl of proteinase K at 55°C overnight. After centrifugation, the supernatant containing DNA was saved. The DNA was precipitated by ethanol and redissolved in Tris-ethylenediaminetetraacetic acid buffer for future use.
T-Cell-Receptor γ Gene Rearrangement
The TCR-γ-chain gene was examined by multiplex nested PCR, as modified from a previous report. 22 In the first multiplex PCR, four primers, VrI, VrII, VrIII, and VrIV, were used to anneal to the variable regions of TCR-γ, and three primers, Jr1/2, Jpr, and Jpr1/2, were used to anneal to the junctional regions of the TCR-γ. The primer sequences were as follows: VrI, 5′-TACATCCACTGGTACCTACACCAG-3′; VrII, 5′-GAAAGGAATCTGGCATTCCG-3′; VrIII, 5′-AAGCAACAAAGTGGAGGCAAGAAAG-3′; VrIV, 5′-CTCACACTCTCACTTC-3′; Jr1/2, 5′-CAAGTGTTGTTCCACTGCC-3′; Jpr, 5′-TTGTTCCGGGACCAAATACC-3′; and Jpr1/2, 5′-GTTACTATGAGCCTAGTC-3′.
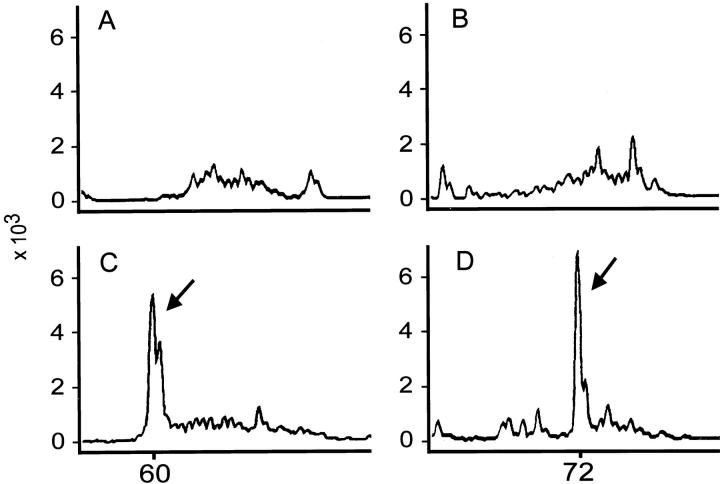
In the second nested PCR, we used primers V, 5′-TCTGGAGTCTATTACTGTGC-3′, and Jr, 5′-(6-FAM)-AGTGTAGTCCCTGTACCAAACATTTT-3′, to anneal to the consensus regions of the variable and junctional regions of TCR-γ. Each 20-μl reaction mixture contained 50 mmol/L Tris-HCl, pH 9.1, 3.5 mmol/L MgCl2, 16 mmol/L ammonium sulfate, 150 μg/ml bovine serum albumin, 200 μmol/L of each dNTP, 0.3 μmol/L of each primer set, 2 μl of DNA from the first PCR reaction, and 1 U of Taq polymerase. The reaction mixture was subjected to 35 cycles of PCR after an initial 2-minute denaturation step at 94°C. Each cycle consisted of denaturation at 94°C for 45 seconds, annealing at 40°C for 45 seconds, and extension at 72°C for 45 seconds. At the end of 35 cycles, a portion of the PCR product was loaded on and separated by a DNA analyzer (ABI377 equipped with GeneScan software, Perkin-Elmer). A typical result is shown in Figure 3 ▶ .
Figure 3.
Examination of TCR-γ gene rearrangement in peripheral T-cell lymphoma and nasal T/NK cell lymphoma. The status of TCR-γ gene rearrangement was determined by PCR-based tests. The products were separated and analyzed by GeneScan. The x axis is the size of the PCR product; the y axis is the size of the peak. A polyclonal pattern was detected for a case of reactive lymphoid hyperplasia (tonsil, A) and a case of sinonasal lymphoma (B). A monoclonal peak (arrows in C and D) was demonstrated for a case of peripheral T-cell lymphoma (C) and a case of sinonasal lymphoma (D).
Immunohistochemistry
Immunohistochemistry was done according to a protocol published from our laboratory. 10 Briefly, immunostaining for cdk6 was performed with an affinity-purified rabbit polyclonal antibody that recognized a peptide corresponding to amino acid residues 306 to 326 mapping at the carboxyl terminus of cdk6 (C-21 antibody; Santa Cruz Biotechnology, Santa Cruz, CA). Immunostaining for CD44 was performed with a monoclonal antibody, anti-human CD44H (2C5/IgG2a subclass; R&D Systems Ltd., Minneapolis, IN), that recognized all CD44 isoforms, including the standard CD44 isoform (CD44s). Additional antibodies, CD3ε (polyclonal) or CD45RO (DAKO SA, Glostrup, Denmark), and CD56 (Novocastra Lab Ltd., Newcastle, UK), were used according to the manufacturer’s recommendation.
Quantitative evaluation of cdk6 expression was performed by counting of the percentage of positively stained cells in high-power (×40) microscopic fields. 10 The percentages of tumor cells expressing nuclear cdk6 were defined as follows: −, <10%; +, 10 to 30%; ++, 30 to 50%; and +++, 50 to 100%.
Results
Patient Characteristics and Pathological Findings
The series of sinonasal T/NK cell lymphomas included 10 male patients with an age distribution from 24 to 67 years. All initially had disease localized to the sinonasal area. Biopsies were taken before initiation of chemotherapy or radiation therapy, and they were classified histologically as pleomorphic medium- and large-cell lymphoma. Coagulative necrosis was seen in nearly all of these cases. Angioinvasion was present to variable degrees. All 10 cases were CD3-positive or CD45RO (UCHL-1)-positive, and most of them were CD56-positive by immunohistochemistry (Table 2) ▶ . In situ hybridization for EBV was positive in all cases. The immunophenotypes of these 10 cases were included in our previous publication. 10 The male predominance and the pathology findings are consistent with other previous reports. 1,2,
KIR Repertoire by Group-Specific RT-PCR
The RT-PCR approach for characterizing the KIR repertoire is well established, and was shown to correlate well with surface expression of KIR receptors. 16,17 For the present study, we modified and developed a group-specific RT-PCR for KIR by amplifying the unique junction that is present only in one of the three groups. A schematic presentation for this approach is shown in Figure 1 ▶ . The PCR product was then separated by a high-resolution polyacrylamide system, the GeneScan, a typical result of which is shown in Figure 2 ▶ .
If any one of the three group-specific RT-PCRs yielded a peak, then a KIR repertoire was detected, and the relative sizes of the three peaks were entered in Table 2 ▶ . If the sizes of the three peaks were all indistinguishable from baseline fluctuations, a minus sign was entered in Table 2 ▶ . In our system, a peak would become undetectable if it was equal to or less than 1% of the size of the actin peak.
Most sinonasal lymphomas listed in Table 2 ▶ had a detectable KIR repertoire (9 of 10). The relative proportions of the three groups of KIR transcripts were shown, and it is apparent that the KIR repertoires for all nine cases were restricted to one or two groups, consistent with monoclonal NK cell proliferation. One case did not have detectable KIR transcripts. Although accidental degradation of RNA and extensive necrosis of the tissue might have caused a false-negative result, this particular case seemed to be true negative, as RT-PCR for β-actin and NKG2A were both positive and served as internal positive controls.
The majority of T-cell lymphomas were negative for KIR (8 of 10), as predicted, but two cases were KIR-positive. Expression of KIR in these two peripheral T-cell lymphomas could be false-positive, or they might represent lymphomas arising from a minor subset of T cells that normally expressed KIR. Although we could not distinguish between the two possibilities, the data showed that a KIR repertoire is much more likely to be associated with a sinonasal lymphoma than with a T-cell lymphoma, and can be used to define a NK-cell lineage in sinonasal lymphomas.
Measurements of the KIR repertoire were done on 10 additional cases of reactive T/NK proliferation. These included two hyperplastic tonsils, two lymph nodes with T-zone hyperplasia, two B-cell lymphomas, mononuclear cells from the peripheral blood of two healthy normal patients, and two gestational endometrial biopsy specimens. An unrestricted pattern of KIR repertoire was found, as expected, from the endometrium and peripheral blood. The hyperplastic tonsils, lymph nodes with T-zone hyperplasia, and one B-cell lymphoma showed no detectable KIR repertoires, which simply meant that the NK cells in the specimens were below the detection limit of our measurement system. The absence of a KIR repertoire in tonsils is consistent with a previous report that the number of NK cells in acute tonsillitis or hypertrophic tonsils is only ∼1%, 23 which is about the detection limit of our system. Interestingly, the second B-cell lymphoma showed a restricted pattern of KIR repertoire. The intensities of KIR transcripts in this case were weak and should be because of infiltrating NK cells reactive to the B-cell lymphoma, but the cause for the restriction deserves further investigation. The KIR repertoire of case 9 was very disturbed because of a low percentage of KIR2DL4 at 2%, which was confirmed by a repeated measurement. This also raised the possibility of monoclonal NK mixed with some reactive NK cells. Although some of these possibilities could not be resolved, the data did show that most reactive conditions had either no KIR repertoires or unrestricted KIR repertoires, and gave further support to the use of restricted KIR repertoires as markers for sinonasal lymphomas.
RT-PCR for NKG2A
We also used RT-PCR to determine NKG2A transcript in sinonasal lymphoma, and we identified its presence in most cases (9 of 10), but only rarely in T-cell lymphomas (2 of 10). For the reactive controls, NKG2A was positive in the endometrium and peripheral blood, but was not found in B-cell lymphoma, hyperplastic tonsils, or lymph nodes with T-zone expansion. The result is consistent with a previous report that four out of four cases of sinonasal lymphoma had NKG2A expression by immunohistochemistry, but this expression was rarely found in other T- or B-cell lymphomas. 15
TCR-γ Gene Rearrangement
A PCR-based TCR-γ-GR study was performed according to a previously published protocol, with slight modifications. 22 The PCR products were separated by the GeneScan system, a typical result of which is shown in Figure 3 ▶ . The data for all of the cases are listed in Table 2 ▶ . We found that most sinonasal lymphomas did not have monoclonal TCR-γ rearrangement (7 of 10) and most T-cell lymphomas were monoclonal (7 of 10). However, there were three cases of sinonasal lymphoma with monoclonal TCR rearrangement and three cases of T-cell lymphoma without TCR rearrangement. The three T-cell lymphomas without a monoclonal TCR-γ-GR were false-negative because monoclonality could be shown by either TCR-β rearrangement or cytogenetics. As for the three sinonasal lymphomas with monoclonal TCR-γ-GR, it is less clear whether they were true or false-positive. Because KIR could be found rarely on a minor subset of T cells, a lymphoid tumor cell might simultaneously express KIR and rearrange TCR.
Immunophenotype
We performed immunostaining for CD3, CD56, CDK6, and CD44. A typical result is shown in Figure 4 ▶ . Sinonasal lymphomas are usually CD3+ and CD56+, and peripheral T-cell lymphomas are usually CD3+ and CD56−. Sinonasal lymphomas have strong nuclear expression of CDK6, but frequent loss of surface CD44, and peripheral T-cell lymphomas are usually CDK6-negative but usually CD44-positive. The two cases of lymphoblastic lymphoma had strong expression of CDK6, which is also consistent with previous reports. 24 The immunophenotyping provides an independent criterion for lineage assignment, and is approximately consistent with lineage assignment based on consideration of KIR repertoires and TCR rearrangements.
Figure 4.
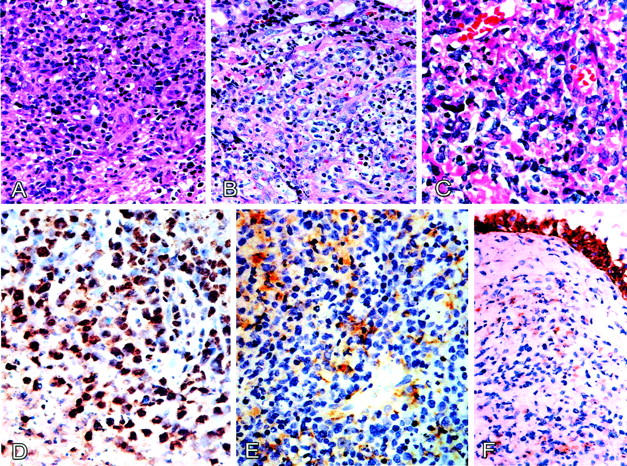
Morphology and expression of cdk6 and CD44 in sinonasal lymphoma of NK cell origin. A: Tumor cells exhibiting an angiocentric growth pattern. B: Focal clear cell changes with increased karyorrhexis. C: Pleomorphic medium and large lymphoma cells. D: Strong nuclear cdk6 immunostaining is present in most lymphoma cells, and some faint cytoplasmic staining can also be noted. E: Most lymphoma cells are CD44-negative with squamous epithelium as internal positive control (F).
Lineage Assignment
Sinonasal lymphoma seems, as shown in Table 2 ▶ , to be heterogeneous both genotypically and phenotypically, and cannot be completely separated from peripheral T-cell lymphomas by any single marker. However, the expression of KIR correlates fairly well with the expression of NKG2a, CD56, and CDK6, and the absence of CD44, and is about as good as the other markers in distinguishing sinonasal lymphomas from peripheral T-cell lymphomas.
We therefore propose to use restriction of the KIR repertoire without TCR-γ rearrangement as a criterion for a true NK cell lineage. When the criterion was applied to the 10 cases listed in Table 2 ▶ , 6 cases could be assigned to the true NK lineage. Three cases had both KIR expression and TCR-γ rearrangement, which probably represents dual differentiations toward both T and NK lineages, although a false-positive TCR-γ rearrangement could not be excluded completely. The lineage in the remaining one case without TCR-γ rearrangement or KIR expression is undetermined.
When the same principle was applied to two lymphoblastic lymphomas and eight peripheral T-cell lymphomas, we found that the two lymphoblastic lymphomas and four of the eight peripheral T-cell lymphomas had TCR-γ rearrangement without KIR expression, consistent with a T lineage. Cases 3 and 6 did not have monoclonal TCR-γ rearrangement and did not express KIR. The lineage was undetermined. Case 9 expressed KIR without a monoclonal TCR-γ rearrangement, consistent with an NK-cell lineage. Case 10 showed both TCR-γ rearrangement and KIR expression, consistent with dual differentiation. Case 6 would have a T-lineage and case 9 a lineage of dual differentiation, if we also considered the status of TCR-β rearrangement. Apparently the test was limited by the low sensitivity for detecting TCR-γ rearrangement, that led to wrong assignments in two cases.
Correlation with Morphology and Response to Chemotherapy and Clinical Outcome
We were unable to find significant morphological differences between the KIR+ and KIR− cases. There seemed to be no correlation between the clinical course and the status of KIR repertoires. A larger sample size and a longer follow-up might be necessary for a significant conclusion.
Discussion
There is uncertainty regarding the true nature of nasal T/NK cell lymphoma of T versus NK cell lineage. 25,26 Lineage assignment is difficult because NK cells are developmentally close and immunophenotypically similar to T cells. 5-8 Most classification schemes, therefore, rely heavily on molecular studies of monoclonal TCR-GR for assignment to a T lineage. So far, there have been no direct criteria for a monoclonal NK-cell neoplasm except for cytogenetic criteria. 27
Monoclonality is probably the single most important test in hematopathology. The classical example is the light-chain restriction in B-cell neoplasms. The rationale for the KIR repertoire restriction is similar to that of light-chain restriction. However, experimentally it is more complicated because each NK cell can express 3 or 4 KIRs out of 12 families, whereas each B cell displays either κ or λ light chain. 16,17 To simplify the experiments, we took advantage of the fact that the 12 families share sequence homology and could be divided into three groups. 18 By group-specific or junction-specific RT-PCR, we demonstrated that, in sinonasal lymphoma, a restricted pattern could be identified in all nine cases with a KIR repertoire.
Strictly speaking, a restricted pattern may be because of a monoclonal or oligoclonal NK proliferation. Further experimental data would be required for finding out how common it is to have an oligoclonal NK proliferation, and how important it is to distinguish between this condition and a true monoclonal proliferation. Similarly, more data would be required for determining whether other NK-cell neoplasms also have restricted KIR repertoires.
Regardless of these limitations, we can use the status of the KIR repertoire restriction in conjunction with TCR-γ-GR to achieve a lineage assignment for sinonasal lymphoma. We find six cases with true NK cell differentiation, three cases with probable dual T/NK differentiation, and one case with no differentiation. This reflects the current model of T- and NK-cell development, in which a common lymphoid precursor gives rise to a bipotent T/NK progenitor, which subsequently develops into T and true NK cells. 28-30
Extensive characterization of the KIR and NKG2 repertoire of normal NK cells in peripheral blood has been reported. 16,17 It has been established that the RT-PCR approach to mRNA transcripts gives the same result as does immunocytochemical staining of surface receptors. 16,17 However, only limited data are available on surface KIR and NKG2 expression in sinonasal lymphoma. In the only report based on immunohistochemical staining, each of four cases stained positively for NKG2A, but only one of four cases stained positively for KIR. 15 Taking the small sample sizes into consideration, the data of Haedicke and colleagues 15 are approximately consistent with the present report that 9 of 10 cases were positive for NKG2A. We, therefore, agree with the view that NKG2A is a general marker in sinonasal lymphoma. In contrast, we found a higher frequency of KIR positive cases (nine of ten). Although multiple antibodies were used in the previous study, the failure to detect KIR raised the possibility of false negativity. Because RT-PCR is more sensitive than immunostaining, the increased sensitivity might account for the discrepancy.
Sinonasal lymphoma is usually TCR-γ-GR-negative, but our data show that some cases (3 of 10) had a monoclonal TCR-γ-GR. Although a false-positive result could not be completely excluded, the monoclonal rearrangement might be true, considering that we used the GeneScan system, which offered a higher resolution than did the agarose gels used in previous studies, including ours. 10 The finding of monoclonal TCR-γ-GR in sinonasal lymphoma is in consistent with a recent publication that demonstrated both NK-cell and gamma-delta T-cell lines established from primary lesions of nasal T/NK-cell lymphomas associated with the Epstein-Barr virus. 31 On the other hand, we unexpectedly found two KIR-positive T-cell lymphomas. Whether these are false-positive or represent dual differentiation requires further investigation. Taken together, our data showed that most sinonasal lymphomas belong to the true NK cell lineage, 7 of 10 T-cell lymphomas belong to the T-lineage if the status of TCR-β rearrangement was also considered, and a minority of sinonasal or T-cell lymphomas might have dual differentiations.
More importantly, the interpretation of TCR-γ-GR itself should be modified now as a corollary to our analysis of the KIR repertoire in sinonasal lymphoma. A polyclonal TCR-GR is no longer equivalent to a reactive T-cell proliferation, because there is still the possibility of a monoclonal process due to NK-cell proliferation. Furthermore, a monoclonal TCR-GR is no longer equivalent to a T-cell lymphoma, because the possibility of a lymphoma with dual T/NK differentiation cannot be excluded. We suggest that an isolated analysis for either the TCR or KIR repertoire is no longer sufficient, but that they could be done together for more accurate lineage assignment than either one alone.
In summary, we present a novel approach to the lineage assignment of sinonasal lymphoma, based on the consideration of a restricted KIR repertoire as a criterion of monoclonal or oligoclonal NK-cell proliferation. Our data showed that most sinonasal lymphomas have a restricted KIR repertoire. This is consistent with and provides preliminary support for the hypothesis. We conclude that a restricted killer cell immunoglobulin-like receptor repertoire without TCR-γ-GR can be used to support a true NK-cell lineage in a subset of sinonasal lymphomas.
Acknowledgments
We thank Drs. M. Albitar, M. S. Lee, J. Medeiros, and C. Ramos-Buesora for helpful discussions.
Footnotes
Address reprint requests to Su-Ming Hsu, M.D., Department of Pathology, National Taiwan University College of Medicine, Jen-Ai Road, Taipei, Taiwan 10016.
Supported, in part, by grants 89-N027, 89-B-FA01-1-4, NSC892320-B-002-276, NHRI-EX90-8704SL, and NHRI-GT-EX89S704L from National Taiwan University Hospital, National Science Council, and National Health Research Institute, Taiwan.
References
- 1.Ho FCS, Todd D, Loke SL, Ng RP, Khoo RKK: Clinico-pathological features of malignant lymphomas in 294 Hong Kong Chinese patients; retrospective study covering an eight-year period. Int J Cancer 1984, 34:143-148 [DOI] [PubMed] [Google Scholar]
- 2.Arber DA, Weiss LM, Albujar RF, Chen YY, Jaffe ES: Nasal lymphoma in Peru, high incidence of T-cell immunophenotype and Epstein-Barr virus infection. Am J Surg Pathol 1993, 17:392-399 [PubMed] [Google Scholar]
- 3.Jaffe ES: Classification of natural killer (NK) cell and NK-like T-cell malignancies. Blood 1996, 87:1207-1210 [PubMed] [Google Scholar]
- 4.Jaffe ES, Chan JKC, Su IJ, Frizzera G, Mori S, Feller AC, Ho FCS: Report on the workshop on nasal and related extranodal angiocentric T/natural killer cell lymphomas. Am J Surg Pathol 1996, 20:103-111 [DOI] [PubMed] [Google Scholar]
- 5.Denning SM, Jones DM, Ware RE, Weinhold KJ, Brenner MB, Haynes BF: Analysis of clones derived from human CD7+CD4−CD8−CD3− thymocytes. Int Immunol 1991, 3:1015-1024 [DOI] [PubMed] [Google Scholar]
- 6.Lanier LL, Chang C, Spits H, Phillips JH: Expression of cytoplasmic CD3ε proteins in activated human adult NK cells and CD3γ,δ,ε complexes in fetal NK cells: implications for the relationship of NK and T lymphocytes. J Immunol 1992, 14:1876-1880 [PubMed] [Google Scholar]
- 7.Phillips JH, Hori T, Nagler A, Bhat H, Spits H, Lanier LL: Ontogeny of human natural killer cells: fetal NK cells mediate cytolytic function and express cytoplasmic CD3ε, δ proteins. J Exp Med 1982, 175:1055-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spits H, Lanier LL, Phillips JH: Development of human T and natural killer cells. Blood 1995, 85:2654-2670 [PubMed] [Google Scholar]
- 9.Suzumiya J, Takeshita M, Kimura N, Kikuchi M, Uchida T, Hisano S, Eura Y, Kozuru M, Nomura Y, Tomita K, Komiyama S, Okumura M: Expression of adult and fetal natural killer cell markers in sinonasal lymphomas. Blood 1994, 83:2255-2260 [PubMed] [Google Scholar]
- 10.Lien HC, Lin CW, Huang PH, Chang ML, Hsu SM: Expression of cyclin-dependent kinase 6 (cdk6) and frequent loss of CD44 in nasal-nasopharyngeal NK/T cell lymphomas: comparison with CD56-negative peripheral T cell lymphomas. Lab Invest 2000, 80:893-900 [DOI] [PubMed] [Google Scholar]
- 11.Long EO, Rajagopalan S: HLA class I recognition by killer cell Ig-like receptors. Semin Immunol 2000, 12:101-108 [DOI] [PubMed] [Google Scholar]
- 12.Moretta A, Botino C, Vitale M, Pende D, Biassoni R, Mingari MC, Moretta L: Receptors for HLA class I molecules in human natural killer cells. Annu Rev Immunol 1996, 14:619-648 [DOI] [PubMed] [Google Scholar]
- 13.Raulet DH: Development and tolerance of natural killer cells. Curr Opin Immunol 2000, 11:129-134 [DOI] [PubMed] [Google Scholar]
- 14.Mingari MC, Vitale C, Cantoni C, Bellomo R, Ponte M, Schiavetti F, Bertone S, Moretta L: Interleukin-15 induced maturation of human natural killer cells from early thymic precursors: selective expression of CD94/NKG2-A as the only HLA class I-specific inhibitory receptor. Eur J Immunol 1997, 27:1374-1380 [DOI] [PubMed] [Google Scholar]
- 15.Haedicke W, Ho FCS, Chott A, Moretta L, Rudiger T, Ott G, Muller-Hermelink HK: Expression of CD94/NKG2A and killer immunoglobulin-like receptors in NK cells and a subset of extranodal cytotoxic T-cell lymphomas. Blood 2000, 95:3628-3630 [PubMed] [Google Scholar]
- 16.Uhrberg M, Valiante NM, Shun BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P: Human diversity in killer cell inhibitory receptor genes. Immunity 1997, 7:753-763 [DOI] [PubMed] [Google Scholar]
- 17.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Philips JH, Lanier LL, Parham P: Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity 1997, 7:739-751 [DOI] [PubMed] [Google Scholar]
- 18.Steffens U, Vyas Y, Dupont B, Selvakkumar A: Nucleotide and amino acid sequence alignment for human killer cell inhibitory receptors (KIR), 1998. Tissue Antigens 1998, 51:386-413 [DOI] [PubMed] [Google Scholar]
- 19.Hiby SE, King A, Sharkey M, Loke YW: Human uterine NK cells have a similar repertoire of killer inhibitory and activatory receptors to those found in blood, as demonstrated by RT-PCR and sequencing. Mol Immunol 1997, 34:419-430 [DOI] [PubMed] [Google Scholar]
- 20.Verma S, King A, Loke YW: Expression of killer cell inhibitory receptors on human uterine natural killer cells. Eur J Immunol 1997, 27:979-983 [DOI] [PubMed] [Google Scholar]
- 21.Nakajima-Iijima S, Hamada H, Reddy P, Kakunaga T: Molecular structure of the human cytoplasmic β-actin gene: interspecies homology of sequence in the introns. Proc Natl Acad Sci USA 1985, 82:6133-6137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Signoretti S, Murphy M, Cangi MG, Puddu P, Kadin ME, Loda M: Detection of clonal T-cell receptor γ gene rearrangement in paraffin-embedded tissue by polymerase chain reaction and nonradioactive single-strand conformational polymorphism analysis. Am J Pathol 1999, 154:67-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez-Gonzalez MA, Sanchez B, Mata F, Delgado F: Tonsillar lymphocyte subsets in recurrent acute tonsillitis and tonsillar hypertrophy. Int J Pediatr Otolaryngol 1998, 43:33-39 [DOI] [PubMed] [Google Scholar]
- 24.Chilosi M, Doglioni C, Yan Z, Lestani M, Menestrina F, Sorio C, Benedetti A, Vinante F, Pizzolo G, Inghirami G: Differential expression of cyclin-dependent kinase 6 in cortical thymocytes and T-cell lymphoblastic lymphoma/leukemia. Am J Pathol 1998, 152:209-217 [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang AKS, Srivastava G, Lau PWF, Ho FCS: Differences in T-cell-receptor gene rearrangement and transcription in nasal lymphomas of natural killer and T-cell types: implications on cellular origin. Hum Pathol 1996, 27:701-707 [DOI] [PubMed] [Google Scholar]
- 26.Chiang AKS, Chan ACL, Srivastava G, Ho FCS: Nasal T/natural killer (NK)-cell lymphomas are derived from Epstein-Barr virus-infected cytotoxic lymphocytes of both NK- and T-cell lineage. Int J Cancer 1997, 73:332-338 [DOI] [PubMed] [Google Scholar]
- 27.Tien HF, Su IJ, Tang JL, Liu MC, Lee FY, Chen YC, Chuang SM: Clonal chromosomal abnormalities as direct evidence for clonality in nasal T/natural killer cell lymphomas. Br J Haematol 1997, 97:721-725 [DOI] [PubMed] [Google Scholar]
- 28.Mingari MC, Schiavetti F, Ponte M, Vitale C, Maggi E, Romagnani S, Demarest J, Pantaleo G, Fauci AS, Moretta L: Human CD8+ T lymphocyte subset that express HLA class I specific inhibitory receptors represent oligoclonally or monoclonally expanded cell populations. Proc Natl Acad Sci USA 1996, 93:2433-2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seki S, Habu Y, Kawamura T, Takeda K, Dobashi H, Ohkawa T, Hiraide H: The liver as a crucial organ in the first line of host defense: the role of Kupffer cells, natural killer (NK) cells and NK 1.1 Ag+ T cells in T helper 1 immune response. Immunol Rev 2000, 174:35-46 [DOI] [PubMed] [Google Scholar]
- 30.Taniguchi M, Makno Y, Chui J, Masuda K, Kawano T, Sato H, Kondo E, Koseki H: Va14+ NK T cells: a novel lymphoid cell lineage with regulatory function. J Allergy Clin Immunol 1996, 98:S263-S269 [DOI] [PubMed] [Google Scholar]
- 31.Nagata H, Konno A, Kimura N, Zhang Y, Kimura M, Demachi A, Sekine T, Yamamoto K, Shimizu N: Characterization of novel natural killer (NK)-cell and gammadelta T-cell lines established from primary lesions of nasal T/NK-cell lymphomas associated with the Epstein-Barr virus. Blood 2001, 97:708-713 [DOI] [PubMed] [Google Scholar]