Abstract
At present, a major challenge in the initial diagnosis of leukemia of large granular lymphocytes (LGLs) is to establish the clonal nature of the expanded population. In the present study we have analyzed by flow cytometry immunophenotyping the TCR-Vβ repertoire of 98 consecutive cases of persistent expansions of CD4+ or CD8+bright CD3+/TCR-αβ+ LGLs and compared the results with those obtained in molecular studies of TCR-β gene rearrangements. Fifty-eight cases were considered to be monoclonal in molecular studies whereas in the remaining 40 cases there was no evidence for monoclonality (11 cases were considered oligoclonal and 29 polyclonal). The TCR-Vβ repertoire was biased to the preferential use of one or more TCR-Vβ families in 96% of cases, a total of 124 TCR-Vβ expansions being diagnosed: one TCR-Vβ expansion in 71 cases and two or more TCR-Vβ expansions in 23 cases. The highest TCR-Vβ expansion observed in each case was higher among monoclonal (74 ± 19%) as compared to nonmonoclonal cases (24 ± 14%) (P = 0.001), as did the fraction of LGLs that exhibited a TCR-Vβ-restricted pattern (86 ± 16% and 42 ± 23%, respectively; P = 0.0001); by contrast, the proportion of cases displaying more than one TCR-Vβ expansion was higher in the latter group: 7% versus 48%, respectively (P = 0.001). Results obtained in oligoclonal cases were intermediate between those obtained in polyclonal and monoclonal cases and similar results were observed for CD4+ as for CD8+bright T-cell expansions. TCR-Vβ familiesexpressed in CD8+bright T-cell-LGL proliferations showed a pattern of distribution that mimics the frequency at which the individual TCR-Vβ families are represented in normal peripheral blood T cells. Assuming that a given proliferation of LGLs is monoclonal whenever there is an expansion of a given TCR-Vβ family of at least 40% of the total CD4+ or CD8+bright T-cell compartment, we were able to predict clonality with a sensitivity of 93% and a specificity of 80%. By increasing the cut-off value to 60%, sensitivity and specificity were of 81% and 100%. In summary, our results suggest that flow cytometry immunophenotypic analysis of the TCR-Vβ repertoire is a powerful screening tool for the assessment of T-cell clonality in persistent expansions of TCR-αβ+ LGLs.
Leukemias of large granular lymphocytes (LGLs) represent a well-recognized group of chronic T- and natural killer (NK)-cell neoplasias. 1 Like their normal counterparts, leukemic LGLs are usually classified into two major groups based on the expression of both CD3 and the T-cell receptor (TCR) molecules: CD3+/TCR+ T- and CD3−/TCR− NK-LGL leukemias. 2 Among the former group, monoclonal expansions of LGLs displaying the TCR-αβ represent the majority of cases, whereas TCR-γδ+ T-LGL leukemias are relatively infrequent. From the clinical point of view whereas most clonal T-LGL leukemias show a benign clinical outcome, in some cases they behave as an aggressive condition. Either transient or persistent expansions of polyclonal, oligoclonal, or even monoclonal LGLs are relatively common in various disease conditions and such expansions of LGLs have also been reported in otherwise normal healthy individuals, 3,4 although their neoplastic nature remains unclear. 5,6 In any case, at present, a major challenge in the initial diagnosis of LGL leukemias is to establish the clonal nature of the expanded population of LGLs, to distinguish between polyclonal, oligoclonal, and monoclonal LGL proliferations.
Molecular investigation into the existence of monoclonal rearrangements of the TCR-β and TCR-γ genes, using Southern blot analysis is the preferred method for investigation of T-cell clonality. 7 Despite the high reliability of the Southern blot, this method has some drawbacks that limit its routine use in diagnostic laboratories: it is a labor-intensive and time-consuming method and large quantities of high-quality DNA are needed to obtain reliable results. In recent years alternative approaches have been developed for the assessment of T-cell clonality. Of them, the immunophenotypic analysis of the repertoire of the variable (V) regions of the TCR-α, -β, -γ, and -δ chains represent one of the most attractive options. In humans, the Vα and Vβ gene segments are estimated to contain ∼46 and 52 different functional members that, based on nucleotide homology, can be grouped into 32 and 25 different families, respectively. 8,9 Polymerase chain reaction using TCR-Vβ- and TCR-Vα-specific primers and, more recently, flow cytometry using TCR-Vβ- or TCR-Vα-specific monoclonal antibodies (mAbs) can be used to investigate the TCR-Vβ and TCR-Vα gene usage. Flow cytometry is not only routinely available in many laboratories but also offers several advantages: 1) it allows both a quantitative and qualitative characterization of the T-cell repertoire; 2) the T-cell repertoire can be specifically evaluated within the population of interest by combining TCR-Vβ and TCR-Vα specific with other mAbs; and 3) a large panel of mAbs against Vβ-region determinants is now commercially available, making it possible to access a large fraction of the T-cell repertoire. Although the analysis of the TCR-V repertoire is now being used to indirectly assess T-cell clonality, the finding of an expansion of T cells restricted to a particular V-region family, does not necessarily mean the presence of underlying monoclonal TCR gene rearrangements. Thus, studies in which immunophenotyping is compared with molecular methods are necessary to establish the value of the TCR-V repertoire analysis in investigating T-cell clonality.
Several studies have already been performed aimed at the characterization of both TCR-Vβ and TCR-Vα T-cell repertoires from normal healthy individuals 10-12 and the T-cell responses occurring in various pathological conditions, including autoimmune 13-15 and infectious diseases, 16 tumors, 17-19 allogeneic transplants, 20 and other disease states. 21 However, only a few reports on the TCR-Vβ or TCR-Vα repertoire in LGL leukemias are available and, furthermore, in the majority of these studies the numbers of TCR-Vβ families analyzed were limited. 22-27 Some of these studies showed that several specific TCR-Vβ and TCR-Jβ genes are randomly used, 23,24,27 whereas others suggested a preferential usage of a few TCR-Vβ regions in LGL leukemias. 22,24 To the best of our knowledge, until now no study has been reported in which the utility of using a relatively broad panel of anti-Vβ mAbs for the assessment of T-cell clonality in a large series of consecutive individuals showing a persistent expansion of peripheral blood (PB) TCR-αβ+ LGLs has been evaluated in comparison to molecular techniques.
The aim of our study was to characterize the TCR-Vβ repertoire from a group of 98 consecutive patients displaying a persistent expansion of either CD4+ or CD8+ TCR-αβ+ T-LGLs in the PB and to establish its utility in the diagnosis of clonality. For that purpose, a large panel of 23 mAbs directed against 24 members of 19 different TCR-Vβ families was used and these results were compared with those obtained with conventional molecular techniques.
Materials and Methods
Patients and Controls
Ninety-eight consecutive patients with persistent TCR-αβ+ T-cell proliferations of LGLs were studied (51 males and 47 females; ages 12 to 90 years; median, 61 years). The diagnosis was established on the basis of a LGL morphology, together with a typical CD3+/TCR-αβ+ LGL phenotype, as defined by high FSC/SSC values, absence of expression of CD28, and reactivity for NK-associated antigens. 2,3 From the phenotypic point of view, LGL proliferations were classified into two groups: TCR-αβ+/CD8+bright/CD4− (n = 72 cases) and TCR-αβ+/CD4+/CD8−/+dim (n = 26 cases). The mean absolute number of LGLs in the PB was 2856 ± 3086 × 106/L (median, 1725 × 106/L) and the mean percentage of LGLs within the CD4+ or CD8+bright T-cell population was of 78 ± 19% (median, 83%). Increased numbers of LGLs were associated with absolute lymphocytosis (>3.5 × 109/L) in 60 cases (61%). Neutropenia (<1.5 × 109/L) was observed in 34 cases (35%), anemia (Hb < 10 g/dl) in 13 cases (13%), and thrombocytopenia (<100 × 109/L) in 11 cases (11%). Organomegalies were rarely detected (<10% of cases). Median follow-up for the patients analyzed was of 29 months. During this period, 37 patients (38%) showed associated conditions, including autoimmune disorders (14 cases), neoplastic diseases (15 cases), or other disease states (8 cases). Ten age- and sex-matched healthy individuals (six males and four females; median age, 47 years) were used as controls.
Immunophenotypic Studies
Ethylenediaminetetraacetic acid-anti-coagulated PB samples were stained using a direct immunofluorescence technique. Briefly, 100 μl of whole PB containing between 0.5 to 2.0 × 10 6 nucleated cells were incubated with saturating concentrations of mAbs for 15 minutes at room temperature in the dark. Then 2 ml of fluorescence-activated cell sorting lysing solution (Becton Dickinson, San Jose, CA) were added to lyse nonnucleated red cells. After another 10 minutes incubation at room temperature in the dark, cells were washed once and resuspended in 0.5 ml of phosphate-buffered saline (PBS).
The repertoire of the Vβ chain of TCR-αβ+/CD8+bright/CD4− and TCR-αβ+/CD4+/CD8−/+dim lymphocytes was analyzed by combining either anti-CD4 or anti-CD8 with the following panel of 23 mAbs specific against 24 members of a total of 19 families of variable regions of the TCR-β chain: BV1S1 (Vβ1.1), BV5S1 (Vβ5.1), BV5S2 (Vβ5.2), BV5S3 (Vβ5.3), BV6S1 (Vβ6.1), BV7S1 (Vβ7.1), BV8S1 + BV8S2 (Vβ8.1 + 8.2), BV9S1 (Vβ9.1), BV11S1 (Vβ11.1), BV12S2 (Vβ12.2), BV13S1 (Vβ13.1), BV13S6 (Vβ13.6), BV14S1 (Vβ14.1), BV16S1 (Vβ16.1), BV17S1 (Vβ17.1), BV18S1 (Vβ18.1), BV20S1 (Vβ20.1), BV21S3 (Vβ21.3), BV22S1 (Vβ22.1), BV23S1 (Vβ23.1) (Beckman-Coulter Immunotech, Marseille, France), BV2S1 (Vβ2.1) (Beckman-Coulter Immunotech or Biodesign International, Kennebunk, ME), BV3S1 (Vβ3.1) (Beckman-Coulter Immunotech or Endogen, Woburn, MA) and BV6S7 (Vβ6.7) (Endogen). All anti-TCR-Vβ reagents were tested in all samples except for the anti-Vβ1.1, -Vβ6.7, -Vβ7.1, and -Vβ9.1 that were tested in only 62%, 42%, 57% and 60% of the cases, respectively. In all cases, isotype-matched fluorochrome-conjugated nonspecific immunoglobulins were used as negative control.
Data acquisition was performed in two FACSCalibur flow cytometers (BD) using the Cell Quest software program (BD). Information on a minimum of 2 × 10 5 events was acquired for each reagent combination. Data analysis was performed using the Paint-a-Gate PRO software program (BD). For each TCR-Vβ family, the proportion of positive cells within the CD8+bright or CD4+ lymphocytes was calculated as the percentage of cells stained above the negative isotype control value.
For those TCR-Vβ families that were assessed with the panel of mAbs used in this study (direct identification), we considered that there is a TCR-Vβ expansion whenever its representation exceeded by at least two standard deviations the mean value observed in CD4+ or CD8+ circulating T cells in normal healthy individuals. 16 For the remaining TCR-Vβ families that were not explored with the panel of mAbs used in this study, criteria used to define a TCR-Vβ expansion (indirect identification) was based on the observation of a relative decrease in the fraction of either CD4+ or CD8+bright circulating T cells that were recognized with the panel of mAbs to values <85% of those observed for the same T-cell subsets in PB samples from normal adult individuals. These criteria rely on previous studies demonstrating that a value of 15% should be considered a T-cell expansion. 28,29
Molecular Biology Studies
Rearrangements of the TCR-β chain genes were evaluated by conventional Southern blotting 30 in all cases. Briefly, mononuclear cells were obtained after fractionation on a Lymphoprep (Nycomed Pharma AS, Oslo, Norway) density gradient, washed twice in PBS, and cryopreserved. DNA was extracted using the phenol/chloroform method and digested with EcoRI and HindIII restriction enzymes. DNA fragments were separated by 0.8% agarose gel electrophoresis and transferred to nitrocellulose membranes by vacuum blotting, UV fixed, and hybridized with 32P-labeled probes for the TCR-β gene region (Cβ, TCRBC, and TCRBJ2; DAKO A/S, Glostrup, Denmark). In those cases (n = 5) in which the CD4+ or CD8+bright LGL population represented <10% of the total nucleated cells present in the sample and because all TCR-αβ+ T-cell malignancies have rearranged TCR-γ genes, 31 the clonality studies were performed by polymerase chain reaction analysis of TCR-γ gene rearrangements, using the strategies and primers previously described. 32
Statistical Methods
For all variables under study, median, mean, SD, and range values were calculated. Comparison between groups was calculated using the Mann-Whitney U and chi-square tests for continuous and dichotomic variables, respectively (SPSS 9.0; SPSS, Chicago, IL). P values <0.05 were considered to be associated with statistically significant differences.
Results
Immunophenotypic Analysis of the TCR-Vβ Repertoire of Both PB CD4+ and CD8+bright T Cells and Molecular Studies in Normal Healthy Individuals
The panel of 23 mAbs used in the present work allowed us to identify 60.3 ± 4.3% (range, 53.3 to 65.5%) and 46.1 ± 5.6% (range, 37.4 to 53.5%) of all CD4+ and CD8+bright T cells in the PB of 10 normal healthy controls. The specific distribution of each of the different TCR-Vβ families on both CD4+ and CD8+bright T cells from normal individuals is displayed in Table 1 ▶ . As shown in this table, certain TCR-Vβ families are represented more than others, either within the CD4+ or the CD8+bright normal T lymphocytes. Overall, the relative distribution of the different TCR-Vβ families within the CD4+ and CD8+ subsets was comparable; however, some TCR-Vβ families were preferentially expressed either within the CD4+ or the CD8+bright subsets, with differences only statistically significant for a preferential expression of TCR-Vβ2.1, -Vβ5.1, -Vβ5.3, -Vβ6.7, -Vβ12.2, -Vβ13.1, -Vβ18.1, -Vβ20.1, and -Vβ22.1 on CD4+ T cells and TCR-Vβ7.1 on CD8+bright T cells. Molecular studies showed a polyclonal pattern for the TCR-β gene rearrangements in all control individuals.
Table 1.
Usage of the TCR V-β Families Tested in Peripheral Blood CD8+bright and CD4+ T Cells from Normal Healthy Adult Individuals (n = 10)
| TCR-Vβ families | % CD8+bright T cells | % CD4+ T cells | P value | ||||
|---|---|---|---|---|---|---|---|
| Vβ1.1 (BV1S1) | 3.5 | 3.5 ± 1.2 | (1.4–5.3) | 3.0 | 3.0 ± 0.5 | (2.1–3.8) | N.S. |
| Vβ2.1 (BV2S1) | 4.4 | 4.7 ± 1.5 | (3.0–6.7) | 9.6 | 9.6 ± 0.9 | (8.1–11.2) | .0001 |
| Vβ3.1 (BV3S1) | 4.3 | 4.4 ± 3.5 | (0.7–11.4) | 4.3 | 4.8 ± 2.3 | (1.3–7.3) | N.S. |
| Vβ5.1 (BV5S1) | 2.7 | 3.0 ± 1.2 | (1.7–5.8) | 6.5 | 6.6 ± 1.0 | (5.0–8.3) | .0001 |
| Vβ5.2 (BV5S2) | 0.3 | 0.3 ± 0.2 | (0.1–0.7) | 0.5 | 0.4 ± 0.1 | (0.3–0.6) | N.S. |
| Vβ5.3 (BV5S3) | 0.8 | 0.9 ± 0.4 | (0.5–1.8) | 1.2 | 1.2 ± 0.3 | (0.8–1.8) | .030 |
| Vβ6.1 (BV6S1) | 0.2 | 0.2 ± 0.2 | (0.0–0.8) | 0.3 | 0.4 ± 0.2 | (0.1–0.9) | N.S. |
| Vβ6.7 (BV6S7) | 1.1 | 1.4 ± 0.9 | (0.7–3.7) | 3.4 | 4.0 ± 1.8 | (1.5–7.0) | .003 |
| Vβ7.1 (BV7S1) | 3.1 | 3.3 ± 1.8 | (0.9–7.5) | 2.0 | 1.9 ± 0.3 | (1.4–2.4) | .030 |
| Vβ8.1+8.2 (BV8S1+S2) | 2.9 | 3.4 ± 1.7 | (1.9–7.6) | 3.8 | 3.8 ± 0.6 | (2.8–4.7) | N.S. |
| Vβ9.1 (BV9S1) | 2.2 | 2.5 ± 1.3 | (1.1–5.4) | 1.9 | 2.0 ± 0.8 | (0.7–3.0) | N.S. |
| Vβ11.1 (BV11S1) | 0.3 | 0.8 ± 1.6 | (0.1–5.3) | 0.6 | 0.5 ± 0.2 | (0.2–0.8) | N.S. |
| Vβ12.2 (BV12S2) | 1.1 | 1.0 ± 0.4 | (0.2–1.4) | 1.6 | 1.5 ± 0.3 | (0.9–2.0) | .008 |
| Vβ13.1 (BV13S1) | 1.3 | 1.2 ± 0.6 | (0.2–1.99) | 3.0 | 2.8 ± 1.2 | (0.6–4.7) | .003 |
| Vβ13.6 (BV13S6) | 1.4 | 1.4 ± 0.6 | (0.6–2.3) | 1.8 | 1.6 ± 0.2 | (1.1–1.8) | N.S. |
| Vβ14.1 (BV14S1) | 1.5 | 1.6 ± 1.1 | (0.4–3.7) | 0.9 | 0.9 ± 0.5 | (0.3–2.0) | N.S. |
| Vβ16.1 (BV16S1) | 0.6 | 1.3 ± 1.4 | (0.4–4.9) | 0.9 | 0.9 ± 0.2 | (0.7–1.2) | N.S. |
| Vβ17.1 (BV17S1) | 3.8 | 4.7 ± 2.4 | (2.1–9.3) | 5.6 | 5.5 ± 0.9 | (4.4–7.1) | N.S. |
| Vβ18.1 (BV18S1) | 0.1 | 0.1 ± 0.1 | (0.0–0.3) | 0.4 | 0.5 ± 0.6 | (0.1–2.3) | .040 |
| Vβ20.1 (BV20S1) | 1.6 | 1.6 ± 0.9 | (0.5–2.8) | 2.9 | 2.6 ± 1.1 | (1.0–4.3) | .048 |
| Vβ21.3 (BV21S3) | 2.7 | 2.7 ± 0.6 | (1.7–3.5) | 2.3 | 2.4 ± 0.5 | (1.8–3.3) | N.S. |
| Vβ22.1 (BV22S1) | 2.5 | 2.4 ± 0.6 | (1.1–3.6) | 2.9 | 3.1 ± 0.9 | (2.3–4.9) | .046 |
| Vβ23.1 (BV23S1) | 0.6 | 0.7 ± 0.3 | (0.4–1.1) | 0.6 | 0.6 ± 0.2 | (0.2–0.8) | N.S. |
| Total | 47.8 | 46.1 ± 5.6 | (37.4–53.5) | 61.3 | 60.3 ± 4.3 | (53.3–65.5) | – |
Results are presented as median values (bold), mean ± standard deviation, and range (minimum-maximum).
Assessment of T-Cell Clonality by Immunophenotypic Analysis of the TCR-Vβ Repertoire of PB CD4+ and CD8+bright T Cells and Molecular Studies in Patients Showing Persistent Expansions of LGLs
From the molecular point of view, 58 of the 98 cases studied (59%) were found to be monoclonal by either Southern blot and/or polymerase chain reaction whereas in the remaining 40 cases there was no evidence of monoclonality. Among these latter 40 cases, 11 were classified as oligoclonal whereas 29 were classified polyclonal.
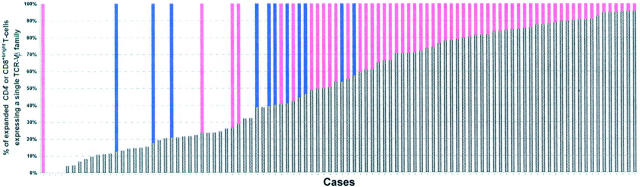
The immunophenotypic analysis of the TCR-Vβ repertoire of both CD4+ and CD8+bright PB T cells from individuals showing LGL expansions revealed the existence of an expansion of at least one Vβ family in all but four cases (n = 94; 96%), with a total of 124 expansions detected. Ninety-one of these TCR-Vβ expansions (73%) were directly identified with the panel of mAbs used whereas in the remaining 33 cases the identification was indirect. Seventy-one patients (72%) showed an expansion of a single TCR-Vβ family whereas 23 cases (23%) displayed expansions of two or more TCR-Vβ families: two TCR-Vβ families were simultaneously expanded in 17 patients, three in 5 patients, and four in 1 case. The highest TCR-Vβ family expansion found in each case was highly variable, ranging from 4 to 96% of the total PB CD4+ and CD8+bright T cells (mean, 55 ± 30%; median, 57%) (Figure 1) ▶ . The other TCR-Vβ expanded families represented 4 to 28% (mean, 9 ± 5%) of the total PB CD4+ and CD8+bright T cells. Interestingly, no significant expansions of TCR-αβ+ T cells not included in the suspected LGL population were detected in cases analyzed here.
Figure 1.
Percentage of TCR-αβ+/CD4+ or TCR-αβ+/CD8+bright T cells from each individual patient displaying a single expanded TCR-Vβ family, as represented by shaded bars superimposed by white bars in polyclonal, blue in oligoclonal, and red in monoclonal expansions of LGLs, as determined by molecular techniques.
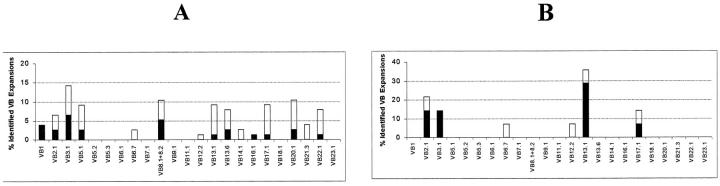
Figure 2 ▶ shows the relative incidence of expansions for each of the TCR-Vβ families within the CD4+ (Figure 2A) ▶ and the CD8+bright (Figure 2B) ▶ T cells. As may be seen, overall, the expanded TCR-Vβ families within the CD8+bright LGLs showed a similar distribution to that observed in normal PB because the must frequently expanded TCR-Vβ families were those usually highly represented in normal circulating CD8+bright T cells (Table 1 ▶ and Figure 2A ▶ ). In contrast, among cases showing an expansion of CD4+ LGLs only a few TCR-Vβ families were found to be expanded: TCR-Vβ2.1, -Vβ3.1, -Vβ6.7, -Vβ12.2, -Vβ13.1, and -Vβ17.1. When considering only monoclonal expansions of CD4+ LGLs, two cases (22%) corresponded to TCR-Vβ2.1, two cases (22%) to TCR-Vβ3.1, and four cases (44%) to TCR-Vβ13.1, a frequency that exceeds the frequency that was expected on the basis of the representation of each of these TCR-Vβ families among normal PB CD4+ T cells (Table 1 ▶ and Figure 2B ▶ ).
Figure 2.
Incidence of expansions of each TCR-Vβ family in proliferations of TCR-αβ+/CD8+ (A) and TCR-αβ+/CD4+ (B) LGLs in the whole series and in cases classified as monoclonal by molecular techniques (black bars).
Correlation between Immunophenotypic and Molecular Clonality Studies
Nonmonoclonal (polyclonal plus oligoclonal) and monoclonal T-LGL expansions differed in a number of aspects including the number of expanded TCR-Vβ families and the magnitude of the highest TCR-Vβ expansion. Table 2 ▶ summarizes the immunophenotypic features of monoclonal versus nonmonoclonal expansions by considering separately the CD4+ and CD8+bright LGL proliferations.
Table 2.
Relative and Absolute Counts of PB Lymphocytes, LGL, and Expanded Vβ-Families in Patients with Persistent Expansions of CD4+ and CD8+bright LGL According to the Presence or Not of a Monoclonal Expansion of PB T Cells by Molecular Techniques
| Expansions of CD8+bright LGL | Expansions of CD4+ LGL | |||||
|---|---|---|---|---|---|---|
| Polyclonal/oligoclonal n = 34 | Monoclonal n = 38 | P value | Polyclonal/oligoclonal n = 6 | Monoclonal n = 20 | P value | |
| Number of PB lymphocytes× 109/L | 3.1± 2.3 (2.4) (1.0–13.4) | 5.9± 3.6 (5.0) (1.1–18.3) | .0001 | 3.2 ± 2.6 (2.7) (0.9–8.3) | 7.1± 4.2 (6.5) (1.4–17.3) | .028 |
| Number of total PB CD8+bright or CD4+ T cells× 106/L | 1398± 1446 (842) (363–8080) | 4178± 3520 (3312) (572–17755) | .0001 | 1791± 1420 (1551) (492–4467) | 5063± 3161 (4633) (491–11479) | .025 |
| Number of total PB CD8+bright or CD4+ LGL× 106/L | 1070± 1288 (607) (225–7111) | 3892± 3517 (3146) (481–17577) | .0001 | 678± 446 (618) (280–1519) | 4455± 3049 (4309) (123–11020) | .003 |
| Percentage of total PB CD8+bright or CD4+ LGL | 68± 15 (67) (31–97) | 90 ± 8 (92) (73–99) | .0001 | 46± 22 (38) (20–76) | 81± 18 (88) (25–96) | .001 |
| Number of cases with expansions of one or more TCR-Vβ families (%) | 31/34 (91%) | 37/38 (97%) | N.S. | 6/6 (100%) | 20/20 (100%) | N.S. |
| Number of cases showing expansions of more than one TCR-Vβ families (%) | 18/34 (53%) | 3/38 (8%) | .001 | 1/6 (17%) | 1/20 (5%) | N.S. |
| Number of cases with at least one Vβ expansion > 40% | 5/34 (15%) | 36/38 (95%) | .0001 | 3/6 (50%) | 18/20 (90%) | .006 |
| Highest Vβ expansion | 22± 12 (21) (4–47) | 74± 17 (79) (27–96) | .0001 | 34± 19 (32) (13–58) | 75± 22 (83) (24–96) | .001 |
| Ratio between the highest Vβ expansion and the % of LGL within CD8+bright or CD4+ T-cells | 0.35± 0.18 (0.33) (0.07–0.66) | 0.82± 0.16 (0.87) (0.31–1.00) | .0001 | 0.75± 0.21 (0.78) (0.38–0.99) | 0.91± 0.14 (0.97) (0.41–1.00) | .028 |
Results are presented as mean ± standard deviation, median (in brackets), and range (italic, in brackets). N.S.: no significant differences.
All but one monoclonal proliferation of LGLs showed expansion of at least one TCR-Vβ family (98%). Furthermore, in most of these cases expansion of a single TCR-Vβ family was detected by immunophenotyping (53 of 57 cases; 93%), with other smaller Vβ expansions being simultaneously detected in only four cases (7%). As a mean the highest TCR-Vβ expansion accounted for 74 ± 19% (range, 24 to 96%) of the expanded T-cell compartment: 74 ± 17% of CD8+bright and 75 ± 22% of CD4+ T cells, for CD8+bright and CD4+ LGLs, respectively, representing >40% of the CD4+ or CD8+bright T-cell populations in all but four patients (93%).
Similarly, all but three LGL proliferations classified as not being monoclonal by molecular techniques displayed expansion of at least one TCR-Vβ family (93%). However, the frequency of cases with expansion of more than one TCR-Vβ family among nonmonoclonal proliferations of LGLs (19 of 40 patients; 48%)—18 of 34 patients (53%) and 1 of 6 patients (17%) for CD8+bright and CD4+ T-LGL, respectively—was much higher than that observed in monoclonal cases (P = 0.001). In addition, the magnitude of the highest TCR-Vβ expansion observed was also much lower (P = 0.001) among these cases as compared to the monoclonal cases, accounting for only 24 ± 14% of the expanded T-cell population: 22 ± 12% of CD8+bright and 34 ± 19% of the CD4+ T cells. Interestingly, the proportion of the TCR-Vβ expanded T cells was higher among the oligoclonal as compared to the polyclonal cases (38 ± 15% versus 18 ± 9%). Also in contrast (P = 0.0001) to what was observed in monoclonal proliferations, the percentage of CD4+ or CD8+bright T cells expressing a single TCR-Vβ family exceeded 40% of the total CD4+ or CD8+bright T cells in only eight cases (20%), all of them being classified as oligoclonal by molecular techniques. Interestingly, the TCR-Vβ expansion did not exceed 60% in any of them. On comparing the proportion of T cells expressing the expanded TCR-Vβ family from either the total CD4+ or CD8+bright T cells that fulfilled the phenotypic criteria for LGLs in monoclonal and in oligoclonal/polyclonal T-LGL expansions (Figure 2) ▶ , significantly higher numbers (P = 0.0001) were observed in monoclonal cases—86 ± 16% versus 42 ± 23%—the lowest values being observed for polyclonal T-LGL expansions (32 ± 17%) whereas oligoclonal LGL expansions showed intermediate values (64 ± 21%).
Taking 40% of the total CD4+ or CD8+bright T cells as the cut-off value for diagnosis of a monoclonal T-cell expansion, immunophenotyping showed a sensitivity of 93% and a negative predictive value of 89%, with a specificity and positive predictive value of 80% and 87%, respectively. By increasing the cut-off value to 60% both the specificity and positive predictive value increased to 100%, although a large proportion of cases displaying a monoclonal expansion of T-LGL proliferations were improperly classified as nonmonoclonal by flow cytometry: sensitivity of 81% and negative predictive value of 78% (Table 3) ▶ .
Table 3.
Sensitivity and Specificity of Immunophenotypic Analysis of the TCR-Vβ Repertoire for the Diagnosis of (Mono)clonal Expansions of TCRαβ+ LGL
| % of the TCR-Vβ expansion | ||
|---|---|---|
| >40% | >60% | |
| Positive predictive value | 87% | 100% |
| Negative predictive value | 89% | 78% |
| Sensitivity | 93% | 81% |
| Specificity | 80% | 100% |
Discussion
The results presented here show that persistent expansions of TCR-αβ+ T-LGLs form a continuum spectrum of polyclonal, oligoclonal, and monoclonal proliferations of TCR-Vβ-restricted T cells that differ both in type and number of expanded TCR-Vβ families, as well as in the magnitude of the TCR-Vβ expansion.
Overall, once cases displaying a monoclonal expansion of TCR-αβ+ T-LGLs based on molecular techniques were compared to the other patients, major differences in the immunophenotypic result were observed: 1) polyclonal and oligoclonal expansions of TCR-αβ+ LGLs frequently showed expansions of more than one TCR-Vβ family; 2) the proportion of the most represented TCR-Vβ family was usually much lower in these cases; and 3) even in the presence of a TCR-Vβ dominance, nonmonoclonal LGL proliferations usually displayed a large fraction of residual LGLs with a highly diversified T-cell repertoire.
Such observations would support the notion that monoclonal LGL proliferations may arise as a consequence of an antigen-driven immune response that would start as a polyclonal reactive T-cell response and could thereafter subsequently evolve into oligoclonal and monoclonal processes. This hypothesis would also be supported by the fact that TCR-Vβ families expressed in CD8+bright LGL proliferations showed a pattern of distribution that mimics the frequency at which individual TCR-Vβ families are represented in normal CD8+bright T cells. This would suggest that CD8+bright T-LGLs are clonally transformed in a random manner; the possibility of a bias to the preferential use of some TCR-Vβ families in cases showing a monoclonal expansion of CD4+ LGLs needs further evaluation using a larger number of cases. The notion that monoclonal expansions of T-LGLs may represent an evolutionary end stage of an antigen-mediated proliferation of LGLs is not new. 5,6 In fact, there is a great deal of evidence supporting this view: 1) clonal dominance with preservation of a polyclonal reservoir is a typical feature in the normal TCR-αβ+ T-cell repertoire; 33 2) the immune TCR-Vβ repertoire is dynamic and can be continuously modulated; 34 3) in normal individuals, TCR-Vβ-restricted T-cell expansions accumulate with age; 10-12 4) TCR-Vβ-restricted T-cell expansions are more frequently found in association with pathological conditions characterized by chronic antigen stimulation; 35,36 5) preliminary reports suggest that a higher incidence of clonality seems to be a frequent finding once TCR-Vβ-restricted T-cell repertoires are observed; 10,37,38 this phenomenon seems to be particularly frequent in the TCR-αβ+/CD8+bright T-cell compartment, although expansions of TCR-αβ+/CD4+ cells, TCR-αβ+/CD4−/CD8− 39 and TCR-γδ+ T cells 40 have also been sporadically observed; 6) similarly to LGL-leukemia cells, polyclonal T-LGLs displaying a restricted usage of TCR-Vβ families are usually CD28−, 41 express NK-associated antigens and NK receptors, 42 suggesting that they represent antigen-driven cytotoxic T cells; and 7) molecular studies provided evidence that TCR-Vβ-restricted T-cell expansions may depend on an antigen-mediated selection process. 25,43,44
If this hypothesis is true, the higher the TCR-Vβ expansion is, the greater the probability for a monoclonal T-cell proliferation, as found in the present study. In turn, this indicates that a large expansion of a TCR-Vβ family would be highly predictive of clonal transformation of a chronic reactive process. Nevertheless, these phenomena do not always necessarily correlate. In fact, some cases displaying LGL expansions in which a single TCR-Vβ family account for more than a half of the CD4+ or CD8+bright T-cell compartment proved to be oligoclonal by molecular techniques. Moreover, we have found a case of monoclonal T-cell proliferation in which a normal distribution of the different TCR-Vβ families was observed in accordance to previous reports. 45,46 It could be argued that such discrepancies could be because of problems related to technical uses such as the different sensitivity of the molecular and immunophenotypic techniques in cases in which minor populations of clonal LGLs are present. 7 Alternatively, these observations could also suggest that monoclonal T-cell rearrangements could theoretically occur at any of the stages of the process of continuous T-cell stimulation. Based on the results presented here and in previous reports indicating that a TCR-Vβ expansion representing >40% of the overall population of CD4+ and CD8+bright T cells is hardly ever found in normal individuals, 10-12 we propose that, for routine purposes, expansions of a single TCR-Vβ family representing >60% of the overall population of CD4+ and CD8+bright T cells could be considered as highly suggestive of monoclonal whereas those of <40% would be nonmonoclonal, pending confirmation by molecular techniques. In those cases in which the TCR-Vβ expansion represents between 40% and 60% of the total CD4+ and CD8+bright T cells, molecular studies are essential to establish clonality. Further studies are necessary to clarify the utility of the immunophenotypic assessment of T-cell clonality based on the analysis of the TCR-Vβ repertoire together with a characterization of the phenotypic profile of the expanded TCR-Vβ family aimed at the identification of specific aberrant phenotypes, not only at the time of diagnosis, but also during the follow-up of patients, in particular if therapy is required.
Acknowledgments
We thank Beckman/Coulter for providing part of the anti-TCR-Vβ monoclonal antibodies used in this study.
Footnotes
Address reprint requests to Margarida Lima, Clinical Hematology, Unit of Flow Cytometry, Hospital Geral de Santo António, Rua D Manuel II, s/n, 4050 Porto, Portugal. E-mail: mmc.lima@clix.pt.
Supported in part by the Comissão de Fomento da Investigação em Cuidados de Saúde, Ministério da Saúde, Portugal (PI 51/99); Acções Integradas Luso-Espanholas do Conselho de Reitores das Universidades Portuguesas (E-31/99); and Acción Integrada Hispano-Portuguesa (HP 1998-0091), Dirección General de Enseñanza Superior e Investigación Científica, Ministerio de Educación y Cultura, Spain.
This work was done in the context of the BIOMED2 program (action BMH4-CT97-2611).
References
- 1.Loughran TP: Clonal diseases of large granular lymphocytes. Blood 1993, 82:1-14 [PubMed] [Google Scholar]
- 2.Scott CS, Richard SJ: Classification of large granular lymphocyte (LGL) and NK-associated (NKa) disorders. Blood Rev 1992, 6:220-233 [DOI] [PubMed] [Google Scholar]
- 3.Semenzato G, Pandolfi F, Chisesi T, De Rossi G, Pizzolo G, Zambello R, Trentim L, Agostini C, Dini E, Vespignani M, Cafaro A, Pasqualetti D, Giubellino MC, Migone N, Foa R: The lymphoproliferative disease of granular lymphocytes. A heterogeneous disease ranging from indolent to aggressive conditions. Cancer 1987, 60:2971-2978 [DOI] [PubMed] [Google Scholar]
- 4.Scott CS, Richards SJ, Sivakumaran M, Short M, Child JÁ, Hunt KM, McEvoy M, Steed AJ, Balfour IC, Parapia LA: Transient and persistent expansions of large granular lymphocytes (LGL) and NK associated (NKs) cells: the Yorkshire Leukaemia Group Study. Br J Haematol 1993, 83:505-517 [DOI] [PubMed] [Google Scholar]
- 5.Semenzato G, Pizzoto G, Rannunci A, Agostini C, Chilosi M, Quinti I, De Sanctis G, Vercelli B, Pandolfi F: Abnormal expansion of polyclonal large to small size granular lymphocytes: reactive or neoplastic process? Blood 1984, 63:1271-1277 [PubMed] [Google Scholar]
- 6.Richards SJ, Short M, Scott CS: Clonal CD3+CD8+ large granular lymphocyte (LGL)/NK-associated (NKa) expansions: primary malignancies or secondary reactive phenomena? Leuk Lymphoma 1995, 17:303-311 [DOI] [PubMed] [Google Scholar]
- 7.Ryan DK, Alexander HD, Morris TC: Routine diagnosis of large granular lymphocytic leukaemia by Southern blot and polymerase chain reaction analysis of clonal T cell receptor gene rearrangement. Mol Pathol 1997, 50:77-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei S, Charmley P, Robinson MA, Concannon P: The extent of the human germline T-cell receptor V beta gene segment repertoire. Immunogenetics 1994, 40:27-36 [DOI] [PubMed] [Google Scholar]
- 9.Wei S, Concannon P: Repertoire and organization of human T-cell receptor alpha region variable genes. Genomics 1996, 38:442-445 [DOI] [PubMed] [Google Scholar]
- 10.Posnett D, Sinha R, Kabak S, Russo C: Clonal populations of T cells in normal elderly humans: the T cell equivalent to “benign monoclonal gammapathy.” Exp Med 1994, 179:609-618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wack A, Cossarizza A, Heltai S, Barbieri D, D’Addato S, Fransceschi C, Dellabona P, Casorati G: Age-related modifications of the human alphabeta T cell repertoire due to different clonal expansions in the CD4+ and CD8+ subsets. Int Immunol 1998, 10:1281-1288 [DOI] [PubMed] [Google Scholar]
- 12.van Den Beemd R, Boor PP, van Lochem EG, Hop WC, Langerak AW, Wolvers-Tettero IL, Hooijkaas H, van Dongen JJ: Flow cytometric analysis of the vbeta repertoire in healthy controls. Cytometry 2000, 40:336-345 [DOI] [PubMed] [Google Scholar]
- 13.Conrad B, Weldmann E, Trucco C, Rudert WA, Behboo R, Ricordi C, Rodriquez-Rilo H, Finegold D, Trucco M: Evidence for superantigen involvement in insulin-dependent diabetes mellitus aetiology. Nature 1994, 371:351-355 [DOI] [PubMed] [Google Scholar]
- 14.Holbrook MR, Tighe PJ, Powell RJ: Restrictions of T cell receptor beta chain repertoire in the peripheral blood of patients with systemic lupus erythematosus. Ann Rheum Dis 1996, 55:627-631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borgato L, Beri R, Biasi D, Testoni R, Cugola L, Ceru S, De Sandre G, Lunardi C: Analysis of the T cell receptor repertoire in rheumatoid arthritis. Clin Exp Rheumatol 1997, 15:475-479 [PubMed] [Google Scholar]
- 16.Roglic M, Macphee RD, Duncan SR, Sattler FR, Theofilopoulos AN: T cell receptor (TCR) BV gene repertoires and clonal expansions of CD4 cells in patients with HIV infections. Clin Exp Immunol 1997, 107:21-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puisieux I, Even J, Pannetier C, Jotereau F, Favrot M, Kourilsky P: Oligoclonality of tumor-infiltrating lymphocytes from human melanomas. J Immunol 1994, 153:2807-2818 [PubMed] [Google Scholar]
- 18.Halapi E, Werner A, Wahlstrom J, Osterborg A, Jeddi-Tehrani M, Yi Q, Janson CH, Wigzell: T cell repertoire in patients with multiple myeloma and monoclonal gammopathy of undetermined significance: clonal CD8+ T cell expansions are found preferentially in patients with a low tumor burden. Eur J Immunol 1997, 27:2245-2252 [DOI] [PubMed] [Google Scholar]
- 19.Goolsby CL, Kuchnio M, Finn WG, Peterson L: Expansions of clonal and oligoclonal T cells in B-cell chronic lymphocytic leukemia are primarily restricted to the CD3(+)CD8(+) T-cell population. Cytometry 2000, 42:188-195 [PubMed] [Google Scholar]
- 20.Gorochov C, Debré P, Leblond V, Sadat-Sowti B, Sigaux F, Autran B: Oligoclonal expansion of CD8+CD57+ T cells with restricted T cell receptor chain variability after bone marrow transplantation. Blood 1994, 83:587-595 [PubMed] [Google Scholar]
- 21.Probert CS, Chott A, Turner JR, Stevens AC, Bodinaku K, Elson CO, Balk SP, Blumberg RS: Persistent clonal expansions of peripheral blood CD4+ lymphocytes in chronic inflammatory bowel disease. J Immunol 1996, 157:3183-3191 [PubMed] [Google Scholar]
- 22.de Totero D, Tazzari PL, DiSanto JP, di Celle PF, Raspadori D, Conte R, Gobbi M, Ferrara GB, Flomenberg N, Lauria F, Foá R: Heterogeneous immunophenotype of large granular lymphocyte expansions: differential expression of the CD8α and CD8β chains. Blood 1992, 80:1765-1773 [PubMed] [Google Scholar]
- 23.Kaneko T, Mizoguchi H, Oshimi K: Expression of T-cell receptor Vβ regions in granular lymphocyte-proliferative disorders. Blood 1993, 81:3482-3483 [PubMed] [Google Scholar]
- 24.Zambello R, Trentin L, Facco M, Cerutti A, Sancetta R, Milani A, Raimondi R, Tassinari C, Agostini C, Semenzato G: Analysis of the T cell receptor in the lymphoproliferative disease of granular lymphocytes: superantigen activation of clonal CD3+ granular lymphocytes. Cancer Res 1995, 55:6140-6145 [PubMed] [Google Scholar]
- 25.Kasten-Sportes C, Zahnoen S, Steis RG, Chan WCC, Winton EF, Waldmann TA: T-cell receptor gene rearrangement in T-cell large granular leukocyte leukemia: preferential Vα but diverse Jα usage in one of five patients. Blood 1994, 83:767-775 [PubMed] [Google Scholar]
- 26.Bowman SJ, Bhavnani M, Geddes GC, Corrigall V, Boylston AW, Panayi GS, Lanchbury JS: Large granular lymphocyte expansions in patients with Felty’s syndrome: analysis using anti-T cell receptor V beta-specific monoclonal antibodies. Clin Exp Immunol 1995, 101:18-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davey MP, Starkebaum G, Loughran TP, Jr: CD3+ leukemic granular lymphocytes utilize diverse T-cell receptor V beta genes. Blood 1995, 51:146-150 [PubMed] [Google Scholar]
- 28.Morley J, Batliwaua F, Hingorani R, Cregersen P: Oligoclonal CD8+ T cells are preferentially expanded in the CD57+ subset. J Immunol 1995, 154:6182-6190 [PubMed] [Google Scholar]
- 29.Halapi E, Werner A, Wahlstrom J, Osterborg A, Jeddi-Tehrani M, Yi Q, Janson CH, Wigzell H, Grunewald J, Mellstedt H: T cell repertoire in patients with multiple myeloma and monoclonal gammopathy of undetermined significance: clonal CD8+ T cell expansions are found preferentially in patients with a low tumor burden. Eur J Immunol 1997, 27:2245-2252 [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Sanz R, Vargas Montero M, Gonzalez Diaz M, del Carmen M, Santos C, Balanzategui Echevarria A, Flores Corral T, Hernandez Martin JM, Caballero Barrigon MD, San Miguel JF: Action of single and associated lesions on the Bcl-l, Bcl-2, Bcl-6, c-Myc, p53, and p16 genes in B-cell non Hodgkin’s lymphomas. Value of molecular analysis for a better assignment of the histologic subtype. Haematologica 1998, 83:209-216 [PubMed] [Google Scholar]
- 31.Van Dongen JJ, Wolvers-Tettero IL: Analysis of immunoglobulin and T cell receptor genes. Part II: possibilities and limitations in the diagnosis and management of lymphoproliferative diseases and related disorders. Clin Chim Acta 1991, 198:93-174 [DOI] [PubMed] [Google Scholar]
- 32.Langerak AW, Szczepanski T, Van der Burg M, Wolvers-Tettero ILM, van Dongen JJM: Heteroduplex PCR analysis of rearranged T cell receptor genes for clonality assessment in suspect T cell proliferations. Leukemia 1997, 1:2192-2199 [DOI] [PubMed] [Google Scholar]
- 33.Lantelme E, Granziero L, Angman L, Giachino C: Clonal predominance, but preservation of a polyclonal reservoir, in the normal alpha beta T-cell repertoire. Hum Immunol 1997, 53:49-56 [DOI] [PubMed] [Google Scholar]
- 34.Blish CA, Gallay BJ, Turk GL, Kline KM, Wheat W, Fink PJ: Chronic modulation of the TCR repertoire in the lymphoid periphery. J Immunol 1999, 162:3131-3140 [PubMed] [Google Scholar]
- 35.Pannetier C, Even J, Kourilsky P: T-cell repertoire diversity and clonal expansions in normal and clinical samples. Immunol Today 1995, 16:176-181 [DOI] [PubMed] [Google Scholar]
- 36.Grunewald J, Wigzell H: T cell receptors in health and disease. Introduction. Springer Semin Immunopathol 1999, 21:1-4 [PubMed] [Google Scholar]
- 37.Fitzgerald J, Ricalton N, Meyer A-C, West S, Kaplan H, Behrendt C, Kotzin B: Analysis of clonal CD8+ T cell expansions in normal individuals and patients with rheumatoid arthritis. J Immunol 1995, 154:3538-3547 [PubMed] [Google Scholar]
- 38.Wang E, Moss P, Frodsham P, Lehner P, Bell J, Borvsiewicz L: CD8+highCD57+ T lymphocytes in normal, healthy individuals are oligoclonal and respond to human cytomegalovirus. J Immunol 1995, 155:5046-5056 [PubMed] [Google Scholar]
- 39.Niehues T, Gulwani-Akolkar B, Akolkar PN, Tax W, Silver J: Unique phenotype and distinct TCR V beta repertoire in human peripheral blood alpha beta TCR+, CD4−, and CD8− double negative T cells. J Immunol 1994, 152:1072-1081 [PubMed] [Google Scholar]
- 40.Giachino C, Granziero L, Modena V, Maiocco V, Lomater C, Fantini F, Lanzavecchia A, Migone N: Clonal expansions of V delta 1+ and V delta 2+ cells increase with age and limit the repertoire of human gamma delta T cells. Eur J Immunol 1994, 24:1914-1918 [DOI] [PubMed] [Google Scholar]
- 41.Mugnaini EN, Egeland T, Spurkland A, Brinchmann JE: The T cell receptor repertoire of CD8+CD28− T lymphocytes is dominated by expanded clones that persist over time. Clin Exp Immunol 1999, 117:298-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mingari MC, Schiavetti F, Ponte M, Vitale C, Maggi E, Romagnani S, Demarest J, Pantaleo G, Fauci AS, Moretta L: Human CD8+ T lymphocyte subsets that express HLA class I-specific inhibitory receptors represent oligoclonally or monoclonally expanded cell populations. Proc Natl Acad Sci USA 1996, 93:12433-12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sottini A, Bettinardi A, Quiros-Roldan E, Plebani A, Airo P, Primi D, Imberti L: Evidence for antigenic selection of large granular lymphocytes in a patient with Wiskott-Aldrich syndrome. Blood 1995, 86:2240-2247 [PubMed] [Google Scholar]
- 44.Quiros-Roldan E, Sottini A, Gulletta M, Stellini R, Puoti M, Primi D, Imberti L: The T-cell receptor repertoires expressed by CD4+ and CD4− large granular lymphocytes derived from the same patients suggest the persistent action of an immune-mediated selection process. Blood 1996, 88:2133-2143 [PubMed] [Google Scholar]
- 45.Hingorani R, Choi I-H, Akolkar P, Gulwani-Akolkar B, Pergolizzi R, Silver, Cregersen P: Clonal predominance of T cell receptors within the CD8+CD45R0+ subset in normal human subjects. J Immunol 1993, 151:5762-5769 [PubMed] [Google Scholar]
- 46.Monteiro J, Flingorani R, Choi I-H, Silver J, Pergolizzi R, Cregersen P: Oligoclonality in the human CD8+ T cell repertoire in normal subjects and monozygotic twins: implications for studies of infectious and autoimmune diseases. Mol Med 1995, 1:614-624 [PMC free article] [PubMed] [Google Scholar]