Abstract
It has been proposed that the regulation of telomerase takes place at the transcriptional level, the expression of the catalytic subunit human telomerase reverse transcriptase (hTERT) being crucial for telomerase activity (TA). Recently, differential splicing of hTERT mRNA has been demonstrated in various tissues during embryonal development, and it has been suggested that only full-length transcripts translate into functionally active telomerase. With this in view, we analyzed the different hTERT transcripts by reverse transcriptase-polymerase chain reaction in neuroblastic tumors and compared the results with the TA, the tumor growth fraction, and the MYCN status. In a series of 38 neuroblastic tumors, high TA and full-length hTERT transcripts were found in nine samples, whereas nine samples showed absence of both enzymatic activity and hTERT transcripts. Interestingly, in another eight samples, low or absent TA coincided with a lack of full-length hTERT transcripts. Eleven samples contained hTERT transcripts with low or undetectable TA and one sample had low TA but no hTERT transcripts. TA correlated with MYCN amplification and was weakly associated with the proliferative activity. Moreover, a significant correlation with tumor progression was observed. Our findings point at a posttranscriptional regulation of TA in a subset of neuroblastic tumors. Because high TA was detected only in tumors with full-length hTERT transcripts, reverse transcriptase-polymerase chain reaction analysis of archival neuroblastic tumor samples might help to appraise the malignant potential in individual cases.
Neuroblastoma is the most common extracranial solid tumor in children. Its biological behavior is notoriously unpredictable and comprises a spectrum ranging from spontaneous regression to rapid metastatic spread. 1 A singular trait of neuroblastic tumors is their capacity of differentiation. Indeed, in a portion of neuroblastic tumors, maturation of neuroblasts to ganglionic cells engenders ganglioneuroblastomas that may further eventuate into benign ganglioneuromas. The degree of cellular maturation and the amount of tumor cells showing signs of differentiation form the basis of various grading schemes, 2-6 but the determinants of the divergent courses of the disease are primarily unknown.
The recently identified enzyme telomerase is considered to play an important role in the development and progression of malignant tumors. 7,8 Because of the inability of the replication machinery to synthesize the uttermost 3′ ends of chromosomes, there is a gradual decrease in telomere length at each cell division, limiting the replicative potential of normal somatic cells. 9 The continuous unrestricted proliferation of most cancer cells would thus be expected to lead to a critical telomere attrition resulting in chromosomal catastrophe and cell death. 10,11 To compensate for the loss of telomeric DNA sequences, the ribonucleoenzyme telomerase is activated under certain circumstances, adding hexameric nucleotide repeats (TTAGGG)n to the telomeres. Accordingly, telomerase activity (TA) is found in renewing tissues and in rapidly dividing cells, ie, in the germline and in malignant tumors. Because TA may be present in nonneoplastic cells and hyperplastic tissues, 12-15 it cannot be regarded as a true tumor marker, and it has even been suggested that TA might be a mere indicator of proliferation. 16 However, recent investigations have shown an association between TA and loss of cell cycle regulators, establishing a new link to malignancy. 17,18 TA has also been described as a prognostic marker in neuroblastic tumor, a high TA correlating with an aggressive biological behavior. 19-21 However, it seems that some neuroblastic tumors contain hTERT transcripts without displaying noticeable TA, which may represent one mechanism limiting the growth potential of these tumors.
Telomerase is likely to constitute a multiprotein complex, of which three essential components have been identified: human telomerase reverse transcriptase (hTERT), an internal RNA strand (hTR), and a RNA-binding protein (hTEP1). The genes for hTR and hTERT have been cloned, 22-25 and although several other telomerase-associated proteins have been identified, 26,27 hTR and hTERT are sufficient to reconstitute TA in vitro. 28,29 Despite intensive research, the regulation of TA is not well understood. Although hTR is detectable in most embryonal and adult human cells, 22,30 the expression of hTERT, which closely correlates to the TA, seems to be tightly controlled, but the underlying mechanism remains elusive. Recently, it has been shown that different transcripts of the hTERT gene are present in human tissues, 23,31 suggesting a posttranscriptional modulation of TA by alternative splicing that may result in truncated, and possibly dysfunctional, protein products. This mechanism would explain the discordant finding of hTERT transcription without detectable TA in some neuroblastic tumors.
With this in view, we sought to elucidate whether there is a relationship between the different hTERT splice variants and TA in neuroblastic tumors, and whether TA correlates with the proliferative status of neuroblastic tumors. Additionally, we performed a search for MYCN amplification, which is regarded as one of the main prognostic factors in neuroblastic tumors. 32
Materials and Methods
Patients
Freshly excised samples of neuroblastic tumors from 38 patients (Table 1) ▶ were snap-frozen in liquid nitrogen and stored at −80°C. One part of each sample was processed for routine histological examination. Only viable tumor tissue, verified by the presence of undegraded RNA, was used for further analysis. Diagnosis, 4 grading, 2,33 and staging 34 were made according to the established criteria.
Table 1.
Tumor and Patient Characteristics
| No. | Age at diagnosis (days) | Hughes’ grade | Pretreatment | Shimada | MYCN | Relative TA | hTERT | Ki-S5 [%] | INSS stage | Status | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | Grade | ||||||||||
| 1 | 96 | 3 | + | — | — | + | 7.2% | + | 31 | 4s | NED |
| 2 | 1048 | 1a | − | GNB i | FH | − | — | — | <1 | 3 | NED |
| 3 | 509 | 1a | + | — | — | − | 0.4% | nfl | <1 | 2a | NED |
| 4 | 3975 | 3 | − | NBL ud | UH | − | — | nfl | 43 | 3 | NED |
| 5 | 22 | 3 | − | NBL ud | UH | − | — | nfl | 33 | 4 | NED |
| 6 | 272 | 2 | − | NBL pd | FH | − | 18.5% | + | 39 | 2a | PD |
| 7 | 22 | 3 | − | NBL ud | UH | − | 0.1% | − | 48 | 4s | NED |
| 8 | 1003 | 3 | − | NBL ud | UH | − | 5.9% | + | 44 | 3 | DOD |
| 9 | 50 | 3 | − | NBL ud | UH | − | 0.4% | nfl | 49 | 4s | NED |
| 10 | 371 | 2 | − | NBL d | FH | − | 0.6% | nfl | 24 | 2b | NED |
| 11 | 2987 | 3 | − | NBL ud | UH | − | — | − | 43 | 4 | DOD |
| 12 | 2847 | 2 | + | — | — | − | 5.1% | + | 40 | 2a | REL |
| 13 | 932 | 2 | − | GNB n | UH | − | 1.2% | + | 47 | 4 | REL |
| 14 | 1014 | 3 | + | — | — | − | — | − | 31 | 4 | REL |
| 15 | 581 | 3 | + | — | — | + | — | − | 39 | 4 | DOD |
| 16 | 1481 | 3 | − | NBL ud | UH | − | — | + | 29 | 4 | DOD |
| 17 | 304 | 3 | − | NBL ud | UH | − | — | + | 45 | 3 | PD |
| 18 | 3395 | GN | − | GN mature | FH | − | — | − | <1 | 2a | NED |
| 19 | 216 | 3 | − | NBL pd | FH | − | — | − | 3 | 1 | NED |
| 20 | 199 | 3 | − | NBL ud | UH | + | 3.1% | + | 56 | 4 | REL |
| 21 | 455 | 3 | − | NBL pd | FH | − | — | − | 23 | 2a | NED |
| 22 | 4630 | 1a | − | GNB i | FH | − | — | − | 1 | 1 | NED |
| 23 | 982 | 3 | − | NBL ud | UH | + | 4.4% | + | 47 | 4 | NED |
| 24 | 2402 | 1a | − | GN mature | FH | − | — | nfl | <1 | na | NED |
| 25 | 512 | 2 | − | NBL d | FH | − | 0.5% | + | 11 | 1 | NED |
| 26 | 91 | 2 | + | — | — | − | 0.1% | + | 14 | 2 | NED |
| 27 | 506 | 3 | − | NBL pd | FH | − | — | − | 9 | 3 | NED |
| 28 | 1152 | 2 | + | — | — | − | 0.1% | + | 13 | 4 | NED |
| 29 | 2831 | 3 | − | NBL ud | UH | − | 1.0% | + | 26 | na | NED |
| 30 | 392 | 2 | + | — | — | − | — | + | 21 | 3 | NED |
| 31 | 416 | 2 | − | NBL pd | FH | − | 0.3% | + | 25 | 3 | NED |
| 32 | 141 | 2 | − | NBL d | FH | − | 0.5% | + | 57 | 1 | NED |
| 33 | 1509 | 2 | − | NBL d | FH | + | 1.0% | + | 16 | 4 | NED |
| 34 | 112 | 2 | − | NBL d | FH | − | — | nfl | 31 | 3 | NED |
| 35 | 215 | 2 | + | — | — | − | — | + | 25 | 4 | NED |
| 36 | 429 | 3 | − | NBL pd | FH | − | 0.2% | + | 27 | 2a | NED |
| 37 | 1110 | 2 | + | — | — | − | — | nfl | 33 | 4 | NED |
| 38 | 1311 | 1a | + | — | — | + | — | + | <1 | 4 | NED |
NBL, neuroblastoma; ud, undifferentiated; pd, poorly differentiated; d, differentiating; GNB, ganglioneuroblastoma; GN, ganglioneuroma; i, intermixed; n, nodular; FH, favorable histology; UH, unfavorable histology; nfl, no full-length hTERT transcripts; NED, no evidence of disease; DOD, died of disease; REL, relapse; PD, progressive disease; na, no information available.
RNA Isolation and Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
RNA was isolated with the chaotropic reagent Trizol (Life Technologies, Inc., Gaithersburg, MD) according to the manufacturer. One μg of total RNA was reverse-transcribed using the RNA PCR kit (Perkin Elmer, Langen, Germany) and subjected to PCR as recommended by the manufacturer. We used 1.25 U/50 μl of Amplitaq Gold (Perkin Elmer) as polymerase, which must be activated by heating to 95°C for 10 minutes. This procedure avoids primer-dependant PCR artifacts. Primer sequences were chosen to amplify a region of the hTERT mRNA containing the motifs for the reverse transcriptase activity. 23,35 Primers were 5′ end labeled with Fam (6-carboxyfluorescein, Perkin Elmer), primer sequences: 2164 5′ gcctgagctgtactttgtcaa 3′ and 2620 5′ cgcaaacagcttgttctccatgtc 3′. 31 After 40 cycles of amplification with an annealing temperature of 68°C, PCR products were analyzed by capillary electrophoresis (ABIprism 310, Perkin Elmer). As a positive control, glyceraldehyde-3-phosphate dehydrogenase mRNA (GAPDH) was amplified in parallel with the primers GAPDH-S: 5′ acgaccactttgtcaagctcat 3′, and GAPDH-A: 5′ ggtactttattgatggtacatg 3′ for 30 cycles with an annealing temperature of 60°C. Validation of PCR products was done by automated sequencing (ABIprism 310, Perkin Elmer).
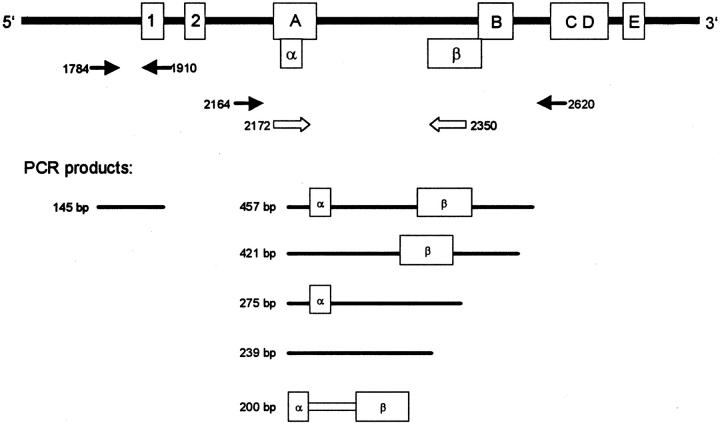
Analysis of matched formalin-fixed, paraffin-embedded tumor tissue was done as follows: five sections, each 10-μm thick, were cut from paraffin blocks and collected in a 1.5-ml sterile polypropylene tube. Paraffin was removed with two changes of xylene and two changes of absolute ethanol (both from Sigma, Deisenhofen, Germany). The remaining tissue pellet was air-dried, overlaid with 1 ml of Trizol, and homogenized with a hand-held mechanical device (Ultra Turrax T8; IKA Labortechnik, Hohenstaufen, Germany). The tissue lysates were left overnight at room temperature and RNA isolation was proceeded as described above. RNA integrity was ascertained by RT-PCR amplification of GAPDH mRNA. Two different regions of hTERT mRNA were amplified in parallel (Figure 1) ▶ : primers hTERT 1784 (5′ cggaagagtgtctggagcaa 3′) and 1910 (5′ ggatgaagcggagtctgga 3′) amplify a 145-bp segment present in all transcripts whereas the primers hTERT 2172 (5′ tgtactttgtcaaggtggatgtg 3′) and hTERT 2350 (5′ gtacggctggaggtctgtcaag 3′) were designed to amplify a 200-bp segment being unique to the full-length hTERT transcript. Annealing temperature was 60°C for hTERT 1784/1910 and 68°C for hTERT 2172/2350.
Figure 1.
Schematic drawing of the hTERT mRNA with the sequences coding for the reverse-transcriptase motifs 1, 2, A, B, C, D, and E. 35 A full-length transcript (457 bp) is shown at the top. Splicing of the α region results in a 36-base abridgement of the mRNA that causes a deletion in the reverse-transcriptase motif A. Splicing of the β region causes a 182-base abridgement of the mRNA, the resulting protein lacks the reverse-transcriptase motifs B, C, D, and E. RT-PCR primers 2164 and 2620 flank the region containing the α and β splice sites, the size of the amplicons being dependent on the type of transcript. Primers 2172 and 2350 bind within the critical splice sites and amplify full-length transcripts only, primers 1784 and 1910 amplify hTERT transcripts irrespective of alternate splicing.
TRAP Assay
For the assessment of TA, a modified version of the telomeric repeat amplification protocol (TRAP) assay was applied as described. 36 Briefly, tumor samples were lysed in ice cold lysis buffer (0.5% CHAPS, (Sigma), 10 mmol/L Tris-HCl, pH 7.5, 1 mmol/L MgCl2, 1 mmol/L EGTA, 5 mmol/L β-mercaptoethanol, 0.1 mmol/L 4-(2-aminoethyl)benzenesulfonylfluoride) (Sigma), 1 U/μl RNase Inhibitor (MBI Fermentas, St. Leon-Rot, Germany), 10% glycerin), incubated for 30 minutes on ice and centrifuged at 20,000 × g for 30 minutes at 4°C. Protein concentration of the supernatant was determined with the Bradford assay (Bio-Rad, München, Germany). One μg of protein was assayed in a 48-μl reaction mix (20 mmol/L Tris-HCl, pH 8.0, 1 mmol/L EGTA, 0.005% Tween 20, 1.5 mmol/L MgCl2, 63 mmol/L KCl, 50 μmol/L of each deoxynucleosidetriphosphate (MBI Fermentas), 2 U Taq DNA-polymerase (MBI Fermentas), 0.1 attomol PCR standard, 10 pmol primer TS (5′-TAMRA-TS; Eurogentec, Seraing, Belgium), 10 pmol CX-ext-primer). After elongation for 30 minutes at 30°C, 30 cycles of PCR (95°C for 30 seconds, 50°C for 30 seconds, and 72°C for 30 seconds) were run. PCR products were analyzed by capillary electrophoresis (ABIprism 310, Perkin Elmer). Integrated values were added up for all telomerase products and adjusted by dividing them by the value obtained for the internal amplification standard. Different dilutions of lysates of highly telomerase-positive L428 cells (Hodgkin’s disease-derived cell line 37 ) were analyzed in the same way and used to generate a calibration curve. The TA values of each tumor were matched to this curve to express the results as a percentage of the TA measured in 1 μg of protein extract from L428 cells.
Immunohistochemistry
The tumor cell-growth fraction was assessed immunohistochemically by means of monoclonal antibody Ki-S5 directed to a formalin-resistant epitope of the Ki-67 protein. 38 Tumor sections were deparaffinized in xylene and rehydrated in graded ethanol. Immunoreactivity was restored by microwaving the slides in 10 mmol/L of sodium citrate, pH 6.0, for 20 minutes at 800 W, and the staining reaction was enhanced with use of the alkaline phosphatase anti-alkaline phosphatase technique. 39 For each tumor sample, a total of 1000 cells were evaluated in different areas of the tumor at high magnification (×350) using a standard light microscope (Zeiss, Oberkochen, Germany). The quantity of positive nuclei was expressed as a percentage of the total tumor cell count.
Fluorescence in Situ Hybridization
The MYCN status was determined by in situ hybridization 40 with a digoxigenin-labeled probe complementary to the MYCN gene (Appligene Oncor, Illkirch, France). Briefly, paraffin-embedded tumor samples were cut as for immunohistochemistry. After deparaffinization and rehydration, tissue slides were treated with 40 μg/ml of proteinase K (Roche Molecular Biochemicals, Mannheim, Germany) at 37°C for 30 minutes and hybridized overnight. Visualization was accomplished with anti-digoxigenin F(ab)2-fragments from sheep coupled to fluorescein (Roche Molecular Biochemicals), diluted 1:200. After repeated washings in phosphate-buffered saline the slides were air-dried, stained with propidium iodide supplemented with antifade [1,4-diazabicyclo(2.2.2)octane; Sigma]. Fluorescence microscopy was performed with use of a Zeiss photomicroscope equipped with fluorescein epifluorescence filters. A tumor was considered MYCN amplified when more than six nuclear signals were seen or when tumor nuclei showed relatively large signals compared with admixed fibroblasts or lymphocytes, which usually showed a pair of distinct nuclear signals. This procedure is supposed to adjust for potential hyperdiploidy, in which case the tumor cells would show more than the expected two signals without a MYCN amplification per se.
Statistical Analysis
The SPSS software package, version 10.0, was used for all calculations. Binary factors were correlated by means of Fisher’s exact test, and categories of continuous variables were compared with the use of the Mann-Whitney U test or the Kruskal-Wallis nonparametric analysis of variance. Statistical significance was assumed at P < 0.05.
Results
Clinicopathological data of the patients are detailed in Table 1 ▶ . Nineteen out of 38 neuroblastic tumors showed TA in the TRAP assay. The relative enzymatic activity ranged from 0.1 to 18.5%, as compared to the L428 cell extracts. Lack of TA was not attributable to failure of the assay (eg, because of the presence of inhibitors), because regular amplification of the internal amplification standard was seen with all tumor cell extracts. Also, previous chemotherapy did not seem to influence TA as two pretreated samples contained high TA. Well-differentiated tumors, ie, Hughes’ grade 1/ganglioneuroblastomas, had very low or absent TA, and the one ganglioneuroma in our series was telomerase-negative. TA was nevertheless very heterogeneous in the less well-differentiated tumors (Hughes’ grades 2 or 3), which displayed either high or low TA levels, or even no activity at all. Thus, no statistically significant correlation could be established between Hughes’ grade and TA, whereas there was a trend toward an correlation of TA with Shimada’s grade (P = 0.06 by Fisher’s exact test). Conversely, an association with tumor stage was not evident. The highest TA was found in a localized (stage 2) neuroblastoma whereas some tumors in advanced stages exhibited no or little TA.
RT-PCR experiments were conducted using primers (2164 and 2620) partly encompassing the reverse transcriptase domain of the hTERT mRNA (Figure 1) ▶ . Within this region, two splice sites have been identified. Splicing at the α site causes a 36-base deletion (bases 2186 to 2221) in the mRNA, and β splicing results in the loss of 182 bases (bases 2342 to 2524) from the transcript. Nucleotide loss at the α splice site causes partial ablation of the conserved reverse transcriptase motif A, whereas splicing at the β site results in loss of the motifs B, C, D, and E. As deletion of one of the conserved reverse transcriptase motifs is sufficient to abrogate the function of the enzyme, intactness of the transcript at both these splice sites is essential for TA.
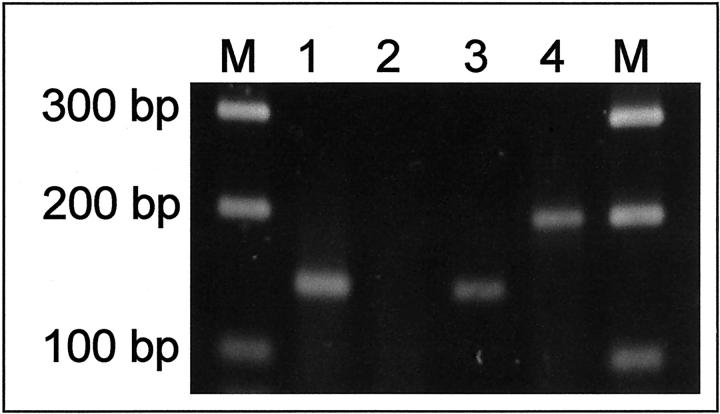
For analysis of formalin-fixed paraffin-embedded tumor samples, two primer pairs were used. Primers 1784 and 1910 amplify every hTERT message whereas primers 2172 and 2350 were designed to amplify full-length transcripts only. This was achieved by choosing primer-binding sites spanning the sequences at which α- and β-splicing occurs, thus preventing amplification of spliced hTERT transcripts. In case of a positive reaction, a 200-bp PCR product was seen whereas primers 1784 and 1910 generated a 145-bp PCR product.
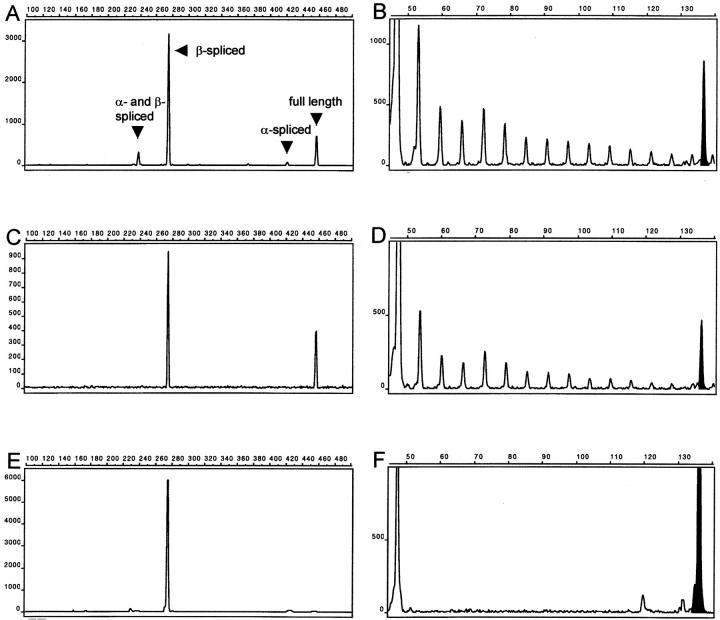
Twenty-eight tumors contained hTERT mRNA, and all samples were positive for the ubiquitously expressed glyceraldehyde-3-phosphate dehydrogenase mRNA used as a positive control (data not shown). Twenty formalin-fixed paraffin-embedded tumor samples showed identical results compared with their fresh-frozen counterparts. RT-PCR with the primers 2164 and 2620 revealed splice variants in virtually all tumors containing hTERT mRNA transcripts and in the cell line L428 (examples are shown in Figures 2 and 3 ▶ ▶ ). Notably, β-spliced transcripts were consistently present, whereas the two other splice variants were randomly distributed. Full-length transcripts were present in 20 tumors, nine of which displayed TA of 1% or higher. Eight tumors that lacked the full-length hTERT transcripts had either low or undetectable TA. Eleven tumors contained full-length transcripts with low TA in six and absence of TA in five cases. By contrast, one tumor displayed very low TA in the absence of hTERT transcripts. The association of any amount of TA with the presence of full-length hTERT transcripts was highly significant (P < 0.0001 by Fisher’s exact test). Also, full-length hTERT transcripts were significantly associated with high TA levels (equal to or greater than 1%, P = 0.001). By contrast, neither the distribution of hTERT splice variants nor TA levels correlated with the patient’s age at diagnosis.
Figure 2.
RT-PCR demonstration of hTERT transcripts (left) and TRAP assay results (right) of the tumor cell line L428 (A and B), patient 1 (C and D), and patient 4 (E and F). The x axis defines the fluorescence intensities and the y axis indicates the length of the PCR fragments in bp. Presence of full-length hTERT transcripts (A and C) correlates with TA (B and D), demonstrated here by the characteristic 6-bp ladder, representing the elongated template primer TS. Lack of full-length transcripts (E) is associated with inactive telomerase and absent TA (F). The amount of protein analyzed in the TRAP assay was 1 μg, except in the case of L428, in which 120 ng were used. The internal amplification standard peak is in black.
Figure 3.
RT-PCR demonstration of hTERT transcripts in formalin-fixed paraffin-embedded neuroblastoma samples. Lanes 1 and 2 correspond to case no. 4 and lanes 3 and 4 correspond to case no. 1. Primers hTERT 1784/1910 amplify a 145-bp PCR product common to all hTERT transcripts (lanes 1 and 3) whereas primers hTERT 2172/2350 amplify a 200-bp region unique to full-length transcripts. M, molecular weight marker.
The proliferative status of the different neuroblastic tumors was highly variable. Ki-S5 immunolabeling revealed a heterogeneous pattern in all tumor samples that was accentuated at the tumor margins with occasional clusters of proliferating cells in the bulk of the tumor. Well-differentiated tumors contained only a few Ki-S5-positive cells (as a rule <3%). The other samples showed labeling indices within a range between 9% and 57%. The proliferative activity showed a tendency toward an association with TA (P = 0.06 according to the Kruskal-Wallis test), and to a lesser extent with the presence of hTERT transcripts (P = 0.16, Kruskal-Wallis test).
MYCN amplification, as shown by multiple nuclear signals revealed by in situ hybridization, was found only in six tumors, four of which were Hughes’ grade 3 whereas only two were of unfavorable histology according to Shimada’s grading. Four contained full-length hTERT transcripts associated with high TA, one showed full-length transcripts without detectable TA, and one lacked both TA and hTERT transcripts. Statistical analysis revealed a significant association of MYCN amplification with high TA (P = 0.019, Fisher’s exact test), and to a lesser extent with the presence of full-length hTERT transcripts (P = 0.08).
We further investigated whether TA or hTERT transcription and splicing may provide prognostic information. High TA was significantly associated with disease progression in terms of either local recurrence, distant metastasis, or tumor-related mortality (P = 0.004, Fisher’s exact test). This corresponds to the significance level of Shimada’s grade determined on this series (P = 0.005). Cases with full-length hTERT transcripts, however, merely showed a tendency toward an adverse prognosis (P = 0.13). As this study was designed as an analysis of fresh tissue with a view toward basic regulatory mechanisms, the follow-up period (median, 12 months) may be too short for a confident estimation of the clinical outcome, and therefore, the results have to be considered with reserve. Our analysis nevertheless suggests that high TA may predict early disease progression. Because of the limited correlation with other prognostic factors, TA might thus be a truly independent indicator of prognosis in neuroblastic tumors.
Discussion
In neuroblastic tumors, the presence of TA is believed to portend an adverse outcome, 19,21 and previous studies have shown a strong statistical correlation between TA and the transcription of hTERT mRNA. 20,21 However, it was found that a subset of neuroblastic tumors exhibited no detectable TA although hTERT transcripts were present, suggesting a regulation at the posttranscriptional level. To address this issue, we performed on a series of 38 neuroblastic tumors a comparative analysis of the TA in relation to hTERT transcription determined by RT-PCR.
Owing to the inclusion of an internal amplification standard, our modified TRAP assay virtually allows us to exclude false-positive results, and a deproteinization step warrants a sufficient sensitivity. 41 Moreover, comparison with a calibration curve generated with stepwise diluted protein extracts from L428 cells enables a reasonable appraisal of the relative TA in tissue samples. 36
Overall, TA values in the samples of neuroblastic tumors were low compared with L428 extracts, possibly reflecting suboptimal growth conditions in the tumor in contrast to an optimum nutrient supply in cell culture. This posed the problem of categorizing high versus low TA values. As we observed a spectrum from 0.1 to 18.5% relative TA, we followed previous reports to assume that very low TA (<1%) is below the threshold required for telomere maintenance or elongation. 42 On this basis, significant TA would be present in 24% of our tumors.
The first publication relating telomerase to prognosis in neuroblastic tumors reported TA in 96% of untreated neuroblastic tumors. 19 The much lower prevalence in our series is nevertheless unlikely to be attributable to the inclusion of 10 patients having received cytotoxic treatment before the diagnostic biopsy, as pretreatment does not seem to affect TA levels. 21 Rather, one may assume that the observations mentioned above are biased by false-positive results as they easily occur with the original TRAP assay because of template slippage on PCR-derived primer dimers. 41 In more recent studies 29% 21 and 80% 20 of neuroblastic tumors contained TA, the former result being well in line with our observations.
It is nevertheless remarkable that the percentage of telomerase-positive neuroblastic tumors is rather small compared with other cancers, notably carcinomas. 7,8 This may be attributable to the propensity of neuroblastic tumors toward differentiation and even spontaneous regression, which might reflect the inability of sustaining tumor cell proliferation in the absence of mechanisms counteracting the attrition of telomeres. In light of these reflections, it is well conceivable that a differential regulation of TA may be a major determinant of the biological behavior of neuroblastic tumors. In our series, there was no clear-cut association between TA and clinical stage or histopathological tumor grade. However, well-differentiated tumors (ganglioneuroblastomas and ganglioneuromas) hardly exhibited any TA, which supports the above given considerations.
Looking for hTERT-transcripts and their splice variants by means of RT-PCR, we found hTERT transcription in 28 out of 38 tumors. Interestingly, eight tumors in our series lacked full-length hTERT transcripts, which is reminiscent of a phenomenon seen in certain tissue types during embryonal development. 31 It seems that, at later stages of gestation, full-length hTERT transcripts are replaced by splice variants with alterations in the region spanning the reverse transcriptase motifs. More recently, analogous alternate transcripts were also observed in normal and neoplastic ovarian tissue, endometrium, and myometrium. 43 These truncated forms failed to produce TA in the absence of full-length transcripts, consistent with in vitro experiments showing that the β-deletion variants are unable to reconstitute a functional telomerase complex. 28 Although the various splice variants are likely to be ubiquitous in hTERT-expressing cells, the lack of full-length transcripts was primarily restricted to nonneoplastic tissues. Hence, the authors concluded that alternate splicing of hTERT mRNA might constitute a regulatory mechanism of TA at the posttranscriptional level, which may be lost with malignant transformation. 43
Although our observations support this idea, they additionally provide evidence for an occasional maintenance of this physiological mechanism in malignant neoplasms. In the eight tumors lacking full-length transcripts TA was absent or at least well below 1% indicating that, beyond transcription, differential splicing of hTERT mRNA has a significant impact on the TA in neuroblastic tumors, which identifies a novel regulatory mechanism in these tumors. It is therefore tempting to speculate that regulatory mechanisms of embryonal life might to some extent be conserved in neuroblastic tumors. Although we could not establish a significant overall correlation between alternative hTERT splicing and patient age, the median age of the patients with truncated hTERT transcripts was 14.4 months. This observation suggests that the negative regulation of TA by alternate splicing of hTERT mRNA might persist through early infancy, and thus might account for the favorable biological behavior of neuroblastic tumors in children younger than 1.5 years of age. 3,44
It emerges from our data that full-length hTERT transcripts are indispensable for the enhancement of significant TA. Thus, the clear-cut correlation between the lack of full-length transcripts and low or absent TA may explain the discrepancy between TA and the presence of hTERT transcripts observed by others. 20,21 Because primers amplifying all types of hTERT transcripts were used in these analyses, variant transcripts inevitably escaped detection. However, five tumors in our series transcribed hTERT mRNA with complete reverse transcriptase motifs while lacking detectable TA. In concert with previous observations, 43 this is suggestive of additional regulatory mechanisms, eg, posttranslational modifications by phosphorylation or dephosphorylation of certain protein subunits, 45-48 or possibly, truncations outside the RT motifs of hTERT. 49
Yet another reason for this apparent discrepancy might be that the expression levels of full-length hTERT, which were not investigated in this study, might modulate the TA. This idea would be supported by the very close association of full-length hTERT transcripts with detectable TA in this series (P < 0.0001). In fact, initial data had shown a good correlation between hTERT mRNA levels and TA. 23,24,50 Also, functional hTERT mRNA was detected in lymphocytes irrespective of the degree of TA 51 as well as in normal, telomerase-negative tissues, such as human brain, liver, prostate, heart and primary fibroblasts. 52 It has therefore been suggested 15 that cells with the ability to proliferate on a certain stimulus, eg, antigen-stimulated lymphocytes, constitutively express a low amount of hTERT mRNA, which would be dramatically up-regulated in malignant cells.
Several studies have shown that TA closely correlates to the proliferative activity in normal tissues and cancers. 16,17,36 We therefore determined the tumor growth fraction by means of a monoclonal antibody (Ki-S5) against the Ki-67 protein, which is expressed in all cycling cells from G1 through M phase of the division cycle. 53 However, despite a weak correlation with TA, a high Ki-S5-labeling index alone was neither necessary nor sufficient for telomerase activation, as illustrated by cases no. 4 and no. 17 with high Ki-S5-labeling indices and absent TA (Table 1) ▶ . This may be explained by the inclusion of G1 phase in Ki-67 labeling, because recent results indicate that telomerase is activated at the G1/S transition. 54 Although our observations argue against telomerase being a mere proliferation marker, several lines of evidence point out a relationship between TA and cell-cycle deregulation. 16,17,55
In this regard, MYCN amplification deserved consideration, as the hTERT promoter contains binding sites for the transcription factor MYCN. 56 In accordance with a recent investigation, 57 we observed a significant correlation between MYCN copy number and TA. Out of six tumors with MYCN amplification, five contained full-length hTERT transcripts, and four exhibited significant TA, indicating that MYCN may actually transactivate hTERT whereas additional factors may ultimately modulate its activity.
Our investigation demonstrates at least one novel regulatory mechanism of TA in neuroblastic tumors, which is likely to carry high biological relevance. Although this study could not possibly be designed to test clinicopathological correlations, our preliminary analysis points out a prognostic impact of TA. However, the assessment of TA requires fresh or well-preserved frozen tissue that is not available to the pathologist in most instances. As high TA apparently requires the presence of full-length hTERT transcripts, the rapid and easy method of RT-PCR, being well suitable for the analysis of archival specimens, may provide a novel basis for assessing the prognostic value of TA in neuroblastic tumors in retrospective studies with large sample sizes and long term follow-up. Given the lack of substantial TA in a portion of specimens with full-length transcripts, quantitative analysis of hTERT transcripts using real-time PCR may be necessary to establish a threshold level above which enzyme activity is detectable.
Footnotes
Address reprint requests to Matthias Krams, M.D., Department of Pathology, University of Kiel, Michaelisstr. 11, 24105 Kiel, Germany. E-mail: mkrams@path.uni-kiel.de.
Supported by the Else Kröner-Fresenius Stiftung, Bad Homburg, Germany, and the Kinder Krebs-Initiative Buchholz Holm-Seppensen, Germany.
References
- 1.Evans AE, Gerson J, Schnaufer L: Spontaneous regression of neuroblastoma. Natl Cancer Inst Monogr 1976, 44:49-54 [PubMed] [Google Scholar]
- 2.Hughes M, Marsden HB, Palmer MK: Histologic patterns of neuroblastoma related to prognosis and clinical staging. Cancer 1974, 34:1706-1711 [DOI] [PubMed] [Google Scholar]
- 3.Shimada H, Chatten J, Newton WAJ, Sachs N, Hamoudi AB, Chiba T, Marsden HB, Misugi K: Histopathologic prognostic factors in neuroblastic tumors: definition of subtypes of ganglioneuroblastoma and an age-linked classification of neuroblastomas. J Natl Cancer Inst 1984, 73:405-416 [DOI] [PubMed] [Google Scholar]
- 4.Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B: Terminology and morphologic criteria of neuroblastic tumors: recommendations by the International Neuroblastoma Pathology Committee. Cancer 1999, 86:349-363 [PubMed] [Google Scholar]
- 5.Joshi VV, Chatten J, Sather HN, Shimada H: Evaluation of the Shimada classification in advanced neuroblastoma with a special reference to the mitosis-karyorrhexis index: a report from the Childrens Cancer Study Group. Mod Pathol 1991, 4:139-147 [PubMed] [Google Scholar]
- 6.Joshi VV: Peripheral neuroblastic tumors: pathologic classification based on recommendations of international neuroblastoma pathology committee (Modification of Shimada classification). Pediatr Dev Pathol 2000, 3:184-199 [DOI] [PubMed] [Google Scholar]
- 7.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW: Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266:2011-2015 [DOI] [PubMed] [Google Scholar]
- 8.Shay JW, Bacchetti S: A survey of telomerase activity in human cancer. Eur J Cancer 1997, 33:787-791 [DOI] [PubMed] [Google Scholar]
- 9.Greider CW: Mammalian telomere dynamics: healing, fragmentation shortening and stabilization. Curr Opin Genet Dev 1994, 4:203-211 [DOI] [PubMed] [Google Scholar]
- 10.Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC: Telomere reduction in human colorectal carcinoma and with ageing. Nature 1990, 346:866-868 [DOI] [PubMed] [Google Scholar]
- 11.Harley CB, Kim NW, Prowse KR, Weinrich SL, Hirsch KS, West MD, Bacchetti S, Hirte HW, Counter CM, Greider CW: Telomerase, cell immortality, and cancer. Cold Spring Harb Symp Quant Biol 1994, 59:307-315 [DOI] [PubMed] [Google Scholar]
- 12.Harle-Bachor C, Boukamp P: Telomerase activity in the regenerative basal layer of the epidermis in human skin and in immortal and carcinoma-derived skin keratinocytes. Proc Natl Acad Sci USA 1996, 93:6476-6481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasumoto S, Kunimura C, Kikuchi K, Tahara H, Ohji H, Yamamoto H, Ide T, Utakoji T: Telomerase activity in normal human epithelial cells. Oncogene 1996, 13:433-439 [PubMed] [Google Scholar]
- 14.Norrback KF, Roos G: Telomeres and telomerase in normal and malignant haematopoietic cells. Eur J Cancer 1997, 33:774-780 [DOI] [PubMed] [Google Scholar]
- 15.Kolquist KA, Ellisen LW, Counter CM, Meyerson M, Tan LK, Weinberg RA, Haber DA, Gerald WL: Expression of TERT in early premalignant lesions and a subset of cells in normal tissues. Nat Genet 1998, 19:182-186 [DOI] [PubMed] [Google Scholar]
- 16.Belair CD, Yeager TR, Lopez PM, Reznikoff CA: Telomerase activity: a biomarker of cell proliferation, not malignant transformation. Proc Natl Acad Sci USA 1997, 94:13677-13682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landberg G, Nielsen NH, Nilsson P, Emdin SO, Cajander J, Roos G: Telomerase activity is associated with cell cycle deregulation in human breast cancer. Cancer Res 1997, 57:549-554 [PubMed] [Google Scholar]
- 18.Roos G, Nilsson P, Cajander S, Nielsen NH, Arnerlov C, Landberg G: Telomerase activity in relation to p53 status and clinicopathological parameters in breast cancer. Int J Cancer 1998, 79:343-348 [DOI] [PubMed] [Google Scholar]
- 19.Hiyama E, Hiyama K, Yokoyama T, Matsuura Y, Piatyszek MA, Shay JW: Correlating telomerase activity levels with human neuroblastoma outcomes. Nat Med 1995, 1:249-255 [DOI] [PubMed] [Google Scholar]
- 20.Hiyama E, Hiyama K, Yokoyama T, Fukuba I, Yamaoka H, Shay JW, Matsuura Y: Rapid detection of MYCN gene amplification and telomerase expression in neuroblastoma. Clin Cancer Res 1999, 5:601-609 [PubMed] [Google Scholar]
- 21.Poremba C, Scheel C, Hero B, Christiansen H, Schaefer KL, Nakayama J, Berthold F, Juergens H, Boecker W, Dockhorn-Dworniczak B: Telomerase activity and telomerase subunits gene expression patterns in neuroblastoma: a molecular and immunohistochemical study establishing prognostic tools for fresh-frozen and paraffin-embedded tissues. J Clin Oncol 2000, 18:2582-2592 [DOI] [PubMed] [Google Scholar]
- 22.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J: The RNA component of human telomerase. Science 1995, 269:1236-1241 [DOI] [PubMed] [Google Scholar]
- 23.Kilian A, Bowtell DD, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA: Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet 1997, 6:2011-2019 [DOI] [PubMed] [Google Scholar]
- 24.Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, Bacchetti S, Haber DA, Weinberg RA: hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 1997, 90:785-795 [DOI] [PubMed] [Google Scholar]
- 25.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR: Telomerase catalytic subunit homologs from fission yeast and human. Science 1997, 277:955-959 [DOI] [PubMed] [Google Scholar]
- 26.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB, Arruda I, Robinson MO: A mammalian telomerase-associated protein. Science 1997, 275:973-977 [DOI] [PubMed] [Google Scholar]
- 27.Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F: TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell 1997, 88:875-884 [DOI] [PubMed] [Google Scholar]
- 28.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB: Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet 1997, 17:498-502 [DOI] [PubMed] [Google Scholar]
- 29.Beattie TL, Zhou W, Robinson MO, Harrington L: Reconstitution of human telomerase activity in vitro. Curr Biol 1998, 8:177-180 [DOI] [PubMed] [Google Scholar]
- 30.Avilion AA, Piatyszek MA, Gupta J, Shay JW, Bacchetti S, Greider CW: Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res 1996, 56:645-650 [PubMed] [Google Scholar]
- 31.Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR: Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res 1998, 58:4168-4172 [PubMed] [Google Scholar]
- 32.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM: Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 1984, 224:1121-1124 [DOI] [PubMed] [Google Scholar]
- 33.Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B, Stram DO, Gerbing RB, Lukens JN, Matthay KK, Castleberry RP: The International Neuroblastoma Pathology Classification (the Shimada system). Cancer 1999, 86:364-372 [PubMed] [Google Scholar]
- 34.Brodeur GM, Pritchard J, Berthold F, Carlsen NL, Castel V, Castelberry RP, De Bernardi B, Evans AE, Favrot M, Hedborg F: Revisions of the international criteria for neuroblastoma diagnosis, staging and response to treatment. Prog Clin Biol Res 1994, 385:363-369 [PubMed] [Google Scholar]
- 35.Eickbush TH: The Evolutionary Biology of Viruses. 1994:pp 121-157 Raven, Edited by SS Morse. New York
- 36.Rudolph P, Schubert C, Tamm S, Heidorn K, Hauschild A, Michalska I, Majewski S, Krupp G, Jablonska S, Parwaresch R: Telomerase activity in melanocytic lesions: a potential marker of tumor biology. Am J Pathol 2000, 156:1425-1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diehl V, Kirchner HH, Burrichter H, Stein H, Fonatsch C, Gerdes J, Schaadt M, Heit W, Uchanska-Ziegler B, Ziegler A, Heintz F, Sueno K: Characteristics of Hodgkin’s disease-derived cell lines. Cancer Treat Rep 1982, 66:615-632 [PubMed] [Google Scholar]
- 38.Rudolph P, Lappe T, Hero B, Berthold F, Parwaresch R, Harms D, Schmidt D: Prognostic significance of the proliferative activity in neuroblastoma. Am J Pathol 1997, 150:133-145 [PMC free article] [PubMed] [Google Scholar]
- 39.Cordell JL, Falini B, Erber WN, Ghosh AK, Abdulaziz Z, MacDonald S, Pulford KA, Stein H, Mason DY: Immunoenzymatic labeling of monoclonal antibodies using immune complexes of alkaline phosphatase and monoclonal anti-alkaline phosphatase (APAAP complexes). J Histochem Cytochem 1984, 32:219-229 [DOI] [PubMed] [Google Scholar]
- 40.Shapiro DN, Valentine MB, Rowe ST, Sinclair AE, Sublett JE, Roberts WM, Look AT: Detection of N-myc gene amplification by fluorescence in situ hybridization. Diagnostic utility for neuroblastoma. Am J Pathol 1993, 142:1339-1346 [PMC free article] [PubMed] [Google Scholar]
- 41.Krupp G, Kuhne K, Tamm S, Klapper W, Heidorn K, Rott A, Parwaresch R: Molecular basis of artifacts in the detection of telomerase activity and a modified primer for a more robust ‘TRAP’ assay. Nucleic Acids Res 1997, 25:919-921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM: Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci USA 1994, 91:9857-9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulaner GA, Hu JF, Vu TH, Oruganti H, Giudice LC, Hoffman AR: Regulation of telomerase by alternate splicing of human telomerase reverse transcriptase (hTERT) in normal and neoplastic ovary, endometrium and myometrium. Int J Cancer 2000, 85:330-335 [PubMed] [Google Scholar]
- 44.Berthold F, Kassenbohmer R, Zieschang J: Multivariate evaluation of prognostic factors in localized neuroblastoma. Am J Pediatr Hematol Oncol 1994, 16:107-115 [PubMed] [Google Scholar]
- 45.Li H, Zhao L, Yang Z, Funder JW, Liu JP: Telomerase is controlled by protein kinase c-alpha in human breast cancer cells. J Biol Chem 1998, 273:33436-33442 [DOI] [PubMed] [Google Scholar]
- 46.Li H, Zhao LL, Funder JW, Liu JP: Protein phosphatase 2A inhibits nuclear telomerase activity in human breast cancer cells. J Biol Chem 1997, 272:16729-16732 [DOI] [PubMed] [Google Scholar]
- 47.Bodnar AG, Kim NW, Effros RB, Chiu CP: Mechanism of telomerase induction during T cell activation. Exp Cell Res 1996, 228:58-64 [DOI] [PubMed] [Google Scholar]
- 48.Ku WC, Cheng AJ, Wang TC: Inhibition of telomerase activity by PKC inhibitors in human nasopharyngeal cancer cells in culture. Biochem Biophys Res Commun 1997, 241:730-736 [DOI] [PubMed] [Google Scholar]
- 49.Beattie TL, Zhou W, Robinson MO, Harrington L: Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol Biol Cell 2000, 11:3329-3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura Y, Tahara E, Tahara H, Yasui W, Ide T: Quantitative reevaluation of telomerase activity in cancerous and noncancerous gastrointestinal tissues. Mol Carcinog 1999, 26:312-320 [PubMed] [Google Scholar]
- 51.Liu K, Schoonmaker MM, Levine BL, June CH, Hodes RJ, Weng NP: Constitutive and regulated expression of telomerase reverse transcriptase (hTERT) in human lymphocytes. Proc Natl Acad Sci USA 1999, 96:5147-5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramakrishnan S, Eppenberger U, Mueller H, Shinkai Y, Narayanan R: Expression profile of the putative catalytic subunit of the telomerase gene. Cancer Res 1998, 58:622-625 [PubMed] [Google Scholar]
- 53.Gerdes J, Li L, Schlueter C, Duchrow M, Wohlenberg C, Gerlach C, Stahmer I, Kloth S, Brandt E, Flad HD: Immunobiochemical and molecular biologic characterization of the cell proliferation-associated nuclear antigen that is defined by monoclonal antibody Ki-67. Am J Pathol 1991, 138:867-873 [PMC free article] [PubMed] [Google Scholar]
- 54.Feist H, Zeidler R, Skrebsky T, Schmatloch S, Kreipe H: Induction of telomerase activity in stimulated human lymphocytes precedes expression of topoisomerase II alpha. Ann Hematol 1998, 76:111-115 [DOI] [PubMed] [Google Scholar]
- 55.Bonatz G, Frahm SO, Klapper W, Helfenstein A, Heidorn K, Jonat W, Krupp G, Parwaresch R, Rudolph P: High telomerase activity is associated with cell cycle deregulation and rapid progression in endometrioid adenocarcinoma of the uterus. Hum Pathol 2001, 32:605-614 [DOI] [PubMed] [Google Scholar]
- 56.Cong YS, Wen J, Bacchetti S: The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet 1999, 8:137-142 [DOI] [PubMed] [Google Scholar]
- 57.Streutker CJ, Thorner P, Fabricius N, Weitzman S, Zielenska M: Telomerase activity as a prognostic factor in neuroblastomas. Pediatr Dev Pathol 2001, 4:62-67 [DOI] [PubMed] [Google Scholar]