Abstract
The role of genes in normal birth-weight variation is poorly understood, and it has been suggested that the genetic component of fetal growth is small. Type 2 diabetes genes may influence birth weight through maternal genotype, by increasing maternal glycemia in pregnancy, or through fetal genotype, by altering fetal insulin secretion. We aimed to assess the role of the recently described type 2 diabetes gene TCF7L2 in birth weight. We genotyped the polymorphism rs7903146 in 15,709 individuals whose birth weight was available from six studies and in 8,344 mothers from three studies. Each fetal copy of the predisposing allele was associated with an 18-g (95% confidence interval [CI] 7–29 g) increase in birth weight (P=.001) and each maternal copy with a 30-g (95% CI 15–45 g) increase in offspring birth weight (P=2.8×10-5). Stratification by fetal genotype suggested that the association was driven by maternal genotype (31-g [95% CI 9–48 g] increase per allele; corrected P=.003). Analysis of diabetes-related traits in 10,314 nondiabetic individuals suggested the most likely mechanism is that the risk allele reduces maternal insulin secretion (disposition index reduced by ∼0.15 standard deviation; P=1×10-4), which results in increased maternal glycemia in pregnancy and hence increased offspring birth weight. We combined information with the other common variant known to alter fetal growth, the −30G→A polymorphism of glucokinase (rs1799884). The 4% of offspring born to mothers carrying three or four risk alleles were 119 g (95% CI 62–172 g) heavier than were the 32% born to mothers with none (for overall trend, P=2×10-7), comparable to the impact of maternal smoking during pregnancy. In conclusion, we have identified the first type 2 diabetes–susceptibility allele to be reproducibly associated with birth weight. Common gene variants can substantially influence normal birth-weight variation.
The role of genes in normal variation in birth weight is poorly understood, and it has been suggested that the genetic component of fetal growth in the general population is small.1–4 In contrast, the maternal intrauterine environment, including maternal glucose tolerance, BMI, and smoking, has a large impact on birth weight.5–8 However, the importance of these environmental factors does not negate the role of common maternal or fetal gene variants as determinants of normal fetal growth. Family, twin, and linkage studies suggest a role for common genetic variants,1,3,4,9–11 but, to date, specific genetic loci remain largely unknown.
Diabetes-susceptibility genes or genes that alter fasting glucose are good candidates for genes that influence birth weight, since they may impact insulin secretion or insulin action in nondiabetic individuals. Altered fetal insulin secretion would alter fetal growth and hence birth weight, since insulin is a key intrauterine growth factor. A diabetes-risk allele in the mother may alter fetal growth indirectly, by altering maternal glycemia during pregnancy and thereby influencing fetal insulin secretion. Alternatively, a diabetes-risk allele in the fetus may act directly on fetal insulin secretion. Reduced birth weight is associated with an increased risk of type 2 diabetes (MIM 125853) later in life,12,13 and it has been proposed under the fetal insulin hypothesis that this association could have a genetic explanation.14 There is some evidence of this from population studies.15–18 Direct evidence that birth weight is altered by fetal and maternal inheritance of diabetes-susceptibility genes is provided by several rare subtypes of diabetes. These include mutations in monogenic diabetes genes that reduce birth weight as a result of reduced fetal insulin secretion in utero19–24 and mutations in other monogenic diabetes genes that increase birth weight as a result of increased fetal insulin secretion.25
The observations in monogenic diabetes and the general population led to the hypothesis that inheritance of type 2 diabetes–susceptibility alleles by the fetus would reduce birth weight, whereas their inheritance by the mother would increase offspring birth weight. Some studies have suggested that inheritance by the fetus of polymorphisms associated with type 2 diabetes is associated with reduced birth weight,26,27 but these results have not been replicated.28–32 A maternal copy of the A allele of the common glucokinase (GCK [MIM 138079]) promoter polymorphism (GCK-30; rs1799884), which is associated with raised fasting glucose at all ages in the general population (P=1×10-9), is also associated with a 32-g (95% CI 11–53 g) increase in offspring birth weight (P=.002). However, there is no evidence of an independent effect of fetal genotype.33,34
The gene encoding transcription factor 7–like 2 (TCF7L2 [MIM 602228]) is the most important type 2 diabetes susceptibility gene found to date.35 Since its discovery, the association has been replicated in subjects of U.K., Amish, Finnish, French, U.S., Polish, Scandinavian, Dutch, Indian, and West African origin.36–45 In the U.K. population, the allelic odds ratio for rs7903146 (risk-allele frequency ∼30%) is 1.36 (95% CI 1.24–1.48; P=1.3×10-11), and individuals carrying two risk (T) alleles are at nearly twice the risk of type 2 diabetes as are those with none.36 Studies of nondiabetic subjects indicate that TCF7L2 diabetes-risk genotypes alter insulin secretion.37,42,46 The impact of this polymorphism on birth weight has not been studied.
In the present study, we hypothesized that fetal TCF7L2 type 2 diabetes–predisposing genotypes at rs7903146 and rs12255372 would be associated with reduced birth weight and that maternal genotypes would be associated with increased offspring birth weight through elevated maternal glucose. We investigated this hypothesis in >24,000 individuals from six population-based studies. In addition, we explored the role of these variants in diabetes-related intermediate traits, including beta-cell function, in >10,000 young (median age ⩽45 years), nondiabetic individuals from five studies.
Subjects and Methods
Subjects for Analysis of Fetal TCF7L2 Genotype with Birth Weight
To assess the association of fetal TCF7L2 genotypes with birth weight, we used subjects from six studies (table 1). All subjects were born at 36 full wk gestation or later and were of white European origin, either from the United Kingdom (Barry Caerphilly Growth Study [BCG],29 Exeter Family Study of Childhood Health [EFSOCH],47 North Cumbria Community Genetics Project [NCCGP],48 British 1958 Birth Cohort [1958BC],36,49 and Avon Longitudinal Study of Parents and Children [ALSPAC]50) or Finland (Northern Finland 1966 Birth Cohort [NFBC1966]).51–53 These studies have been described in depth elsewhere, but brief details are as follows. BCG is a longitudinal study of individuals born between 1972 and 1974 whose growth was monitored from birth to age 5 years. Data for analysis of the association of rs7903146 with birth weight were available for 571 subjects. EFSOCH is a prospective study of children, born between 2000 and 2004, and their parents from a geographically defined region of Exeter, United Kingdom. Data for 792 EFSOCH babies were available for the present analysis. Maternal fasting glucose, assessed at 26–28 wk gestation, was available for these subjects. Maternal smoking status was also assessed at that time. NCCGP is a community-based DNA-banking project. Data were available for 1,096 babies born between April 1999 and March 2002. Maternal smoking status was assessed during the first 12 wk of pregnancy. The 1958BC is a national cohort of U.K. subjects born during the same week in March 1958. The 1,779 subjects included in the present analysis are from the first group of subjects from whom DNA was extracted during 2003–2005. Information about smoking during pregnancy was reported by the mothers after the birth of the child and was coded as smoking continuing after the 4th mo of pregnancy (“yes/no”). ALSPAC is a prospective study, which recruited pregnant women from Bristol, United Kingdom, with expected delivery dates between April 1991 and December 1992. We were able to include 6,893 of the ALSPAC children in the present analysis. Maternal smoking status was assessed during the first 12 wk of gestation. NFBC1966 is a study of offspring born in the two northernmost provinces of Finland to mothers with expected dates of delivery in 1966. The 4,578 subjects included in the present analysis are from a subset of individuals who had anthropometric data taken and DNA extracted at age 31 years. Maternal smoking status was assessed throughout the pregnancy and was classified as “yes/no” after the 8th gestational wk.
Table 1. .
Clinical Characteristics of Subjects Included in the Analysis of the TCF7L2 rs7903146 Genotype with Birth Weight[Note]
| Characteristics by Study |
||||||
| Characteristic | BCG | EFSOCH | NCCGP | 1958BC | NFBC1966 | ALSPAC |
| Na (% male) | 571 (52.2) | 792 (52.9) | 1,096 (49.9) | 1,779 (49.5) | 4,578 (47.7) | 6,893 (51.2) |
| Mean (SD) birth weight, in g | 3,387 (496) | 3,504 (478) | 3,441 (485) | 3,351 (494) | 3,536 (490) | 3,489 (474) |
| Median (IQR) gestation, in wk | 40.0 (39.0–41.0) | 40.3 (39.3–41.1) | 40.0 (39.0-41.0) | 40.3 (39.4–41.3) | 40.0 (38.0–42.0) | 40.0 (39.0–41.0) |
| Median (IQR) maternal age, in years | NA | 31.0 (27.0–34.0) | 28.8 (24.0–32.8) | 27.0 (23.0–31.0) | 27.2 (17.4–36.9) | 29.0 (26.0–32.0) |
| Median (IQR) maternal prepregnancy BMIb | NA | 23.0 (21.1–25.7) | NA | 22.1 (20.4–24.5) | 22.7 (18.9–26.5) | 22.2 (20.5–24.4) |
| Primiparous births (%) | NA | 55.3 | 66.2 | 35.2 | 30.6 | 43.3 |
| Maternal smoking during pregnancy (%) | NA | 13.8 | 26.0 | 32.9 | 13.4 | 19.9 |
Note.— The clinical characteristics of offspring with maternal rs7903146 genotype data available (917, 1,133, and 6,294 individuals from EFSOCH, NCCGP, and ALSPAC, respectively) were very similar (data not shown). In some cases, offspring birth weight and maternal genotype were available, whereas offspring genotype was unavailable. Therefore, for EFSOCH and NCCGP, the number of subjects in the maternal genotype analysis was larger than that included in the fetal genotype analysis. NA=not available; IQR=interquartile range.
Includes white, singleton individuals, genotyped for rs7903146, with birth weight available, born at minimum gestation of 36 wk.
Calculated as weight in kilograms divided by the square of height in meters.
Only singleton pregnancies were included in the analyses. In all studies, birth weight was obtained from hospital records, and gestation was inferred from the last menstrual period or ultrasound scan. In most studies, data were available on parity (dichotomized as first child or subsequent), maternal prepregnancy BMI, and maternal smoking status (smoking or nonsmoking during pregnancy). All subjects involved gave their informed consent, and ethical approval was obtained from the local institutional review board for each study.
Subjects for Analysis of Maternal TCF7L2 Genotype with Offspring Birth Weight
To assess the association of maternal TCF7L2 genotypes with offspring birth weight, we used a total of 8,344 genotyped mothers from the three studies in which maternal DNA was available (917 from EFSOCH, 1,133 from NCCGP, and 6,294 from ALSPAC). Altogether, there were 6,044 mother-offspring pairs with both mother's and child’s rs7903146 genotype and offspring birth weight available (754 from EFSOCH, 1,024 from NCCGP, and 4,266 from ALSPAC). Maternal DNA was not available for BCG, 1958BC, or NFBC1966 subjects.
Subjects for Analysis of TCF7L2 Genotype with Diabetes-Related Intermediate Trait Data
We investigated possible associations of TCF7L2 genotypes with diabetes-related intermediate traits in the BCG, EFSOCH, 1958BC, NFBC1966, and ALSPAC studies. Since the BCG, 1958BC, and NFBC1966 studies are long-term follow-up cohorts, detailed quantitative trait data were available for the individuals as young adults, in addition to their birth and early-life data. In the case of ALSPAC, diabetes-related intermediate traits were measured in subsets of children at ages 7 and 8 years. For EFSOCH, we used data from the mothers and fathers. Again, analysis was restricted to individuals of white European origin. In addition, when information was available, subjects with diabetes, fasting plasma glucose >6 mmol/liter, or glycated hemoglobin (HbA1c) >6% were excluded. The clinical characteristics of these subjects are shown in table 2.
Table 2. .
Clinical Characteristics of Individuals Included in the Diabetes-Related Trait Analyses for the TCF7L2 rs7903146 Genotype[Note]
| Characteristics by Study |
|||||
| Characteristic | BCG | EFSOCH | 1958BC | NFBC1966 | ALSPAC |
| Na (% male) | 619 (52.8) | 1,810 (49.1) | 1,813 (49.7) | 4,486 (46.3) | 697 (53.8) |
| Median (IQRb) age, in years | 25.0 (24.5–25.6) | 31.0 (28.0–35.0) | 45.0 | 31.0 | 8.0 |
| Median (IQR) BMIc | 24.3 (22.0–27.3) | 24.8 (22.2–27.7)d | 26.5 (23.9–29.8) | 24.0 (19.2–28.8) | 16.2 (15.2–17.6) |
| Median (IQR) fasting plasma: | |||||
| Glucose, in mmol/liter | 4.6 (4.3–4.8) | 4.5 (4.2–4.8) | NA | 5.0 (4.5–5.5) | 4.9 (4.7–5.1) |
| Insulin, in pmol/liter | 39.6 (29.4–55.8) | 54.8 (40.4–79.0) | NA | 52.1 (29.2–75.0) | 33.3 (21.5–54.9) |
| HbA1c (IQR) [%] | NA | 5.1 (4.8–5.3) | 5.2 (5.0–5.5) | NA | 4.9e (4.7–5.1) |
Note.— NA=not available; IQR=interquartile range.
Includes white, unrelated individuals genotyped for rs7901346. When information was available, subjects with diabetes, HbA1c >6%, or fasting plasma glucose >6 mmol/liter were excluded.
IQR not applicable for 1958BC, NFBC1966, and ALSPAC, since participants were all studied at the same age.
Calculated as weight in kilograms divided by the square of height in meters.
Females were pregnant (28 wk gestation) at time of study, but BMI is the prepregnancy value.
HbA1c was measured in 1,280 subjects at age 7 years (50.6% male), whereas all other data in this column were gathered 1 year later. A total of 391 ALSPAC children were included at both times.
Oral glucose-tolerance test data were available for 619 BCG adults and 697 ALSPAC children. These consisted of plasma glucose and insulin measures taken after fasting and then repeated 30 min after administration of an oral glucose load. We calculated early insulin response to glucose54
 |
and insulin disposition index (insulinogenic index × homeostasis model assessment of insulin resistance [HOMA-IR]), where
 |
and42
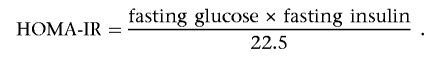 |
Whereas early insulin response is a measure of insulin secretion, disposition index is a corrected measure, accounting for an individual’s insulin resistance. Fasting-glucose and insulin data were also available for 1,810 EFSOCH parents and 4,486 NFBC1966 adults. Using the online HOMA Calculator version 2.2 (available from the Diabetes Trials Unit Web site) we obtained model-derived estimates of HOMA-IR for these subjects. HbA1c was available for 1,280 ALSPAC children, 1,442 EFSOCH parents, and 1,813 subjects from 1958BC. Finally, BMI was available for all subjects and for 5,817 ALSPAC mothers.
Genotyping and Quality Control
The rs7903146 and rs12255372 polymorphisms were genotyped in all cohorts. Genotyping was performed on the BCG, 1958BC, EFSOCH, NFBC1966, and ALSPAC samples by KBiosciences, with use of their own novel system of fluorescence-based competitive allele-specific PCR (KASPar). Details of assay design are available from the KBiosciences Web site. Genotyping of the NCCGP samples was performed in-house (Peninsula Medical School), with use of TaqMan SNP genotyping assay (Applied Biosystems) according to the manufacturer’s protocol. Additional EFSOCH samples (70 children and 104 parents) were genotyped in this way for rs7903146. These genotypes were added to the genotypes from KBiosciences, since the concordance for samples genotyped by both centers (n=1,706) was 98.9%.
The percentage of successful genotype calls was >90% in all groups, apart from NCCGP children (88% rs7903146; 89% rs12255372). The percentage of duplicate samples included for genotyping in each study group was 1% (1958BC), 3% (ALSPAC children), 6% (ALSPAC mothers and NFBC1966), 10% (BCG and EFSOCH), and 21% (NCCGP). Concordance between duplicate samples was ⩾99% in all groups except NFBC1966 (98.1% rs7903146) and ALSPAC children (98.4% rs7903146). No deviation from Hardy-Weinberg equilibrium was observed (all P>.05). The minor-allele frequency (MAF) of rs7903146 was 19.1% in NFBC1966 and had a range of 29.0%–30.4% in the U.K. samples. The MAF of rs12255372 was 16.8% in NFBC1966 and had a range of 28.4%–29.8% in the U.K. samples. These figures are consistent with those observed elsewhere in Finnish38 and U.K.36 subjects, and the difference is therefore likely to reflect underlying differences in population allele frequencies than differences due to our sampling or genotyping. The SNPs were in linkage disequilibrium in all study groups (r2 range 0.74–0.79 in U.K. samples; 0.69 in NFBC1966).
Statistical Analysis
We performed linear regression analysis within each study, with birth weight as the dependent and SNP genotype (coded as zero, one, or two risk alleles) as the independent variable and with sex and gestation as covariates. We performed additional analyses correcting also for parity, maternal smoking, maternal prepregnancy BMI, and maternal fasting plasma–glucose concentration, when these variables were available. All regression analyses were performed using Stata SE version 9 (StataCorp) or SPSS version 11.5. We looked for evidence of deviation from an additive genetic model in ALSPAC by comparing the full genotype model with the additive model, using the likelihood-ratio test. We found no evidence to reject the additive model (P>.05), so this was assumed for all regression analyses. Inverse variance meta-analysis (fixed effects) statistics and plots were generated using StatsDirect version 2.5.6. The I2 (inconsistency) statistic was used to estimate the proportion of variance attributable to between-study heterogeneity.55 By performing a meta-analysis of summary data from the separate studies, we were able to avoid the potential confounding effect of the difference between the NFBC1966 and U.K. allele frequencies. To investigate the relative contributions to birth weight of maternal and fetal genotypes, we generated standardized residuals from a regression of birth weight on sex and gestation within the EFSOCH, NCCGP, and ALSPAC studies. We combined these sex- and gestational age–corrected birth weight Z scores (mean=0; SD=1) into one data set. We then performed a regression analysis with birth-weight Z score as the dependent and both maternal and fetal genotypes as independent variables. Since there is a degree of colinearity between maternal and fetal genotype (r≈0.5), we performed stratified analyses to verify this result; within each of the three strata defined by maternal genotype, birth-weight Z score was regressed on fetal genotype, then the three regression coefficients were combined using a meta-analysis approach (fixed effects, inverse-variance method). The same analysis was performed for maternal genotype with data stratified by fetal genotype.
All diabetes-related trait variables, apart from fasting glucose, required log transformation to obtain a normal distribution before analysis. Linear regression analyses were performed for each trait against genotype (additive model). When appropriate, we included age, sex, and ln(BMI) as covariates.
To investigate the combined effects of maternal TCF7L2 rs7903146 and GCK rs1799884 genotype on offspring birth weight, we combined sex- and gestational age–corrected birth-weight Z scores from the 6,122 ALSPAC and 737 EFSOCH subjects for whom genotypes at both loci were available. We performed a regression analysis with Z score as the independent variable and “TCF7L2-GCK genotype” (values of zero, one, two, and three or four maternal risk alleles) as the dependent variable. We combined individuals with three or four risk alleles, because of small numbers in the final category. We looked for evidence of deviation from an additive genetic model in the separate analyses of birth-weight Z score versus TCF7L2 rs7903146 or GCK rs1799884 and, in the combined analysis, by comparing the full genotype model with the additive model with use of the likelihood-ratio test. We found no evidence to reject the additive model (all P>.4), so this was assumed for the combined analysis. The mean birth-weight Z score of offspring born to mothers with no risk alleles was compared with those born to mothers carrying three or four risk alleles, with use of an independent-samples t test.
Power calculations were performed using Lenth’s Java Applets for Power and Sample Size (available from the Russ Lenth’s Power and Sample Size Web site), with the assumption of an MAF of 30%, birth-weight SD of 480 g, and a two-tailed P<.05. Our sample of 15,709 subjects with available birth weight gave us 80% power to detect differences of 39 g in birth weight between homozygotes. In assessment of the effect of maternal genotype, our sample of 8,344 genotyped mothers gave us 80% power to detect differences of 53 g between homozygotes.
Results
Association of Fetal TCF7L2 Genotype with Birth Weight
Since SNPs rs7903146 and rs12255372 are highly correlated (r2≈0.75), the results obtained for each were similar. In this work, we focus on rs7903146, since that SNP is associated more strongly with type 2 diabetes.36,45 Detailed data for rs12255372 are available from the authors.
Meta-analysis of the association between birth weight (corrected for sex and gestation) and fetal rs7903146 genotype showed an 18-g (95% CI 7–29 g) increase in birth weight per T allele (P=.001) (fig. 1 and table 3). Heterogeneity between studies was low (I2=21.0%). The association observed with rs12255372 was similar (combined per-allele difference=19 g [95% CI 8–30 g]; P=.0009; I2=0%). The largest contribution to the overall effect size was made by ALSPAC, the largest individual study. In that study, fetal rs7903146 genotype was associated with increased birth weight (per–T allele increase 23 g [95% CI 7–39 g]; P=.004). Correction for other covariates of birth weight made little difference to the results (data available from the authors).
Figure 1. .
Meta-analysis of the association of birth weight (corrected for sex and gestation) with fetal rs7903146 genotype across six studies, arranged in order of increasing size. The effect size (in grams) and 95% CI per risk allele is presented. Combined per-allele difference=18 g (95% CI 7–29 g); P=.001; I2=21.0%.
Table 3. .
Analysis of Birth Weight by Fetal TCF7L2 rs7903146 Genotype[Note]
| CC |
CT |
TT |
P |
||||||||
| Study | Total N | Mean Birth Weighta (95% CI) | n | Mean Birth Weighta (95% CI) | n | Mean Birth Weighta (95% CI) | n | Per–T Allele Birth-Weighta Difference (SE)b | Uncorrected | Corrected for Sex and Gestation |
Corrected for Additional Covariatesc |
| BCG | 571 | 3,355 (3,301–3,409) | 281 | 3,411 (3,352–3,471) | 233 | 3,444 (3,324–3,564) | 57 | 49 (29) | .13 | .10 | NA |
| EFSOCH | 792 | 3,475 (3,432–3,517) | 399 | 3,523 (3,476–3,570) | 316 | 3,583 (3,487–3,679) | 77 | 52 (23) | .11 | .03 | .05 |
| NCCGP | 1,096 | 3,455 (3,419–3,491) | 542 | 3,412 (3,374–3,451) | 467 | 3,505 (3,416–3,596) | 87 | −4 (20) | .38 | .83 | NA |
| 1958BC | 1,605 | 3,359 (3,327–3,392) | 764 | 3,358 (3,324–3,392) | 700 | 3,365 (3,290–3,440) | 141 | 1 (18) | .76 | .96 | .87 |
| NFBC1966 | 4,578 | 3,534 (3,516–3,551) | 2,978 | 3,541 (3,516–3,566) | 1,444 | 3,540 (3,466–3,615) | 156 | 9 (12) | .66 | .47 | .52 |
| ALSPAC | 6,893 | 3,474 (3,460–3,489) | 3,372 | 3,499 (3,483–3,515) | 2,876 | 3,517 (3,483–3,550) | 645 | 23 (8) | .03 | .004 | .01 |
Note.— All weights are given in grams.
Corrected for sex and gestation.
Rounded to the nearest 1 g.
Corrected for sex, gestation, parity (first child/subsequent), maternal smoking (yes/no), and maternal prepregnancy BMI. NA=not available.
Association of Maternal TCF7L2 Genotype with Offspring Birth Weight
Meta-analysis of the association between birth weight (corrected for sex and gestation) and maternal rs7903146 genotype showed a 30-g (95% CI 15–45 g) increase in birth weight per T allele (P=2.8×10-5) (fig. 2 and table 4). Again, heterogeneity between studies was low (I2=14.3%). The association observed with rs12255372 was similar (combined per-allele difference=29 g [95% CI 14–43 g]; P=4.5×10-5; I2=0%). Again, the largest individual study, ALSPAC, made the largest contribution to the combined association. In ALSPAC, the maternal rs7903146 genotype was associated with a per–T allele increase in offspring birth weight of 35 g (95% CI 17–53 g; P=4×10-5). Correction for other covariates of birth weight again made little difference to the meta-analysis result (data available from the authors).
Figure 2. .
Meta-analysis of the association of birth weight (corrected for sex and gestation) with maternal rs7903146 genotype across three studies, arranged in order of increasing size. The effect size (in grams) and 95% CI per risk allele is presented. Combined per-allele difference=30 g (95% CI 15–45 g); P=2.8×10-5. I2=14.3%.
Table 4. .
Analysis of Offspring Birth Weight by Maternal TCF7L2 rs7903146 Genotype[Note]
| CC |
CT |
TT |
P |
||||||||
| Study | Total N | Mean Birth Weighta (95% CI) | n | Mean Birth Weighta (95% CI) | n | Mean Birth Weighta (95% CI) | n | Per–T Allele Birth-Weighta Difference (SE)b |
Uncorrected | Corrected for Sex and Gestation |
Corrected for Additional Covariatesc |
| EFSOCH | 917 | 3,471 (3,432–3,509) | 460 | 3,488 (3,446–3,530) | 384 | 3,549 (3,452–3,646) | 73 | 30 (22) | .25 | .18 | .34 |
| NCCGP | 1,133 | 3,439 (3,403–3,475) | 563 | 3,448 (3,409–3,487) | 470 | 3,434 (3,349–3,519) | 100 | 2 (20) | .39 | .91 | NAd |
| ALSPAC | 6,294 | 3,443 (3,426–3,460) | 3,051 | 3,491 (3,473–3,509) | 2,702 | 3,494 (3,453–3,534) | 541 | 35 (9) | .0002 | .00004 | .00002 |
Note.— All weights are given in grams.
Corrected for sex and gestation.
Rounded to the nearest 1 g.
Corrected for sex, gestation, parity (first child/subsequent), maternal smoking (yes/no), and maternal prepregnancy BMI.
NA=not available.
Data on maternal diabetes status were variable and incomplete across the cohorts, so no subject was excluded from the main analysis on that basis. Data on diabetes status were available for 96% of the ALSPAC mothers. These mothers had been asked whether they had ever had diabetes. Removal of the 64 mothers who had answered “yes” from the analysis of offspring birth weight versus maternal rs7903146 genotype, sex, and gestation did not alter the results.
Adjustment of Fetal and Maternal Genotype Effects for One Another
Maternal and fetal genotypes are 50% correlated. To establish whether maternal or fetal genotypes were driving the association, we used 6,044 mother-offspring pairs for whom both maternal and fetal rs7903146 genotypes were available. Regression of birth-weight Z score on fetal and maternal genotype revealed an association of maternal (per–T allele increase 0.07 [95% CI 0.02–0.11]; P=.003) but not fetal (per–T allele increase 0.02 [95% CI −0.02 to 0.07]; P=.29) genotype with birth weight. Stratified analyses confirmed this result. With use of the SD of birth weight (440 g), corrected for sex and gestation, in ALSPAC children with genotyped mothers, the effect size of 0.07 (95% CI 0.02–0.11) Z scores is equivalent to an increase of 31 g (9–48 g) per maternal T allele. The results of this analysis for rs12255372 were similar: there was an association of maternal (per–T allele increase 0.05 [95% CI 0.01–0.10]; P=.025) but not fetal (per–T allele increase 0.03 [95% CI −0.01 to 0.08]; P=.16) genotype with birth weight.
Combined Analysis of Maternal TCF7L2 and GCK Genotype with Offspring Birth Weight
A previous study showed that maternal genotypes of the glucokinase (GCK) variant rs1799884 (GCK-30) are reproducibly associated with offspring birth weight.33,34 To assess the combined effect of maternal TCF7L2 rs7903146 and GCK rs1799884 variants on birth weight, we used the 737 EFSOCH and 6,122 ALSPAC subjects for whom maternal genotype was available at both loci. In this data set, maternal GCK rs1799884 genotype was associated with a per–A allele increase in offspring birth-weight Z score of 0.06 (95% CI 0.02–0.10; P=.006; ∼27 g [95% CI 8–46 g]). This is similar to the effect size observed in the present study for TCF7L2 rs7903146. We analyzed sex- and gestational age–corrected birth-weight Z score against the number of TCF7L2 rs7903146 and GCK rs1799884 maternal risk alleles (zero, one, two, and three or four) in 6,859 individuals (fig. 3). We confirmed that it was valid to combine data from the two polymorphisms in this way, because there was no deviation from an additive model (P=.53). The addition of each maternal allele (or alleles) was associated with an increase in Z score of 0.08 (95% CI 0.05–0.10; P=2×10-7). This equates to a per-allele increase of 35 g (95% CI 22–44 g). Offspring born to the 4% of mothers with three or four risk alleles had a 0.27 (95% CI 0.14–0.39) higher birth-weight Z score than those born to the 32% of mothers with no risk alleles (P=3×10-5), which equates to a difference of 119 g (95% CI 62–172 g).
Figure 3. .
Bar graph showing sex- and gestational age–corrected birth-weight Z score plotted against the number of maternal risk alleles for TCF7L2 rs7903146 and GCK rs1799884. Error bars show 95% CIs.
Association of TCF7L2 Genotypes with Diabetes-Related Intermediate Traits
To help establish the mechanism through which maternal TCF7L2 genotype may be altering birth weight, we analyzed a number of type 2 diabetes–related continuous traits in 10,314 subjects. The results of the intermediate trait analysis for rs7903146 are shown in table 5. The type 2 diabetes–risk allele (T) was associated with a reduction in insulin secretion in the BCG and ALSPAC studies, as measured by early-insulin response (combined per–T allele decrease [logged data] −0.067 [95% CI −0.118 to −0.016]; P=.02; n=1,235) and insulin-disposition index (combined per–T allele decrease [logged data] −0.192 [95% CI −0.288 to −0.096]; P=1×10-4; n=1,184). An association was also seen with increased fasting plasma glucose across the BCG, EFSOCH, ALSPAC, and NFBC1966 studies (combined per–T allele increase 0.020 mmol/liter [95% CI 0.005–0.035 mmol/liter]; P=.01; n=7,612; I2=9.2%). After a Bonferroni correction for the six studied traits was applied, the early-insulin response and fasting-glucose associations did not remain at P<.05. However, the correlation between these traits means that this is a conservative correction, and there is insufficient statistical power to rule out the associations observed. Whereas the rs7903146 T allele was associated with reduced fasting insulin and HOMA-IR in the BCG cohort (P<.05), this association was not seen in the two larger cohorts (P>.1). No association was observed between the TCF7L2 genotype and HbA1c (P>.05). There was no evidence of association of TCF7L2 genotype with BMI in any cohort (all P>.05) (data not shown). The results of the analyses for rs12255372 were similar (data available from the authors).
Table 5. .
Type 2 Diabetes–Related Intermediate Trait Analyses for the TCF7L2 rs7903146 Genotype[Note]
| Uncorrected Mean (95% CI) |
P |
||||||
| Trait (units) and Study | N | CC | CT | TT | Per–T Allele Trait Difference (SE) |
Uncorrected | Correcteda |
| Fasting plasma glucose (mmol/liter): | |||||||
| BCG | 619 | 4.61 (4.57–4.65) | 4.61 (4.56–4.66) | 4.56 (4.47–4.66) | −.016 (.023) | .48 | .72 |
| ALSPAC | 697 | 4.92 (4.88–4.96) | 4.92 (4.90–4.95) | 4.96 (4.88–5.03) | .012 (.019) | .52 | .53 |
| EFSOCH | 1,810 | 4.49 (4.46–4.52) | 4.55 (4.52–4.58) | 4.52 (4.44–4.59) | .031 (.016) | .06 | .08 |
| NFBC1966 | 4,486 |
4.98 (4.96–4.99) | 4.99 (4.97–4.99) | 5.03 (4.97–5.09) | .025 (.011) | .03 | .06 |
| Combined | 7,612 | .020 (.008) | .01 | .03 | |||
| Fasting plasma insulin (pmol/liter): | |||||||
| BCG | 619 | 42.8 (40.4–45.4) | 40.9 (38.3–43.5) | 35.7 (31.5–40.4) | −.076 (.031) | .02 | .002 |
| ALSPAC | 697 | 37.2 (34.5–40.0) | 34.8 (33.5–37.6) | 35.8 (31.0–41.4) | −.052 (.038) | .17 | .26 |
| EFSOCH | 1,780 | 56.1 (54.2–58.1) | 59.7 (57.5–62.0) | 55.3 (50.8–60.2) | .023 (.020) | .25 | .16 |
| NFBC1966 | 4,453 |
53.6 (52.9–54.3) | 53.7 (52.7–54.7) | 55.7 (52.5–59.0) | .007 (.010) | .45 | .99 |
| Combined | 7,549 | .0009 (.008) | .92 | .75 | |||
| HOMA-IR: | |||||||
| BCG | 619 | .89 (.84–1.06) | .85 (.80–.91) | .75 (.66–.84) | −.076 (.031) | .01 | .002 |
| EFSOCH | 1,780 | 1.15 (1.11–1.19) | 1.23 (1.18–1.23) | 1.14 (1.05–1.24) | .025 (.019) | .21 | .14 |
| NFBC1966 | 4,452 |
1.00 (.99–1.01) | 1.00 (.99–1.01) | 1.02 (.99–1.04) | .004 (.004) | .41 | .95 |
| Combined | 6,851 | .004 (.004) | .35 | 1.00 | |||
| Early insulin response (pmol/mmol/liter): | |||||||
| BCG | 604 | 42.8 (40.0–45.9) | 41.6 (38.6–44.8) | 34.8 (30.1–40.3) | −.078 (.036) | .03 | .02 |
| ALSPAC | 631 |
33.0 (30.4–35.8) | 30.4 (28.2–32..8) | 31.6 (27.7–36.1) | −.055 (.039) | .16 | .27 |
| Combined | 1,235 | −.067 (.026) | .01 | .02 | |||
| Disposition index (pmol2/liter2): | |||||||
| BCG | 593 | 1,151.7 (1,011.3–1,310.3) | 1,028.6 (894.3–1,182.0) | 709.8 (541.9–929.8) | −.199 (.068) | .004 | .001 |
| ALSPAC | 591 |
849.8 (737.3–979.5) | 648.1 (561.7–747.7) | 639.7 (477.7–856.6) | −.184 (.072) | .01 | .03 |
| Combined | 1,184 | −.192 (.049) | .0001 | .0001 | |||
| HbA1c (%): | |||||||
| ALSPAC | 1,280 | 4.90 (4.87–4.92) | 4.90 (4.87–4.92) | 4.87 (4.81–4.92) | −.002 (.002) | .41 | .14 |
| EFSOCH | 1,442 | 5.06 (5.03–5.08) | 5.07 (5.04–5.10) | 5.04 (4.97–5.10) | .0001 (.003) | .97 | .77 |
| 1958BC | 1,813 |
5.20 (5.18–5.22) | 5.21 (5.19–5.24) | 5.25 (5.19–5.31) | .004 (.003) | .10 | .07 |
| Combined | 4,535 | −.0001 (.001) | .95 | .35 | |||
Note.— All variables apart from fasting glucose were log-transformed before analysis. The per–T allele trait difference is presented as the logged value, but means and 95% CIs were back-transformed. The combined values for each trait are from fixed-effects inverse-variance meta-analysis. There was a large amount of heterogeneity among studies in the fasting insulin and HOMA-IR analyses (I2=68% and 75%, respectively).
Corrected for age, sex, and ln(BMI), except in ALSPAC, 1958BC, and NFBC1966, which were corrected for sex and BMI only, since subjects do not differ in age.
Discussion
Using a total of 24,053 subjects from six studies, we have shown that TCF7L2 is the first type 2 diabetes gene to be reproducibly associated with altered birth weight. Each maternal copy of the T allele at rs7903146 increased offspring birth weight by 30 g (95% CI 15–45 g), and our data suggest that the most likely mechanism is through reduced maternal-insulin secretion resulting in maternal hyperglycemia and increased insulin-mediated fetal growth.
The understanding of the regulation of birth weight is important, not only because of direct effects of high or low birth weight on perinatal mortality and morbidity, but also because of the association of altered birth weight with altered adult phenotypes. It has been proposed under the fetal-insulin hypothesis that diabetes-susceptibility alleles might reduce birth weight by reducing fetal insulin secretion or action.14 Our results do not support this hypothesis for TCF7L2, since we found an increase and not a reduction in birth weight with fetal inheritance of the type 2 diabetes risk allele. Our analysis of mother-child pairs suggests that the increase seen in the offspring is not a direct effect of the fetal risk allele but, rather, is a reflection of its presence in the mother.
There was clear evidence in support of the hypothesis that the presence of a maternal type 2 diabetes–susceptibility allele would result in increased offspring birth weight. Maternal TCF7L2 risk genotypes increase birth weight by 30 g (95% CI 15–45 g) per risk allele. This result was seen in a meta-analysis of three U.K. studies that included 8,344 subjects and was independent of any fetal genotype effect.
Studies that have analyzed birth weight in relation to the established type 2 diabetes gene variants—E23K in KCNJ11 (MIM 600937) and Pro12Ala in PPARG (MIM 601487)—have found no evidence of association,56,57 although, in the first of these studies, maternal genotype data were not reported. These variants may genuinely have no effect on birth weight. However, it is possible that, with the smaller type 2 diabetes risk conferred by these variants (odds ratio ∼1.2) compared with TCF7L2 (odds ratio ∼1.4), any effect on birth weight via alterations in insulin secretion and action would be concordantly smaller and would require a sample size of >10,000 individuals for detection. Other studies have investigated the role of common variation in genes that represent good biological candidates for diabetes and related traits in relation to birth weight.58 Positive associations with both diabetes-related traits and birth weight have been shown for the insulin gene (INS [MIM 176730]) variable number tandem repeats (VNTR) locus,59–61 a microsatellite polymorphism in the insulin-like growth factor 1 gene (IGF1 [MIM 147440]),27,62 and a variant in the H19 gene (MIM 103280),63 but the former two have not been consistently replicated,26,28–32,64 and the latter is currently the only study of this gene in relation to common birth-weight variation. Taken together, these results highlight the need for large sample sizes and replication in the study of genetic associations between diabetes genes and fetal growth.
We previously studied the −30G→A (rs1799884) polymorphism in the glucokinase gene in two of the studies with maternal DNA available, including the largest individual study, ALSPAC.33,34 Presence of the A allele confers a 0.06-mmol/liter increase in fasting glucose (95% CI 0.04–0.09 mmol/liter) across all ages in the normal population and, when carried by the mother, is associated with an increase in offspring birth weight of 32 g (95% CI 11–53 g).34 Combining information from these two confirmed common variants, we have shown that the 4% of offspring born to mothers carrying three or four risk alleles were 119 g (95% CI 62–172 g) heavier than the 32% born to mothers with no risk alleles. This effect is similar to the effect of mothers smoking 4–5 cigarettes per day in the third trimester of pregnancy.65,66 It confirms that common genetic variation can have a substantial effect on birth weight.
To understand the mechanism that leads to the increase in offspring birth weight, we studied >10,000 subjects, which represents the largest study to date of the effect of TCF7L2 genotypes on continuous traits related to type 2 diabetes. We found that the T allele at rs7903146 is associated with reduced insulin secretion when correcting for insulin resistance (as measured by disposition index). This supports the findings of previous studies of nondiabetic subjects.37,40,42,46 We also found a weaker association with increased fasting glucose (per–T allele increase 0.020 [95% CI 0.005–0.035] mmol/liter). Results of analyses of glucose concentrations from previous studies show either an increase in fasting glucose or no change in glucose.37,42,44,46 Our data suggest that the increase in birth weight is likely to result from the risk allele reducing maternal insulin secretion, which results in an increase in the pregnant maternal glycemia to which the fetus is exposed. However, further studies in very large cohorts of pregnant women will be needed to confirm this, since most of our data come from young adults, with only 921 females pregnant at the time of study.
To conclude, we have shown that maternal TCF7L2 type 2 diabetes–risk genotypes at rs7903146 are associated with increased offspring birth weight, probably through impaired maternal insulin secretion. Together with the common GCK rs1799884 variant, TFC7L2 rs7903146 is associated with differences in normal birth weight that are comparable to those conferred by smoking, suggesting an important role for genes in this complex trait.
Acknowledgments
R.M.F. holds a Diabetes UK research studentship. A.T.H. is a Wellcome Trust Research Leave Fellow, and M.N.W. is a Vandervell Foundation Research Fellow. The majority of the work was supported by Medical Research Council grant G0500070/73468. We are extremely grateful to all the families who took part in this study and to the midwives who helped recruit them. We are also grateful to the various study teams, which include interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, nurses, receptionists, and managers. The U.K. Medical Research Council, the Wellcome Trust, and the University of Bristol provide core support for ALSPAC. The NFBC1966 study is supported by Wellcome Trust grant GR069224MA and the Academy of Finland. We acknowledge use of DNA from the 1958BC collection, funded by Medical Research Council grant G0000934 and Wellcome Trust grant 068545/Z/02. E.H. is a Department of Health (United Kingdom) Public Health Career Scientist, and research at the Institute of Child Health and Great Ormond Street Hospital for Children National Health Service (NHS) Trust benefits from research and development funding received from the NHS executive.
Web Resources
The URLs for data presented herein are as follows:
- Diabetes Trials Unit, http://www.dtu.ox.ac.uk/
- KBiosciences, http://www.kbioscience.co.uk/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for type 2 diabetes, GCK, TCF7L2, KCNJ11, PPARG, INS, IGF1, and H19)
- Russ Lenth’s Power and Sample Size, http://www.stat.uiowa.edu/~rlenth/Power/
References
- 1.van Baal CG, Boomsma DI (1998) Etiology of individual differences in birth weight of twins as a function of maternal smoking during pregnancy. Twin Res 1:123–130 10.1375/136905298320566258 [DOI] [PubMed] [Google Scholar]
- 2.Baird J, Osmond C, MacGregor A, Snieder H, Hales CN, Phillips DI (2001) Testing the fetal origins hypothesis in twins: the Birmingham twin study. Diabetologia 44:33–39 10.1007/s001250051577 [DOI] [PubMed] [Google Scholar]
- 3.Clausson B, Lichtenstein P, Cnattingius S (2000) Genetic influence on birthweight and gestational length determined by studies in offspring of twins. BJOG 107:375–381 10.1111/j.1471-0528.2000.tb13234.x [DOI] [PubMed] [Google Scholar]
- 4.Magnus P, Gjessing HK, Skrondal A, Skjaerven R (2001) Paternal contribution to birth weight. J Epidemiol Community Health 55:873–877 10.1136/jech.55.12.873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farmer G, Russell G, Hamilton-Nicol DR, Ogenbede HO, Ross IS, Pearson DW, Thom H, Kerridge DF, Sutherland HW (1988) The influence of maternal glucose metabolism on fetal growth, development and morbidity in 917 singleton pregnancies in nondiabetic women. Diabetologia 31:134–141 10.1007/BF00276845 [DOI] [PubMed] [Google Scholar]
- 6.Breschi MC, Seghieri G, Bartolomei G, Gironi A, Baldi S, Ferrannini E (1993) Relation of birthweight to maternal plasma glucose and insulin concentrations during normal pregnancy. Diabetologia 36:1315–1321 10.1007/BF00400812 [DOI] [PubMed] [Google Scholar]
- 7.Kieffer EC, Tabaei BP, Carman WJ, Nolan GH, Guzman JR, Herman WH (2006) The influence of maternal weight and glucose tolerance on infant birthweight in Latino mother-infant pairs. Am J Public Health 96:2201–2208 10.2105/AJPH.2005.065953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindley AA, Gray RH, Herman AA, Becker S (2000) Maternal cigarette smoking during pregnancy and infant ponderal index at birth in the Swedish Medical Birth Register, 1991–1992. Am J Public Health 90:420–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindsay RS, Kobes S, Knowler WC, Hanson RL (2002) Genome-wide linkage analysis assessing parent-of-origin effects in the inheritance of birth weight. Hum Genet 110:503–509 10.1007/s00439-002-0718-2 [DOI] [PubMed] [Google Scholar]
- 10.Arya R, Demerath E, Jenkinson CP, Goring HH, Puppala S, Farook V, Fowler S, Schneider J, Granato R, Resendez RG, et al (2006) A quantitative trait locus (QTL) on chromosome 6q influences birth weight in two independent family studies. Hum Mol Genet 15:1569–1579 10.1093/hmg/ddl076 [DOI] [PubMed] [Google Scholar]
- 11.Fradin D, Heath S, Lepercq J, Lathrop M, Bougneres P (2006) Identification of distinct quantitative trait loci affecting length or weight variability at birth in humans. J Clin Endocrinol Metab 91:4164–4170 10.1210/jc.2006-0529 [DOI] [PubMed] [Google Scholar]
- 12.Hales CN, Barker DJ, Clark PM, Cox LJ, Fall C, Osmond C, Winter PD (1991) Fetal and infant growth and impaired glucose tolerance at age 64. BMJ 303:1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM (1993) Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia 36:62–67 10.1007/BF00399095 [DOI] [PubMed] [Google Scholar]
- 14.Hattersley AT, Tooke JE (1999) The fetal insulin hypothesis: an alternative explanation of the association of low birthweight with diabetes and vascular disease. Lancet 353:1789–1792 10.1016/S0140-6736(98)07546-1 [DOI] [PubMed] [Google Scholar]
- 15.Lindsay RS, Dabelea D, Roumain J, Hanson RL, Bennett PH, Knowler WC (2000) Type 2 diabetes and low birth weight: the role of paternal inheritance in the association of low birth weight and diabetes. Diabetes 49:445–449 10.2337/diabetes.49.3.445 [DOI] [PubMed] [Google Scholar]
- 16.Hypponen E, Smith GD, Power C (2003) Parental diabetes and birth weight of offspring: intergenerational cohort study. BMJ 326:19–20 10.1136/bmj.326.7379.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davey Smith G, Sterne JA, Tynelius P, Rasmussen F (2004) Birth characteristics of offspring and parental diabetes: evidence for the fetal insulin hypothesis. J Epidemiol Community Health 58:126–128 10.1136/jech.58.2.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wannamethee SG, Lawlor DA, Whincup PH, Walker M, Ebrahim S, Davey-Smith G (2004) Birthweight of offspring and paternal insulin resistance and paternal diabetes in late adulthood: cross sectional survey. Diabetologia 47:12–18 10.1007/s00125-003-1270-x [DOI] [PubMed] [Google Scholar]
- 19.Hattersley AT, Beards F, Ballantyne E, Appleton M, Harvey R, Ellard S (1998) Mutations in the glucokinase gene of the fetus result in reduced birth weight. Nat Genet 19:268–270 10.1038/953 [DOI] [PubMed] [Google Scholar]
- 20.Edghill EL, Bingham C, Slingerland AS, Minton JA, Noordam C, Ellard S, Hattersley AT (2006) Hepatocyte nuclear factor-1 beta mutations cause neonatal diabetes and intrauterine growth retardation: support for a critical role of HNF-1β in human pancreatic development. Diabet Med 23:1301–1306 10.1111/j.1464-5491.2006.01999.x [DOI] [PubMed] [Google Scholar]
- 21.Wright NM, Metzger DL, Borowitz SM, Clarke WL (1993) Permanent neonatal diabetes mellitus and pancreatic exocrine insufficiency resulting from congenital pancreatic agenesis. Am J Dis Child 147:607–609 [DOI] [PubMed] [Google Scholar]
- 22.Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF (1997) Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet 15:106–110 10.1038/ng0197-106 [DOI] [PubMed] [Google Scholar]
- 23.Gloyn AL, Pearson ER, Antcliff JF, Proks P, Bruining GJ, Slingerland AS, Howard N, Srinivasan S, Silva JM, Molnes J, et al (2004) Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med 350:1838–1849 10.1056/NEJMoa032922 [DOI] [PubMed] [Google Scholar]
- 24.Slingerland AS, Hattersley AT (2006) Activating mutations in the gene encoding Kir6.2 alter fetal and postnatal growth and also cause neonatal diabetes. J Clin Endocrinol Metab 91:2782–2788 10.1210/jc.2006-0201 [DOI] [PubMed] [Google Scholar]
- 25.Pearson ER, Boj SF, Steele AM, Barrett T, Stals K, Shield JP, Ellard S, Ferrer J, Hattersley AT (2007) Macrosomia and hyperinsulinaemic hypoglycaemia in patients with heterozygous mutations in the HNF4A gene. PLOS Med 4:e118 10.1371/journal.pmed.0040118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindsay RS, Hanson RL, Wiedrich C, Knowler WC, Bennett PH, Baier LJ (2003) The insulin gene variable number tandem repeat class I/III polymorphism is in linkage disequilibrium with birth weight but not type 2 diabetes in the Pima population. Diabetes 52:187–193 10.2337/diabetes.52.1.187 [DOI] [PubMed] [Google Scholar]
- 27.Vaessen N, Janssen JA, Heutink P, Hofman A, Lamberts SW, Oostra BA, Pols HA, van Duijn CM (2002) Association between genetic variation in the gene for insulin-like growth factor-I and low birthweight. Lancet 359:1036–1037 10.1016/S0140-6736(02)08067-4 [DOI] [PubMed] [Google Scholar]
- 28.Day IN, King TH, Chen XH, Voropanov AM, Ye S, Syddall HE, Sayer AA, Cooper C, Barker DJ, Phillips DI (2002) Insulin-like growth factor-I genotype and birthweight. Lancet 360:945 (author reply 945–946) 10.1016/S0140-6736(02)11044-0 [DOI] [PubMed] [Google Scholar]
- 29.Frayling TM, Hattersley AT, McCarthy A, Holly J, Mitchell SM, Gloyn AL, Owen K, Davies D, Davey Smith G, Ben-Shlomo Y (2002) A putative functional polymorphism in the IGF-I gene: association studies with type 2 diabetes, adult height, glucose tolerance, and fetal growth in UK populations. Diabetes 51:2313–2316 10.2337/diabetes.51.7.2313 [DOI] [PubMed] [Google Scholar]
- 30.Mitchell SM, Hattersley AT, Knight B, Turner T, Metcalf BS, Voss LD, Davies D, McCarthy A, Wilkin TJ, Davey Smith G, et al (2004) Lack of support for a role of the insulin gene variable number of tandem repeats minisatellite (INS-VNTR) locus in fetal growth or type 2 diabetes-related intermediate traits in United Kingdom populations. J Clin Endocrinol Metab 89:310–317 10.1210/jc.2003-030605 [DOI] [PubMed] [Google Scholar]
- 31.Hansen SK, Gjesing AP, Rasmussen SK, Glumer C, Urhammer SA, Andersen G, Rose CS, Drivsholm T, Torekov SK, Jensen DP, et al (2004) Large-scale studies of the HphI insulin gene variable-number-of-tandem-repeats polymorphism in relation to type 2 diabetes mellitus and insulin release. Diabetologia 47:1079–1087 10.1007/s00125-004-1418-3 [DOI] [PubMed] [Google Scholar]
- 32.Bennett AJ, Sovio U, Ruokonen A, Martikainen H, Pouta A, Taponen S, Hartikainen AL, King VJ, Elliott P, Jarvelin MR, et al (2004) Variation at the insulin gene VNTR (variable number tandem repeat) polymorphism and early growth: studies in a large Finnish birth cohort. Diabetes 53:2126–2131 10.2337/diabetes.53.8.2126 [DOI] [PubMed] [Google Scholar]
- 33.Weedon MN, Frayling TM, Shields B, Knight B, Turner T, Metcalf BS, Voss L, Wilkin TJ, McCarthy A, Ben-Shlomo Y, et al (2005) Genetic regulation of birth weight and fasting glucose by a common polymorphism in the islet cell promoter of the glucokinase gene. Diabetes 54:576–581 10.2337/diabetes.54.2.576 [DOI] [PubMed] [Google Scholar]
- 34.Weedon MN, Clark VJ, Qian Y, Ben-Shlomo Y, Timpson N, Ebrahim S, Lawlor DA, Pembrey ME, Ring S, Wilkin TJ, et al (2006) A common haplotype of the glucokinase gene alters fasting glucose and birth weight: association in six studies and population-genetics analyses. Am J Hum Genet 79:991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al (2006) Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38:320–323 10.1038/ng1732 [DOI] [PubMed] [Google Scholar]
- 36.Groves CJ, Zeggini E, Minton J, Frayling TM, Weedon MN, Rayner NW, Hitman GA, Walker M, Wiltshire S, Hattersley AT, et al (2006) Association analysis of 6,736 U.K. subjects provides replication and confirms TCF7L2 as a type 2 diabetes susceptibility gene with a substantial effect on individual risk. Diabetes 55:2640–2644 10.2337/db06-0355 [DOI] [PubMed] [Google Scholar]
- 37.Damcott CM, Pollin TI, Reinhart LJ, Ott SH, Shen H, Silver KD, Mitchell BD, Shuldiner AR (2006) Polymorphisms in the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in the Amish: replication and evidence for a role in both insulin secretion and insulin resistance. Diabetes 55:2654–2659 10.2337/db06-0338 [DOI] [PubMed] [Google Scholar]
- 38.Scott LJ, Bonnycastle LL, Willer CJ, Sprau AG, Jackson AU, Narisu N, Duren WL, Chines PS, Stringham HM, Erdos MR, et al (2006) Association of transcription factor 7-like 2 (TCF7L2) variants with type 2 diabetes in a Finnish sample. Diabetes 55:2649–2653 10.2337/db06-0341 [DOI] [PubMed] [Google Scholar]
- 39.Cauchi S, Meyre D, Dina C, Choquet H, Samson C, Gallina S, Balkau B, Charpentier G, Pattou F, Stetsyuk V, et al (2006) Transcription factor TCF7L2 genetic study in the French population: expression in human beta-cells and adipose tissue and strong association with type 2 diabetes. Diabetes 55:2903–2908 10.2337/db06-0474 [DOI] [PubMed] [Google Scholar]
- 40.Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D (2006) TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 355:241–250 10.1056/NEJMoa062418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C, Qi L, Hunter DJ, Meigs JB, Manson JE, van Dam RM, Hu FB (2006) Variant of transcription factor 7-like 2 (TCF7L2) gene and the risk of type 2 diabetes in large cohorts of U.S. women and men. Diabetes 55:2645–2648 10.2337/db06-0643 [DOI] [PubMed] [Google Scholar]
- 42.Saxena R, Gianniny L, Burtt NP, Lyssenko V, Giuducci C, Sjogren M, Florez JC, Almgren P, Isomaa B, Orho-Melander M, et al (2006) Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes 55:2890–2895 10.2337/db06-0381 [DOI] [PubMed] [Google Scholar]
- 43.van Vliet-Ostaptchouk JV, Shiri-Sverdlov R, Zhernakova A, Strengman E, van Haeften TW, Hofker MH, Wijmenga C (2007) Association of variants of transcription factor 7-like 2 (TCF7L2) with susceptibility to type 2 diabetes in the Dutch Breda cohort. Diabetologia 50:59–62 10.1007/s00125-006-0477-z [DOI] [PubMed] [Google Scholar]
- 44.Chandak GR, Janipalli CS, Bhaskar S, Kulkarni SR, Mohankrishna P, Hattersley AT, Frayling TM, Yajnik CS (2007) Common variants in the TCF7L2 gene are strongly associated with type 2 diabetes mellitus in the Indian population. Diabetologia 50:63–67 10.1007/s00125-006-0502-2 [DOI] [PubMed] [Google Scholar]
- 45.Helgason A, Palsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, Adeyemo A, Chen Y, Chen G, Reynisdottir I, et al (2007) Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 39:218–225 10.1038/ng1960 [DOI] [PubMed] [Google Scholar]
- 46.Munoz J, Lok KH, Gower BA, Fernandez JR, Hunter GR, Lara-Castro C, De Luca M, Garvey WT (2006) Polymorphism in the transcription factor 7-like 2 (TCF7L2) gene is associated with reduced insulin secretion in nondiabetic women. Diabetes 55:3630–3634 10.2337/db06-0574 [DOI] [PubMed] [Google Scholar]
- 47.Knight B, Shields BM, Hattersley AT (2006) The Exeter Family Study of Childhood Health (EFSOCH): study protocol and methodology. Paediatr Perinat Epidemiol 20:172–179 10.1111/j.1365-3016.2006.00701.x [DOI] [PubMed] [Google Scholar]
- 48.Chase DS, Tawn EJ, Parker L, Jonas P, Parker CO, Burn J (1998) The North Cumbria Community Genetics Project. J Med Genet 35:413–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Power C, Elliott J (2006) Cohort profile: 1958 British Birth Cohort (National Child Development Study). Int J Epidemiol 35:34–41 10.1093/ije/dyi183 [DOI] [PubMed] [Google Scholar]
- 50.Golding J, Pembrey M, Jones R (2001) ALSPAC—the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr Perinat Epidemiol 15:74–87 10.1046/j.1365-3016.2001.00325.x [DOI] [PubMed] [Google Scholar]
- 51.Rantakallio P (1988) The longitudinal study of the northern Finland birth cohort of 1966. Paediatr Perinat Epidemiol 2:59–88 [DOI] [PubMed] [Google Scholar]
- 52.Jarvelin MR, Sovio U, King V, Lauren L, Xu B, McCarthy MI, Hartikainen AL, Laitinen J, Zitting P, Rantakallio P, et al (2004) Early life factors and blood pressure at age 31 years in the 1966 Northern Finland Birth Cohort. Hypertension 44:838–846 10.1161/01.HYP.0000148304.33869.ee [DOI] [PubMed] [Google Scholar]
- 53.Bennett A, Sovio U, Ruokonen A, Martikainen H, Pouta A, Taponen S, Hartikainen AL, Franks S, Peltonen L, Elliott P, et al (2005) No association between insulin gene variation and adult metabolic phenotypes in a large Finnish birth cohort. Diabetologia 48:886–891 10.1007/s00125-005-1737-z [DOI] [PubMed] [Google Scholar]
- 54.Wareham NJ, Phillips DI, Byrne CD, Hales CN (1995) The 30 minute insulin incremental response in an oral glucose tolerance test as a measure of insulin secretion. Diabet Med 12:931 [DOI] [PubMed] [Google Scholar]
- 55.Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327:557–560 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weedon MN, Gloyn AL, Frayling TM, Hattersley AT, Davey Smith G, Ben-Shlomo Y (2003) Quantitative traits associated with the type 2 diabetes susceptibility allele in Kir6.2. Diabetologia 46:1021–1023 10.1007/s00125-003-1135-3 [DOI] [PubMed] [Google Scholar]
- 57.Pfab T, Poralla C, Richter CM, Godes M, Slowinski T, Priem F, Halle H, Hocher B (2006) Fetal and maternal peroxisome proliferator-activated receptor γ2 Pro12Ala does not influence birth weight. Obesity (Silver Spring) 14:1880–1885 [DOI] [PubMed] [Google Scholar]
- 58.Rasmussen SK, Urhammer SA, Hansen T, Almind K, Moller AM, Borch-Johnsen K, Pedersen O (2000) Variability of the insulin receptor substrate-1, hepatocyte nuclear factor-1α (HNF-1α), HNF-4α, and HNF-6 genes and size at birth in a population-based sample of young Danish subjects. J Clin Endocrinol Metab 85:2951–2953 10.1210/jc.85.8.2951 [DOI] [PubMed] [Google Scholar]
- 59.Dunger DB, Ong KK, Huxtable SJ, Sherriff A, Woods KA, Ahmed ML, Golding J, Pembrey ME, Ring S, Bennett ST, et al (1998) Association of the INS VNTR with size at birth: ALSPAC Study Team. Nat Genet 19:98–100 10.1038/ng0598-98 [DOI] [PubMed] [Google Scholar]
- 60.Ong KK, Phillips DI, Fall C, Poulton J, Bennett ST, Golding J, Todd JA, Dunger DB (1999) The insulin gene VNTR, type 2 diabetes and birth weight. Nat Genet 21:262–263 10.1038/6775 [DOI] [PubMed] [Google Scholar]
- 61.Le Stunff C, Fallin D, Schork NJ, Bougneres P (2000) The insulin gene VNTR is associated with fasting insulin levels and development of juvenile obesity. Nat Genet 26:444–446 10.1038/82579 [DOI] [PubMed] [Google Scholar]
- 62.Vaessen N, Heutink P, Janssen JA, Witteman JC, Testers L, Hofman A, Lamberts SW, Oostra BA, Pols HA, van Duijn CM (2001) A polymorphism in the gene for IGF-I: functional properties and risk for type 2 diabetes and myocardial infarction. Diabetes 50:637–642 10.2337/diabetes.50.3.637 [DOI] [PubMed] [Google Scholar]
- 63.Petry CJ, Ong KK, Barratt BJ, Wingate D, Cordell HJ, Ring SM, Pembrey ME, Reik W, Todd JA, Dunger DB (2005) Common polymorphism in H19 associated with birthweight and cord blood IGF-II levels in humans. BMC Genet 6:22 10.1186/1471-2156-6-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ong KK, Petry CJ, Barratt BJ, Ring S, Cordell HJ, Wingate DL, Pembrey ME, Todd JA, Dunger DB (2004) Maternal-fetal interactions and birth order influence insulin variable number of tandem repeats allele class associations with head size at birth and childhood weight gain. Diabetes 53:1128–1133 10.2337/diabetes.53.4.1128 [DOI] [PubMed] [Google Scholar]
- 65.Bernstein IM, Mongeon JA, Badger GJ, Solomon L, Heil SH, Higgins ST (2005) Maternal smoking and its association with birth weight. Obstet Gynecol 106:986–991 [DOI] [PubMed] [Google Scholar]
- 66.Jarvelin MR, Elliott P, Kleinschmidt I, Martuzzi M, Grundy C, Hartikainen AL, Rantakallio P (1997) Ecological and individual predictors of birthweight in a northern Finland birth cohort 1986. Paediatr Perinat Epidemiol 11:298–312 [DOI] [PubMed] [Google Scholar]





