Abstract
Using the National Center for Biotechnology Information Serial Analysis of Gene Expression database, we found that S100A4, a calcium-binding protein previously implicated in metastasis, was expressed in five of seven pancreatic carcinoma libraries but not in the two normal pancreatic duct libraries. We confirmed the overexpression of S100A4 using reverse transcriptase-polymerase chain reaction, which demonstrated that 18 of 19 (95%) pancreatic carcinoma cell lines expressed S100A4. Using immunohistochemistry, we found that 57 of 61 invasive pancreatic carcinomas (93%), 3 of 18 high-grade pancreatic intraepithelial neoplasia lesions (17%), and 0 of the 69 low-grade pancreatic intraepithelial neoplasia lesions expressed S100A4 protein, whereas normal pancreatic tissue and tissue affected by chronic pancreatitis did not label. Expression of S100A4 was associated with poor differentiation of the pancreatic adenocarcinomas (P = 0.001). We found that three CpG sites in the first intron of the S100A4 gene were ∼90% methylated in microdissected normal pancreatic duct cells using bisulfite-modified sequencing and in two cell lines and three primary pancreatic carcinomas with a reduced or absent expression of S100A4. In contrast, these CpGs were 100% hypomethylated in 11 of 12 pancreatic cancer cell lines by methylation-specific polymerase chain reaction. The association between the expression of S100A4 and hypomethylation of the first intron of S100A4 was statistically significant (P = 0.002). These data suggest that the majority of pancreatic carcinomas undergo selection for hypomethylation and overexpression of S100A4. Because most pancreatic carcinomas express S100A4, it may be a useful target for early detection strategies.
S100A4 (also called mts1, p9Ka, calvasculin, CAPL, pEL98) is a member of a family of sixteen S100 calcium-binding proteins, that all have in common a functional EF-hand domain that mediates their activity. 1 The S100A4 gene was originally cloned by differential screening experiments in which cDNAs were compared between cells before and after growth stimulation or transformation by oncogenes. 2-4 S100A4 is thought to promote metastasis. Nonmetastatic tumor cell lines transfected with S100A4 have a higher incidence of metastases and increased motility. 5 S100A4 transcripts are increased in tumor rat cell lines with metastatic properties compared to their nonmetastatic counterparts. 6
The mechanisms by which S100A4 is overexpressed in cancer cell lines has been studied and hypomethylation of CpG sites in the S100A4 gene has been associated with overexpression in colorectal and lymphoma carcinoma cell lines. 7,8
In an initial survey of gene expression analysis of pancreatic cancer using serial analysis of gene expression (SAGE), we identified 47 tags that were overexpressed in pancreatic cancer cell lines compared to normal pancreatic ductal cells (Ryu B, Jones J, Hollingsworth MA, Hruban RH, Kern SG, submitted). 9 Among these tags, one corresponded to S100A4 mRNA. Two-thousand three-hundred and nine S100A4 tags per million were found in the pancreatic cancer cell lines compared to no tags per million in the normal duct epithelial cells. We therefore evaluated the expression of the S100A4 protein in a panel of 61 pancreatic adenocarcinomas and determined the relationship between the methylation status and expression of the S100A4 gene in cancer cell lines and primary pancreatic adenocarcinomas.
Materials and Methods
Analysis of On-Line SAGE Database
The SAGE analysis of pancreatic cancer has been performed by Ryu and colleagues (submitted) and is now available on-line (http://www.ncbi.nlm.nih.gov/SAGE). The SAGE database currently contains SAGE libraries from 143 samples including 4 pancreatic cancer cell lines (CAPAN1, CAPAN2, Panc1, and Hs766T), 2 pancreatic primary adenocarcinomas (Panc 91-16113 and Panc 96-6252), and 2 short term nonneoplastic pancreatic ductal cells (HX and H126). 10 As reported in detail in a previous study, 9 we used the Student’s t-test tool of the database to identify tags that were significantly (P < 0.02) overexpressed in the pancreatic cancer cell lines versus the normal ductal cells, with a >10-fold difference, and excluding tags with an expression of <12 per million. The tag corresponding to the S100A4 calcium-binding protein (Hs.81256) was present among the most highly overexpressed tags in the pancreatic cancer cell lines versus the normal pancreatic ductal cells.
The “virtual Northern” tool on the SAGE website displays the expression level of a specified SAGE tag in all of the 95 SAGE libraries available at the time of analysis. Results are normalized as number of tags per million.
Cell Lines and Tissues
Human cell lines AsPC1, BxPC3, CAPAN1, CAPAN2, CFPAC1, Hs766T, MiaPaca2, Panc1, and SW480 were obtained from the American Type Culture Collection (Rockville, MD). RKO was a gift from Dr. Michael Brattain. Eleven low-passage pancreatic carcinoma cell lines (PL1-6, PL8-11, and PL14) were generously provided by Dr. Elizabeth Jaffee (Johns Hopkins University, Baltimore, MD).
Four normal pancreatic tissues were obtained from surgical resections at the Johns Hopkins Hospital. The institutional review committee on clinical investigation reviewed and approved the collection of tissue samples for genetic analysis. Normal ducts were microdissected using laser capture microdissection (Arcturus Engineering, Santa Clara, CA). A series of 61 well-characterized primary invasive ductal pancreatic adenocarcinomas resected at the Johns Hopkins Hospital were selected solely on the basis of tissue availability for immunostaining. Eighteen pancreatic adenocarcinomas were poorly differentiated, 20 were moderately differentiated, and 23 were well differentiated. Pancreatic tissues from six patients with chronic pancreatitis were also selected for immunostaining.
Reverse-Transcriptase-Polymerase Chain Reaction (RT-PCR)
RNA from 19 pancreatic cancer cell lines and 2 colorectal cancer cell lines was isolated by using Trizol Reagent (Life Technologies, Rockville, MD). One μg of each total RNA was reverse-transcribed using the Superscript II kit (Life Technologies). PCR primers were 5′-AGCTTCTTGGGGAAAAGGAC-3′ (sense) and 5′-CCCCAACCACATCAGAGG-3′ (antisense). A 200-bp PCR product was then amplified simultaneously with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) according to the following conditions: 95°C for 3 minutes; 30 cycles of amplification (95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 20 seconds); 4 minutes at 72°C. The PCR reaction products were resolved by electrophoresis in a 2% agarose gel and stained with ethidium bromide.
Immunohistochemistry
A representative formalin-fixed paraffin-embedded tissue block containing invasive adenocarcinoma was chosen for each of the 61 invasive pancreatic ductal adenocarcinomas selected for immunolabeling. Fifty-four of these cases also contained normal pancreatic tissue. A block from each of the six chronic pancreatitis cases containing reactive ductal changes was also selected. Four-μm sections mounted on positively charged slides were incubated for 30 minutes at 60°C, deparaffinized by standardized methods, and placed in Tris-buffered saline buffer. Antigen retrieval was performed for 20 minutes in 10 mmol/L of sodium citrate buffer (pH 6.0) heated at 95°C in a steamer, followed by cooling for 20 minutes. After blocking endogenous peroxidase activity with a 3% aqueous H2O2 solution for 5 minutes, the primary polyclonal rabbit anti-S100A4 antibody (DAKO, Carpinteria, CA) was incubated with the sections at a final dilution of 2 μg/ml for 30 minutes in a DAKO automatic immunostainer. For each case, a control slide was incubated with Tris-buffered saline buffer substituted for the primary antibody. The EnVision+ DAB+ detection kit (DAKO) was used for the detection of the immunostaining. Sections were counterstained with hematoxylin. The immunolabeling was evaluated jointly by two authors (CR and RHH). The extent of immunolabeling was categorized in four groups: 0%, negative; 1 to 25%, focal; and 26 to 75% or 76 to 100%, diffuse. For statistical analysis, all focally labeled cases were categorized as focal and all cases showing ≥26% labeling were categorized as positive.
Genomic Bisulfite Sequencing and Methylation-Specific PCR (MSP)
Genomic DNA was isolated from the cell lines and frozen normal pancreatic samples by using a tissue DNA isolation kit (Qiagen, Valencia, CA). Pancreatic carcinoma cells from six paraffin-embedded blocks were microdissected before DNA isolation. DNA was modified by sodium bisulfite as previously described. 11
The S100A4 gene does not contain any CpG island. We chose primers to amplify bisulfite-modified DNA in a region of the first intron of the S100A4 gene. This region contains three CpG sites (position + 315, +331, and +387) and one site for HhaI (position +386), a methylation-sensitive restriction enzyme. From a previous study on lymphoma cell lines, the methylation status of this region has been found to correlate with the S100A4 gene expression. 8 Bisulfite-modified DNA was amplified with the S100A4 gene-specific primers 5′-TGTTTTTGAGATGTGGGTTTG-3′ (sense) and 5′-CACAATTACCTTCTACCTTTC-3′ (antisense). PCR conditions were as follows: 95°C for 3 minutes; 35 cycles of amplification (95°C for 20 seconds, 60°C for 20 seconds, and 72°C for 40 seconds); 4 minutes at 72°C. After incubation with exonuclease I and shrimp alkaline phosphatase (Amersham, Arlington Heights, IL), both strands of amplified products were sequenced using the Sequitherm Excel kit, as recommended by the manufacturer (Epicentre Technologies, Madison, WI). The methylation status of each sequence was evaluated visually by determining the percentage of the intensity of the cytosine band versus thymine band for each CpG site.
The methylation status of the first intron of S100A4 was also determined by MSP as described by Herman and colleagues. 11 Primers sequences, available on request, contained the three CpG sites analyzed by bisulfite sequencing.
Results
Analysis of On-Line SAGE Database
A differential analysis of pancreatic cancer samples in the on-line National Center for Biotechnology Information SAGE database identified 47 different tags that were overexpressed in the four available pancreatic cancer cell lines as compared to the two short-term normal ductal cell lines. Among the top 10 overexpressed tags, the tag matching the S100 calcium-binding protein A4 (ATGTGTAACG), Hs.81256, was identified 71 times in the pancreatic cancer lines versus 0 in the normal duct cells. Three of the four pancreatic cancer cell line libraries contained the Hs.81256 tag: CAPAN1, CAPAN2, and Panc1 contained 1133, 775, and 401 Hs.81256 tags per million total tags, respectively. The pancreatic cancer cell line Hs766T library did not contain the Hs.81256 tag.
Using the “virtual Northern” tool from the on-line SAGE program, the S100A4 tag was present in 45 SAGE libraries with a total count of 454 (ranging from 1 to 78) among 3,888,724 tags in 95 SAGE libraries available at the time of the analysis in July 2001. The primary pancreatic adenocarcinoma libraries also were found to contain the Hs.81256 with 530 and 755 Hs.81256 tags per million tags for Panc 91-16113 and Panc 96-6252, respectively. Remarkably, 5 of the 11 SAGE libraries with the highest number of Hs.81256 tags (normalized per million) were pancreatic cancer libraries. The other libraries that contained high counts of Hs.81256 were derived from glioblastoma multiforme cell lines, a breast cancer cell line and a vascular endothelial cell line.
Reverse-Transcriptase PCR
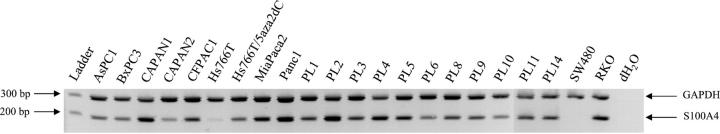
To confirm SAGE results on pancreatic cancer cell lines, we used RT-PCR to evaluate the mRNA expression of S100A4 in 19 pancreatic cancer lines and 2 colorectal cancer cell lines. PCR primers were chosen to overlap an exon-intron boundary and to be specific for the A4 member of the S100 family. The S100A4 transcript was present in 18 of the 19 pancreatic cancer cell lines (95%) (Figure 1) ▶ . For Hs766T, there was a weak amplification of the cDNA at 35 cycles that parallels the SAGE analysis results. Among the colorectal cancer cell lines, S100A4 was expressed in SW480 but not in RKO (Figure 1) ▶ .
Figure 1.
RT-PCR analysis of 19 pancreatic carcinoma cell lines and 2 colorectal carcinoma cell lines (SW480 and RKO). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) serves as a RNA control. All cancer cell lines but Hs766T and RKO strongly express S100A4. Treatment of Hs766T with 5-aza-2-deoxycytidine restores the expression of S100A4.
Immunohistochemistry
With 2 μg/μl as a final dilution of the purified polyclonal rabbit anti-S100A4 antibody, the normal pancreatic ducts and acini did not label in any of the 54 cases that contained normal tissue. Fifteen reactive ductal change lesions have been identified among the six chronic pancreatitis cases; none of them labeled for S100A4. Strong labeling was present in lymphocytes, and in fibroblast-like cells of the stroma of cancer and blood vessels, which served as an internal positive control, as previously reported. 12 Overall, 57 pancreatic carcinomas (93%) expressed S100A4 (Table 1) ▶ . In 28 of the 57 cancer cases, the labeling was present in the majority of tumor cells (76 to 100%), whereas in 17 cases, 26 to 75% of the tumor cells labeled. Labeling was present but focal and weaker in 12 cases, involving <25% of the tumor cells. Four cases did not label at all. All of the 18 poorly differentiated tumors labeled for S100A4. The staining for S100A4 was predominantly cytoplasmic, heterogeneous in some tumors with sometimes a clear demarcation between poorly differentiated tumor cells that were strongly labeled, and well-differentiated areas where the labeling was weaker or negative (Figure 2) ▶ . There was no significant association between S100A4 labeling and overall TNM stage or lymph node status. There was a significant association between tumor differentiation and the extent of labeling for S100A4, with tumors with a uniform expression (>25% of tumor cells) of S100A4 associated with poor differentiation (P = 0.0015, Fisher’s exact test). A range of pancreatic intraepithelial neoplasia (PanIN) lesions in the background of pancreatic adenocarcinoma was identified among 25 sections studied. A total of 87 PanIN lesions have been evaluated for S100A4 labeling. None of the PanIN-1A (0 of 30), PanIN-1B (0 of 26), and PanIN-2 (0 of 13) lesions labeled for S100A4. Among 18 PanIN-3 lesions identified, 3 (17%) labeled.
Table 1.
S100A4 Immunolabeling in Pancreatic Tissues
| S100A4 labeling | Normal pancreatic ducts (n = 54) | Chronic pancreatitis (n = 6) | Low-grade PanINs (n = 69) | High-grade PanINs (n = 18) | Invasive pancreatic ductal adenocarcinomas | |||
|---|---|---|---|---|---|---|---|---|
| Well (n = 23) | Moderately (n = 20) | Poorly (n = 18) | Total (n = 61) | |||||
| Negative | 54 (100%) | 6 (100%) | 0 | 15 (83%) | 3 (14%) | 1 (5%) | 0 | 4 (7%) |
| Focal (1 to 25%) | 0 | 0 | 0 | 0 | 10 (43%) | 2 (10%) | 0 | 12 (20%) |
| Positive (26 to 100%) | 0 | 0 | 0 | 3 (17%) | 10 (43%) | 17 (85%) | 18 (100%) | 45 (73%) |
Figure 2.
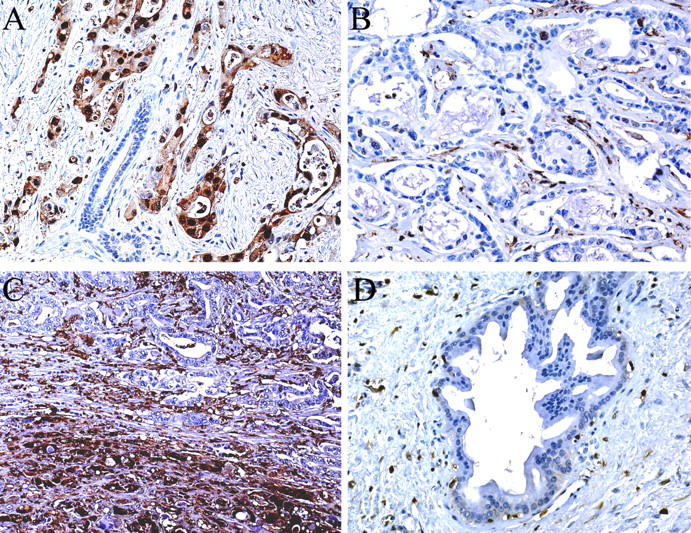
Immunostaining of pancreatic tissue with anti-S100A4 polyclonal antibody. A: Strong labeling of pancreatic adenocarcinoma cells with an absence of labeling of the normal pancreatic cells. B: Pancreatic adenocarcinoma not labeled for S100A4. C: Pancreatic adenocarcinoma with strong labeling of the poorly differentiated area (bottom) whereas the well-differentiated area (top) does not label. D: Reactive ductal change in chronic pancreatitis showing an absence of labeling of duct cells. Original magnifications: ×160 (A, B, D); ×100 (C).
DNA Methylation Analysis
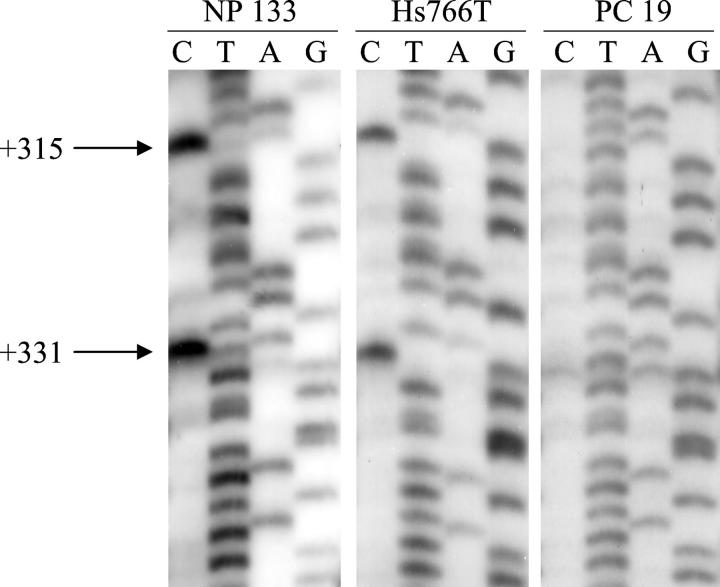
To investigate the relationship between DNA methylation and transcriptional regulation of the S100A4 gene, we sequenced bisulfite-modified DNA from four normal pancreata, one microdissected normal duct cell sample, and nine cancer cell lines (seven pancreatic and two colorectal cancer cell lines). Three CpG sites in the first intron of S100A4 were analyzed by sequencing both strands of a 140-bp PCR product. Overall, 67% (six of nine) of the cancer cell lines analyzed harbored 0% methylation of the S100A4 gene, whereas 100% (four of four) of normal pancreas samples and microdissected duct cells were primarily methylated (60 to 90%). Five of seven pancreatic cancer cell lines (AsPC1, BxPC3, CAPAN1, colo357, and MiaPaCa2) were 0% methylated. For CAPAN2, there was a partial conversion of the cytosines with ∼70% C versus T at the +315 and +331 CpG sites, and ∼30% C versus T at the +387 CpG site. In contrast, Hs766T was 100% methylated (Figure 3) ▶ . For the colorectal cancer cell lines, SW480, which expressed S100A4, was 0% methylated whereas RKO, which did not express S100A4, was completely methylated. Among the four nonneoplastic pancreas samples, S100A4 was almost completely methylated (range from 60 to 90%, Figure 3 ▶ ). Cases for which there was a lower percentage of methylation were those with a low proportion of normal pancreatic tissue that contained inflammatory cells and fibrosis. To minimize contamination with nonpancreatic cells, we microdissected normal duct cells by laser capture microdissection. The S100A4 gene was 90% methylated in the microdissected duct cells.
Figure 3.
Bisulfite-modified sequencing of the first intron of the S100A4 gene showing the methylation status of two CpG sites (+315, +331). NP 133 is a normal pancreas sample with 90% methylation; Hs766T pancreatic cancer cell line is completely methylated; and PC 19 is a primary pancreatic adenocarcinoma, labeled for S100A4 on immunohistochemistry, showing a 90% conversion of the cytosine residues to thymine.
To confirm that the hypomethylation observed in pancreatic cancer cell lines occurred in the primary tumors, we isolated DNA from six paraffin-embedded pancreatic carcinomas. In three cases in which there was no labeling for S100A4 in tumor cells by immunohistochemistry, S100A4 was ∼80% methylated. In three cases with a labeling for S100A4, S100A4 was 0 to 10% methylated for two cases and partially (60%) methylated for one case (Figure 3) ▶ .
Both cell lines methylated for S100A4 had reduced (Hs766T) or complete lack (RKO) of expression of S100A4 at the RT-PCR level, whereas all cell lines with lacking S100A4 methylation expressed S100A4 (Figure 1) ▶ . The association between expression of S100A4 and methylation status in pancreatic carcinoma cell lines and primary tumors was statistically significant (P = 0.002, Fisher’s exact test). Moreover, after treatment of Hs766T with the demethylating agent 5-aza-2-deoxycytidine for 5 days at 2 μmol/L, we observed a re-expression of the S100A4 gene using RT-PCR (Figure 1) ▶ . These results suggest that the low expression of S100A4 in Hs766T is related to the methylation status of the S100A4 gene.
To determine whether cancer cell lines are clonal with respect to S100A4 hypomethylation, we designed MSP primers specific for the methylated and unmethylated versions of the first intron of the gene. We found that 11 of 12 (92%) pancreatic cancer cell lines that express S100A4 were 0% methylated whereas Hs766T was 100% methylated (data not shown). Only CAPAN2 harbored both unmethylated and methylated templates by MSP, which is in agreement with bisulfite sequencing results. These data suggest that the majority of pancreatic cancer cell lines are clonal with respect to S100A4 hypomethylation.
Discussion
With ∼28,000 individuals diagnosed with pancreatic cancer each year in the United States, there is a great need to find better early detection methods that will increase the ability to diagnose this deadly cancer. The discovery of new biomarkers, sensitive and specific for pancreatic cancer, could lead to improved detection of smaller and potentially curative tumors. To discover such new biomarkers, we searched for genes overexpressed in pancreatic cancer by SAGE. This approach has already led to the identification of prostate stem cell antigen as a gene overexpressed in pancreatic cancer. 9 We found that S100A4 was overexpressed by SAGE analysis and RT-PCR. Immunohistochemistry demonstrated that 93% of pancreatic adenocarcinomas expressed S100A4.
Previous immunohistochemical studies of S100A4 showed an overexpression in 41% of breast carcinomas, 12 55% of gastric carcinomas, 13 94% of colorectal adenocarcinomas, 14 and 25% of esophageal squamous cell carcinomas. 15 Our data support the putative role of S100A4 in cell motility and invasion, 5,6 as S100A4 expression was limited to high-grade PanIN lesions and invasive carcinomas, with a correlation between poor tumor differentiation and S100A4 overexpression.
We examined the role of DNA hypomethylation in S100A4 gene expression. We found that expression of S100A4 was associated with hypomethylation of the first intron of the S100A4 gene in both the cell lines and in the primary pancreatic carcinomas. Additional evidence of the role of hypomethylation inducing S100A4 gene expression was provided by the re-expression of S100A4 after treatment of the methylated pancreatic cancer cell line Hs766T with 5-aza-2-deoxycytidine. Global DNA hypomethylation has been previously reported in carcinomas. 16 Only a few studies examined the relationship between aberrant hypomethylation and overexpression of specific genes in carcinomas. Cho and colleagues 17 recently reported an association between expression and hypomethylation of MN/CA9 in renal cell carcinomas. Hypomethylation and overexpression of oncogenes c-jun and c-myc have been reported in chemical-induced mouse liver tumors. 18 It is not known whether hypomethylation of S100A4 in pancreatic carcinomas is related to selection of S100A4-expressing cells or a byproduct of genome-wide DNA hypomethylation. The fact that the majority of pancreatic cancer cell lines expressing S100A4 were not methylated at the first intron of the gene by MSP-PCR suggests that the evolving neoplasm may have undergone selection for hypomethylation of S100A4.
Our findings of S100A4 overexpression in pancreatic adenocarcinoma has several potential clinical applications. First, because S100A4 was specifically found in the cancer cells and not in the normal pancreas cells, detection of S100A4 transcripts or proteins could be used as an aid in the diagnosis of pancreatic cancer in biopsies of suspicious pancreatic lesions or fine needle aspirates. Second, measurement of soluble S100A4 in biological fluids, such as pancreatic juice or serum, could potentially be used for early detection to detect a small amount of pancreatic cancer cells. Because lymphocytes express S100A4, a quantitative technique, such as real-time PCR or enzyme-linked immunosorbent assay would be more suitable for early detection strategies. Third, S100A4 is a potential therapeutic target. In vitro and in vivo studies have demonstrated that blocking the expression of S100A4 inhibits the metastatic spread of cancer cells. 19,20
In conclusion, we found that S100A4 is overexpressed in the majority of pancreatic adenocarcinomas, and that its expression in pancreatic cancer is associated with hypomethylation of the S100A4 gene.
Footnotes
Address reprint requests to Michael Goggins, M.D., Department of Pathology, 632 Ross Building, The Johns Hopkins Medical Institutions, 720 Rutland Ave., Baltimore, MD 21205-2196. E-mail: mgoggins@jhmi.edu.
Supported by The Lustgarten Foundation for Pancreatic Cancer Research (http://www.lustgartenfoundation.org/), the National Institutes of Health Specialized Program in Research Excellence in Gastrointestinal Cancer (grant p50-CA62924), the Michael Rolfe Foundation, and the Pancreatic Cancer Action Network (http://www.pancan.org/).
Dr. Rosty’s current address is Institut Curie Laboratoire de Pathologie 26 Rue d’Ulm 75005 Paris, France.
References
- 1.Schafer BW, Heizmann CW: The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem Sci 1996, 21:134-140 [DOI] [PubMed] [Google Scholar]
- 2.Linzer DI, Nathans D: Growth-related changes in specific mRNAs of cultured mouse cells. Proc Natl Acad Sci USA 1983, 80:4271-4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Vouge MW, Mukherjee BB: Transformation of normal rat kidney cells by v-K-ras enhances expression of transin 2 and an S-100-related calcium-binding protein. Oncogene 1992, 7:109-119 [PubMed] [Google Scholar]
- 4.Goto K, Endo H, Fujiyoshi T: Cloning of the sequences expressed abundantly in established cell lines: identification of a cDNA clone highly homologous to S-100, a calcium binding protein. J Biochem 1988, 103:48-53 [DOI] [PubMed] [Google Scholar]
- 5.Davies BR, Davies MP, Gibbs FE, Barraclough R, Rudland PS: Induction of the metastatic phenotype by transfection of a benign rat mammary epithelial cell line with the gene for p9Ka, a rat calcium-binding protein, but not with the oncogene EJ-ras-1. Oncogene 1993, 8:999-1008 [PubMed] [Google Scholar]
- 6.Ebralidze A, Tulchinsky E, Grigorian M, Afanasyeva A, Senin V, Revazova E, Lukanidin E: Isolation and characterization of a gene specifically expressed in different metastatic cells and whose deduced gene product has a high degree of homology to a Ca2+-binding protein family. Genes Dev 1989, 3:1086-1093 [DOI] [PubMed] [Google Scholar]
- 7.Nakamura N, Takenaga K: Hypomethylation of the metastasis-associated S100A4 gene correlates with gene activation in human colon adenocarcinoma cell lines. Clin Exp Metastasis 1998, 16:471-479 [DOI] [PubMed] [Google Scholar]
- 8.Tulchinsky E, Grigorian M, Tkatch T, Georgiev G, Lukanidin E: Transcriptional regulation of the mts1 gene in human lymphoma cells: the role of DNA-methylation. Biochim Biophys Acta 1995, 1261:243-248 [DOI] [PubMed] [Google Scholar]
- 9.Argani P, Rosty C, Reiter RE, Wilentz RE, Murugesan SR, Leach SD, Ryu B, Skinner HG, Goggins M, Jaffee EM, Yeo CJ, Cameron JL, Kern SE, Hruban RH: Discovery of new markers of cancer through serial analysis of gene expression: prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res 2001, 61:4320-4324 [PubMed] [Google Scholar]
- 10.Lal A, Lash AE, Altschul SF, Velculescu V, Zhang L, McLendon RE, Marra MA, Prange C, Morin PJ, Polyak K, Papadopoulos N, Vogelstein B, Kinzler KW, Strausberg RL, Riggins GJ: A public database for gene expression in human cancers. Cancer Res 1999, 59:5403-5407 [PubMed] [Google Scholar]
- 11.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB: Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996, 93:9821-9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudland PS, Platt-Higgins A, Renshaw C, West CR, Winstanley JH, Robertson L, Barraclough R: Prognostic significance of the metastasis-inducing protein S100A4 (p9Ka) in human breast cancer. Cancer Res 2000, 60:1595-1603 [PubMed] [Google Scholar]
- 13.Yonemura Y, Endou Y, Kimura K, Fushida S, Bandou E, Taniguchi K, Kinoshita K, Ninomiya I, Sugiyama K, Heizmann CW, Schafer BW, Sasaki T: Inverse expression of S100A4 and E-cadherin is associated with metastatic potential in gastric cancer. Clin Cancer Res 2000, 6:4234-4242 [PubMed] [Google Scholar]
- 14.Takenaga K, Nakanishi H, Wada K, Suzuki M, Matsuzaki O, Matsuura A, Endo H: Increased expression of S100A4, a metastasis-associated gene, in human colorectal adenocarcinomas. Clin Cancer Res 1997, 3:2309-2316 [PubMed] [Google Scholar]
- 15.Ninomiya I, Ohta T, Fushida S, Endo Y, Hashimoto T, Yagi M, Fujimura T, Nishimura G, Tani T, Shimizu K, Yonemura Y, Heizmann CW, Schafer BW, Sasaki T, Miwa K: Increased expression of S100A4 and its prognostic significance in esophageal squamous cell carcinoma. Int J Oncol 2001, 18:715-720 [DOI] [PubMed] [Google Scholar]
- 16.Feinberg AP, Vogelstein B: Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983, 301:89-92 [DOI] [PubMed] [Google Scholar]
- 17.Cho M, Uemura H, Kim SC, Kawada Y, Yoshida K, Hirao Y, Konishi N, Saga S, Yoshikawa K: Hypomethylation of the MN/CA9 promoter and upregulated MN/CA9 expression in human renal cell carcinoma. Br J Cancer 2001, 85:563-567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao L, Yang S, Xie M, Kramer PM, Pereira MA: Hypomethylation and overexpression of c-jun and c-myc protooncogenes and increased DNA methyltransferase activity in dichloroacetic and trichloroacetic acid-promoted mouse liver tumors. Cancer Lett 2000, 158:185-193 [DOI] [PubMed] [Google Scholar]
- 19.Maelandsmo GM, Hovig E, Skrede M, Engebraaten O, Florenes VA, Myklebost O, Grigorian M, Lukanidin E, Scanlon KJ, Fodstad O: Reversal of the in vivo metastatic phenotype of human tumor cells by an anti-CAPL (mts1) ribozyme. Cancer Res 1996, 56:5490-5498 [PubMed] [Google Scholar]
- 20.Takenaga K, Nakamura Y, Sakiyama S: Expression of antisense RNA to S100A4 gene encoding an S100-related calcium-binding protein suppresses metastatic potential of high-metastatic Lewis lung carcinoma cells. Oncogene 1997, 14:331-337 [DOI] [PubMed] [Google Scholar]