Abstract
Epigenetic mechanisms of gene silencing, including promoter hypermethylation of tumor suppressor genes, have been shown to contribute to tumorigenesis. PTEN is an important tumor suppressor implicated in the pathogenesis of a number of familial and sporadic cancers. Germline mutations of PTEN predispose to dominantly inherited hamartomatous disorders Cowden syndrome and Bannayan-Zonana syndrome. Somatic PTEN mutations commonly occur in endometrial, breast, prostate, and thyroid tumors. Several investigators have speculated on PTEN promoter hypermethylation as a possible mechanism of PTEN inactivation but data supporting such observations is not forthcoming. The genomic sequence of PTEN is 98% identical to a highly conserved processed PTEN pseudogene (psiPTEN) and this sequence identity extends 841 base pairs into the promoter region. This high degree of homology has made analysis of the methylation status of the PTEN promoter quite challenging. We have investigated the methylation profiles of the promoter region of PTEN in endometrial, breast, and colon cancer cell lines, as well as in a panel of primary endometrial tumors using a combination of methylation-specific polymerase chain reaction, methylation-sensitive restriction analysis, and bisulfite sequencing. Our results show that the pseudogene, and not PTEN, is predominantly methylated in cell lines and tumors. Without careful consideration of the critical nucleotide differences between the two sequences, results obtained from PTEN analysis may not necessarily represent the methylation status of PTEN.
Tumor suppressor gene PTEN, also know as MMAC1/TEP1, has been implicated in a number of familial and sporadic cancers. Germline mutations of PTEN are responsible for Cowden syndrome 1 and BannayanZonana 2 syndrome, dominantly inherited conditions, causing predisposition to a variety of cancers. Inactivation of PTEN is observed in a variety of sporadic cancers including glioblastomas, 3 melanomas, 4 and carcinomas of the endometrium, 5 ovary, 6 thyroid, 7 breast, 8 and prostate. 9 Among these cancers, PTEN is considered to be a “gatekeeper” in the endometrium. 10 PTEN mutations occur in 50 to 80% of endometrial adenocarcinomas. 5,11 Furthermore, PTEN alterations are observed in up to 30% of complex atypical hyperplasias, which are considered to be the direct precursor lesions of endometrial adenocarcinomas and even in early lesions that are intermediate between normal endometrium and benign hyperplasias. 12 This progressive accumulation of PTEN mutations may contribute to the transition from premalignant to malignant disease. In a study by Mutter et al, 11 computerized morphometric analysis and selective UV irradiation were used to identify and obtain endometrial adenocarcinomas and associated hyperplasias. Mutations of PTEN were shown to occur in 83% of endometrial adenocarcinomas and 55% of hyperplasias. Most of these mutations occurred in only one allele. However, complete loss of PTEN expression, as seen from immunohistochemical analysis, was observed in 61% of tumors and 97% showed at least some degree of diminished expression. 11 These observations suggest that other mechanisms, including the epigenetic regulation of PTEN by differential methylation may contribute to the loss of PTEN expression. 4
Differential methylation is an important epigenetic control mechanism, which has been implicated in the development of a variety of cancers. Research has shown that methylation of promoter regions of normally unmethylated genes may lead to transcriptional inactivation and ultimately to tumorigenesis. Hypermethylation of several tumor suppressor genes has been associated with the development of various cancers. For example, methylation of p16 has been implicated in the development of gliomas, 13 while methylation of APC has been observed in the development of colon 14 and breast 15 tumors. Methylation of MLH1, a mismatch repair gene, has been shown to occur in sporadic colon and endometrial tumors and is associated with the development of the microsatellite instability (MSI) phenotype. 16 Approximately 90% of endometrial carcinomas with MSI have been shown to display MLH1 methylation. Whether hypermethylation of other tumor suppressor genes including PTEN contribute to endometrial cancer development is not yet known.
PTEN (GenBank accession number AF143312) is localized to chromosome 10q23. 17-19 A highly conserved processed PTEN pseudogene (GenBank accession number AF040103, PTEN pseudogene; AF029308, Homo sapiens chromosome 9 duplication of the T cell receptor β locus and trypsinogen gene families) is located on chromosome 9p21. 20 Examination of the two sequences indicates that the sequence identity extends −841 nucleotides upstream of the PTEN translational start site. Because of the high degree of homology between PTEN and its pseudogene, analysis of PTEN has proven to be difficult. We have examined the methylation status of PTEN in a panel of carcinoma cell lines and endometrial tumors. In this study, we report the technical challenges and possible pitfalls faced when undertaking the analysis of methylation profiles of PTEN.
Materials and Methods
Tissues and Cell Lines
All cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). RL95–2 cells were maintained in a 1:1 mixture of Ham’s F-12 and Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Invitrogen Corp., Carlsbad, CA). HEC-1B, AN3CA and MCF-7 cell lines were maintained in modified Eagle’s medium (MEM) supplemented with Earle’s salts (Sigma Genosys, Oakville, Canada) and 10% FBS (Invitrogen). MDA-MB231, MDA-MB468, SW-48, and SW-480 cells were maintained in Leibovitz’s L-15 media with 10% FBS (Invitrogen). HT-29 cells were maintained in McCoys’s 5A media with 10% FBS (Invitrogen). DNA was extracted from cells using the Dneasy Tissue Kit (Qiagen Inc., Mississauga, Canada).
Formalin-fixed, paraffin-embedded tissue sections of endometrial adenocarcinomas and matched normal specimens (n = 20) were obtained from patients according to a protocol approved by the Ethics Committee, University of Toronto. Tissue was microdissected from 10-μm cut sections, then transferred into lysis buffer (10 mmol/L Tris-Cl, pH 8; 100 mmol/L KCl; 2.5 mmol/L MgCl2; 0.45% Tween 20) with 6 μl of Proteinase K (20 mg/ml) and incubated at 65°C for 16 hours to obtain genomic DNA.
Methylation-Specific Polymerase Chain Reaction (MSP)
Genomic DNA (18 μl) obtained from tissues was treated with 5 mol/L sodium metabisulfite (Fisher Scientific Ltd., Ottawa, Canada) for 16 hours (protocol kindly provided by Peter Laird, University of Southern California), while cell line DNA (1 μg) was modified using the CpGenome DNA Modification Kit (Intergen Company, Purchase, NY). Three primer sets were designed as outlined in Table 1 ▶ . For sets I and II, the polymerase chain reaction (PCR) mixture contained 1X PCR buffer, 21 dNTPs (each at 1.28 mmol/L; Invitrogen), primers (4 ng/μl; Sigma Genosys), Platinum Taq polymerase (1.5 units; Invitrogen), and bisulfite-modified DNA (∼100 ng) or unmodified DNA (50 to 100 ng) in a final volume of 25 μl. The reactions were carried out in a DNA Engine Dyad Peltier Thermal Cycler (MJ Research Inc., Waltham, MA) at 95°C for 5 minutes, cycled at 30 seconds at 95°C, 30 seconds at the annealing temperature listed in Table 1, and 1 ▶ minute at 72°C (35 cycles), followed by a 4 minute extension at 72°C. For set III, the PCR mixture contained 1X PCR buffer, 21 dNTPs (each at 1.28 mmol/L; Invitrogen), primers (4 ng/μl; Sigma Genosys), HotStarTaq DNA polymerase (1.5U; Qiagen), and bisulfite-modified DNA (∼100 ng) or unmodified DNA (50 to 100 ng) in a final volume of 25 μl. The reactions were carried out in a Dyad thermal cycler at 95°C for 15 minutes, cycled at 30 seconds at 94°C, 30 seconds at the annealing temperature listed in Table 1, and 1 ▶ minute at 72°C (35 cycles), followed by a 10 minute extension at 72°C. Normal lymphocyte DNA supermethylated with SssI methyltransferase (New England Biolabs, Beverly, MA), and subsequently treated with sodium metabisulfite served as the methylated control. Normal lymphocyte DNA treated with sodium metabisulfite alone was included as an unmethylated control. DNA not treated with sodium metabisulfite was also included as a negative control. Ten microliters of PCR product was visualized on a 2% agarose gel.
Table 1.
PCR Primers Used for MSP and MSRA
| Primer set | Forward primer 5′ to 3′ | Reverse primer 5′ to 3′ | Size (bp) | Genomic position | Anneal temperature (°C) |
|---|---|---|---|---|---|
| MSP | |||||
| Set I-M | TTTTTTTTCGGTTTTTCGAGGC | CAATCGCGTCCCAACGCCG | 134 | −984 | 59 |
| Set I-U | TTTTGAGGTGTTTGGGTTTTTGGT | ACACAATCACATCCCAACACCA | 124 | −971 | 59 |
| Set II-M | TGTTTTTAATACGGCGGCGGC | AAACGAATAATCCTCCGAACG | 143 | −392 | 62 |
| Set II-U | TTTTTATTGTTTTTAATATGGTGGT | ACCAACAACCTCCAAAACCCA | 120 | −399 | 57 |
| Set III-M | GGTTTCGGAGGTCGTCGGC | CAACCGAATAATAACTACTACGACG | 155 | −298 | 58 |
| Set III-U | TGGGTTTTGGAGGTTGTTGGT | ACTTAACTCTAAACCACAACCA | 173 | −300 | 56 |
| MSRA | |||||
| Set IV-A | TCCGGAGGCCGCCGGCGGAA | CCGGGTAATGGCTGCTGCGGCG | 151 | −295 | 65 |
| Set IV-B | TCCGGAGGCCGCTGGAGGAG | GGGTAATGGCTGCGGCAGCA | 149 | −295 | 72 |
| Set IV-C | CAGCCGTTCGGAGGATTATT | GTTTCTGCTAACGATCTCTTTG | 310 | −273 | 56 |
Sequence differences between the bisulfite modified and untreated genomic DNA are shown in bold. Differences between methylated and unmethylated sequences are underlined. Genomic position is defined by the location of the 5′ nucleotide of the sense primer from the translational start site of PTEN (Genbank accession number AF040103). PCR primers and annealing temperatures are indicated.
Set IV-A and B were used to amplify PTEN and the pseudogene, respectively, following AciI digestion. Nucleotide differences between PTEN gene and pseudogene are underlined. Set IV-C primers are the same as reported by Salvesen et al. 23 Genomic position is defined by the location of the 5′ nucleotide of the sense primer from the translational start site of PTEN (Genbank accession number AF040103).
Methylation-Sensitive Restriction Analysis (MSRA)
Three hundred nanograms of genomic cell line DNA was incubated with 10 units of AciI (New England Biolabs) at 37°C for 16 hours. AciI digests only unmethylated DNA at its recognition sequence (5′- C↓CGC- 3′), leaving methylated sites intact. Two microliters of digested DNA was then used for PCR. Primers were designed as outlined in Table 1 ▶ . Set IV-A and -B primers encompass 6 AciI sites, while set IV-C primers flank 7 sites. For set IV-A primers, the PCR mixture contained 1X PCR buffer (Qiagen), dNTPs (each at 0.2 mmol/L; Invitrogen), primers (4 ng/μl; Sigma Genosys), HotStarTaq DNA polymerase (1.5 units; Qiagen), and digested DNA (∼30 ng) or undigested DNA (∼100 ng) in a final volume of 25 μl. The reactions were carried out as described above for MSP set III primers. For sets IV-B and -C, the PCR mixture contained 1X PCR buffer (Qiagen), dNTPs (each at 0.2 mmol/L; Invitrogen), primers (4 ng/μl; Sigma Genosys), Platinum Taq polymerase (1.5U; Invitrogen), and digested DNA (∼30 ng) or undigested DNA (∼100 ng) in a final volume of 25 μl. The reactions were carried out as described above for MSP set I primers. Normal lymphocyte DNA alone or treated with SssI methyltransferase (New England Biolabs) followed by AciI digestion served as unmethylated and methylated controls, respectively. Ten microliters of PCR product was visualized on a 2% agarose gel.
Sequencing of Bisulfite-Modified DNA and MSRA Products
Manual sequencing of unmethylated and methylated bisulfite-modified DNA products (sets I and II) was conducted using the ThermoSequenase radiolabeled terminator cycle sequencing kit (Amersham Pharmacia Biotech, Inc., Piscataway, NJ) to distinguish between PTEN and the pseudogene. Manual sequencing of PCR products using set-IV-A and -B primers was conducted to verify primer specificity.
Results and Discussion
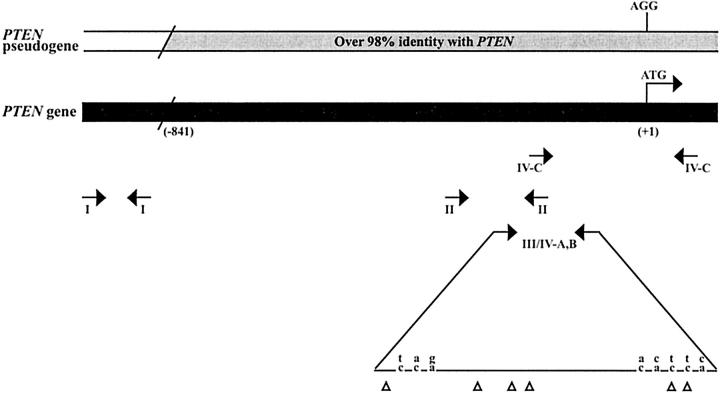
The role of methylation in PTEN silencing has been suggested by a number of researchers. 4,22 However, little information has been published on the methylation status of PTEN. Salvesen et al 23 recently reported that 26 of 138 endometrial carcinomas displayed PTEN promoter methylation. To our knowledge, this is the only published study of its kind. However, these researchers did not address the issue of the PTEN pseudogene and its relevance to determining PTEN methylation status. We analyzed PTEN methylation status in a panel of cancer cell lines (endometrial: RL95–2, AN3CA, HEC-1B; breast: MCF-7, MDA-MB231, MDA-MB468; colon: HT-29, SW-48, SW-480) and in primary endometrial adenocarcinomas using a combination strategy of methylation-specific PCR (MSP), methylation-sensitive restriction analysis (MSRA) and bisulfite genomic sequencing. We designed three primer sets for MSP analysis of the PTEN promoter region (Figure 1 ▶ , Table 1 ▶ ). Set I primers (nucleotides −984 to −848) amplified a region of the promoter specific to PTEN that is not homologous to the pseudogene. Set II and III primers (nucleotides −399 to −250; nucleotides −300 to −128, respectively) were downstream of set I primers and were localized to the PTEN sequence which shares over 98% identity with the pseudogene. Set II primers did not discriminate between PTEN and the pseudogene while set III primers exclusively amplified PTEN and were designed by taking advantage of critical nucleotide differences between the two sequences. MSP analysis using PTEN gene specific set I and III primers (Table 2 ▶ ; Figure 2a, 3a ▶ ▶ ) showed that none of the cell lines were methylated. However, using set II primers we found that these same cell lines exhibited methylation (Figure 3b) ▶ with the exception of the MCF-7 cell line. Furthermore, analysis of endometrial tumors was consistent with these results. For set I and III primers, no tumors were found to exhibit methylation, while all tumor and matched normal tissues displayed methylation for set II primers (Figure 3, a and b) ▶ .
Figure 1.
Alignment of the PTEN gene (lower) and pseudogene (upper). The region upstream of nucleotide (−841) represents a divergent sequence. MSP primer sets I, II, and III are shown. Set IV-A, -B primers were used for MSRA to amplify the PTEN gene and pseudogene, respectively. AciI restriction sites are indicated with triangles. Differences between the two sequences are shown with the pseudogene sequence on top. Set IV-C are primers reported by Salvesen et al 23 for MSRA are shown. Genomic position is defined by the location relative to the translational start site of PTEN (GenBank accession number AF143312).
Table 2.
MSP and MSRA Results for Cell Lines
| Tissue type | Cell line | MSP | MSRA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Set I | Set II | Set III | Set IV-A M | Set IV-B M | Set IV-C M | |||||
| M | U | M | U | M | U | |||||
| Endometrial | RL95-2 | − | + | + | + | − | + | − | + | + |
| Endometrial | AN3CA | − | + | + | + | − | + | − | + | + |
| Endometrial | HEC-1B | − | + | + | + | − | + | − | + | + |
| Breast | MCF-7 | − | + | − | + | − | + | − | − | − |
| Breast | MDA-MB231 | − | + | + | + | − | + | − | + | + |
| Breast | MDA-MB468 | − | + | + | + | − | + | − | + | + |
| Colon | HT-29 | − | + | + | + | − | + | − | + | + |
| Colon | SW-48 | − | + | + | + | − | + | − | + | + |
| Colon | SW-480 | − | + | + | + | − | + | − | + | + |
MSP (primer sets I, II, III) and MSRA (primer sets IV-A, B, and C) analysis of cancer cell lines. U, unmethylated; M, methylated.
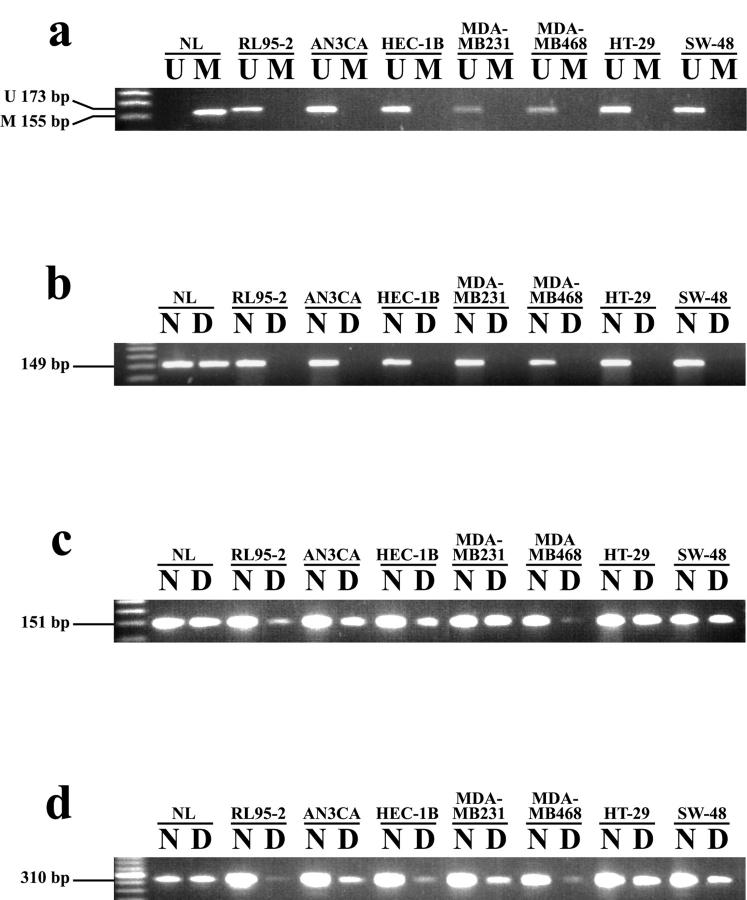
Figure 2.
Methylation-specific PCR using set III PTEN specific primers (a) and methylation-sensitive restriction analysis (b, c, d) using primer set IV-A (PTEN-specific), IV-B (pseudogene-specific), and IV-C (reported by Salvesen et al 23 ) respectively for cell lines. MSP analysis, as well as MSRA using primer set IV-A (PTEN), show lack of methylation in cell lines (a, b). Results for MSRA using set IV-B and -C primers (c, d) show methylation positive products in the same cell lines. NL treated with SssI methyltransferase was used as a methylated positive control. Following AciI digestion, equal amounts of cell line DNA were used for PCR analysis. U, unmethylated; M, methylated; N, undigested DNA; D, digested DNA; NL, normal lymphocyte DNA.
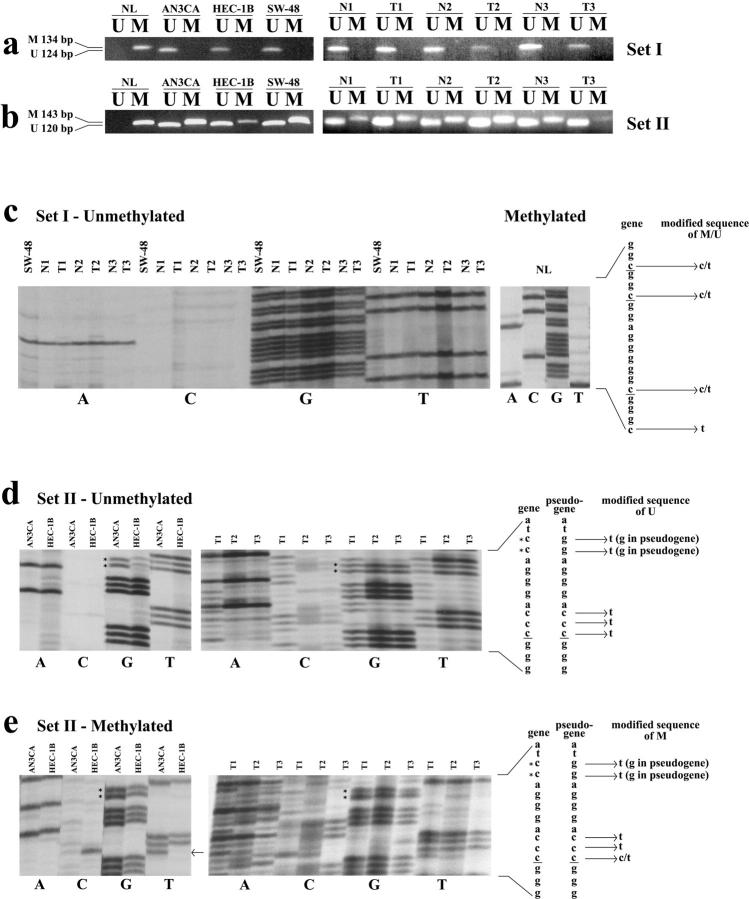
Figure 3.
MSP and sodium bisulfite sequencing for set I and II primers. a and b: MSP results for PTEN primer sets I and II respectively. Panels on the left show results for cell lines while those on the right are representative of matched normal (N) and tumor (T) samples. PTEN set I sodium bisulfite sequencing of unmethylated cell lines and matched normal and tumors, and methylated NL is shown (c). Sequencing of unmethylated (d) and methylated (e) cell lines and tumors using set II primers are shown. PTEN and pseudogene sequences are shown with critical differences between the 2 sequences identified with an * and differences between the genomic and sodium bisulfite modified DNA are indicated. Note the presence of a T instead of a C (arrow) in the AN3CA cell line indicating that this C of a CpG dinucleotide is not methylated, unlike in HEC-1B (e). Cytosine residues of CpG dinucleotides are underlined. U, unmethylated; M, methylated; NL, normal lymphocyte DNA.
We verified these results by performing MSRA on the same cancer cell line DNA. MSRA primers were designed (Table 1) ▶ in the same region as the MSP set III primers to amplify either PTEN (set IV-A; nucleotides −295 to −147) or the PTEN pseudogene (set IV-B; nucleotides −295 to −149). This was important so that results from MSP could be correlated with those of MSRA based on the same CpG sites. In the study published by Salvesen et al, 23 PTEN promoter methylation status was analyzed by performing MSRA using primers that were essentially located in the same region as our set IV primers, but extended approximately 150 base pairs (bp) further downstream. We found that none of the cell lines showed PTEN methylation using set IV-A (Figure 2b) ▶ following AciI digestion, while 8 of 9 cell lines showed methylation of the pseudogene using set IV-B (Figure 2c) ▶ . When we performed MSRA using the same primers as reported by Salvesen et al, 23 (primers IV-C) we again found that all cell lines, with the exception of MCF-7, exhibited methylation (Figure 2d) ▶ .
To further confirm the specific contributions of PTEN and the pseudogene to these observations, we sequenced MSP and MSRA products. Sequencing of set I unmethylated tumor products and control supermethylated NL DNA, confirmed the PTEN-specific sequence (Figure 3c) ▶ . As expected, sequencing of set II unmethylated cell line and tumor PCR products demonstrated nucleotide sequences corresponding to both PTEN and the pseudogene. This was evident by the presence of both a TT from the PTEN gene (CC modified to TT following bisulfite treatment) and GG from the pseudogene (Figure 3d) ▶ . While for methylated PCR products, only the pseudogene was amplified as determined from the presence of only GG which is encoded by the pseudogene sequence (Figure 3e ▶ ; note the absence of a TT at this location as compared to the unmethylated sequencing products).
From our results, we conclude that PTEN methylation is likely to be a rare event since set I and III primers, which were specific to the PTEN gene, showed absence of methylation in all cell lines and endometrial tumors examined. However, methylation was observed in 8 of 9 cell lines and in all endometrial tumors tested using set II primers which could not discriminate between PTEN and the pseudogene. Taken together, our findings confirm an absence of PTEN methylation. Our results demonstrate the extreme difficulties in determining the methylation status of the promoter/5′ untranslated region of PTEN due to the high homology in this region with that of the PTEN pseudogene. It is important that researchers pay particular attention to the few critical residues that differ between the PTEN and pseudogene sequences. Methylation detection methods which do not make the distinction between PTEN and the pseudogene sequences, may lead to false positives and may suggest a higher degree of PTEN promoter methylation than truly exists. Specific contribution of PTEN promoter methylation should be determined by conducting different assessment methods in parallel and should include sequencing to check the identity of the critical nucleotides that differ between the gene and its pseudogene. Studies to examine the PTEN expression including immunohistochemical analysis and RT-PCR should also be used. Besides promoter methylation, other genetic events such as loss of allelic expression or dysregulation of control elements may explain the high frequency of loss of PTEN expression observed by immunohistochemical analysis. Determining the effects of such molecular mechanisms on the regulation of PTEN warrants further study.
Acknowledgments
We thank Dr. P. Laird for the bisulfite-modification protocol, A. Saka and S. Esufali for technical support, A. Firestone for assistance with preparation of figures, and the members of our lab for helpful discussions in the preparation of this manuscript.
Footnotes
Supported in part by funds provided by Concern Foundation (B.B.) and American Institute for Cancer Research Grant 99BO55 (B.B.). M.Z. is a recipient of a University of Toronto Open Fellowship.
References
- 1.Nelen MR, Padberg GW, Peeters EA, Lin AY, van den Helm B, Frants RR, Coulon V, Goldstein AM, van Reen MM, Easton DF, Eeles RA, Hodgsen S, Mulvihill JJ, Murday VA, Tucker MA, Mariman EC, Starink TM, Ponder BA, Ropers HH, Kremer H, Longy M, Eng C: Localization of the gene for Cowden disease to chromosome 10q22–23. Nat Genet 1996, 13:114-116 [DOI] [PubMed] [Google Scholar]
- 2.Marsh DJ, Dahia PL, Zheng Z, Liaw D, Parsons R, Gorlin RJ, Eng C: Germ-line mutations in PTEN are present in Bannayan-Zonana syndrome. Nat Genet 1997, 16:333-334 [DOI] [PubMed] [Google Scholar]
- 3.Rasheed BK, Stenzel TT, McLendon RE, Parsons R, Friedman AH, Friedman HS, Bigner DD, Bigner SH: PTEN gene mutations are seen in high-grade but not in low-grade gliomas. Cancer Res 1997, 57:4187-4190 [PubMed] [Google Scholar]
- 4.Zhou XP, Gimm O, Hampel H, Niemann T, Walker MJ, Eng C: Epigenetic PTEN silencing in malignant melanomas without PTEN mutation. Am J Pathol 2000, 157:1123-1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tashiro H, Blazes MS, Wu R, Cho KR, Bose S, Wang SI, Li J, Parsons R, Ellenson LH: Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res 1997, 57:3935–3940 [PubMed]
- 6.Kurose K, Zhou XP, Araki T, Cannistra SA, Maher ER, Eng C: Frequent loss of PTEN expression is linked to elevated phosphorylated Akt levels, but not associated with p27 and cyclin D1 expression, in primary epithelial ovarian carcinomas. Am J Pathol 2001, 158:2097-2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahia PL, Marsh DJ, Zheng Z, Zedenius J, Komminoth P, Frisk T, Wallin G, Parsons R, Longy M, Larsson C, Eng C: Somatic deletions and mutations in the Cowden disease gene, PTEN, in sporadic thyroid tumors. Cancer Res 1997, 57:4710-4713 [PubMed] [Google Scholar]
- 8.Perren A, Weng LP, Boag AH, Ziebold U, Thakore K, Dahia PL, Komminoth P, Lees JA, Mulligan LM, Mutter GL, Eng C: Immunohistochemical evidence of loss of PTEN expression in primary ductal adenocarcinomas of the breast. Am J Pathol 1999, 155:1253-1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cairns P, Okami K, Halachmi S, Halachmi N, Esteller M, Herman JG, Jen J, Isaacs WB, Bova GS, Sidransky D: Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res 1997, 57:4997-5000 [PubMed] [Google Scholar]
- 10.Ali IU: Gatekeeper for endometrium: the PTEN tumor suppressor gene. J Natl Cancer Inst 2000, 9211:861-863 [DOI] [PubMed] [Google Scholar]
- 11.Mutter GL, Lin MC, Fitzgerald JT, Kum JB, Baak JP, Lees JA, Weng LP, Eng C: Altered PTEN expression as a diagnostic marker for the earliest endometrial precancers. J Natl Cancer Inst 2000, 92:924-930 [DOI] [PubMed] [Google Scholar]
- 12.Levine RL, Cargile CB, Blazes MS, van Rees B, Kurman RJ, Ellenson LH: PTEN mutations and microsatellite instability in complex atypical hyperplasia, a precursor lesion to uterine endometrioid carcinoma. Cancer Res 1998, 58:3254-3258 [PubMed] [Google Scholar]
- 13.Costello JF, Berger MS, Huang HS, Cavenee WK: Silencing of p16/CDKN2 expression in human gliomas by methylation and chromatin condensation. Cancer Res 1996, 56:2405-2410 [PubMed] [Google Scholar]
- 14.Esteller M, Sparks A, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Gonzalez S, Tarafa G, Sidransky D, Meltzer SJ, Baylin SB, Herman JG: Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res 2000, 60:4366-4371 [PubMed] [Google Scholar]
- 15.Virmani AK, Rathi A, Sathyanarayana UG, Padar A, Huang CX, Cunnigham HT, Farinas AJ, Milchgrub S, Euhus DM, Gilcrease M, Herman J, Minna JD, Gazdar AF: Aberrant methylation of the adenomatous polyposis coli (APC) gene promoter 1A in breast and lung carcinomas. Clin Cancer Res 2001, 7:1998-2004 [PubMed] [Google Scholar]
- 16.Esteller M, Catasus L, Matias-Guiu X, Mutter GL, Prat J, Baylin SB, Herman JG: hMLH1 promoter hypermethylation is an early event in human endometrial tumorigenesis. Am J Pathol 1999, 155:1767–1772 [DOI] [PMC free article] [PubMed]
- 17.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R: PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997, 275:1943-1947 [DOI] [PubMed] [Google Scholar]
- 18.Li DM, Sun H: TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res 1997, 57:2124-2129 [PubMed] [Google Scholar]
- 19.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV: Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet 1997, 15:356-362 [DOI] [PubMed] [Google Scholar]
- 20.Dahia PL, FitzGerald MG, Zhang X, Marsh DJ, Zheng Z, Pietsch T, von Deimling A, Haluska FG, Haber DA, Eng C: A highly conserved processed PTEN pseudogene is located on chromosome band 9p21. Oncogene 1998, 16:2403-2406 [DOI] [PubMed] [Google Scholar]
- 21.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB: Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996, 93:9821-9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mutter GL: Pten, a protean tumor suppressor. Am J Pathol 2001, 158:1895-1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salvesen HB, MacDonald N, Ryan A, Jacobs IJ, Lynch ED, Akslen LA, Das S: PTEN methylation is associated with advanced stage and microsatellite instability in endometrial carcinoma. Int J Cancer 2001, 91:22-26 [DOI] [PubMed] [Google Scholar]