Abstract
The detection of t(14;18) translocations is widely used for the diagnosis and monitoring of follicular lymphomas displaying a high prevalence for this aberration. Cytogenetics, Southern blotting, and polymerase chain reaction (PCR) are commonly used techniques. It is generally believed that the vast majority of the breakpoints occurs on chromosome 18 in the major breakpoint region (mbr) and the minor cluster region (mcr). Yet, by improving long-distance PCR protocols we identified half of the breakpoints outside of these clusters. Our study included biopsies from 59 patients with follicular lymphoma. Seventy-one percent carried translocations detectable with our long-distance PCR protocol. The novel primer sets were derived from the hitherto uncharacterized 25-kb-long stretch between mbr and mcr that we have sequenced for this purpose. Sequence analysis of the novel breakpoints reveals a wide distribution between mbr and mcr displaying some clustering 16 kb downstream from the BCL2 gene. By including a primer for this intermediate cluster region in standard PCRs we could also improve the detection of t(14;18) translocations in formalin-fixed and paraffin-embedded biopsies. Our new PCRs are highly sensitive, easy to perform, and thus well suited for routine analysis of t(14;18) translocations for the primary diagnosis of follicular lymphoma and surveillance of minimal residual disease.
Follicular lymphoma, one of the most common non-Hodgkin’s lymphomas of B-cell type in the Western world, is closely associated with a t(14;18) translocation. 1 This chromosomal translocation juxtaposes the BCL2 locus on chromosome 18q21.3 to the immunoglobulin heavy chain gene (IGH) locus on chromosome 14q32.33. 2,3 Consequently, the BCL2 gene is subjected to the control of the IGH Eμ-enhancer leading to the overexpression of the anti-apoptotic protein Bcl-2. 4-6 The resulting suppression of apoptosis probably represents the initial step of the malignant transformation that facilitates the accumulation of further genomic lesions eventually leading to uncontrolled cell proliferation and progression to a follicular lymphoma. 7
Hence, the demonstration of the t(14;18) translocations in biopsy samples has been widely used as an accessory diagnostic tool for the identification of follicular lymphomas. Because of its relative simplicity, polymerase chain reaction (PCR) is nowadays the most commonly applied technique to detect this translocation. Other more laborious approaches include Southern blotting, conventional karyotyping, fluorescence in situ hybridization, and fiber fluorescence in situ hybridization. In the past, the standard PCR detection methods have been mainly based on observations that the breakpoints on chromosome 18 cluster at two sites: in a 150-bp-long region located in the untranslated 3′-portion of the third exon of the BCL2 gene 2 and in a 500-bp DNA stretch ∼30 kb downstream of the BCL2 locus. 8 These clusters have been named major breakpoint region (mbr) and minor cluster region (mcr), respectively. On chromosome 14, breakpoints are predominantly found within the joining elements (JH) of the immunoglobulin heavy chain gene suggesting an aberrant recombination process as the primary cause for the translocation. Because of these apparently restricted breakpoint localizations, primers immediately 5′ to the mbr and the mcr clusters on chromosome 18 and in conserved portions of the JH elements on chromosome 14 have been widely used for standard PCR assays. The combination of these mbr or mcr primers with a JH consensus primer gives rise to PCR products in the range of 150 and 500 bp, respectively, allowing a successful amplification even starting from formalin-fixed and paraffin-embedded tissues.
In recent years, a large number of studies have analyzed t(14;18) translocations in follicular lymphoma. 9 Surprisingly, the detection ratios of BCL2-IGH rearrangements varied greatly among these reports, likely being explained by the different methodologies used. From recent studies, 10,11 using either a long-distance or an inverse PCR approach, it has become evident that the breakpoints on chromosome 18 are not only occurring within the mbr and mcr loci, but are often scattered between these two cluster regions. In consequence, these breakpoints escape the detection by conventional PCR techniques leading to a large number of false-negative results.
Although the long-distance PCR (LD-PCR) protocol of Akasaka and co-workers 10 significantly enhances the detection rate of t(14;18) translocations in follicular lymphomas, it is not easily included in routine laboratory assays. Depending on the breakpoint localization, fragments up to 23 kb have to be amplified. In this size range the sensitivity of the assay is greatly affected by the quality of the DNA and minimal tissue infiltrates of lymphoma cells may be missed. Furthermore, exact breakpoint localization is often impossible because of the lack of sequence data covering the stretch between mbr and mcr.
We therefore isolated and sequenced the hitherto unknown genomic region between these two cluster regions, designed sets of new primer pairs for shorter t(14;18) target amplifications, and compared our improved PCR assay with the widely used standard PCR approaches.
Materials and Methods
Tumor Specimens
For this study, we have analyzed frozen tissues from 59 patients with follicular lymphoma admitted to University Hospital, Zürich, Switzerland. The tissues were subjected to standard histochemical and immunohistochemical analysis, including Bcl-2 immunostaining. Histological classification of follicular non-Hodgkin’s lymphoma was performed according to the World Health Organization classification.
DNA Extraction
DNA was isolated from snap-frozen tumor specimens using standard protocols. Briefly, four to six 20-μm-thick cryostat sections were digested overnight at 56°C in extraction buffer (100 mmol/L NaCl, 10 mmol/L Tris buffer, pH 8, 25 mmol/L ethylenediaminetetraacetic acid, pH 8, and 0.5% sodium dodecyl sulfate) containing 100 μg/ml proteinase K (Roche Biochemicals, Rotkreuz, Switzerland). Remaining protein debris was removed by phenol-chloroform extraction. Phase separation was done in Phase Lock light tubes (Eppendorf, Basel, Switzerland) according to the manufacturer’s instruction. Precipitation of the purified DNA was performed with half a volume of 7.5 mol/L NH4OAc and 3 volumes of ice cold ethanol. The DNA was dissolved in TE buffer (10 mmol/L Tris buffer, pH 8, and 1 mmol/L ethylenediaminetetraacetic acid, pH 8) and stored at 4°C until use. The DNA concentration was measured by UV absorbance at 260 nm. The integrity of the extracted DNA was tested by performing control LD-PCRs with two different primer sets (A2-up/A2-low and B4-up/B4-low, Table 1 ▶ ).
Table 1.
Primers for PCR and Breakpoint Identification
| Primer | Nucleotide sequence 5′ > 3′ | Application | Reference |
|---|---|---|---|
| s-MBR | GAGAGTTGCTTTACGTGGCC | Standard PCR t(14;18) | Liu et al. 21 |
| s-mcr | CGCTTGACTCCTTTACGTGC | Standard PCR t(14;18) | Liu et al. 21 |
| s-icr | TCGTTCTCAGTAAGTGAGAGTGC | Standard PCR t(14;18) | This study |
| s-JH | ACCTGAGGAGACGGTGACC | Standard PCR t(14;18) | Liu et al. 21 |
| A1-up | CACAAGTGAAGTCAACATGCCTGCCCCAAACAAAT | Long distance PCR t(14;18) | Akasaka et al. 10 (MBR01) |
| A2-up | TTATGGTTATGTGTCTTTCTGGGGATGG | Long distance PCR t(14;18) | This study |
| A3-up | GGTAGAGGTGAATACCCCAGGGCTGAGCAGGAAGG | Long distance PCR t(14;18) | Akasaka et al. 10 (mcr01) |
| A4-up | TGTTGGTTGACATTTGATGGCTTTGCTGAGAGGTA | Long distance PCR t(14;18) | Akasaka et al. 10 (mcr02) |
| B1-up | GTCTCAATGGGTTTTGCTCCTTCA | Long distance PCR t(14;18) | This study |
| B2-up | GCCACCATGCTAGGGTAGTTTTTGTA | Long distance PCR t(14;18) | This study |
| B3-up | TTGCTAATGACATCCTCTAAGTCCTCCTCT | Long distance PCR t(14;18) | This study |
| B5-up | CAGAGAGCCAGAAGCGTCAGGTC | Long distance PCR t(14;18) | This study |
| Eμ-low | TTGTCCAGGTGTTGTTTTGCTCAGTAG | Long distance PCR t(14;18) | This study |
| A2-up | TTATGGTTATGTGTCTTTCTGGGGATGG | LD-PCR (3′-BCL2 cloning) | This study |
| A2-low | GGCTGCTCTGTTATTTCTCGTCTGATG | LD-PCR (3′-BCL2 cloning) | This study |
| B4-up | TGAAAGAAACGAAAGCAACAGGAACA | LD-PCR (3′-BCL2 cloning) | This study |
| B4-low | GGGAAGGAAGCCAGGGAAACATT | LD-PCR (3′-BCL2 cloning) | This study |
| Eμ-B | AACCAGCTTCAAGGCA | Sequencing (breakpoint identification | This study |
| 3′ JH-6 | AAAGGCCCTAGAGTGC | Sequencing (breakpoint identification | This study |
| 3′ JH-5 | GACCCTGGCAAGCTG | Sequencing (breakpoint identification | This study |
| 3′ JH-4 | GGAGAGAGGTTGTGAGGA | Sequencing (breakpoint identification | This study |
| 3′ JH-2 | GGGGGCTGCAGTGGGA | Sequencing (breakpoint identification | This study |
| 3′ JH-1 | ACCTGAGGAGACGGTGACC | Sequencing (breakpoint identification | This study |
Isolation and Sequencing of the DNA Region between mbr and mcr
Two genomic fragments covering the entire stretch between mbr and mcr were amplified using two LD-PCRs (primer pairs A2-up/A2-low and B4-up/B4-low, Table 1 ▶ ). These primer pairs were designed on the basis of previously published sequences (M14745, AB010948, AB010949, sequence from Ngan and colleagues 12 ). The cycling conditions included an initial denaturation for 1 minute at 94°C followed by 14 cycles with 98°C denaturing for 20 seconds and 20 minutes annealing/primer extension at 68°C plus 16 cycles consisting of 20 seconds of denaturing at 98°C and annealing/primer extension at 68°C for 20 minutes with 15-second increments per cycle; final extension 10 minutes at 72°C. Each reaction mixture (50 μl) contained 100 to 200 ng DNA in 1× PCR reaction buffer (TaKaRa) with 0.4 μmol/L primers, 0.4 mmol/L dNTPs, 2.5 mmol/L MgCl2, and 2.5 U LA Taq-Polymerase (TaKaRa, Otsu, Japan). The reactions were overlaid with one drop of mineral oil. PCR amplification was performed in a Cetus/Perkin-Elmer thermocycler 480.
The LD-PCR products were either directly ligated into the pGEM-T-easy vector (Promega, Madison, WI) (13.3-kb 5′-fragment) or subcloned into the pBluescript SK+ vector (Stratagene, La Jolla, CA) after digestion with either EcoRI, SacI, or PstI restriction enzymes (11.5-kb 3′-fragment). For all subcloning experiments the recombination-deficient Escherichia coli strain Sure (Stratagene) was used as host. Direct sequencing of the PCR products and/or the subcloned fragments was done on an ABI 377 sequencer using Big Dye terminator kits (Applied Biosystems, Rotkreuz, Switzerland) and custom primers.
t(14;18)-Specific Polymerase Chain Reactions for Standard and Long Target Amplification
Conventional PCRs were essentially done according to the protocol of Liu and co-workers. 21 To obtain optimal primer annealing (primer analysis with Oligo 5.0 software), the mbr and mcr standard primers were shortened at their 3′ end by two nucleotides. PCR conditions for standard PCR were as follows: initial Taq activation/DNA denaturation for 3 minutes at 96°C, followed by 35 cycles consisting of 2 minutes of denaturation at 94°C, 1 minute annealing at 58°C, and 2 minutes of primer extension at 72°C. The 50-μl reactions contained 100 ng DNA, 1× reaction buffer II (Perkin Elmer, Rotkreuz, Switzerland), 0.2 μmol/L each primer, 0.2 mmol/L dNTPs, 2.5 mmol/L MgCl2, and 2.5 U Amplitaq Gold (Perkin Elmer). PCR amplification was performed in a Perkin-Elmer thermocycler 9600.
For LD-PCR new primer pairs were designed with the Oligo 5.0 software (MedProbe, Oslo, Norway) on the basis of our novel sequence data (Table 1) ▶ . These primer sets were supplemented with previously published LD-PCR primers. 10 Identical LD-PCR-cycling conditions as described before were applied. Each DNA sample was analyzed at least two times in independent experiments.
To avoid contamination, all PCRs were prepared in a laminar flow hood and pipette tips with aerosol filters were used. Each run included a negative control, in which the DNA solution was replaced by water, and a positive control amplification of a patient’s sample with confirmed t(14;18) translocation in the corresponding region. Furthermore, the DNA quality of each extract was tested in an internal control amplification using primers specific for the versican 13 (vExon3up: CAACGATGCCTACTTTGCCACCC/vExon3low: ATCCCGTACATGACGTCACAGCG; standard PCR) or the BCL2-gene (A2-up/A2-low and B4-up/B4-low; LD-PCR).
t(14;18) Breakpoint Identification
Ten-μl aliquots of the PCR reactions were analyzed by agarose gel electrophoresis (1% agarose gels for standard PCR products and 0.5% for LD-PCR products) followed by ethidium bromide staining. PCR products were excised from the gels and isolated with an agarose gel extraction kit (Qiagen, Basel, Switzerland).
The presence of a t(14;18) translocation was verified by sequencing each patient’s sample with the corresponding PCR primers and JH-specific sequencing primers for exact breakpoint identification. The amplification products were either sequenced directly or after subcloning into pGEM-T vector (Promega). Sure bacteria (Stratagene) were used as host strain. The resulting sequences were aligned with the IGH gene locus (accession number X97051), with the BCL2 (M14745), mcr, 12 and the novel DNA sequence between these two clusters (AF325194, AF325195) using Blast2 (www.ncbi.nlm.nih.gov/BLAST/).
Results
Isolation and Sequence Determination of the Genomic Region Downstream of the BCL2 Locus between Major Breakpoint and Minor Cluster Region
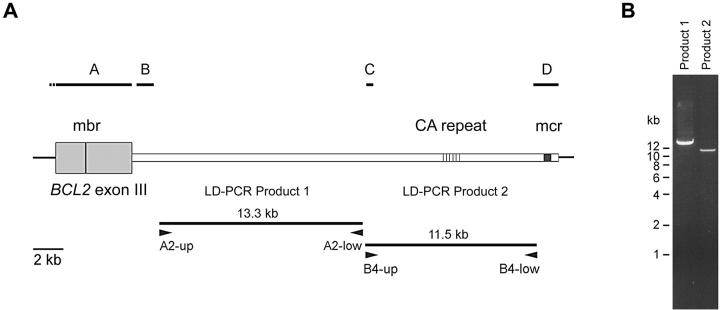
To improve PCR-based t(14;18) translocation detection assays and to allow an unambiguous identification of specific breakpoints between mbr and mcr, we have isolated and sequenced an ∼25-kb-long hitherto uncharacterized DNA stretch downstream of the BCL2 gene locus. For this purpose, we performed two LD-PCRs with primer pairs (A2-up/A2-low and B4-up/B4-low; Figure 1A ▶ ) designed on the basis of the previously published sequences in this region (GenBank accession numbers M14745, AB010948, AB010949, and from Ngan and colleagues 12 ). In this way we obtained a 13.3 kb- (A2-up/A2-low) and an 11.5-kb (B4-up/B4-low) amplification product, respectively, bridging the entire unknown sequence between mbr and mcr (Figure 1B) ▶ .
Figure 1.
PCR cloning of the genomic region spanning mbr and mcr. A: Schematic representation of the 3′BCL2 gene locus. Black bars on top indicate the localization of previously determined sequences (A, M14745; B, AB010948; C, AB010949; D, sequence from Ngan et al 12 ). Two large fragments containing the unknown sequences were amplified with LD-PCR (primer pairs A2-up/A2-low and B4-up/B4-low, respectively). B: Gel electrophoresis of LD-PCR products derived from human genomic DNA. Two percent of the reaction volumes were run on a 0.5% agarose gel and stained with ethidium bromide.
Sequence determination of these two products either by direct sequencing or after subcloning into a plasmid vector revealed a genomic sequence with a large number of repetitive elements between mbr and mcr (detected by RepeatMasker, ftp.genome.washington.edu/cgi-bin/RepeatMasker). This includes 15 SINEs (5 Alu and 10 MIR), 6 LINEs, 12 DNA transposons, 6 LTR elements, and at least 3 simple repeats. One of the latter, an ∼1-kb-long CA microsatellite-containing element 22 kb downstream of the mbr locus, was inaccessible to exact sequence analysis. Hence, the novel genomic sequence downstream of the BCL2 was submitted to GenBank in two parts: 1) the sequence stretch between mbr and the CA dinucleotide repeat (accession number AF325194), and 2) the stretch between the CA repeat and the mcr locus (accession number AF 325195). No evidence for the presence of a gene in this DNA stretch immediately downstream of the BCL2 locus could be found.
Detection of t(14;18) Translocation Using Standard PCR Analysis
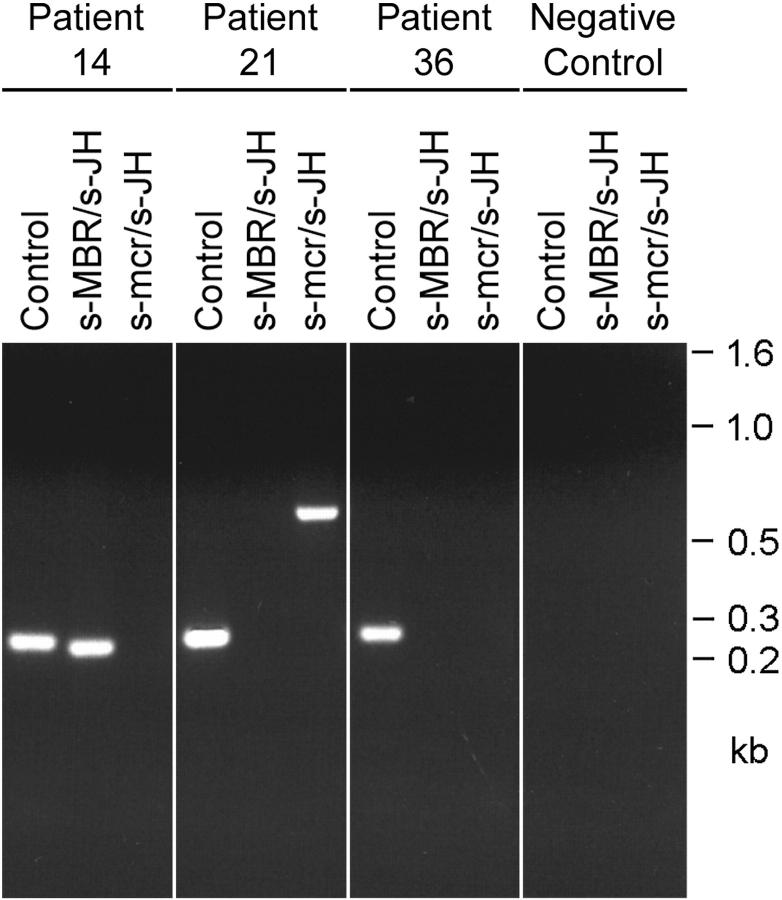
In a study of 59 patients with histologically confirmed follicular lymphoma we initially screened for the presence of a t(14;18) translocation applying the widely used standard PCRs with mbr/JH and mcr/JH primer pairs (Table 1) ▶ . In this way we could demonstrate a translocation in 36% of the samples (21 patients) including 19 samples (32.2%) with breakpoints within the mbr cluster and 2 samples (3.4%) with involvement of the mcr locus. The size of the PCR products containing breakpoints in the mbr ranged from 180 to 275 bp. The amplicons of the two mcr samples were 580-bp and 630-bp long. Representative results from the standard t(14;18) PCRs are shown in Figure 2 ▶ . In addition an ∼500-bp amplification product was obtained from patient 40 in both the mbr and the mcr reaction. Further examination of this sample revealed that the product resulted from an amplification with JH consensus primer alone (for exact breakpoint identification see separate paragraph below).
Figure 2.
Detection of t(14;18) translocations with standard PCR analysis. Representative examples of the initial screening of patients’ samples with standard PCRs using s-MBR/s-JH and s-mcr/s-JH primer pairs, respectively. Twenty percent of the amplification volumes were loaded on a 2% agarose gel stained with ethidium bromide. Direct sequencing of the PCR products confirmed breakpoints in the mbr (patient 14) and mcr (patient 21). No amplification product was obtained from the sample of patient 36 despite the presence of a t(14;18) translocation (for breakpoint determination see Table 2 ▶ and Figure 4 ▶ ). A control amplification of a 268-bp genomic fragment was included in each analysis to verify the quality of the DNA extracts. Furthermore, negative reagent controls were performed each time to exclude contamination.
Improved Detection of the t(14;18) Translocation with Newly Designed LD-PCR Assays
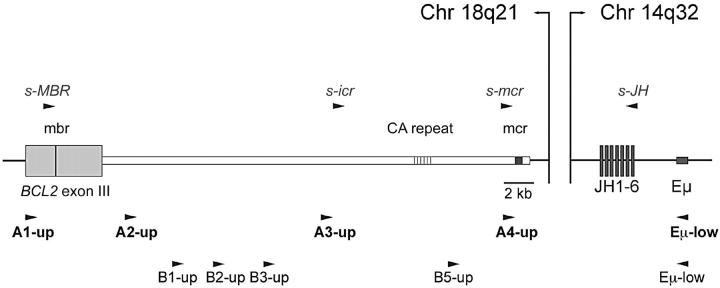
Because the standard PCRs detect only breakpoints within the two cluster regions, we have performed a second screening round using a novel LD-PCR approach. Based on our new sequence data from the genomic locus 3′ of the BCL2 gene, we established two sets of LD-PCR assays (Figure 3 ▶ , set A and B). With these assays, we covered the entire region between the third exon of the BCL2 gene and mcr allowing the detection of breakpoints outside of mbr and mcr. The new primers were positioned in regular intervals along the ∼31-kb-long sequence stretch to limit the size of the PCR products to less than 15 kb (set A) or 9 kb (set A and B), respectively.
Figure 3.
Localization of primers used in long-distance and modified standard PCRs for the detection of t(14;18) translocations. Position of upper primers in close vicinity to the BCL2 gene locus on chromosome 18 are shown together with their lower strand primer partners localized in the IGH Eμ enhancer region or the JH elements on chromosome 14. Long-distance PCR screening was initially done with primer set A and extended with primer set B (lower primer: Eμ-low). Improved standard PCR analysis included s-MBR, s-mcr, and the new s-icr upper primer together with a JH consensus primer (s-JH).
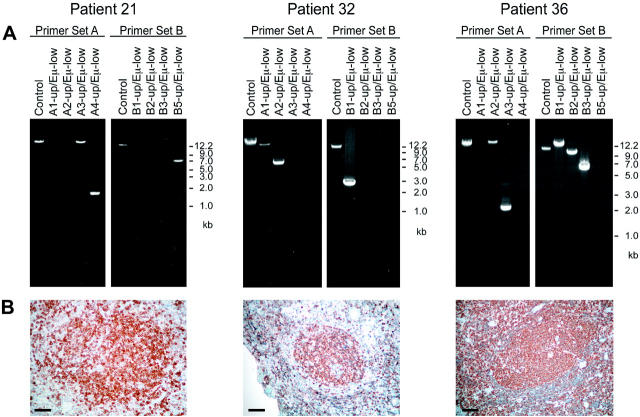
Depending on the breakpoint localization, we observed specific amplification patterns. In case the breakpoint on chromosome 18 was localized within the exon III of the BCL2 gene or immediately 3′ thereof only the LD-PCR with the A1-up/Eμ primer combination gave rise to an amplification product. In all other samples a specific set of PCR products was observed (examples in Figure 4 ▶ ), allowing an approximate positioning of the translocation breakpoint between mbr and mcr.
Figure 4.
Representative examples of t(14;18) LD-PCR analysis. A: Products of LD-PCRs with primer sets A and B were run on 0.5% agarose gels and stained with ethidium bromide. The DNA quality of each extract was tested in a reaction amplifying a 13.3-kb (set A) or a 11.5-kb control fragment (set B). Depending on breakpoint location a unique amplification pattern was observed and the approximate position of the breakpoint on chromosome 18 was established. Direct sequencing of the LD-PCR products revealed in patient 21 a breakpoint in the mcr region, in patient 32 a localization 9.5 kb downstream from MBR, and in patient 36 a translocation with involvement of the icr. B: The neoplastic cells of all three patients expressed Bcl-2 as revealed by immunohistochemistry. Scale bar, 50 μm.
Using these two primer sets, we detected another 21 t(14;18) translocations in the 38 samples that were negative in the standard PCR analysis. Furthermore, all samples being positive in the standard assays could also be detected with the LD-PCR technique. In total, we identified with this modified LD-PCR approach t(14;18) translocations in 71% (42 samples) of the follicular non-Hodgkin’s lymphomas in our study. Because the fresh-frozen samples yielded rather high-quality genomic DNA (>20 kb), no difference in sensitivity between the LD-PCR assays with primer set A alone or set A plus set B was observed.
Exact Characterization of Translocation Breakpoints by Sequence Determination
Exact breakpoint localization through sequence determination and comparison confirmed in 19 samples a translocation involving the mbr region (mbr as defined by Bakhshi and colleagues 14 ) and in 2 samples a breakpoint within the mcr (determined by Ngan and colleagues 12 ) (Figure 5 ▶ , Table 2 ▶ ). Twenty patients with a follicular lymphoma displayed breakpoints scattered throughout the 30-kb region spanning mbr and mcr and in one case (patient 42) a breakpoint ∼800-bp 5′ from the mbr, but 1.5 kb downstream from the bcl-2 translation stop codon was detected.
Figure 5.
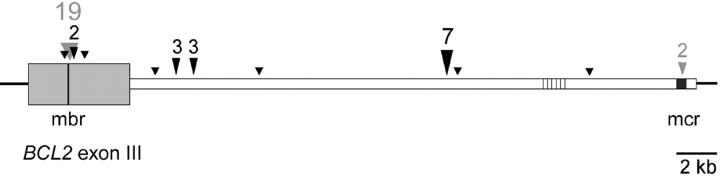
Distribution of t(14;18) breakpoints on chromosome 18. The exact positions of the breakpoints on the 3′BCL2 genomic locus were determined by sequencing of the PCR products. Black arrowheads indicate breakpoints being only detectable by LD-PCR. Breakpoints within the MBR and mcr clusters, which were also amplified by standard PCRs, are shown with gray arrowheads. In certain regions clustering of breakpoints was observed. In those cases, the numbers of breakpoint in close vicinity to each other are indicate above the arrowheads.
Table 2.
t(14;18) Breakpoint Detection
| Patient | Bcl-2 IHC | BCL2 BrP | JH (D) | Standard/ LD-PCR | BCL2 sequence | de novo sequence (+D) | JH sequence | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A: t(14;18) mbr breakpoints detected with standard and LD-PCR | ||||||||||||||
| 1 | pos | mbr | 5 | +/+ | ..CTCCTGCCCTCCTTC> | GCTT> | CAACTGGTTCGACCCCT… | |||||||
| 2 | pos | mbr | 6 | +/+ | ..CTCCTGCCCTCCTTC> | TGACTCTTCGGTGATT | ||||||||
| CGACTCAAAAAGTGGG | ||||||||||||||
| TACCCCG> | ACTACGGTATGGACGT… | |||||||||||||
| 3 | pos | mbr | 4 | +/+ | ..CTCCTGCCCTCCTTC> | GATAGCAACCGT> | GGCCAGGGAACCCTGG… | |||||||
| 4 | pos | mbr | 6 | +/+ | ..CCTGCCCTCCTTCCG> | AGA> | GGACGTCTGGGGCCAA… | |||||||
| 5 | pos | mbr | 6 | +/+ | ..CCTTCAGGGTCTTCC> | GACTAGGCGCGGTG> | CTACTACTACGGTATGG.. | |||||||
| 6 | pos | mbr | 5 | +/+ | ..GGGTCTTCCTGAAA> | GGGAGAACCTTTT> | TTCGACCCCTGGGGCCA… | |||||||
| 7 | nd | mbr | 6 | +/+ | ..TGAAATGCAGTGGT> | CCCAATGC> | CTACTACTACGGTATGG… | |||||||
| 8 | pos | mbr | 4 | +/+ | ..AAATGCAGTGGTG> | AC> | GACTACTGGGGCCAGG… | |||||||
| 9 | pos | mbr | 6 | +/+ | ..AAATGCAGTGGTGC> | AAATCACGGTCCACC> | TTACTACTACTACTACG… | |||||||
| 10 | pos | mbr | 6 | +/+ | ..TGCAGTGGTGCTTA> | GGCCCCGGGGTACGG | ||||||||
| TATTCC> | CTACTACTACTACTACA… | |||||||||||||
| 11 | pos | mbr | 6 | +/+ | ..TGCAGTGGTGCTTA> | TCCTCCCTTTCCCAGC | ||||||||
| TGCTAAGTGTGGTC> | CTACTACTACTACTACA… | |||||||||||||
| 12 | pos | mbr | 6 | +/+ | ..GCAGTGGTGCTTAC> | TATGGCGTCTG> | TACTACTACGGTATGG… | |||||||
| 13 | pos | mbr | 4 | +/+ | ..GCAGTGGTGCTTAC> | CCGCCGGTCA> | ACTACTGGGGCCAGGG… | |||||||
| 14 | pos | mbr | 6 | +/+ | ..TCCACCAAGAAAGC> | CCCAAAGAGGTGAGAA | ||||||||
| (3–10) | AATATGGTTCGGGGAGTT | |||||||||||||
| ATTGCTCGAGGCATATG | ||||||||||||||
| GGGATCATCATCATCA | ||||||||||||||
| TCATC> | ATGGACGTCTGGGGCA… | |||||||||||||
| 15 | pos | mbr | 6 | +/+ | ..TCCACCAAGAAAGC> | CCCGACGAGATAC> | TACTACTACTACTACGG… | |||||||
| 16 | nd | mbr | 4 | +/+ | ..TGGTATGAAGCCAG> | GACCGATTCCCGACG | ||||||||
| TT> | CTTTGACTACTGGGGCC… | |||||||||||||
| 17 | pos | mbr | 5 | +/+ | ..TGAAGCCAGACCTC> | GGGTGAGTTTCCCCC | ||||||||
| GCTCTGTA> | AACTGGTTCGACCCCTG… | |||||||||||||
| 18 | pos | mbr | 6 | +/+ | ..GCCAGACCTCCCCG> | CCGTATAGTT> | GTATGGACGTCTGGGG… | |||||||
| 19 | pos | mbr | 5 | +/+ | ..CAGACCTCCCCGGC> | TCC> | ACTGGTTCGACCCCTGG… | |||||||
| B: t(14;18) mcr breakpoints detected with standard and LD-PCR | ||||||||||||||
| 20 | pos | mcr | 6 | +/+ | ..GGACAATCTCCTGC> | CGCGCGACGCGAGCG | ||||||||
| GTCGGTTTGGAGGGT | ||||||||||||||
| CTATTATTCGAATAGG | ||||||||||||||
| CATCGGCCGAGCC> | CTACGGTATGGACGTCT… | |||||||||||||
| 21 | pos | mcr | 6 | +/+ | ..ATCTCATGCTGCAA> | > | CTACTACTAC… | |||||||
| C: t(14;18) intermediate breakpoints detected with LD-PCR only | ||||||||||||||
| 22 | pos | 3′-mbr | inv. | −/+ | ..GAGTCAGTTTGCAG> | GCGCCAGCCGGATGT | ||||||||
| 5′-5* | GGGAGATGGTA> | CACATTGTGACAACAA… | ||||||||||||
| 23 | pos | 3′-mbr | 4 | −/+ | ..CAGTTTGCAGTATG> | TCTTCTCCTTTTCTAC | ||||||||
| CCCTCCGTGGGTAAT> | TTTGACTACTGGGGCCA… | |||||||||||||
| 24 | nd | 3′-mbr | 6 | −/+ | ..GCCAGGGTCAGAG> | CTCGTTACTAC> | ATGGACGTGTGGGGCA… | |||||||
| 25 | pos | Far 3′- | 6 | −/+ | ..TTATACCATGTAAA> | ACACGAAAACCCAGTA | ||||||||
| mbr | (6-6) | TAGCAGCTCGGCGACG | ||||||||||||
| GTGGGCCGATAC> | CTACTACTACTACGGTA… | |||||||||||||
| 26 | pos | Far 3-mbr | 6 | −/+ | ..AAATGTGAATTCTT> | C> | CTACGGTATGGACGTC… | |||||||
| 27 | pos | Far 3′-mbr | 5 | −/+ | ..TTCAGACTGGCCTC> | TCT> | AACTGGTTCGACCCCTG… | |||||||
| 28 | pos | Far 3′-mbr | 4 | −/+ | ..GAAGAACAGCTCG> | GCCC> | ACTACTGGGGCCAGGG… | |||||||
| 29 | pos | Far 3′-mbr | 6 | −/+ | ..ATCAAACCATTGTG> | ATGGCAGACC> | CTACTACTACTACGGTA… | |||||||
| 30 | pos | Far 3′- | 4 | −/+ | ..ATCAAACCATTGTG> | TAGAATCGGCACAGTG | ||||||||
| mbr | (3–10) | TCACAGAGTTCGGGG | ||||||||||||
| RSS† | AGTTATCAAGTCTT> | CTTTGACTACTGGGGCC… | ||||||||||||
| (Continued) | ||||||||||||||
| Patient | Bcl-2 IHC | BCL2 BrP | JH (D) | Standard/ LD-PCR | BCL2 sequence | de novo sequence (+D) | JH sequence | |||||||
| 31 | pos | Far 3′- | 6 | −/+ | ..TCAAACCATTGTGG> | ACACAGTGTCACAGAGT | ||||||||
| mbr | (3–10) | …TGGTTCGGGGAG | ||||||||||||
| RSS† | TCCCGCAGCGAGAT> | ATTACTACTACTACTAC… | ||||||||||||
| 32 | pos | Very Far 3′-mbr | 6 | −/+ | ..TTAAGTCCTCTTTTT> | CGAAGTGGTGG> | CGGTATGGACGTCTGG… | |||||||
| 33 | pos | 5′-mcr | 6 | (+)§/+ | ..GTGAGAGTGCAGA> | TCCGGG> | CTACTACGGTATGGAC… | |||||||
| 34 | pos | 5′-mcr | 6 | (+)§/+ | ..AGTGCAGAATCTGA> | TCCTGGGGGGTTACGG> | TACTACTACTACGGTAT… | |||||||
| 35 | pos | 5′-mcr | ؇ | − /+ | ..CAAATGAGTAACTC> | GTTGGAG> | CTGTCGGAGTGGGTGA… | |||||||
| 36 | pos | 5′-mcr | 6 | (+)§/+ | ..AAATGAGTAACTCC> | ACCTAGGGGTGCTAC | ||||||||
| AATAC> | ATGGACGTCTGGGGCA… | |||||||||||||
| 37 | pos | 5′-mcr | 6 | (+)§/+ | ..AAATGAGTAACTCC> | CACGCCACC> | ACTACTACTACGGTAG… | |||||||
| 38 | pos | 5′-mcr | 6 | (+)§/+ | ..AAATGAGTAACTCC> | TGGCCC> | TTACTACTACTACTACG… | |||||||
| 39 | pos | 5′-mcr | 6 | (+)§/+ | ..AGGTAACTTTTTAG> | GAGGGATAAGGAATA | ||||||||
| CTTGGAAGAATATCAA | ||||||||||||||
| TCCGCCA> | TACTACTACTACGGTAT… | |||||||||||||
| 40 | pos | 5′-mcr | 5* | − /+ | ..GTAGCTAGTAGCTA> | TTATCTACG> | GACAACAATGCCAGGA… | |||||||
| 41 | pos | 3′-Sat | 6 | − /+ | ..CCTTCACCTCCTCTT> | CTCCTCCC> | GTATGGACGTCTGGGG… | |||||||
| D: t(14;18) 5′-mbr breakpoint detected with LD-PCR only | ||||||||||||||
| 42 | nd | 5′-mbr | ؇ | − /+ | ..TGGTTTTATTTGAA> | > | TTTCAAGTATATTAATT… | |||||||
IHC, Immunohistochemistry; BrP, breakpoint; pos, positive; nd, not determined.
*t14;18 translocation plus partial inversion of the JH segment.
†Contains sequence 5′ to D3–10 including the complete 3′-recombination signal sequence of D3-9.
‡BCL2 gene locus directly joined to region immediately upstream of IGH Eμ-enhancer.
§In standard PCR, this translocation can only be amplified with the new intermediate cluster primer pair (s-icr/s-JH).
Apart from mbr and mcr, some degree of clustering was observed ∼5.3 kb and 6.2 kb downstream of the mbr locus (three samples each). In addition, a very high prevalence for chromosome 18 breakpoints was observed ∼19-kb 3′ from the mbr. Seventeen percent of the (t14;18) translocations (seven samples) could be detected in an ∼200-bp-long sequence portion, we now call the “intermediate cluster region” (icr). In comparison, we observed in our study an involvement of mbr or mcr in 45% and 5% of the translocations, respectively. Interestingly, the breakpoints in this new cluster were identical in patients 36, 37, and 38 and differed only by 1 bp in patient 35. Despite the similarities on the 3′-BCL2 side, N-nucleotide additions and breakpoints in the IGH locus on chromosome 14 were unique in each of these samples. A representative LD-PCR analysis of a patient with a breakpoint in this icr is shown in Figure 4 ▶ (sample 36).
On chromosome 14 the translocation involved in six samples the IGH joining element JH4 (14%), five times JH5 (12%), and in 23 patients the JH6 segment (55%). In an additional two samples (4.75%) the translocation occurred further downstream lacking all JH segments on chromosome 14+, but leaving the Eμ enhancer intact. Twice we observed a link between BCL2 or 3′ sequences thereof to partially rearranged (D-J) diversity elements (4.75%) and another two times the 3′-recombination signal sequence (3′-RSS) of D3-9 participated in the joint at the breakpoint (4.75%). Finally, more complex rearrangements including a t(14;18) translocation and a partial inversion of the JH region were found in two instances (4.75%). At these breakpoints some of the joining elements were aberrantly arranged in opposing directions. In consequence a conventional PCR with JH consensus primers alone yielded in one analysis a short amplification product (patient 40).
All except for one breakpoint fusion sequences included on chromosome 14+ variable numbers of N-nucleotide additions. No such modifications were present in a rather unusual translocation joining a sequence 5′ to the mbr directly with a DNA stretch 3′ to the JH segments (patient 42).
Improved Detection of t(14;18) Translocations in Formalin-Fixed Tissues
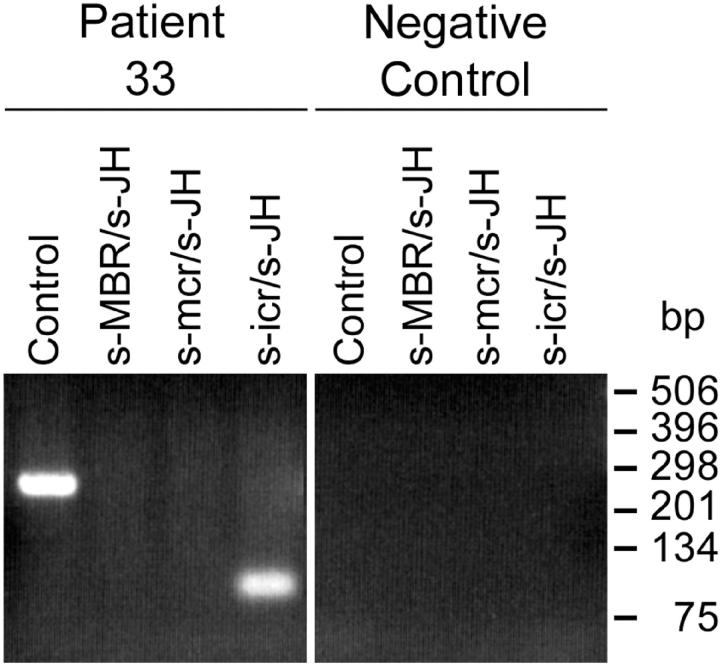
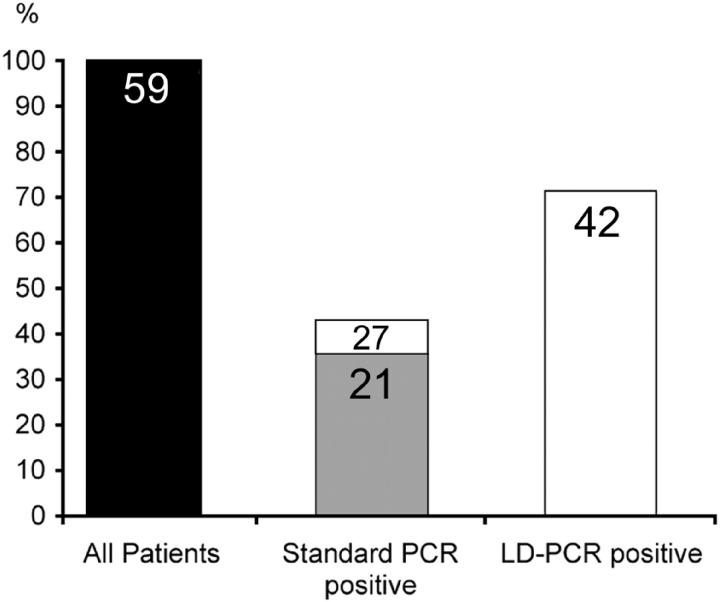
Our follicular lymphoma study demonstrates that breakpoints occur more frequently within the icr relative to translocations involving the mcr. Nevertheless, neither the widely used protocols for standard PCRs nor our LD-PCRs are suitable to detect breakpoints in the icr in DNA extracts from tissues that have been formalin-fixed and paraffin-embedded. To make this novel cluster accessible to such analysis, we have designed a new primer (s-icr) to supplement the widely used standard PCRs with s-MBR and s-mcr primers (Table 1) ▶ . The combination of s-icr with the JH consensus primer detected in six instances breakpoints located in the icr also starting from formalin-fixed tissue samples (for example see Figure 6 ▶ ). Only in one case with a breakpoint downstream from JH6 on chromosome 14 (patient 35), the PCR with the s-icr/JH primer pair failed to detect a translocation because of the absence of the JH primer-annealing site. Hence, by including the s-icr/JH primer pair in the standard PCR assay we could significantly increase the overall t(14;18) detection rate in DNA extracts from formalin-fixed tissues from 36 to 46% (Figure 7) ▶ .
Figure 6.
Improved standard PCR analysis for the detection of t(14;18) translocation in formalin-fixed tissues. By supplementing the standard MBR- and mcr-specific PCR analysis with the s-icr/JH primer pair, frequently occurring breakpoints within the icr can be detected in extracts even from formalin-fixed and paraffin-embedded biopsies (example: patient 33; 2% agarose gel electrophoresis, ethidium bromide staining).
Figure 7.
Frequency of t(14;18) translocations detected in follicular lymphomas by standard and LD-PCR assays. In comparison to the standard PCR protocols using MBR and mcr primers, our LD-PCR assay greatly increases the detection rate of t(14;18) translocations in follicular lymphoma samples from European patients (from 36 to 71%). Some improvement of the standard PCR can be achieved by including mbr-, mcr-, and icr-specific primers in the analysis (from 36 to 46%). Numbers in the columns indicate number of patients.
Discussion
The large variance in detection frequencies of t(14;18) translocations in follicular lymphomas has in the past often been attributed to ethnic and geographic differences and only partly to methodological limitations. Whereas a high prevalence for the occurrence of t(14;18) has been reported for studies in the United States (71 to 84%), the detection rates were significantly lower in Europe (41 to 61%) and in the Far East (32 to 39%). 9 Nonetheless, primarily discrepant frequencies were also described in studies from different laboratories in the same geographic region, even when similar techniques were applied. 15 This confusion may to some extent be caused by differences in histological diagnosis, but seems more likely to originate from the absence of tools to independently validate the results obtained by cytogenetic or molecular analysis. This gap is recently being filled by the analysis of t(14;18) translocations with new technologies such as fiber fluorescence in situ hybridization, 16 inverse and LD-PCR 10,11 supplementing conventional cytogenetics, Southern blotting, and standard PCR assays. Our characterization of an extended sequence portion 3′ to the BCL2 gene and the design of new LD-PCR approaches has not only allowed us to detect a high number of novel breakpoints on chromosome 18 but also has enabled us to verify these t(14;18) translocations by direct sequence comparison.
Our study makes evident that only half of the breakpoints localize within the commonly analyzed clusters mbr and mcr. The other breakpoints are widely spread throughout the sequence stretch between mbr and mcr, nevertheless displaying some degree of clustering. This is most pronounced in the icr, where we have observed more than three times more breakpoints than in the mcr. These data are supported by the findings of Willis and colleagues 11 and Akasaka and colleagues, 10 who also identified a few breakpoints in this region.
The observation that a significant proportion of the chromosome 18 breakpoints localize far distant from mbr and mcr explains why standard PCR assays detect fewer t(14;18) translocations than other techniques. Although the sensitivity of these assays can be improved by including primers for the icr, approximately a third of the t(14;18) translocations remain inaccessible to these conventional PCR methods (Figure 7) ▶ . Nevertheless, in situations in which only formalin-fixed and paraffin-embedded biopsies are available, standard PCRs with s-MBR/JH, s-mcr/JH and s-icr/JH primer combinations may be the only detection technique available, because of extensive DNA degradation in fixed samples.
The 71% proportion of t(14;18) translocations we detect with our LD-PCR protocol in follicular lymphomas correlates well with results from cytogenetic and Southern blot studies from the United States and is significantly higher than similar data from Europe and the Far East. 9 Because we found in each of the LD-PCR-positive samples distinct translocation sequences, false-positive results because of unspecific amplification and contamination can be excluded. We assume that the difference in sensitivity between European and North American Southern blot studies may rather arise from methodological differences than from ethnic and/or geographic peculiarities. Theoretically, all breakpoints in our study could have been detected in Southern blots using an mbr and an mcr probe together with a BamHI restriction, despite the fact that BamHI does not cover the entire stretch between mbr and mcr. However, some of the breakpoints would have given rise to fragments larger than 30 kb being well beyond the separation capacity of conventional agarose gel electrophoresis. Even more significant, 8 and 16 breakpoints of our study would have been missed in protocols using HindIII and EcoRI digestions, respectively. Because not all of the European studies included BamHI in the Southern blot analysis, a major proportion of the breakpoints may have remained undiscovered.
Exact t(14;18) breakpoint characterization by directly sequencing the PCR products not only allows the verification of the amplification specificity, it may also give insight into the mechanisms that lead to these aberrant gene rearrangement events. Various models have been presented, all proposing an involvement of the VDJ recombination machinery in the translocation. Whereas a participation of RAG-1 and RAG-2 in the formation of double-strand breaks in the IGH gene on chromosome 14 is undisputed, there are still doubts about the mechanisms generating the breakpoints on chromosome 18. Cryptic recombination signal sequences (RSS) 17 within or near the BCL2 gene locus or RAG-mediated transposition events 18 may be involved in the translocation process. In our study, we have found only one breakpoint localization with a potentially cryptic RSS ∼3.3 kb downstream of the BCL2 gene. It contains a putative nonamer and heptamer sequence with a 22-bp spacer still conforming to the 12/23 rule. 19 At this site, the chromosome 18 sequence of three patients either ended directly after the heptamer (patients 29 and 30) or 1-bp 3′ thereof (patient 31). Recently, the same breakpoint location has been identified in a study by Vandraager and colleagues. 20 In patients 30 and 31 of our study, these putative RSS were linked to the 3′-RSS of the D3-9 element in the IGH locus. Hence, these translocations may have resulted from an aberrant signal joining, despite the unusual presence of N-nucleotides in both joints. In one instance (patient 22) we found also a breakpoint that partly conforms to the transposition model, in which one signal end attacks the double-stranded DNA of another chromosome. The addition of N-nucleotides as present in our sample is, however, rather uncharacteristic for such a mechanism. The high variability of breakpoint localization and junctional sequences observed in the rest of the samples argues against a single aberrant process causing all t(14;18) translocations. It seems rather likely, that different defects in the RAG-driven immunoglobulin rearrangement and eventually even joining of randomly broken chromosomal ends may lead to similar translocations becoming only manifest as a lymphoma after an extended cellular selection.
In conclusion, our protocol has greatly improved the detection rate of t(14;18) translocations in follicular lymphomas using a relatively simple PCR technique. This method is suitable for routine diagnosis in a standard molecular pathology laboratory. Our LD-PCR and sequencing approach is rapid, highly specific, and sensitive and could eventually replace Southern blotting analysis commonly used for samples in which high-molecular weight DNA is available. By determining a large sequence stretch downstream from the BCL2 gene locus, we have furthermore provided the basis for the exact characterization of a series of novel breakpoints distant from mbr and mcr, finally allowing the development of sensitive patient-specific PCR assays for the monitoring of therapeutic success and the detection of minimal residual disease.
Acknowledgments
We thank Karin Camiu and Regula Städeli-Brodbeck for the preparation of the sequencing reactions and Rita Moos for cutting the cryostat sections.
Footnotes
Address reprint requests to Dieter R. Zimmermann, Laboratory of Molecular Biology, Institute of Clinical Pathology, Department of Pathology, Schmelzbergstr. 12, CH-8091 Zürich, Switzerland. E-mail: dieterzi@pathol.unizh.ch.
Present addresses of A. A.-H.: HNO-Universitätsklinik Freiburg, Killianstr. 5, 79106 Freiburg, Germany; and of B. H.: Medizinische Abteilung, Spital Limmattal, Urdorferstr. 100, 8952 Schlieren, Switzerland.
References
- 1.Fukuhara S, Rowley JD, Variakojis D, Golomb HM: Chromosome abnormalities in poorly differentiated lymphocytic lymphoma. Cancer Res 1979, 39:3119-3128 [PubMed] [Google Scholar]
- 2.Cleary ML, Sklar J: Nucleotide sequence of a t(14;18) chromosomal breakpoint in follicular lymphoma and demonstration of a breakpoint-cluster region near a transcriptionally active locus on chromosome 18. Proc Natl Acad Sci USA 1985, 82:7439-7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM: Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science 1984, 226:1097-1099 [DOI] [PubMed] [Google Scholar]
- 4.Korsmeyer SJ: BCL-2 gene family and the regulation of programmed cell death. Cancer Res 1999, 59:1693s-1700s [PubMed] [Google Scholar]
- 5.Kroemer G: The proto-oncogene Bcl-2 and its role in regulating apoptosis. Nat Med 1997, 3:614-620 [DOI] [PubMed] [Google Scholar]
- 6.Strasser A: Dr. Josef Steiner Cancer Res Prize Lecture: the role of physiological cell death in neoplastic transformation and in anti-cancer therapy. Int J Cancer 1999, 81:505-511 [DOI] [PubMed] [Google Scholar]
- 7.Vaux DL, Cory S, Adams JM: Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 1988, 335:440-442 [DOI] [PubMed] [Google Scholar]
- 8.Cleary ML, Galili N, Sklar J: Detection of a second t(14;18) breakpoint cluster region in human follicular lymphomas. J Exp Med 1986, 164:315-320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segel MJ, Paltiel O, Zimran A, Gottschalk-Sabag S, Schibi G, Krichevski S, Ludkovski O, Yehuda DB: Geographic variance in the frequency of the t(14;18) translocation in follicular lymphoma: an Israeli series compared to the world. Blood Cells Mol Dis 1998, 24:62-72 [DOI] [PubMed] [Google Scholar]
- 10.Akasaka T, Akasaka H, Yonetani N, Ohno H, Yamabe H, Fukuhara S, Okuma M: Refinement of the BCL2/immunoglobulin heavy chain fusion gene in t(14;18)(q32;q21) by polymerase chain reaction amplification for long targets. Genes Chromosom Cancer 1998, 21:17-29 [DOI] [PubMed] [Google Scholar]
- 11.Willis TG, Jadayel DM, Coignet LJ, Abdul-Rauf M, Treleaven JG, Catovsky D, Dyer MJ: Rapid molecular cloning of rearrangements of the IGHJ locus using long-distance inverse polymerase chain reaction. Blood 1997, 90:2456-2464 [PubMed] [Google Scholar]
- 12.Ngan BY, Nourse J, Cleary ML: Detection of chromosomal translocation t(14;18) within the minor cluster region of bcl-2 by polymerase chain reaction and direct genomic sequencing of the enzymatically amplified DNA in follicular lymphomas. Blood 1989, 73:1759-1762 [PubMed] [Google Scholar]
- 13.Zimmermann DR, Ruoslahti E: Multiple domains of the large fibroblast proteoglycan, versican. EMBO J 1989, 8:2975-2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakhshi A, Wright JJ, Graninger W, Seto M, Owens J, Cossman J, Jensen JP, Goldman P, Korsmeyer SJ: Mechanism of the t(14;18) chromosomal translocation: structural analysis of both derivative 14 and 18 reciprocal partners. Proc Natl Acad Sci USA 1987, 84:2396-2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson PW, Swinbank K, MacLennan S, Colomer D, Debuire B, Diss T, Gabert J, Gupta RK, Haynes A, Kneba M, Lee MS, Macintyre E, Mensink E, Moos M, Morgan GJ, Neri A, Johnson A, Reato G, Salles G, van’t Veer MB, Zehnder JL, Zucca E, Selby PJ, Cotter FE: Variability of polymerase chain reaction detection of the bcl-2-IgH translocation in an international multicentre study. Ann Oncol 1999, 10:1349–1354 [DOI] [PubMed]
- 16.Vaandrager JW, Schuuring E, Raap T, Philippo K, Kleiverda K, Kluin P: Interphase FISH detection of BCL2 rearrangement in follicular lymphoma using breakpoint-flanking probes. Genes Chromosom Cancer 2000, 27:85-94 [PubMed] [Google Scholar]
- 17.Tsujimoto Y, Gorham J, Cossman J, Jaffe E, Croce CM: The t(14;18) chromosome translocations involved in B-cell neoplasms result from mistakes in VDJ joining. Science 1985, 229:1390-1393 [DOI] [PubMed] [Google Scholar]
- 18.Hiom K, Melek M, Gellert M: DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 1998, 94:463-470 [DOI] [PubMed] [Google Scholar]
- 19.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG: The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol 2000, 18:495-527 [DOI] [PubMed] [Google Scholar]
- 20.Vaandrager JW, Schuuring E, Philippo K, Kluin PM: V(D)J recombinase-mediated transposition of the BCL2 gene to the IGH locus in follicular lymphoma. Blood 2000, 96:1947-1952 [PubMed] [Google Scholar]
- 21.Liu J, Johnson RM, Traweek ST: Rearrangement of the BCL-2 gene in follicular lymphoma. Detection by PCR in both fresh and fixed tissue samples. Diagn Mol Pathol 1993, 2:241-247 [PubMed] [Google Scholar]