Abstract
It is generally assumed that follicular lymphomas (FL) not only morphologically resemble normal germinal centers but have retained some functional characteristics of their non-neoplastic counterparts as well. Recent IgV gene analyses on a panel of FLs however, strongly suggested that FLs do not retain the capacity of somatic hypermutation and are not being selected on basis of the quality of their mIgV regions. To extend these findings, we investigated the follicular organization and class switching in a FL that consisted of both IgM- and IgG-expressing tumor cells with a high somatic mutation load and significant intraclonal VH gene diversity. VH-Cμ and VH-Cγ gene transcripts were amplified and sequenced from samples of approximately 50 tumor cells, isolated from frozen tissue sections by laser microdissection. We identified many different subclones and obtained limited evidence of subclone dominance in individual follicles. Remarkably, several subclones were found scattered over different follicles. All samples contained IgM- and IgG-expressing tumor cells with, in general, non-identical mutation patterns, which is not in support of ongoing class switching. Accordingly, no switch circle recombination products were found. The findings indicate that the neoplastic follicles lack the organization and functions typical of reactive germinal centers.
During normal B cell maturation, the developing cells are continuously tested for expression and quality of their B cell antigen receptors (BCR). 1,2 This principle most obviously rules the germinal center (GC) reaction, where B cells compete to obtain survival signals by binding antigen that is presented at the surface of follicular dendritic cells (FDCs). 3 This competition, combined with changes that are introduced in the immunoglobulin variable (IgV) region genes due to somatic hypermutation, 4 results in clonal evolution of the BCRs and forms the molecular basis of affinity maturation of the humoral immune response.
Several in situ studies have provided insight into the selection and differentiation processes that take place within germinal centers. 5-7 Both anatomically and functionally, germinal centers seem rather closed compartments. It has been shown that, although a single B cell clone can be seeded into multiple GCs, clonal evolution thereafter occurs independently in these GCs. 7,8 The stringency of selection within this environment was demonstrated by the fact that the B cell populations of individual GCs, initially being highly polyclonal, become oligoclonal or even monoclonal within a relatively short time span. 5,6,9
Follicular lymphoma (FL) is considered as the prototype of a germinal-center cell-derived B cell non-Hodgkin’s lymphoma, 10 as the centroblast- and centrocyte-like tumor cells proliferate in nodular networks of non-neoplastic FDCs. Many investigators have demonstrated that FLs carry heavily mutated IgV genes. 11-13 Moreover, based on the findings of intraclonal IgV gene diversity and the co-presence of different heavy (H) chain switch variants in some FLs, it was concluded that the processes of somatic hypermutation 14-17 and H chain isotype switching 18,19 remain active in these neoplasms. Finally, as the mutation patterns in the IgV genes of FLs were found to be highly comparable to those in normal antigen-selected B cells, it was proposed that FL cells are, during their malignant growth, still being clonally selected on basis of the antigen-binding capacity of their BCRs. 15,20-22
We have recently studied a large panel of FLs. 13 These analyses indicated that FLs may not retain the capacity to somatically mutate. Additionally, we obtained evidence for subclone selection instead of clonal BCR evolution over time. 23 These findings challenged the concept of antigen-driven lymphomagenesis.
We here analyze an interesting case of a FL that harbored, in all tumor follicles, IgM+ and IgG+ tumor cells of the same clonal origin. By in situ analyses using laser microdissection we aimed to obtain insight in the composition of the neoplastic follicles, the process of isotype switching and the repertoire of infiltrating T cells within the tumor compartment. The results again indicate that FL cells may have retained less properties of normal GC B cells than is generally assumed.
Materials and Methods
Patient Material
Fresh tissue of FL no. 8 was obtained from surgically removed lymph nodes from the departments of pathology of the Westeinde Hospital, The Hague, the Netherlands and was kindly provided by Dr. E.C.M. Ooms. The patient, a 48-year-old woman, presented with a FL in 1983 (FL 8-‘83). Complete remission was achieved after chemotherapy. After irradiation of a local relapse, in 1988, a systemic relapse developed in 1992 (FL 8-‘92). As control tissue, fresh human tonsil was used. All tissues had been snap-frozen and stored in liquid nitrogen until usage.
Immunohistochemistry
Cryostat sections were immunohistochemically stained as described, 24 using monoclonal antibodies specific for human CD21L (DRC-1, Dako, Glostrup, Denmark), Bcl-2 (clone 124, DAKO), and for Ig heavy and light chain isotypes (all from Dako except for anti-IgM, -κ, and -λ, which were obtained from Becton Dickinson, Erembodegem-Aalst, Belgium). Heavy chain isotype expression of the lymphoma of patient 8 was also assessed by immunofluorescence. Briefly, acetone-fixed tissue sections were incubated at 37°C with the primary antibody (anti-IgM, -IgG, -IgA, -IgD, -κ or -λ; Dako), washed three times in phosphate-buffered saline (PBS), and incubated for 30 minutes with swine-anti-rabbit, labeled with FITC (Dako).
Microdissection of Samples
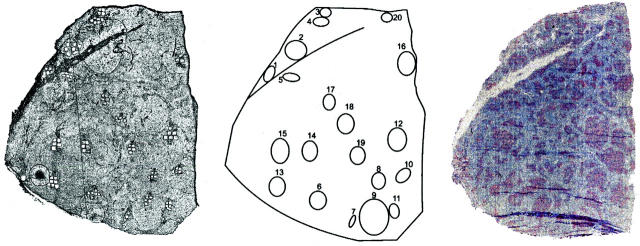
Microdissection out of frozen sections was performed with a laser-microbeam system (PALM GmbH, Bernried, Germany). The potential risk of RNA carry-over by cutting was evaluated using slides with membranes on which tissue of two distinct lymphomas was mounted together. These analyses indicated that cell-specific results were obtained and that out of microdissected membrane located adjacently to the tissue no polymerase chain reaction (PCR) products were amplifiable by reverse transcriptase (RT)-PCR (data not shown). Similar results were reported by others even after a brief fixation and staining procedure. 24 If, however, microdissection is possible using polarized light only, like in the present study, we use untreated sections to exclude the risk of RNA carry-over. Thus, here 10-μm frozen sections were dried but were left unfixed and unstained before use. For RNA analyses, samples of approximately 50 cells were dissected from lymphoma 8-’83 (Figure 3) ▶ , catapulted into 3 μl of cDNA reaction mixture (see below), and stored on ice until cDNA synthesis. For DNA samples, approximately 100 cells were dissected from follicles of hematoxylin-stained tissue sections of lymphoma 8-‘83 and its relapse, 8-‘92 and from normal tonsil. Tissue samples were “catapulted” into 5 μl of water.
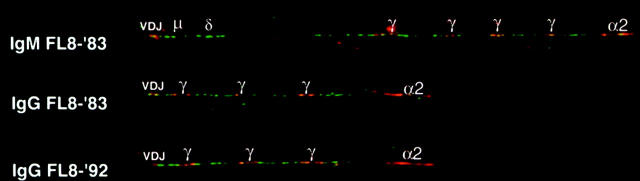
Figure 3.
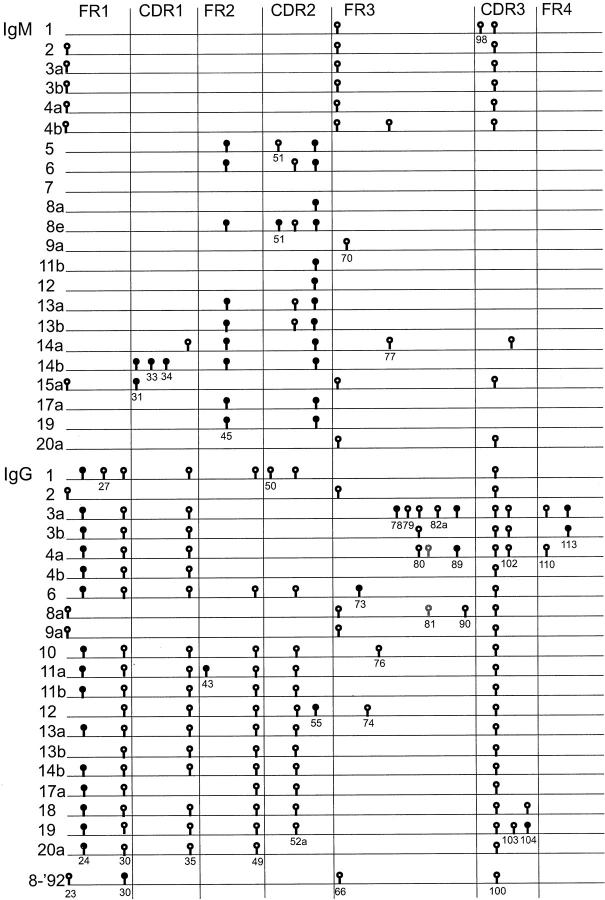
Schematic representation of IgM- and IgG-derived sequences from FL. no 8-‘83 (upper part and lower part, respectively) and its relapse 8-‘92 (bottom line). Only mutations that differ from the IgM consensus sequence, as determined on crude tissue, 13 are shown. The numbers represent the different follicles from which samples were taken, as indicated in Figure 2 ▶ . Different samples from one follicle are designated with a different letter. Replacement and silent mutations are represented as closed and open circles, respectively. Codon numbers are indicated underneath the symbols. Whereas in the consensus sequence at position 81 a mutation was present as compared to the germ-line gene, the gray symbols at this site in sequence 8aγ and 4aγ represent the germ-line nucleotide.
cDNA Synthesis
RNA of bulk material was isolated from frozen tissue sections using the TRIzol reagent (Life Technologies, Breda, the Netherlands) and cDNA was synthesized as described. 25 From the microdissected samples, cDNA was synthesized without prior RNA isolation. The microdissected samples were incubated in a total volume of 20 μl of cDNA reaction mixture. 25 The reaction was performed for 15 minutes at 37°C. Subsequently, the enzyme was inactivated during 10 minutes at 95°C. After cDNA synthesis 20 μl of water was added.
DNA Preparation
For DNA samples, 15 μl of 0.25 mol/L proteinase K in 1X Taq buffer was added (20 mmol/L Tris-HCl, 50 mmol/L KCl, pH 8.4). After incubation overnight at 56°C, the enzyme was inactivated for 10 minutes at 95°C. Bulk DNA was isolated from frozen tissue sections using the DNAzol reagent, according to the manufacturer’s instructions (Life Technologies).
Amplification of the VH Gene
In the first rounds of amplification 1 μl of cDNA reaction mixture was used in a 25 μl PCR reaction volume using a forward primer with specificity for the leader of the VH3 gene family in combination with a reverse primer specific for Cμ (Cμ1-: 5′-CGTATCCGACGGGGAATTCTC-3′), or Cγ. 25 Next, a nested PCR was performed using 2.5 μl of the first PCR product in a 25-μl reaction. PCR reactions were performed with a VH3 primer that anneals in the FRI region (VH3FR: 5′-TCCCTGAGACTCTCCTGTG-3′) combined with the appropriate reverse primer either Cμ or Cγ2. 25 The PCR consisted of 50 cycles under conditions that were the same as described previously for the CDR3-specific PCR. 25 PCR products were analyzed on a 1% standard agarose gel (Sigma, St. Louis, MO). All PCR reactions and the subsequent sequencing reactions were performed in duplicate to obtain a reliable consensus sequence of a sample. Specificity of the PCR results was further controlled using many buffer-only samples to which no cDNA was added. These controls were in all instances negative.
Amplification of Switch Circle Fragments
Quantification and subsequent stratification of the amount of amplifiable DNA was performed using a PCR on the β2 microglobulin gene (β2m). The primers used were 5′-AGCATTCAGACTTGTCTTTCAG-3′ and 5′-GATGCTGCTTACATGTCTCG-3′, which yield a product of 776 base pairs from DNA. The PCR protocol for this β2m-PCR was the same as for the amplification of the VH gene. Sγ-Sμ switch circle fragments were amplified in 25-μl reaction mixtures from 250 ng or less DNA, using the upstream Iγ1/2 primer, in combination with the downstream Sμ primer. 26 As positive controls, 10 copies of artificial construct plasmids 27 were amplified. All plasmids (Sγ1-Sμ, Sγ2-Sμ, Sγ3-Sμ, and Sγ4-Sμ) could be amplified using this PCR approach (data not shown). The PCR samples were first incubated at 95°C for 4 minutes, then 40 cycles of 1 minute at 95°C, 1 minute of 68°C and 1 minute of 72°C were performed, followed by 10 minutes at 72°C. Next, a nested PCR was performed with 2 μl of PCR product of the first PCR in a 25-μl PCR, using the Sγ and Sμ primers. 26 The same PCR mixtures were used as for the amplification of the VH gene, except that 100 nmol/L of each primer was used and Taq platinum polymerase (Life Technologies). With this PCR, one switch circle copy in at least 1000 cells is detectable. The PCR products were analyzed on a 1% agarose gel (Sigma) and subsequently sequenced to confirm their origin.
DNA Fiber Fluorescence in Situ Hybridization
The configuration of the constant heavy chain gene locus of both time points of FL 8 was analyzed by DNA fiber fluorescence in situ hybridization (FISH) as described. 28 Probes for the J region, the different constant region genes and the BCL-2 locus were used. At least 20 fibers representative for each configuration were analyzed out of tissue of each time point.
Amplification and Analysis of Clonality of the Variable Genes of the TCR-γ Locus
The T cell receptor-γ (TCRG) gene rearrangements were amplified from DNA of microdissected samples. The first PCR was performed with the PCR primers as described, 29 except that Taq platinum polymerase (Life Technologies) was used instead of AmpliTaq Gold polymerase (PE Biosystems, Foster City, CT). For the second, nested PCR, the sequencing primers 29 were used with the same PCR protocol, with exception that 2.0 mmol/L MgCl2 was used and 0.25 μmol/L of each primer. The products were first analyzed on a 1% agarose gel and subsequently by heteroduplex analysis. 30 Briefly, the PCR products were denatured for 5 minutes at 95°C, then they were allowed to reanneal at 4°C for at least 1 hour. The resulting homo- and/or heteroduplexes were analyzed on a 6% non-denaturing polyacrylamide gel and visualized by staining with ethidium-bromide.
Cloning and Sequencing of PCR Products
Both strands of the PCR products were either directly sequenced with an ABI sequencer (Perkin Elmer Corporation, Norwalk, CT) using the dye-terminator cycle-sequencing kit (Perkin Elmer), or first cloned into pCR2.1-TOPO vectors (Invitrogen, Groningen, the Netherlands) and transformed into TOP10 bacteria (Invitrogen). Subsequently, both strands of the inserts of a clone were sequenced.
Results
Immunoglobulin Analyses of FL 8
We previously analyzed the VH and variable light (VL) chain genes of a large panel of FLs of different Ig isotypes. Among these, two biopsies of FL 8 were included. 13 Interestingly, in all follicles of the presentation biopsy of 1983 (FL 8-‘83) both IgM/κ and IgG/κ tumor cells were found by immunofluorescence as well as immunohistochemically (Figure 1) ▶ . Molecular analyses revealed that these assumed isotype switch variants indeed carried the same VDJ rearrangement which contained 30 and 35 somatic mutations compared to the germ-line gene V3–23, respectively. This FL expressed the L16 Vκ chain gene, which harbored 20 somatic mutations (data not shown). At the second time point (FL 8-‘92), 9 years later, only IgG-expressing tumor cells were found of the same clonal origin (data not shown). In the V3–23 gene, 35 somatic mutations were found compared to the germ-line gene, 30 of which were shared with the IgM and IgG sequences of the first time point. In the κ light chain one extra replacement mutation was present (data not shown). At both time points, significant sequence variation was found between cloned VH products: the means of this intraclonal variation were 5.0, 4.0, and 1.3 mutations/clone compared respectively to the consensus IgM and IgG sequences of FL 8-‘83 and the consensus IgG sequence of FL 8-’92, as determined on crude tissue. 13
Figure 1.
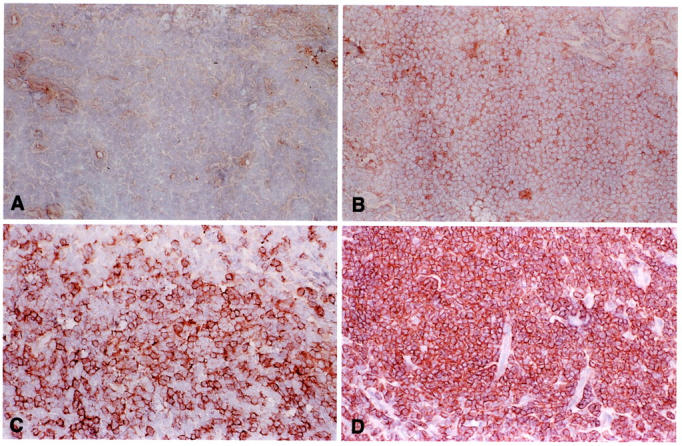
FL 8-‘83 harbors both IgM- and IgG-expressing tumor cells. Frozen section of FL 8-‘83 stained with monoclonal antibodies specific for human IgA (A), IgG (B), IgM (C), and BCL-2 (D) (magnification, ×100). Whereas all cells are IgA-negative, a faint but significant expression of IgG is found on the majority of cells in all areas. In between these cells, scattered tumor cells with relatively strong IgM expression are found.
Distribution of IgM and IgG Subclones in FL 8
To assay the distribution of the subclones within the tissue of FL 8, we microdissected a total of 47 samples from 19 neoplastic follicles from frozen tissue sections (Figure 2) ▶ . Cells were microdissected from unstained tissue sections. Of each sample, containing approximately 50 cells, the VH genes were amplified and sequenced. All PCR and sequence reactions were carried out in duplicate and consensus sequences thus obtained of each sample were compared. The overall PCR efficiency was 86%. All VH PCR products contained the known VDJ rearrangement of this FL. From 5 of 47 samples the duplicates were not reproducible (the IgM-derived sequences of sample 11a and 17b, and the IgG-derived sequences of sample 9c, 15b, and 20b), ie, point mutation differences were observed, most likely due to the presence of too many different subclones without clear dominance of one of them. The sequences that could be reproduced are depicted in Figure 3 ▶ . The variation found within the IgM and IgG sequences of the samples was 2.7 and 3.1 somatic mutations per IgVH sequence, compared to the IgM- or IgG-derived consensus sequences established in crude tissue analyses, respectively. 13
Figure 2.
Tissue sections of FL 8-‘83. A: The unstained tissue section of FL 8-‘83 from which samples were microdissected. B: A schematic drawing of the tissue section. The circles represent the numbered follicles from which samples were taken. C: Tissue section of FL 8-‘83 stained with CD21L antibodies and counterstained with hematoxylin.
In 9 instances (concerning 5 IgM and 4 IgG sequences), a second sample derived from the same follicle was assayed. In two of the five paired IgM samples, the obtained VH sequences were identical (3a and b, 13a and b), suggesting that these follicles were dominated by a specific subclone. However, in the three other sample pairs non-corresponding subclones were identified with respectively 1 (samples 4a and b), 3 (8a and e) and 6 (14a and b) nucleotide differences. All four paired IgG-derived sequences yielded non-identical sequences with either five (samples 3a and b and samples 4a and b) or one (samples 11a and b and samples 13 a and b) nucleotide differences (Figure 3) ▶ .
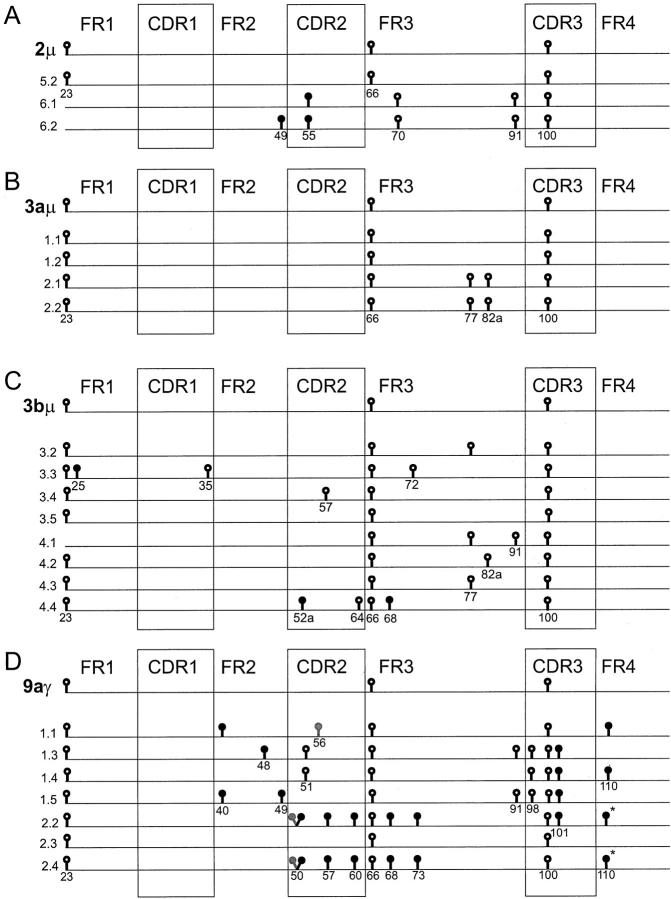
To get more insight into (sub)clonal diversity, three to eight bacterial clones were made from the PCR products of samples 2μ, 3aμ, 3bμ, and 9aγ, and the respective inserts were sequenced (Figure 4) ▶ . Significant intraclonal variation was found, which ranged from 1 mutation per clone (sample 3aμ) to 4.6 mutations per clone (sample 9aγ). It must be emphasized that in these calculations Taq errors may be present. We determined the frequency of Taq polymerase errors to be 0.14% in our experimental setup, which would amount to an average of 0.4 mutations per sequence for these clones. 25 In sample 3aμ and 3bμ, these clonal analyses were concordant with the consensus sequences obtained by directly sequencing the PCR products derived from these samples (Figure 4) ▶ . However, in sample 2μ and 9aγ, the consensus sequence derived from the clones differed from the directly sequenced product. This may be erroneous, due to the low number of analyzed clones, or be a reflection of polyclonality.
Figure 4.
A-D: Intraclonal variation within cloned PCR products. The sequences are depicted as lines. Only differences with the IgM-derived consensus sequence of FL 8-‘83 are shown. The upper line of each figure represents the sequence that was obtained by directly sequencing two independent PCR products from the same sample. The other lines are the sequences of individual bacterial clones made from the PCR products. Replacement and silent mutations are represented as closed and open circles, respectively. Codon numbers are indicated underneath the symbols. Whereas the consensus sequence of 9aγ contains mutations at positions 50 and 56 as compared to the germ-line gene, the gray symbols at these sites in sequences 1.1, 2.2, and 2.4 represent the germ-line nucleotides. *The mutations at position 110 of clones 2.2 and 2.4 (9aγ) are different from the mutations in the same codon of clone 1.1 and 1.4.
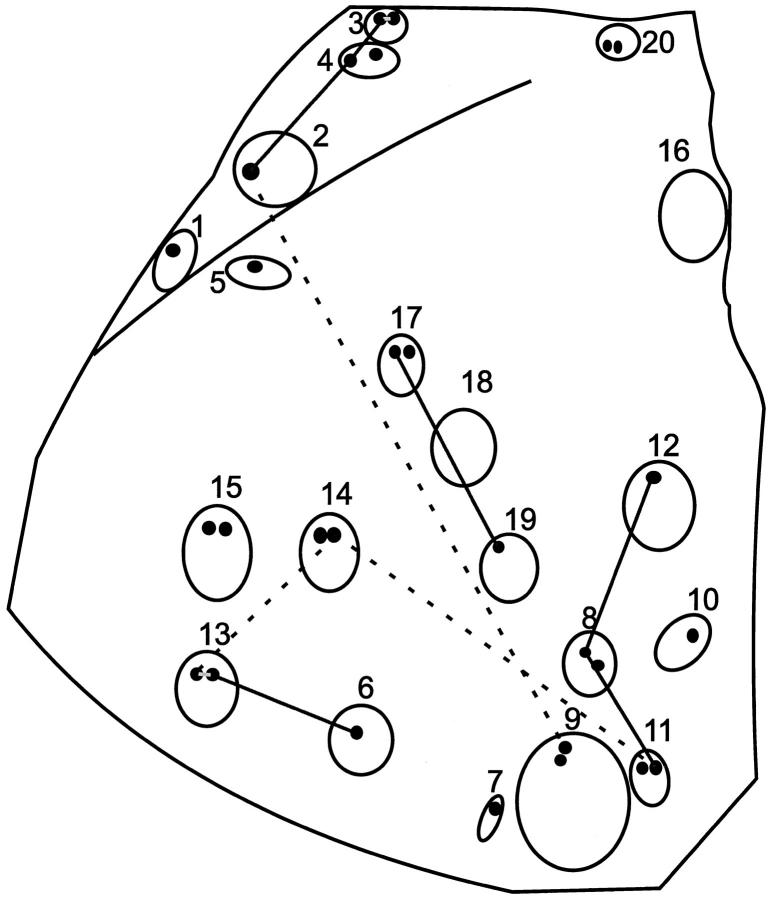
It was striking that several subclones were found in more than one follicle (Figures 3 and 5) ▶ ▶ . The IgM+ subclone of sample 3a and 3b was also found in samples 2 and 4a and closely resembled the subclones of sample 1, 4b and 20a (one nucleotide difference). These follicles are located adjacently (Figures 3 and 5) ▶ ▶ . However, the same samples yielded quite different IgG sequences, not or less clearly related to each other. Furthermore, the IgM sequences of sample 8a, 11b, and 12 were the same, and closely resembled the IgM sequences of 7 and 9a (one nucleotide difference). The IgM-sequences of 13a and 13b were the same as the IgM sequence from 6, and the 17a IgM-sequence was the same as the IgM sequence from sample 19. Similarly, an identical IgG+ subclone was found in samples 11b, 13a, and 14b and in samples 2 and 9a. In all these cases no clear zonation was apparent (Figures 3 and 5) ▶ ▶ .
Figure 5.
A schematic drawing of the unstained tissue section of FL 8-‘83. The open circles represent the numbered follicles from which samples (smaller closed circles) were taken. Labeling of samples with letters was done from left to right, from top to bottom. Follicles in which IgM- or IgG-expressing subclones with identical mutation patterns were found are connected by solid and dashed lines respectively.
Isotype-Switch Variants Do Not Seem to Result from Active Class Switching
Immunohistochemically, no clear compartmentalization was found of the IgM- and IgG-expressing cells. Accordingly, PCR analyses revealed that all dissected samples contained both IgM and IgG transcripts (data not shown). In all samples, except one, nucleotide differences between the isotype-switch variants were observed. Only sample 2 contained an identical IgM- and IgG-expressing subclone. The average number of point mutation differences between the class switch variants within the individual samples was 7.2, ranging from 0 (sample 2) to 13 (sample 3a) differences. These findings are not in support of recent isotype switching of the tumor cells. Therefore, to search for signs of active switching, tissues of both time points of FL 8 were analyzed for the presence of Sγ-Sμ switch circles by PCR and sequencing (Figure 6) ▶ . Whereas switch circle products were abundantly amplified from DNA of three different tonsils, from neither time points of FL 8 were specific switch circle products obtained (Figure 6) ▶ .
Figure 6.
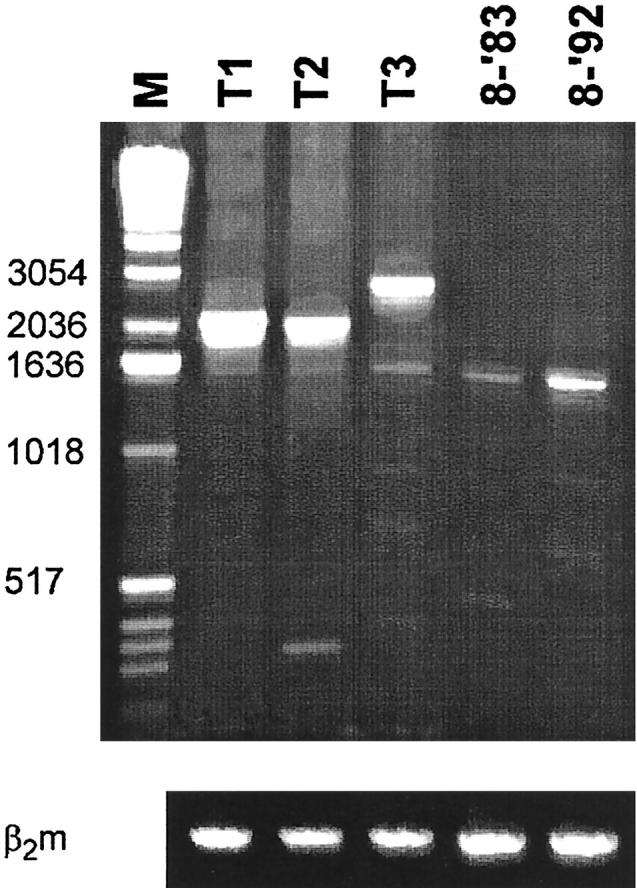
PCR analysis to detect Sγ-Sμ switch circle recombination products. The first lane is the 1 kb DNA ladder (M) with the fragment sizes indicated on the left. Lanes T1-T3 represent the PCR products obtained out of crude tonsil tissue, whereas lanes 8-‘83 and 8-‘92 show the PCR results obtained on crude tissue of FL 8. The amount of genomic DNA (approximately 250 ng) had been stratified using a β2-microglobulin PCR (lower panel). Sequencing proved that the PCR products obtained out of tonsil DNA (T1-T3) were indeed switch circle recombination products, whereas the faint bands found in lanes 8-‘83 and 8-‘92 proved to represent non-specific products (data not shown).
Fiber FISH Analysis of CH Gene Rearrangements in FL 8
To further address the isotype switching process in this FL, the configuration of the constant heavy chain gene locus of both time points of FL 8 was analyzed by DNA fiber fluorescence in situ hybridization (FISH). 28 Both the functional allele and the translocated allele were detected with probes specific for the J region, the different CH genes and the BCL-2 locus, respectively. Except for one translocated allele, two different functional alleles were detected in the sample from FL 8-‘83, indicating that the isotype-switch variants were derived from different tumor cells (Figure 7 ▶ and Table 1 ▶ ). The first functional allele was a VDJ rearrangement adjacent to Cμ and 4 Cγ genes of the 3′ CH cluster, with a downstream deletion of 2 Cγ and 1 Cα gene. The second allele showed a VDJ rearrangement adjacent to a Cγ gene of the 3′ CH cluster. By fiber FISH analysis of FL 8-‘92, we detected that the translocated and the functional VDJ-Cγ allele were identical to those of FL 8-‘83. In accordance with immunohistochemistry and PCR analyses, no VDJ rearrangement adjacent to Cμ was found in FL 8-‘92 (Figure 7 ▶ and Table 1 ▶ ).
Figure 7.
Fiber FISH detection of CH rearrangements in FL 8-‘83 and FL 8-‘92. Shown are typical examples of the consistently observed bar codes representative for the functional Ig alleles of IgM+ (upper panel) and IgG+ (centerpanel) tumor cells of FL 8-‘83 and the IgG+ tumor cells of FL 8-‘92 (lower panel). The VDJ as well as the different CH regions, as detected by the FISH probes described previously, 28 are indicated by white symbols above each fiber.
Table 1.
Schematic Overview of the Configuration of the Constant Ig Heavy Chain Gene Loci Found at Both Timepoints of FL 8 as Assessed by Fiber FISH
| Alleles | CH configuration |
|---|---|
| Germline allele 1 | Cμ Cδ Cγ3 Cγ1 Cα1 Cγ Cγ Cα2 |
| Germline allele 2 | Cμ Cδ Cγ3 Cγ1 Cα1 Cγ Cγ Cγ Cγ Cα2 |
| Translocated allele* | bcl-2 Cγ1 Cα1 Cγ Cγ Cα2 |
| Functional IgM allele 8-’83 | VDJ Cμ Cδ Cγ Cγ Cγ Cγ Cα2 |
| Functional IgG allele 8-’83 | VDJ Cγ Cγ Cγ Cα2 |
| Functional allele 8-’92 | VDJ Cγ Cγ Cγ Cα2 |
Cγ and Cα genes are numbered when possible.
*The translocated allele was identical at both time points and was derived from germ-line allele 1.
TCRG Analyses of Intrafollicular T Cells
The proposed role for BCR ligands in lymphomagenesis could imply that the tumor cells function as antigen presenting cells for the intrafollicular T cells that might in turn provide essential growth signals. To acquire more insight into the T cell repertoire of this FL, we analyzed T cell receptor-γ (TCRG) variable genes of areas microdissected from the biopsies of both time points of FL 8. Since the TCRG locus, which has a limited repertoire, is rearranged early in T cell development and remains unaltered even in TCRαβ+ cells, 31 TCRG rearrangements can be used as clonality markers. Samples of approximately 100 cells were dissected from follicles of FL 8-‘83, its relapse 8-‘92, as well as from germinal centers of a tonsil tissue section. From the microdissected samples of FL 8 and tonsil, PCR products were obtained of the four different Vγ families. The frequencies of occurrence of these families were strikingly similar between the FL and the tonsil (Table 2) ▶ , with the exception that in FL 8-‘83 VγIV family products were detected in 11 of 33 samples (33%), compared to 11% and 13% of the samples originating from the FL 8-’92 and tonsil, respectively. All PCR products found in FL 8, which appeared as a sharp band in heteroduplex analyses, were sequenced. Finally, of 13 VγI PCR products (8 from 8-‘83 and 5 from 8-‘92), 6 VγII products (4 from 8-‘83 and 2 from 8-‘92), 10 VγIII products (4 from 8-‘83 and 6 from 8-‘92) and 5 VγIV products (3 from 8-‘83 and 2 from 8-‘92), readable sequences were obtained (data not shown). In these analyses, not a single Vγ-Jγ rearrangement was found more than once, strongly suggesting there is no dominating T cell clone in FL 8. It must be realized that in contrast to reactive germinal centers in tonsil or lymph node tissue, in which also polyclonal T cells have been described, 32,33 a FL consists of cells of a single antigen-receptor specificity. By consequence the potential repertoire of peptides presented by MHC class II molecules is restricted. Thus, despite this uniformity of “antigen presenting” tumor cells no indication for oligoclonality of T cells was obtained.
Table 2.
TCRG V Gene Family Products Found in Samples Microdissected from FL 8 and Tonsil Tissue
| Tissue | Samples tested | VγI | VγII | VγIII | VγIV |
|---|---|---|---|---|---|
| 8-’83 | 33 | 21 (64%) | 24 (73%) | 9 (27%) | 11 (33%) |
| 8-’92 | 19 | 12 (63%) | 12 (63%) | 6 (32%) | 2 (11%) |
| Tonsil | 16 | 10 (63%) | 14 (88%) | 3 (19%) | 2 (13%) |
Discussion
We studied clonal distribution and isotype switching in a FL, which harbored IgM and IgG expressing tumor cells of the same clonal origin. Cell samples from neoplastic follicles were microdissected and VH-Cμ and VH-Cγ transcripts present herein were analyzed. We identified many subclones scattered over the different follicles. Four out of 13 identified IgM subclones and 2 of the 16 IgG subclones were found in more than one follicle, some at relatively large distances of each other (Figure 5) ▶ . This is not what would be expected if the neoplastic follicles were anatomically and functionally sequestered compartments, like normal GCs. 7 Additionally, we observed no clear subclone dominance within individual follicles: in 5 of 47 samples too many nucleotide differences were found between the duplicate analyses to be able to assign a consensus sequence. Moreover, in only 2 of the 9 follicles (nos. 3 and 13) of which two samples were taken from different areas (and led to unambiguous results), the same VH sequences were obtained (Figure 3) ▶ , compatible with local outgrowth of individual subclones. The overall picture however is more indicative of a more or less random distribution of subclones. Of note, the VH sequence which was found most frequently in different follicles, both as μ (samples 2, 3, and 4a) and γ (samples 2 and 9a) transcripts, was virtually identical to that of the dominant IgG clone in the relapse tumor 8-‘92, 9 years later. This remarkable evidence for selection of pre-existent subclones was reported previously by us. 23 Also in longitudinal studies on B cell lymphomas reported by others, 16,17,21,22 successive IgV gene patterns were compatible with subclone selection. Apparently, such subclones can finally obtain growth advantage most likely on basis of additional genetic alterations unrelated to the configuration of the BCR.
Our analyses further showed that in all microdissected tissue samples, both IgM and IgG transcripts are present (Figure 3) ▶ . The fact that by fiber FISH, one translocated allele and two distinct functional alleles were found in FL 8-‘83 (data not shown) indicates that these transcripts are not the result of trans-splicing but are indeed derived from different cells. In general, non-identical mutation patterns were found in the isotype-switch variants of individual samples, except for sample 2. The average number of point mutation differences between the IgM- and IgG-derived VH sequences of individual samples was 7.2. Moreover, despite significant intraclonal variation in both IgM- and IgG-derived sequences, the IgM sequences resembled each other more than they resembled the IgG sequences, and vice versa. These findings argue against active class switching. The switch circle-specific PCR analyses support this notion. Abundant switch circle recombination products were amplified from tonsil tissue DNA (Figure 6) ▶ , in which only a minor cell fraction is likely to have undergone a class switch. Among these class-switched cells, a minority will still harbor the excision circles. By contrast, from DNA of the FL, consisting of an almost pure population of isotype switch variants of a single clone, no specific switch circle products were amplified by this sensitive PCR method. This and the fact that we found only one type of VDJ-Cγ allele by fiber FISH, present in identical configuration in FL 8-’92 (Figure 7 ▶ and Table 1 ▶ ), strongly argues against active class switching. Although we can formally not rule out that class switching occurs at very low frequencies in this FL, our findings demonstrate that the tumor cells are not frozen in a stage in which the IgM+ cells are all on the verge of H chain isotype switching. Most likely, the isotype switch took place early after initial transformation, but not before a substantial amount of somatic mutations had been introduced. In the literature, change of isotype expression in time has been documented in some FLs, in one case concurrent with transformation to a diffuse lymphoma. 18,19 In these reports however, no control PCR analyses were performed to exclude that the isotype-switched clones had already been present at low frequencies at the early time points.
It is questionable whether and to what extent the principle molecular and cellular processes occurring in normal GCs, ie, somatic diversification, H chain isotype switching and BCR-based clonal selection, occur in FLs. The concept of antigen-driven lymphomagenesis relies strongly on the capacity of the tumor cells to somatically mutate their IgV genes together with the observed non-random IgV gene mutation patterns. Many investigators consider intraclonal V gene sequence diversity of lymphomas as an indicator of an active mutation machinery. We previously pointed out that, considering the significant tumor volume of FLs in general and thus the number of cell divisions that must have occurred, the degree of intraclonal sequence diversity in this and other FLs is in fact remarkably low and indicates that, like the class switching activity, the mutational activity in advanced stage FLs must be very low or even absent. 13 This contention was supported by the finding that in general the degree of intraclonal sequence diversity within FLs decreases instead of increases over time 13,16,17,21,34 Stringent selection processes must therefore occur which, based on the available evidence for selection of pre-existent subclones, 23 are not necessarily BCR-guided. As we found no indication of T cell oligoclonality within this FL either, we obtained no circumstantial arguments for a role of BCR ligands in lymphomagenesis via antigen-specific T cell help, as has been proposed for low-grade gastric mucosa-associated tissue (MALT) lymphomas. 35-37
The notion that FLs are functionally more crippled than generally assumed seems not inconsistent with their histology. In most FLs, the neoplastic follicles are irregular and in part ill-defined. This can be well appreciated in immunohistochemical stainings with the DRC-1 (anti-CD21L) antibody, in which the FDC networks, unlike normal germinal centers, often have a non-homogeneous, “moth-eaten” appearance. Based on these architectural characteristics, it can be conceived that the formation of new follicles is a process of budding off from pre-existent tumor nodules rather than the result of seeding by founder cells. On the other hand, the fact that individual tumor clones are also found at relatively large distances and that most FLs are systemic indicates that the cells must be actively trafficking and, on recirculation via lymph and blood, home to distant follicles. 38 Hence, whereas normal GCs are rather closed compartments 7 that allow for independent clonal evolution of the included B cells, the neoplastic follicles appear to permit interfollicular tumor cell trafficking, as has previously also been proposed by Dogan et al. 39 This lack of spatial organization in FLs is likely to interfere with strict BCR-based selection processes.
Acknowledgments
We thank H.B.P.M. Dijkman for technical assistance with the PALM and J.B.G. Mulder for assistance with immunohistochemistry. We thank I.L.M. Wolvers-Tettero and E.J. van Gastel-Mol for advice on the TCRG analyses. The artificial switch circle constructs were generously provided by Dr. E.E. Max. We also thank Dr. E.C.M. Ooms and C.C.H. Vellema for providing tissue material.
Footnotes
Address reprint requests to C. J. M. van Noesel, M.D., Ph.D., Department of Pathology, Academic Medical Center, Meibergdreef 9, 1105 AZ, Amsterdam, the Netherlands. E-mail: c.j.vannoesel@amc.uva.nl.
Supported by grant AMC 95–957 from the Dutch Cancer Society. C. J. M. van Noesel is a fellow of the Netherlands Royal Academy of Arts and Sciences.
References
- 1.Rajewsky K: Clonal selection and learning in the antibody system. Nature 1996, 381:751-758 [DOI] [PubMed] [Google Scholar]
- 2.Lam K-P, Kühn R, Rajewsky K: In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell 1997, 90:1073-1083 [DOI] [PubMed] [Google Scholar]
- 3.Lindhout E, Koopman G, Pals ST, de Groot C: Triple check for antigen specificity of B cells during germinal center reactions. Immunol Today 1997, 18:573-577 [DOI] [PubMed] [Google Scholar]
- 4.Kocks C, Rajewsky K: Stable expression and somatic hypermutation of antibody V regions in B cell developmental pathways. Annu Rev Immunol 1989, 7:537-559 [DOI] [PubMed] [Google Scholar]
- 5.Küppers R, Zhao M, Hansmann M-L, Rajewsky K: Tracing B cell development in human germinal centers by molecular analysis of single cells picked from histological sections. EMBO J 1993, 12:4955-4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacob J, Przylepa J, Miller C, Kelsoe G: In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J Exp Med 1993, 178:1293-1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vora KA, Tumas-Brundage K, Manser T: Contrasting the in situ behavior of a memory B cell clone during primary and secondary immune responses. J Immunol 1999, 163:4315-4327 [PubMed] [Google Scholar]
- 8.Jacob J, Kelsoe G: In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. II. A common clonal origin for periarteriolar lymphoid sheath-associated foci and germinal centers. J Exp Med 1992, 176:679-687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroese FG, Wubbena AS, Seijen HG, Nieuwenhuis P: Germinal centers develop oligoclonally. Eur J Immunol 1987, 17:1069-1072 [DOI] [PubMed] [Google Scholar]
- 10.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD: World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the clinical advisory committee meeting-Airlie House, Virginia, November, 1997. J Clin Oncol 1999, 17:3835-3849 [DOI] [PubMed] [Google Scholar]
- 11.Stamatopoulos K, Kosmas C, Papadaki T, Pouliou E, Belessi C, Afendaki S, Anagnostou D, Loukopoulos D: Follicular lymphoma immunoglobulin κ light chains are affected by the antigen selection process, but to a lesser degree than their partner heavy chains. Br J Haematol 1997, 96:132-146 [DOI] [PubMed] [Google Scholar]
- 12.Noppe SM, Heirman C, Bakkus MHC, Brissinck J, Schots R, Thielemans K: The genetic variability of the VH genes in follicular lymphoma: the impact of the hypermutation mechanism. Br J Haematol 1999, 107:625-640 [DOI] [PubMed] [Google Scholar]
- 13.Aarts WM, Bende RJ, Steenbergen EJ, Kluin PM, Ooms ECM, Pals ST, van Noesel CJM: Variable heavy chain gene analysis of follicular lymphomas: correlation between heavy chain isotype expression and somatic mutation load. Blood 2000, 95:2922-2929 [PubMed] [Google Scholar]
- 14.Levy S, Mendel E, Kon S, Avnur Z, Levy R: Mutational hot spots in Ig V region genes of human follicular lymphomas. J Exp Med 1988, 168:475-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zelenetz AD, Chen TT, Levy R: Clonal expansion in follicular lymphoma occurs subsequent to antigen selection. J Exp Med 1992, 176:1137-1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu D, Hawkins RE, Hamblin TJ, Stevenson FK: Clonal history of a human follicular lymphoma as revealed in the immunoglobulin variable region genes. Br J Haematol 1994, 86:505-512 [DOI] [PubMed] [Google Scholar]
- 17.Ottensmeier CH, Thompsett AR, Zhu D, Wilkins BS, Sweetenham JW, Stevenson FK: Analysis of VH genes in follicular and diffuse lymphoma shows ongoing somatic mutation and multiple isotype transcripts in early disease with changes during disease progression. Blood 1998, 91:4292-4299 [PubMed] [Google Scholar]
- 18.Zelenetz AD, Chen TT, Levy R: Histologic transformation of follicular lymphoma to diffuse lymphoma represents tumor progression by a single malignant B cell. J Exp Med 1991, 173:197-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raghoebier S, Broos L, Kramer MHH, van Krieken JHJM, Kluin-Nelemans JC, van Ommen GJB, Kluin PM: Histological conversion of follicular lymphoma with structural alterations of t(14;18) and immunoglobulin genes. Leukemia 1995, 9:1748-1755 [PubMed] [Google Scholar]
- 20.Bahler DW, Zelenetz AD, Chen TT, Levy R: Antigen selection in human lymphomagenesis. Cancer Res 1992, 52:5547s-5551s [PubMed] [Google Scholar]
- 21.Bahler DW, Levy R: Clonal evolution of a follicular lymphoma: evidence for antigen selection. Proc Natl Acad Sci USA 1992, 89:6770-6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matolcsy A, Schattner EJ, Knowles DM, Casali P: Clonal evolution of B cells in transformation from low- to high-grade lymphoma. Eur J Immunol 1999, 29:1253-1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aarts WM, Bende RJ, Bossenbroek JG, Pals ST, van Noesel CJM: Variable heavy chain gene analysis of follicular lymphomas: subclone selection rather than clonal evolution over time. Blood 2001, 98:238-240 [DOI] [PubMed] [Google Scholar]
- 24.Bernsen MR, Dijkman HBPM, De Vries E, Figdor CG, Ruiter DJ, Adema GJ, van Muijen GNP: Identification of multiple mRNA and DNA sequences from small tissue samples isolated by laser-assisted microdissection. Lab Invest 1998, 78:1267-1273 [PubMed] [Google Scholar]
- 25.Aarts WM, Willemze R, Bende RJ, Meijer CJLM, Pals ST, van Noesel CJM: VH gene analysis of primary cutaneous B cell lymphomas: evidence for ongoing somatic hypermutation and isotype switching. Blood 1998, 92:3857-3864 [PubMed] [Google Scholar]
- 26.Cerutti A, Zan H, Schaffer A, Bergsagel L, Harindranath N, Max EE, Casali P: CD40 ligand and appropriate cytokines induce switching to IgG, IgA, and IgE and coordinated germinal center and plamacytoid phenotypic differentiation in a human monoclonal IgM+IgD+ B cell line. J Immunol 1998, 160:2145-2157 [PMC free article] [PubMed] [Google Scholar]
- 27.Malisan F, Brière F, Bridon J-M, Harindranath N, Mills FC, Max EE, Banchereau J, Martinez-Valdez H: Interleukin-10 induces immunoglobulin G isotype switch recombination in human CD40-activated naive B lymphocytes. J Exp Med 1996, 183:937-947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaandrager J-W, Schuuring E, Kluin-Nelemans JC, Dyer MJS, Raap AK, Kluin PM: DNA fiber fluorescence in situ hybridization analysis of immunoglobulin class switching in B cell neoplasia: aberrant CH gene rearrangements in follicle center-cell lymphoma. Blood 1998, 92:2871-2878 [PubMed] [Google Scholar]
- 29.Pongers-Willemse MJ, Seriu T, Stolz F, d’Aniello E, Gameiro P, Pisa P, Gonzalez M, Bartram CR, Panzer-Grumayer ER, Biondi A, San Miguel JF, van Dongen JM: Primers and protocols for standardized detection of minimal residual disease in acute lymphoblastic leukemia using immunoglobulin and T cell receptor gene rearrangements and TAL1 deletions as PCR targets. Report of the BIOMED-1 CONCERTED ACTION: investigation of minimal residual disease in acute leukemia. Leukemia 1999, 13:110–118 [DOI] [PubMed]
- 30.Langerak AW, Szczepanski T, van der Burg M, Wolvers-Tettero ILM, van Dongen JJM: Heteroduplex PCR analysis of rearranged T cell receptor genes for clonality assessment in suspect T cell proliferations. Leukemia 1997, 11:2192-2199 [DOI] [PubMed] [Google Scholar]
- 31.van Dongen JJM, Comans-Bitter WM, Wolvers-Tettero ILM, Borst J: Development of human T lymphocytes and their thymus-dependency. Thymus 1990, 16:207-234 [PubMed] [Google Scholar]
- 32.Golby SJC, Dunn-Walters DK, Spencer J: Human tonsillar germinal center T cells are diverse and widely dissiminated population. Eur J Immunol 1999, 29:3729-3736 [DOI] [PubMed] [Google Scholar]
- 33.Roers A, Hansmann ML, Rajewsky K, Küppers R: Single-cell PCR analysis of T helper cells in human lymph node germinal centers. Am J Pathol 2000, 156:1067-1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fenton JAL, Vaandrager J-W, Aarts WM, Bende RJ, Heering K, van Dijk M, Morgan G, van Noesel CJM, Schuuring E, Kluin PM: Follicular lymphoma with a novel t(14;18) breakpoint involving the immunoglobulin heavy chain switch mu region indicates an origin from germinal center B cells. Blood 2002, 99:716-718 [DOI] [PubMed] [Google Scholar]
- 35.Hussell T, Isaacson PG, Crabtree JE, Dogan A, Spencer J: Immunoglobulin specificity of low grade B cell gastrointestinal lymphoma of mucosa-associated lymphoid tissue (MALT) type. Am J Pathol 1993, 142:285-292 [PMC free article] [PubMed] [Google Scholar]
- 36.Hussell T, Isaacson PG, Crabtree JE, Spencer J: Helicobacter pylori-specific tumor-infiltrating T cells provide contact dependent help for the growth of malignant B cells in low-grade gastric lymphoma of mucosa-associated lymphoid tissue. J Pathol 1996, 178:122-127 [DOI] [PubMed] [Google Scholar]
- 37.Yumoto N, Araki A, Sumida T, Saito T, Taniguchi M, Mikata A: Restricted Vγ gene usage of tumor-infiltrating T lymphocytes in primary gastric malignant B cell lymphoma. Virchows Arch 1995, 426:11-18 [DOI] [PubMed] [Google Scholar]
- 38.Drillenburg P, Pals ST: Cell adhesion receptors in lymphoma dissemination. Blood 2000, 95:1900-1910 [PubMed] [Google Scholar]
- 39.Dogan A, Du M-Q, Aiello A, Diss TC, Ye H-T, Pan L-X, Isaacson PG: Follicular lymphomas contain a clonally linked but phenotypically distinct neoplastic B cell population in the interfollicular zone. Blood 1998, 91:4708–4714 [PubMed]