Abstract
This study was undertaken to analyze genetic alterations in 108 sporadic serous ovarian neoplasms to elucidate ovarian serous carcinogenesis. Our results demonstrate that K-ras mutations occur in approximately 50% of serous borderline tumors (SBTs), non-invasive micropapillary serous carcinomas (MPSCs), and invasive micropapillary serous carcinomas, which represent a morphological continuum of tumor progression. Moreover, progressive increase in the degree of allelic imbalance of chromosomes 1p, 5q, 8p, 18q, 22q, and Xp was observed comparing serous borderline tumors to noninvasive and invasive micropapillary serous carcinomas. In contrast, high-grade (conventional serous carcinoma) tumors contained wild-type K-ras in all 23 cases studied and a high frequency of allelic imbalance even in small (early) primary tumors similar to that found in advanced stage tumors. Based on these findings, we propose a dualistic model for ovarian serous carcinogenesis. One pathway involves a stepwise progression from SBT to noninvasive and then invasive MPSC. The other pathway is characterized by rapid progression from the ovarian surface epithelium or inclusion cysts to a conventional (high-grade) serous carcinoma.
Serous carcinoma is the most common type of ovarian cancer and is the most lethal gynecologic malignancy. Delineation of the molecular pathways involved in the evolution of ovarian serous carcinoma would have profound impact on our understanding of its pathogenesis thereby providing a rational basis for the development of new diagnostic tests and therapeutic strategies. Despite considerable efforts aimed at elucidating the molecular mechanisms of ovarian serous carcinoma, its pathogenesis is still poorly understood 1 largely because of the lack of an established model for its development. At present, the most widely held view is that ovarian serous carcinoma consists of a relatively homogeneous group of neoplasms that arise directly from transformation of the ovarian surface epithelium or inclusion cysts through a de novo process 2 , since definitive precursor lesions have not been detected. Our recent clinical and histopathological studies of a large series of serous neoplasms 3-5 have led to the recognition of a variant of serous carcinoma, designated “micropapillary serous carcinoma” (MPSC) with distinctive histopathological and clinical features. Most MPSCs are noninvasive and are frequently associated with serous borderline tumors (SBTs) also referred to as atypical proliferative serous tumors, a benign form of serous neoplasms. 5 Histological transitions from SBTs to noninvasive MPSCs can be observed as well as areas of infiltrative growth (stromal invasion) immediately adjacent to the MPSC component of these neoplasms (Figure 1A) ▶ . The morphology of the invasive component resembles that of the noninvasive MPSC and can also be seen in frankly invasive low-grade serous carcinomas. We have designated such tumors as invasive MPSCs. 3 Thus, these neoplasms appear to represent a morphological spectrum ranging from a benign proliferative tumor (SBT or atypical proliferative serous tumor) through a noninvasive carcinoma (noninvasive MPSC) to a low-grade invasive carcinoma (invasive MPSC). Our preliminary clinical data indicate that MPSCs (both noninvasive and invasive) generally pursue an indolent course. The frequency of MPSC in the general population is not known, but data from our referral material and a population-based study of noninvasive MPSCs 6 suggest that the prevalence is around 20 to 25% of all ovarian serous tumors. In contrast to invasive MPSCs, conventional serous carcinomas present as high-grade, aggressive neoplasms that evolve rapidly (Figure 1B) ▶ . The aim of this study was to analyze the molecular genetic changes including K-ras mutation and allelic status of different chromosomes in these morphologically distinct ovarian serous neoplasms.
Figure 1.
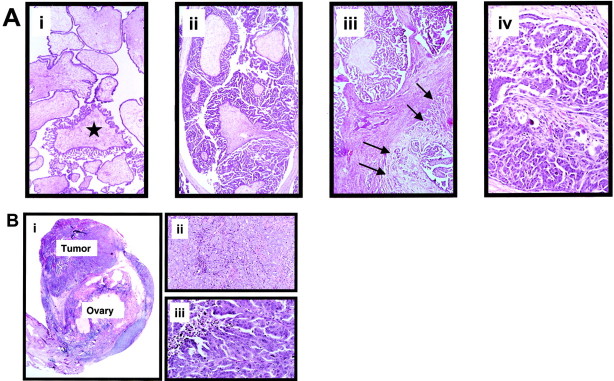
A: Morphological features of the different ovarian serous neoplasms. i: SBT with focal transition to a micropapillary area (asterisk). These focal micropapillary areas are not infrequent in SBTs. ii: Noninvasive MPSC completely replaces an ovary. The tumor is characterized by broad fibrotic papillae from which numerous long delicate micropapillary projections emanate. iii: Noninvasive MPSC (in the upper field) with an area of early invasion (invasive MPSC; right lower field with arrows). iv: Invasive MPSC. The low-grade nuclear features in the invasive carcinoma are the same as in the noninvasive counterpart. Original magnification, ×250. B: Morphological features of conventional serous carcinoma. i: Small (early) conventional serous carcinoma. The tumor measures 0.7 cm and appears to arise from the overlying ovarian surface epithelium. ii: Conventional serous carcinoma. In this area the tumor is composed of solid masses. iii: Conventional serous carcinoma. Spaces within the masses create a pseudopapillary structure as a result of cellular necrosis and processing artifact. The tumor cells contain highly atypical nuclei. Original magnification, ×400.
There are a variety of problems associated with traditional mutational analysis and determination of allelic status in ovarian serous tumors. These include abundant stromal contamination in tumors which can obscure tumor-associated genetic changes (Figure 1A) ▶ , artifactual enrichment for one allele due to limited amounts of DNA purified from microdissected lesions, and DNA degradation of the larger microsatellite alleles which can confound the analysis of allelic status when microsatellite markers and paraffin tissue are used. 7 To overcome these problems, we used a newly developed technique termed digital polymerase chain reaction (PCR) analysis, in which alleles (wild-type/mutant alleles or maternal/paternal alleles) are directly and precisely counted, one by one. 8-11 A rigorous statistical method is then used to conclude whether mutation or allelic imbalance is present in the background of normal DNA. 8-11
Materials and Methods
Tissues and Tumor DNA Samples
Formalin-fixed, paraffin-embedded tissue samples of 108 ovarian serous tumors were used for molecular genetic analysis. These cases were randomly retrieved from the surgical pathology files of The Johns Hopkins Hospital, Baltimore, Maryland and the consultation files of one of the authors (R.J.K.). All of the cases were re-reviewed by three gynecological pathologists who concurred with the diagnoses before microdissection. We did not identify “well differentiated” non-MPSC among the serous carcinomas in this study. So called “moderately differentiated” serous carcinomas showed high-grade nuclear features and were included with “poorly differentiated” carcinomas as conventional serous carcinomas. The specimens included 24 SBTs (5 stage I, 8 stage II, and 11 stage III), 39 noninvasive MPSCs (16 stage I, 8 stage II, and 15 stage III), 22 invasive MPSCs (1 stage I, 1 stage II, 19 stage III, and 1 stage IV) and 23 conventional high-grade serous carcinomas (1 stage I, 2 stage II, 16 stage III, and 4 stage IV). The tumor areas and adjacent normal tissues were microdissected under an inverted microscope with the contamination from non-neoplastic cells estimated at 20 to 50% in the microdissected tumor component. DNA was purified and analyzed for mutational status of K-ras gene and allelic imbalance using digital PCR-based techniques.
Digital Single Nucleotide Polymorphism Analysis for Allelic Imbalance
We used digital single nucleotide polymorphism (SNP) analysis to assess allelic status in tumors since this new method provides a reliable and quantitative measure of the proportion of variant sequences within a mixed DNA sample as always occurs in serous tumors. To perform digital SNP analysis, SNP markers on the chromosomes 1p, 5q, 8p, 18q, 22q and Xp were retrieved from the National Cancer Institute SNP map (http://lpg.nci.nih.gov/html-snp/imagemaps.html). These chromosomal arms were selected based on their frequent losses in serous carcinomas as previously reported. 12-15 SNP markers within a 10 centiMorgan interval were selected from each chromosomal arm. Using these markers, we were able to find at least one heterozygous SNP for each chromosomal arm in most specimens studied.
Digital SNP analysis was performed as previously described 9-11 with modification. In brief, DNA concentrations in the samples were first measured by the PicoGreen dsDNA quantitation kit (Molecular Probes, Eugene, OR) following the manufacturer’s instructions to determine the amount of DNA to be included. DNA samples were diluted and distributed in the wells of a 384-well plate at approximately one genomic equivalent per two wells. In addition to all essential PCR reagents, the PCR cocktail contained a pair of molecular beacons (Gene Link, Thornwood, NY) along with an excess of reverse primer that allowed the generation of single-stranded DNA complementary to the molecular beacons. PCR was performed in a single step using the following protocol: 94°C (1 minute); 4 cycles of 94°C (15 seconds), 64°C (15 seconds), 70°C (15 seconds); 4 cycles of 94°C (15 seconds), 61°C (15 seconds), 70°C (15 seconds); 4 cycles of 94°C (15 seconds), 58°C (15 seconds), 70°C (15 seconds); 60 cycles of 94°C (15 seconds), 55°C (15 seconds), 70°C (15 seconds); 94°C (1 minute) and 60°C (5 minutes). The fluorescence intensity in each well was then measured in a Galaxy FLUOstar fluorometer (BMG Lab Technologies, Durham, NC) and the number of specific alleles in each sample was directly determined from the fluorescence measurements.
Digital PCR Analysis for K-ras Mutations
K-ras mutations at codon 12 and 13 were analyzed using digital PCR and molecular beacons as described in previous reports. 8,16
Statistical Analysis
To determine whether there was statistical significance for allelic imbalance, we used the Sequential Probability Ratio test. 9,10 An allelic imbalance index was determined for each tumor as the number of chromosomal arms with allelic imbalance divided by the total number of chromosomal arms with informative markers. Differences between the allelic imbalance index in different groups and the percentage of allelic imbalance in individual chromosomal arms in different groups were tested using the Student’s t-test and the Mann-Whitney rank-sum test as appropriate. The correlation between tumor size in different groups and allelic imbalance index was assessed using Spearman’s rank-order correlation.
Results
K-ras mutations in codon 12 or 13 were found in 50% of SBTs, 36% of noninvasive, and 54% of invasive MPSCs (Table 1) ▶ . In contrast, K-ras mutations were not found in any of the 23 conventional (high-grade) serous carcinomas examined.
Table 1.
Analysis of K-Ras Mutations and Allelic Imbalance in Serous Ovarian Neoplasms
| Histological pattern | K-ras | Chromosome | |||||
|---|---|---|---|---|---|---|---|
| 1p | 5q | 8p | 18q | 22q | Xp | ||
| SBTs (n = 24) | 12/24 (50) | 1 /22 (4) | 4 /24 (17)* | 2 /19 (10) | 7 /24 (29) | 9 /20 (45) | 9 /15 (60) |
| MPSCs (n = 39) | 14/39 (36) | 5 /38 (13)† | 21 /38 (55)* | 10 /37 (27) | 17 /39 (44) | 15 /30 (50) | 14 /27 (52) |
| Invasive MPSCs (n = 22) | 12/22 (54) | 11 /20 (55)† | 11 /17 (65) | 8 /21 (38) | 8 /19 (42) | 10 /16 (62) | 10 /17 (59) |
| CSCs (n = 23) | 0/23 (0) | 15 /21 (71) | 20 /23 (87) | 18 /22 (81) | 14 /22 (64) | 13 /19 (68) | 7 /16 (43) |
The frequency of K-ras mutations (%) and allelic imbalance (%) in SBTs, noninvasive MPSCs, invasive MPSCs, and CSCs are shown.
*, †, Differences with statistical significance (*, P < 0.02 and †, P < 0.01; Student’s t test and Mann-Whitney Rank-Sum test).
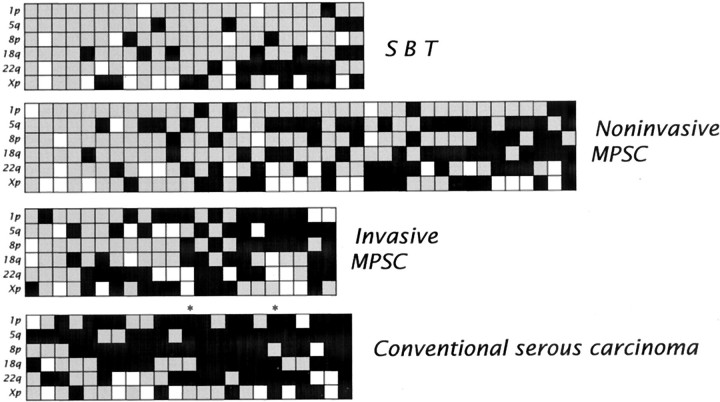
Comparing SBTs to noninvasive and invasive MPSCs (Table 1 ▶ and Figure 2 ▶ ), revealed an increased allelic imbalance index in the progression from SBT to noninvasive MPSC (P <0.01) and to invasive MPSC (P <0.02). In particular, allelic imbalance of chromosome 5q was more frequently observed in noninvasive MPSCs compared to SBTs (P <0.02) and allelic imbalance of chromosome 1p was more frequently found in invasive MPSCs compared to noninvasive MPSCs (P <0.01). Identical allelic imbalance patterns and K-ras mutations were found in the areas of SBT and noninvasive MPSC, or in the areas of noninvasive and invasive MPSC in the 34 tumors containing two components representing stages in progression (SBTs and noninvasive MPSCs or noninvasive and invasive MPSCs). There was no correlation between the allelic imbalance index and the size of the tumors in the different groups. Conventional serous carcinomas showed a high level of allelic imbalance in almost all of the investigated tumors irrespective of their size (Figure 2) ▶ .
Figure 2.
Summary of the results of allelic status in ovarian serous tumors. Each panel represents a group of serous tumors: SBT, noninvasive MPSC, invasive MPSC, and conventional serous carcinoma. Chromosomal arms in which the SNP markers are located are indicated on the left of each panel. In the vertical columns, each column represents one case. Black squares represent chromosomal arms in which allelic imbalance is identified based on the digital SNP analysis, while gray squares represent chromosomal arms in which both alleles were in balance. Blank squares indicate chromosomal arms that cannot be evaluated because the allelic ratio in the SPRT analysis does not achieve a statistical significant difference or because all SNP markers tested are uninformative in the normal tissue. The very small conventional serous carcinomas (maximal dimension 0.6 and 0.7 cm) are marked with asterisks.
Sixteen patients with noninvasive MPSCs (cases 1 to 11) and invasive MPSCs (cases 12 to 16) presented with tumors in both ovaries. These bilateral tumors were of similar size and had a similar gross and microscopic appearance. Comparison of the tumors involving both ovaries in these patients revealed that 15 (94%) of 16 had a discordant pattern of K-ras mutation or allelic imbalance (Table 2) ▶ .
Table 2.
K-ras Mutational Status and Allelic Imbalance in Bilateral Noninvasive and Invasive MPSCs
| Case no. | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1R | 1L | 2R | 2L | 3R | 3L | 4R | 4L | 5R | 5L | 6R | 6L | 7R | 7L | 8R | 8L | ||||||||||||||||
| K-ras status (codon | wt | GCT | GAT | TGC | wt | GAT | TGT | wt | wt | wt | AGC | GAT | wt | wt | wt | wt | |||||||||||||||
| with mutation) | (12) | (12) | (13) | (12) | (12) | (13) | (12) | ||||||||||||||||||||||||
| Allelic imbalance | 5q | 5q | 1p | 8p | 5q | 5q | 5q | 5q | 22q | Xp | 5q | 5q | 22q | 18q | 1p | 5q | |||||||||||||||
| 18q | 18q | 5q | 18q | 8p | 18q | 18q | 18q | Xp | 22q | 18q | Xp | 5q | 8p | ||||||||||||||||||
| Xp | 22q | 18q | Xp | 18q | |||||||||||||||||||||||||||
| 22q | Xp | ||||||||||||||||||||||||||||||
| Xp | |||||||||||||||||||||||||||||||
| Case no. | |||||||||||||||||||||||||||||||
| 9R | 9L | 10R | 10L | 11R | 11L | 12R | 12L | 13R | 13L | 14R | 14L | 15R | 15L | 16R | 16L | ||||||||||||||||
| K-ras status | wt | wt | TGT | wt | AGC | wt | GAC | wt | GAC | GAC | wt | GAC | wt | wt | wt | wt | |||||||||||||||
| (12) | (13) | (13) | (13) | (13) | (13) | ||||||||||||||||||||||||||
| Allelic imbalance | No | 8p | 1p | 18q | 5q | 5q | 5q | 5q | 1p | 1p | 1p | 1p | 22q | 5q | 5q | 5q | |||||||||||||||
| 22q | 8p | 22q | 18q | 8p | 18q | 8p | Xp | Xp | 8p | Xp | 22q | 18q | 22q | ||||||||||||||||||
| 22q | Xp | 22q | 22q | 22q | 18q | 22q | Xp | 22q | Xp | ||||||||||||||||||||||
| Xp | 22q | Xp | |||||||||||||||||||||||||||||
Corresponding case numbers indicate the same patients. Noninvasive MPSCs are the cases 1 to 11, invasive MPSCs are the cases 12 to 16. R, right; L, left; no, no allelic imbalance.
Discussion
By stratifying ovarian serous carcinomas into two histopathologically distinct groups, a low-grade carcinoma designated invasive micropapillary serous carcinoma with its putative precursors (SBT and noninvasive MPSC), and a high-grade carcinoma (conventional serous carcinoma), we were able to demonstrate that these neoplasms displayed very different and characteristic molecular genetic alterations.
First, K-ras mutations were found in nearly half of the invasive MPSCs and their putative precursors, but not in conventional serous carcinoma, suggesting that aberration in the K-ras signaling pathway may play an important role in the development of invasive MPSC. Previous studies of K-ras mutations in SBTs and ovarian serous carcinomas have differed in their findings and interpretation. Some have detected K-ras mutations in SBTs but not in carcinoma and concluded that they are unrelated 17 whereas others have detected them in nearly 40% of SBTs and 30% of serous carcinomas and concluded that SBTs may be precursors of serous carcinoma. 18 Since MPSC (noninvasive and invasive) was not recognized as a distinct entity in these studies, their results cannot be directly compared to ours. Second, we found that the allelic imbalance index gradually increased from SBTs to noninvasive and then to invasive MPSCs. In contrast, all conventional serous carcinomas including the very earliest (tumors less than 0.8 cm confined to one ovary) showed high levels of allelic imbalance. Since the alterations on chromosomes 5q and 1p were not exclusively observed in noninvasive and invasive MPSCs, respectively, and can rarely be demonstrated in SBTs, it is likely that critical genetic alterations may precede the morphological changes. This view is further supported by the identical allelic imbalance patterns and K-ras mutations in the tumors containing different morphological stages of progression (SBTs and noninvasive MPSC or noninvasive and invasive MPSC). Third, our findings that nearly 95% of bilateral ovarian MPSCs have discordant patterns of K-ras mutation or allelic imbalance suggest that they develop independently, although divergent progression from the same early neoplastic lesion cannot be entirely excluded. This contrasts with conventional serous carcinomas in which bilateral tumors have been reported to be monoclonal in most cases. 16
Clear-cut morphologically recognizable precursor lesions of conventional serous carcinomas are rarely observed. In our study, conventional serous carcinomas (including two tumors measuring 0.6 and 0.7 cm), showed massive, clonal allelic imbalance among the different chromosomal arms (Figure 2) ▶ . This finding together with the morphological observations that early conventional serous carcinomas are high-grade 19 underlies the notion that they arise “de novo.” It must be acknowledged, however, that the absence of morphologically established intermediate steps may be due to a higher rate of cellular proliferation resulting in rapid evolution to conventional serous carcinoma, obscuring discrete morphological intermediate stages. This is supported by a substantially higher Ki-67 nuclear labeling (proliferative) index in early conventional serous carcinoma as compared with SBT, noninvasive and invasive MPSC, 20 (and our unpublished data). Thus, the rapid progression of conventional serous carcinoma suggests that a profound loss of cell cycle regulation occurs very early in its development. This interpretation is supported by the finding of p53 mutations in small conventional serous carcinomas confined to the ovary and in adjacent “dysplastic” epithelium. 21 In contrast, p53 mutations have as yet not been detected in MPSC. 4 However, it should be noted that a comprehensive analysis of the pathogenesis of conventional serous carcinoma will require a large collaborative study since early tumors are rarely encountered.
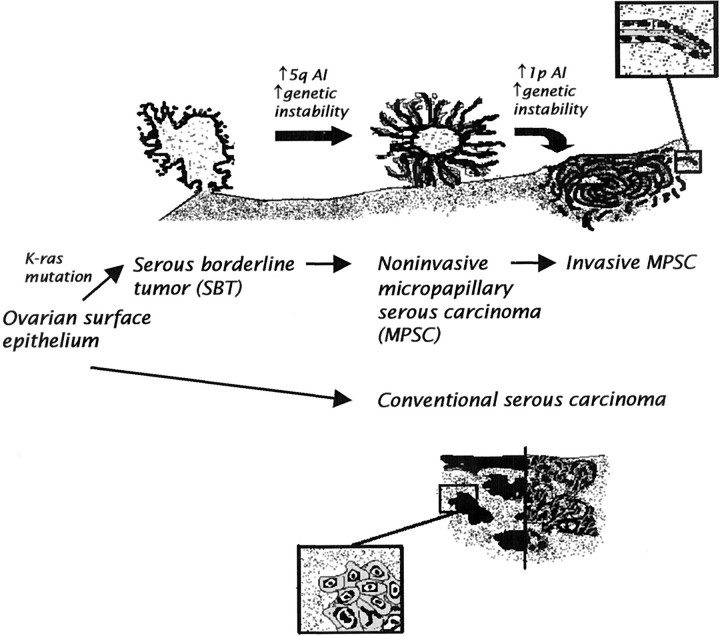
In summary, the molecular findings in this study in conjunction with morphological data support the stratification of ovarian serous carcinomas into two distinct groups with two different pathways of tumorigenesis (Figure 3) ▶ . In one pathway, a low-grade carcinoma (invasive MPSC) develops in a stepwise fashion from a SBT (atypical proliferative serous tumor) and then a noninvasive MPSC. This tumor and its precursors exhibit frequent K-ras mutations. As the precursors evolve into invasive MPSC they gradually acquire more genetic abnormalities. In the second pathway, a high-grade carcinoma (conventional serous carcinoma) develops by transformation from the ovarian surface epithelium or inclusion cysts without morphologically recognizable intermediate stages. These tumors, even early in their development, demonstrate wild-type K-ras and frequent allelic imbalance. This proposed dualistic model is the first step in an attempt to elucidate the pathogenesis of serous ovarian carcinoma, but should not be construed as implying that other pathways of tumorigenesis do not exist. Future studies focusing on gene expression profiles and the early molecular genetic alterations of these two types of serous carcinomas will be necessary to further elucidate the molecular pathogenesis of ovarian serous carcinoma.
Figure 3.
Schematic representation of the dualistic model depicting the development of ovarian serous carcinomas, the most common type of ovarian cancer. In one pathway invasive MPSC develops in a stepwise fashion from a SBT through a noninvasive stage of MPSC before becoming invasive. These tumors are associated with frequent K-ras mutations. Increased allelic imbalance of chromosome 5q is associated with the progression from SBT to MPSC and increased allelic imbalance of chromosome 1p with the progression from noninvasive to invasive MPSC. In the second pathway, conventional serous carcinoma, a high-grade neoplasm, exhibits a solid and/or pseudopapillary morphology and develops from the ovarian surface epithelium without morphologically recognizable intermediate stages. K-ras mutations have not been found in all these neoplasms tested.
Acknowledgments
We thank Drs. Bert Vogelstein and Tian-Li Wang at The Johns Hopkins Oncology Center for the critical comments.
Footnotes
Address reprint requests to Ie-Ming Shih, M.D., Ph.D., 418 N. Bond St., B-315, Baltimore, MD 21231. E-mail: ishih@jhmi.edu.
Supported by the Richard TeLinde Research Endowment from the Department of Gynecology and Obstetrics and the American Cancer Society, and The Johns Hopkins University School of Medicine. Gad Singer was supported by the Swiss National Science Foundation.
References
- 1.Dubeau L: Ovarian cancer. Scriver CR Beaudet AL Sly WS Valle D Childs B Kinzler KW Vogelstein B eds. The Metabolic and Molecular Bases of Inherited Disease. 2001, :pp 1091-1096 McGraw-Hill, New York [Google Scholar]
- 2.Auersperg N, Edelson MI, Mok SC, Johnson SW, Hamilton TC: The biology of ovarian cancer. Semin Oncol 1998, 25:281-304 [PubMed] [Google Scholar]
- 3.Seidman JD, Kurman RJ: Subclassification of serous borderline tumors of the ovary into benign and malignant types: a clinicopathologic study of 65 advanced stage cases. Am J Surg Pathol 1996, 20:1331-1345 [DOI] [PubMed] [Google Scholar]
- 4.Katabuchi H, Tashiro H, Cho KR, Kurman RJ, Hedrick Ellenson L: Micropapillary serous carcinoma of the ovary: an immunohistochemical and mutational analysis of p53. Int J Gynecol Pathol 1998, 17:54–60 [DOI] [PubMed]
- 5.Burks RT, Sherman ME, Kurman RJ: Micropapillary serous carcinoma of the ovary: a distinctive low-grade carcinoma related to serous borderline tumors. Am J Surg Pathol 1996, 20:1319-1330 [DOI] [PubMed] [Google Scholar]
- 6.Slomovitz BM, Caputo TA, Economos K, Gretz H: Non-invasive ovarian proliferations with micropapillary architecture are part of the spectrum of serous borderline tumor. Mod Pathol 2001, 14:146A [Google Scholar]
- 7.Liu J, Zabarovska VI, Braga E, Alimov A, Klien G, Zabarovsky ER: Loss of heterozygosity in tumor cells requires re-evaluation: the data are biased by the size-dependent differential sensitivity of allele detection. FEBS Lett 1999, 462:121-128 [DOI] [PubMed] [Google Scholar]
- 8.Vogelstein B, Kinzler KW: Digital PCR. Proc Natl Acad Sci USA 1999, 96:9236-9241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih IM, Zhou W, Goodman SN, Lengauer C, Kinzler KW, Vogelstein B: Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res 2001, 61:818-822 [PubMed] [Google Scholar]
- 10.Zhou W, Galizia G, Goodman SN, Romans KE, Kinzler KW, Vogelstein B, Choti MA, Montgomery EA: Counting alleles reveals a connection between chromosome 18q loss and vascular invasion. Nature Biotechnol 2001, 19:78-81 [DOI] [PubMed] [Google Scholar]
- 11.Shih IM, Wang TL, Traverso G, Romans K, Hamilton SR, Ben-Sasson S, Kinzler KW, Vogelstein B: Top-down morphogenesis of colorectal tumors. Proc Natl Acad Sci USA 2001, 98:2640-2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zborovskaya I, Gasparian A, Karseladze A, Elcheva I, Trofimova E, Driouch K, Trassard M, Tatosyan A, Lidereau R: Somatic genetic alterations (LOH) in benign, borderline and invasive ovarian tumours: intratumoral molecular heterogeneity. Int J Cancer 1999, 82:822-826 [DOI] [PubMed] [Google Scholar]
- 13.Cliby W, Ritland S, Hartmann L, Dodson M, Halling KC, Keeney G, Podratz KC, Jenkins RB: Human epithelial ovarian cancer allelotype. Cancer Res 1993, 53:2393-2398 [PubMed] [Google Scholar]
- 14.Dodson MK, Hartmann LC, Cliby WA, DeLacey KA, Keeney GL, Ritland SR, Su JQ, Podratz KC, Jenkins RB: Comparison of loss of heterozygosity patterns in invasive low-grade and high-grade epithelial ovarian carcinomas. Cancer Res 1993, 53:4456-4460 [PubMed] [Google Scholar]
- 15.Iwabuchi H, Sakamoto M, Sakunaga H, Ma YY, Carcangiu ML, Pinkel D, Yang-Feng TL, Gray JW: Genetic analysis of benign, low-grade, and high-grade ovarian tumors. Cancer Res 1995, 55:6172-6180 [PubMed] [Google Scholar]
- 16.Shih IM, Yan H, Speyrer D, Shmookler BM, Sugarbaker PH, Ronnett BM: Molecular genetic analysis of appendiceal mucinous adenomas in identical twins, including one with pseudomyxoma peritonei. Am J Surg Pathol 2001, 25:1095-1099 [DOI] [PubMed] [Google Scholar]
- 17.Caduff RF, Svoboda-Newman SM, Ferguson AW, Johnston CM, Frank TS: Comparison of mutations of Ki-RAS and p53 immunoreactivity in borderline and malignant epithelial ovarian tumors. Am J Surg Pathol 1999, 23:323-328 [DOI] [PubMed] [Google Scholar]
- 18.Mok SC, Bell DA, Knapp RC, Fishbaugh PM, Welch WR, Muto MG, Berkowitz RS, Tsao SW: Mutation of K-ras protooncogene in human ovarian epithelial tumors of borderline malignancy. Cancer Res 1993, 53:1489-1492 [PubMed] [Google Scholar]
- 19.Bell DA, Scully RE: Early de novo ovarian carcinoma: a study of fourteen cases. Cancer 1994, 73:1859-1864 [DOI] [PubMed] [Google Scholar]
- 20.Garzetti GG, Ciavattini A, Goteri G, De Nictolis M, Stramazzotti D, Lucarini G, Biagini G: Ki67 antigen immunostaining (MIB 1 monoclonal antibody) in serous ovarian tumors: index of proliferative activity with prognostic significance. Gynecol Oncol 1995, 56:169-174 [DOI] [PubMed] [Google Scholar]
- 21.Pothuri B, Leitao M, Barakat R, Akram M, Bogomolniy F, Olvera N, Lin O: Genetic analysis of ovarian carcinoma histogenesis. Gynecol Oncol 2001, 80:277 [Google Scholar]