Abstract
Both Helicobacter pylori (HP) and Epstein-Barr virus (EBV) have been implicated in carcinogenesis of the stomach. Fifty-seven gastric carcinomas were tested for microsatellite instability and allelic loss at several tumor suppressor loci using 21 polymorphic microsatellite markers. Furthermore, immunohistochemistry for p53 and DPC4/SMAD4 was performed. Results were analyzed according to HP and EBV status of the tumors, as assessed by immunohistochemistry and RNA in situ hybridization, respectively. Fractional allelic loss was lower in EBV-positive carcinomas (n = 15) when compared to EBV-negative carcinomas (P < 0.001). EBV positivity was inversely associated with allelic loss at specific markers on chromosomal arms 5q (APC), 17p (TP53), and 18q (DPC4/SMAD4). Allelic loss at the TP53 locus was not encountered in EBV-positive carcinomas, but occurred in 51% of EBV-negative carcinomas (P < 0.005). Moreover, none of the EBV-positive carcinomas showed unequivocal p53 immunopositivity in contrast to 39% of the EBV-negative carcinomas (P < 0.01). EBV-status was not related to microsatellite instability. There was no correlation between HP-status and any of the molecular alterations tested. In conclusion, EBV-positive gastric carcinomas follow a distinct pathogenesis at the molecular level, in which p53 is not, or differently inactivated.
Despite its declining incidence in the western world, gastric cancer remains one of the most frequent and lethal malignancies worldwide. 1 The natural history of gastric cancer is complex and incompletely understood but diet, infections, and genetic factors are involved. More than 90% of gastric cancers are adenocarcinomas, which can be divided into two major histological types (intestinal and diffuse) by the Laurén classification. 2
Of these two types, the tumorigenesis of the intestinal type of gastric cancer is best understood. It is thought to be governed by environmental factors and is characterized by precursor lesions of the gastric mucosa. 1,3 These precursor lesions are the morphological substrates of a stepwise neoplastic process in which genetic changes have accumulated gradually with tumor progression, similar to the adenoma-carcinoma sequence in colorectal cancer. 4,5
Infectious agents are important factors in carcinogenesis of the stomach. Helicobacter pylori (HP) is a well-known risk factor and it is now considered a first class carcinogen for stomach cancer. 6,7 Epstein-Barr virus (EBV) is encountered in a subset of tumors but its role in gastric carcinogenesis is less well understood. 8 The latency type in gastric carcinomas is different from the known EBV latency types as described for Burkitt’s lymphoma and nasopharyngeal carcinoma. 9,10 For example, the latent membrane protein-1 (LMP-1) is not expressed in EBV-positive stomach cancers. 10 Recent in vitro work by Subramanian and colleagues 11 suggests that the EBV nuclear protein EBNA-3C may functionally inactivate the human metastatic suppressor protein Nm23-H1. Hypermethylation of CpG islands as a mechanism of tumor suppressor gene silencing in EBV carrying gastric cancers has also been mentioned, 12 and expression of RUNX3, a gene causally related to stomach cancer is induced by the EBV transcription factor EBNA-2. 13,14
In the present study, using a variety of molecular markers, we investigated gastric cancers for loss of heterozygosity (LOH) at tumor suppressor loci known to be involved in carcinogenesis of the gastrointestinal tract 15-17 and near tumor suppressor genes involved in syndromes that include gastric cancer in their phenotype. Furthermore, the presence of microsatellite instability (MSI), a hallmark of a defective DNA mismatch repair system, was assessed. The results of these analyses were evaluated with respect to the HP and EBV status of the tumors to evaluate the possible role of these infectious agents in carcinogenesis of the stomach.
Materials and Methods
Patient Material
Formalin-fixed, paraffin-embedded tissue of 57 gastric carcinomas was retrieved from the archives of the pathology departments of the Academic Medical Center (Amsterdam, The Netherlands), the Lublin Medical Academy (Lublin, Poland), and the Johns Hopkins Hospital (Baltimore, MD). DNA was isolated from these tumors and polymerase chain reaction (PCR) was performed using several microsatellite primers as described below. Of these 57 gastric carcinomas the tumors consisted of 28 gastric stump carcinomas (GSCs) and 29 gastric carcinomas of the intact stomach. The tumors were classified according to the Laurén classification by an experienced gastrointestinal pathologist (GJAO). None of the tumors had a lymphoepithelioma-like histology. Patient and tumor characteristics of the GSC and gastric carcinoma of the intact stomach were comparable and not significantly different. GSCs were used in this study because remote partial gastrectomy is a premalignant condition that has our interest, 18 and EBV is relatively common in GSC. 19 The prevalence of HP and EBV positivity was not significantly different in GSCs and gastric carcinoma of the intact stomach in this series. Baseline characteristics according to EBV status are summarized in Table 2 ▶ .
Table 2.
Patient and Tumor Characteristics
| EBV-positive (n = 15) | EBV-negative (n = 42) | |
|---|---|---|
| Sex* | ||
| Male | 13 | 21 |
| Female | 2 | 21 |
| Age† | ||
| Mean | 64.5 | 70.2 |
| SD | 7.37 | 9.49 |
| Median | 63 | 73 |
| Origin | ||
| Netherlands | 7 | 28 |
| Poland | 6 | 9 |
| U.S.A. | 2 | 5 |
| Stomach type | ||
| Primary (GC-IS) | 5 | 24 |
| Stump (GSC) | 10 | 18 |
| Histology | ||
| Intestinal | 14 | 32 |
| Mixed | 0 | 5 |
| Diffuse | 1 | 5 |
| Stage | ||
| Early | 4 | 11 |
| Advanced | 11 | 31 |
| HP | ||
| Positive | 2 | 16 |
| Negative | 13 | 26 |
*P = 0.014; Fisher’s exact test.
†P = 0.015; Mann-Whitney U-test.
Detection of Epstein-Barr Virus and H. pylori
In situ hybridization for EBER1 nuclear RNA transcripts was performed as previously described. 19 HP status was assessed initially by histopathological examination of the hematoxylin and eosin-stained sections. Cases that were negative for HP were subsequently tested by immunohistochemistry using the B471 polyclonal rabbit anti-HP antibody (DAKO, Glostrup, Denmark), as described previously. 19
Microdissection and DNA Isolation
Tumor tissue was carefully microdissected from deparaffinized hematoxylin-stained 5-μm tissue sections. The percentage of cancer cells had to be at least 50 to 60%. For each case, matching nontumorous tissue was obtained from either a tumor-free lymph node or, when this was not available, from duodenum or smooth muscle cells. The tissue was incubated overnight in 50 to 100 μl of PK1 buffer (10 mmol/L Tris, pH 8.3, 50 mmol/L KCl, 2.5 mmol/L MgCl2, 0.45% Nonidet P-40, 0.45% Tween 20, 0.01% gelatin) containing 5% Chelex resin (Chelex 100; Bio-Rad Laboratories, Hercules, CA) and 3 to 5 μl of Proteinase-K (10 mg/ml) at 56°C followed by a 10-minute incubation at 95°C to inactivate Proteinase-K.
Microsatellite Analysis
Microsatellite analysis was performed by PCR with 21 microsatellite primer pairs. Markers were selected either because of their location at tumor suppressor loci known to be involved in gastric carcinogenesis, near (or in) genes involved in syndromes that contain gastric cancer in their phenotype or because of their inclusion in marker panels used for the determination of MSI. The markers used are listed in Table 1 ▶ . The sequences and their corresponding locations on the chromosomes were obtained from the Genome Data Base (http://www.gdb.org), the Cooperative Human Linkage Center (Chttp://lpg.nci.nih.gov/CHLC), or Genéthon (http://www.genethon.fr). One of the primers of each marker was fluorescently labeled. Optimal MgCl2 and dNTP concentrations were obtained for each primer pair at an annealing temperature of 55°C using control human DNA. PCR was performed in a PTC-100 thermal cycler (MJ Research, Inc., Waltham, MA) during 40 cycles in a total reaction volume of 20 μl, containing 40 ng of each primer, 0.1 mg/ml bovine serum albumin, and 1.0 U of Platinum Taq (Life Technologies, Inc., Rockville, MD) in the buffer supplied by the manufacturer. The PCR products were analyzed using an automated ABI 377 sequencer and Genescan 2.1 software (PE Biosystems, Foster City, CA).
Table 1.
Markers for LOH and MSI Analysis
| Marker | Chromosomal location | Repeat type | Putative tumor suppressor gene(s)/remarks |
|---|---|---|---|
| D2S123 | 2p16-21 | Dinucleotide | hMSH2; MSI consensus marker |
| D3S1478 | 3p21 | Dinucleotide | FHIT; hMLH1 |
| D3S2456 | 3p | Tetranucleotide | FHIT; hMLH1 |
| D5S346 | 5q21 | Dinucleotide | APC; MSI consensus marker |
| D5S107 | 5q11.2-q13.3 | Dinucleotide | APC |
| D9S171 | 9p21 | Dinucleotide | p14ARF; p16INK4A; p15INK4B |
| D9S932 | 9p | Tetranucleotide | p14ARF; p16INK4A; p15INK4B |
| D10S2491 | 10q23 | Dinucleotide | PTEN (intragenic marker) |
| D14S68 | 14q24.3-q | Dinucleotide | frequently deleted region in Barrett carcinomas |
| D16S2624 | 16q22.1 | Tetranucleotide | E-cadherin |
| P53 Alu | 17p | Alu repeat | p53 (intragenic marker) |
| TP53 | 17p13.1 | Dinucleotide | p53 |
| D17S250 | 17q11.2-q12 | Dinucleotide | BRCA1; MSI consensus marker |
| D18S64 | 18q21.32 | Dinucleotide | DCC; DPC4/SMAD4; SMAD2 |
| D18S474 | 18q | Dinucleotide | DCC; DPC4/SMAD4; SMAD2 |
| D19S565 | 19p13.3 | Dinucleotide | STK11/LKB1 |
| D19S886 | 19p13.3 | Dinucleotide | STK11/LKB1 |
| D21S49 | 21q22.3 | Dinucleotide | TFF1 |
| BAT25 | 4q12 | Mononucleotide | MSI consensus marker |
| BAT26 | 2p16 | Mononucleotide | MSI consensus marker |
| BAT40 | 1p13.1 | Mononucleotide | MSI consensus marker |
Scoring of LOH and MSI
Normal samples with two distinctly sized alleles at a particular marker were called “informative.” For all informative markers the allelic imbalance factor was calculated essentially as described by Cawkwell and colleagues. 20 A tumor was considered to show LOH at a particular marker if the allelic imbalance factor was >1.6 or <0.63. A finding of LOH had to be confirmed at least once to ensure reproducibility. For each individual the fractional allelic loss (FAL) was calculated as the ratio of LOH-positive markers to the total number of informative markers of that case. The FAL value therefore served as an overall measure of genetic instability at the tested loci.
Cases with an additional peak in the tumor DNA compared with their respective normal sample were scored as “microsatellite instable” (MSI) for a given marker. Tumors that exhibited MSI or that showed inconsistent results in repeated experiments at a given locus were excluded for analysis of LOH at that locus. With respect to MSI, tumors were classified according to international criteria. 21 Tumors were scored as stable (MSS) when no shifts were observed, as MSI-low (MSI-L) when shifts were seen in <40% of the markers and as MSI-high (MSI-H) with instability in ≥40% of the markers. MSI had to be confirmed at least once, to ensure reproducibility.
Immunohistochemistry
Immunohistochemistry for p53 and DPC4 was performed using the monoclonal antibodies DO-7 (DAKO) and clone B8 (Santa Cruz Biotechnology Inc., Santa Cruz, CA), respectively. Briefly, paraffin-embedded specimens were sectioned (5 μm), deparaffinized, and heat treated in 0.01 mol/L of Na-citrate buffer (pH 6.0) for 10 minutes in a Prestige Medical Series 2100 clinical autoclave (Prestige Medical, Blackburn, UK). Subsequently the slides were immersed in 0.3% hydrogen peroxide in methanol for 30 minutes. Nonspecific binding sites were blocked in 5% normal goat serum/phosphate-buffered saline (PBS) for 1 hour at room temperature after which the slides were incubated with the respective primary antibody in 5% normal goat serum/PBS for 1 hour. The Ultravision anti-polyvalent HRP detection system (Lab Vision Corp., Fremont, CA) was used to visualize antibody-binding sites with 3,3′-diaminobenzidine as a chromogen. Sections were counterstained with hematoxylin.
p53 immunoreactivity was scored as negative, weak (with weak to moderate staining in <30% of the tumor cells), or positive (with moderate to strong staining in >30% of the tumor cells). DPC4/SMAD4 immunoreactivity was scored as either negative or positive.
Statistical Analysis
For statistical analysis the Fisher exact test, and the Mann-Whitney U-test were used where indicated. A two-sided P value <0.05 was considered statistically significant.
Results
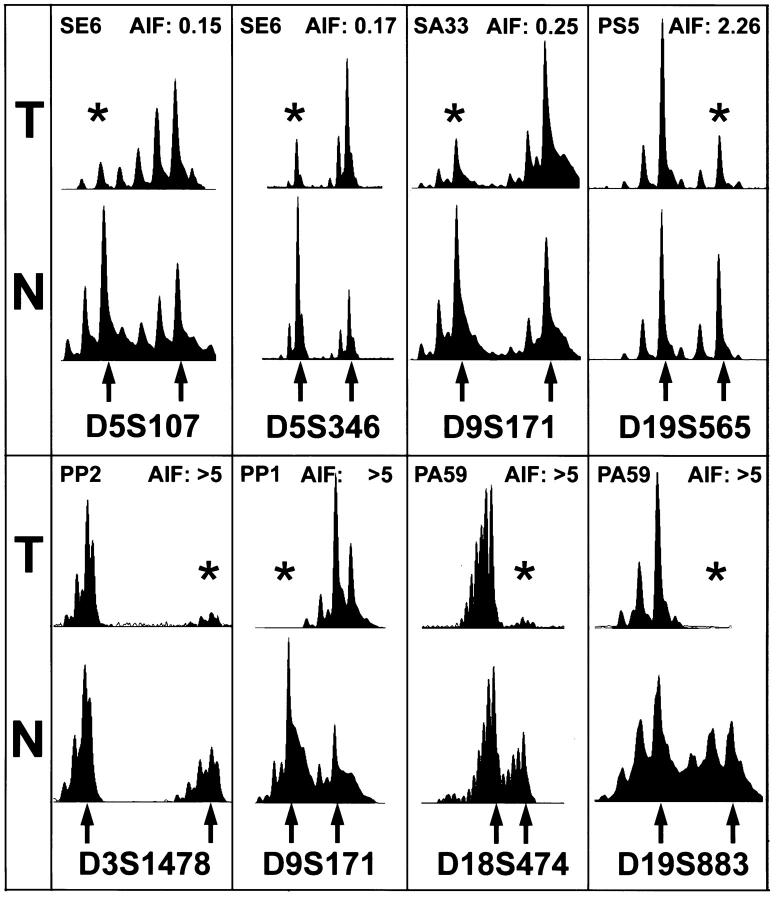
A total of 57 tumors was studied for the presence of HP and EBV and molecular alterations using polymorphic microsatellite markers and immunohistochemistry. The overall frequency of LOH markers as indicated by the mean FAL value was 0.278 for the complete study group. Representative examples of LOH are shown in Figure 1 ▶ . Markers that showed relatively frequent LOH (>30%) were on chromosomal arms 3p (31%), 9p (37%), 17p (40%), 18q (42%), and 19p (48%).
Figure 1.
Representative examples of LOH. Electropherograms of labeled PCR products of paired tumor (T) and normal (N) DNA are shown. The tumor number and the allelic imbalance factor are shown at the top of each frame and the lost alleles are marked by asterisks (see Materials and Methods for calculation of the allelic imbalance factor and scoring of LOH). The tested marker is indicated at the bottom of each frame and individual alleles are marked by arrows.
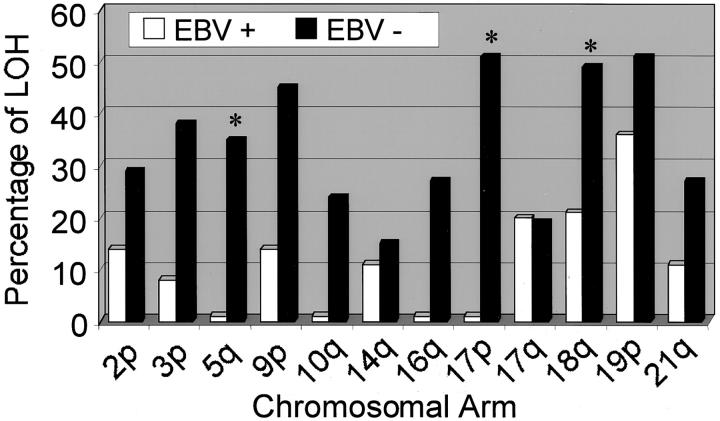
Of all 57 tumors, 15 tumors (26%) were positive for EBV as tested by EBER RNA in situ hybridization. Patients with EBV-positive tumors were predominantly male and on average 5.7 years younger when compared to patients with EBV-negative tumors. Other baseline characteristics were not significantly different between these two groups (Table 2) ▶ . A frequency distribution of allelic loss according to EBV status at the chromosomal arms tested is depicted in Figure 2 ▶ . There was a significant inverse relationship between positivity for EBV and the mean FAL value (Table 3) ▶ . When stratifying LOH results according to EBV status a significant inverse association was found between EBV-positivity and LOH at chromosomal arms 5q (APC), 17p (TP53), and 18q (DPC4/SMAD4) (Table 4 ▶ ; typical results are shown in Figure 3 ▶ ).
Figure 2.
Frequency distribution of allelic loss of EBV-positive and EBV-negative gastric carcinomas at the chromosomal arms tested. Asterisks indicate statistically significant differences at specific chromosomal arms.
Table 3.
Fractional Allelic Loss and MSI Relative to HP and EBV Status
| FAL | MSI-H | MSI-L | |
|---|---|---|---|
| EBV-positive | 0.097* | 2/15 (13%) | 3/15 (20%) |
| EBV-negative | 0.341* | 4/42 (10%) | 8/42 (19%) |
| HP-positive | 0.295 | 1/18 (6%) | 2/18 (11%) |
| HP-negative | 0.268 | 5/39 (13%) | 9/39 (23%) |
*EBV-positive versus EBV-negative; P < 0.001, Mann-Whitney U-test.
Table 4.
Results of LOH Analysis: EBV-Positive Carcinomas Versus EBV-Negative Carcinomas
| Chr. Arm | Marker | EBV positive (n = 15) | EBV negative (n = 42) | P value* | ||
|---|---|---|---|---|---|---|
| No. of LOH/informative cases | %LOH | No. of LOH/informative cases | %LOH | |||
| 2p | D2S123 | 1/7 | 14% | 7/24 | 29% | 0.641 |
| 3p | D3S1478 | 1/8 | 13% | 11/34 | 32% | 0.402 |
| D3S2456 | 0/9 | 0% | 6/25 | 24% | 0.162 | |
| Combined | 1/12 | 8% | 14/37 | 38% | 0.075 | |
| 5q | D5S346 | 0/12 | 0% | 11/32 | 34% | 0.021 |
| D5S107 | 0/12 | 0% | 9/34 | 26% | 0.086 | |
| Combined | 0/15 | 0% | 14/40 | 35% | 0.006 | |
| 9p | D9S171 | 1/10 | 10% | 12/29 | 41% | 0.120 |
| D9S932 | 2/12 | 17% | 11/29 | 38% | 0.275 | |
| Combined | 2/14 | 14% | 17/38 | 45% | 0.055 | |
| 10q | D10S2491 | 0/11 | 0% | 8/33 | 24% | 0.165 |
| 14q | D14S68 | 1/9 | 11% | 4/27 | 15% | 0.999 |
| 16q | D16S2624 | 0/9 | 0% | 8/30 | 27% | 0.160 |
| 17p | p53 Alu | 0/6 | 0% | 14/24 | 58% | 0.019 |
| TP53 | 0/10 | 0% | 12/27 | 44% | 0.016 | |
| Combined | 0/10 | 0% | 19/37 | 51% | 0.003 | |
| 17q | D17S250 | 2/10 | 20% | 4/21 | 19% | 0.999 |
| 18q | D18S64 | 1/11 | 9% | 12/23 | 52% | 0.024 |
| D18S474 | 2/11 | 18% | 12/31 | 39% | 0.282 | |
| Combined | 3/14 | 21% | 19/39 | 49% | 0.115 | |
| 19p | D19S565 | 3/7 | 42% | 15/27 | 56% | 0.681 |
| D19S883 | 1/9 | 11% | 7/30 | 23% | 0.625 | |
| Combined | 4/11 | 36% | 18/35 | 51% | 0.497 | |
| 21q | D21S49 | 1/9 | 11% | 7/26 | 27% | 0.647 |
*All, Fisher’s exact test.
Statistical significant differences are highlighted in bold.
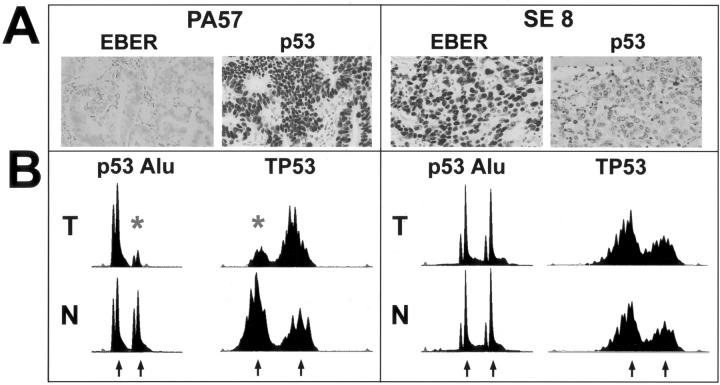
Figure 3.
Relation of EBV and p53 alterations, illustrated by two representative tumors (PA57 and SE8). A: Epstein-Barr virus was detected by in situ hybridization for EBER1 nuclear RNA transcripts. PA57 is negative whereas SE8 shows strong nuclear positivity in the vast majority of tumor cells. Immunohistochemistry for p53 is strongly positive in PA57 but negative in SE8. B: Electropherograms of labeled PCR products of paired tumor (T) and normal (N) DNA. Two microsatellite markers at the TP53 locus are shown (p53 Alu and TP53). Both PA57 and SE8 are informative for each marker. The individual alleles are indicated by arrows. PA57 (EBV-negative, p53-immunopositive) shows LOH at both markers. The lost alleles are marked by asterisks. SE8 (EBV-positive, p53-immunonegative) has retained both alleles in the tumor.
Using immunohistochemistry, we analyzed the tumors for positivity for p53 (17p) and DPC4/SMAD4 (18q). Immunohistochemistry for p53 was evaluated in 54 carcinomas and 33 cases were scored positive (61%). Unequivocal positivity, defined as moderate to strong positivity in more than 30% of the carcinoma cells, was observed in 16 cases (29%). None of the EBV-positive carcinomas were scored positive for p53 immunoreactivity in contrast to 39% of the EBV-negative carcinomas (Figure 3 ▶ , Table 5 ▶ ). Loss of DPC4/SMAD4 expression was observed in eight cases (15%). No association was found between EBV positivity and loss of DPC4/SMAD4 expression. In addition, no association was found between EBV positivity and MSI (Table 3) ▶ . MSI was found in 17 tumors (30%), 6 of which showed MSI-H and 11 of which were MSI-L.
Table 5.
EBV and p53 Immunohistochemistry
| p53 immunohistochemistry negative OR weak | p53 immunohistochemistry positive | |
|---|---|---|
| EBV-positive | 14 | 0 |
| EBV-negative | 25 | 16 |
P < 0.01, Fisher’s exact test.
Of the 57 tumors analyzed, 18 tumors (32%) were positive for HP, as tested by histopathological evaluation and immunohistochemistry of the tissue sections. There was no association between HP positivity and mean FAL value (Table 3) ▶ . HP status was neither significantly associated with LOH at specific markers nor with MSI (Table 3) ▶ .
Discussion
In the present study we compared the prevalence of LOH at several tumor suppressor loci and MSI in 57 gastric carcinomas using 21 polymorphic microsatellite markers. The LOH data for the total number of carcinomas were comparable to those reported in other allelotype studies of gastric cancer. 15-17
The overall prevalence of LOH at the tested loci, as measured by the mean FAL value, was not associated with HP status. In addition HP status was not significantly associated with any specific molecular changes, including MSI, which is in line with several previous reports on HP and molecular alterations. 22,23
In contrast, a significantly lower FAL value appeared to be associated with the presence of EBV in the carcinoma cells, as tested by in situ hybridization for EBER1 nuclear RNA transcripts. The pattern of LOH was also different in the EBV-positive and EBV-negative tumors. These results strongly suggest that EBV-positive gastric carcinomas follow a different pathogenetic pathway, at least on the genetic level, a notion that is in line with recent reports on EBV-positive gastric carcinomas. 12,24,25
LOH is thought to contribute to tumor suppressor gene inactivation. Methylation is currently regarded as an alternative mechanism for silencing tumor suppressor genes. In a recent publication more CpG islands were found to be methylated in EBV-positive gastric cancers when compared with cancers negative for EBV. 12 It would be conceivable that the lower FAL in the present study among the EBV-positive cancers might be compensated for by a higher frequency of gene inactivation through promotor hypermethylation. Frequent targets of hypermethylation are p16 on chromosomal arm 9p, STK11/LKB1 on 19p, and the mismatch repair genes. 26 However, LOH of 9p and 19p or MSI were not significantly different among the EBV-positive and EBV-negative tumors in the current investigation making it somewhat less likely that methylation, as an alternative mechanism for tumor suppressor gene inactivation, could explain the current observations.
Also, LOH was measured at a limited number of specific loci, and it may not be legitimate to generalize this finding and to conclude that EBV infection is accompanied by a genome-wide reduction in genetic instability. The microsatellite markers in this study were chosen based on the reported frequency of LOH at their respective loci in gastric cancer in general and EBV-positive carcinomas comprise only a minority (∼8 to 10%) of conventional gastric adenocarcinomas. Therefore, these results may reflect the fact that the LOH markers were in some way selected for EBV negativity of the tumors.
When LOH at specific markers was assessed, a strong inverse correlation was seen between EBV positivity and LOH at specific markers at chromosomal arms 5q, 17p, and 18q. Particularly, the inverse relation between EBV and LOH at chromosomal arm 17p suggests a difference with regard to the p53 tumor suppressor pathway. For example, it has been reported that the EBV-encoded EBNA-5 protein (alternatively designed EBNA-LP) can form a molecular complex in vitro with both the p53 and retinoblastoma (RB) proteins. 27 It is conceivable that binding with EBNA-5 may lead to an accelerated degradation of either one or both of these tumor suppressor proteins. This would imply a mechanism sharing analogy to that reported for the E6 and E7 proteins of certain human papillomaviruses in the pathogenesis of squamous cell carcinoma of the cervix, resulting in an abrogated tumor suppressor pathway without the need of genetic alteration of the involved gene itself. In line with this, immunohistochemical analysis for p53 protein revealed an inverse correlation of EBV positivity and p53 positivity.
It is more difficult to speculate about the possible mechanisms that are involved in the observed negative association between EBV positivity and LOH at 5q and 18q. Putative targets of LOH on these chromosomal arms may be APC on 5q and DCC, DPC4/SMAD4, or JV18 on 18q. However, there is little evidence for interaction of any of these tumor suppressor gene products and EBV.
LOH at chromosomal arm 18q21, the location of the DPC4/SMAD4 tumor suppressor gene was observed frequently. Inactivation of this gene at the genetic level is strongly correlated to loss of expression of DPC4/SMAD4 protein in pancreatic cancer. 28 We observed loss of expression of DPC4/SMAD4 protein in only 15% of all gastric carcinomas examined and this was not correlated with LOH at 18q21. Furthermore, genetic inactivation of DPC4/SMAD4 is rare in gastric carcinomas, 29 suggesting that DPC4/SMAD4 is not the target of LOH at this locus. In view of the above, also hypermethylation as a potential phenomenon that could explain the observed differences in LOH at 5q, 17p, and 18 is unlikely.
In a previous study using comparative genomic hybridization, no association was found between DNA ploidy and the EBV status and also loss of chromosomal arm 17p was not different. 25 These somewhat contradictory results are not easily explained. Differences in study materials and technicalities because of different methodology provide the most likely explanation. In general, allelic loss measured by specific microsatellite markers will be considered more sensitive and provide more accurate results.
The role of EBV in carcinogenesis of the stomach is not completely understood. The latency type of EBV in gastric adenocarcinomas is distinct from the known EBV latency types, eg, in Burkitt’s lymphomas and nasopharyngeal carcinomas. 9,10 This is mainly because of the expression of the latent membrane protein 2A (LMP2A) and the absence of LMP1 in gastric adenocarcinomas. The transforming BARF1 gene is frequently expressed in EBV-positive gastric carcinomas. 10 Sharing homology with the cellular proto-oncogene c-fms, BARF1 may provide an alternative way for the pathogenesis of EBV-associated epithelial cancers, ie, gastric adenocarcinoma and nasopharyngeal carcinoma, independent of LMP-1 expression. In this manner, EBV could provide a surrogate for further accumulation of genetic instability once the cells are infected and this may also explain our findings in EBV-positive tumors.
An alternative explanation for our results could be that susceptibility for EBV infection is determined by a specific molecular genetic route that involves other genetic changes than those reported frequently for conventional gastric carcinomas. This would be in line with our unpublished observations (Zur Hausen and colleagues, submitted) that EBV infection occurs most likely at a relatively late stage of carcinogenesis in the stomach, ie, at the transition of high-grade dysplasia into invasive carcinoma. EBV positivity would then rather be a consequence of the different molecular pathway.
In conclusion, EBV-positive carcinomas should be regarded as a separate entity with a distinct pathogenesis at the molecular level, when compared to EBV-negative carcinomas. Whether EBV positivity is a cause or merely a consequence of this difference remains to be elucidated. Likewise, the exact mechanism of a possible oncogenic role of EBV in gastric epithelial cells needs further study.
Acknowledgments
We thank F. H. Morsink, A. Musler, and M. J. Clement for excellent technical assistance; Dr. A. M. Cleton-Jansen for helpful advice regarding the allelotype analysis; and Dr. P. Sipponen (Jorvi Hospital, Espoo, Finland), Dr. W. Polkowski, Dr. D. Chibowski (Lublin Medical Academy, Lublin, Poland), and Dr. R. H. Hruban (The Johns Hopkins Hospital, Baltimore, MD) for kindly providing patient material.
Footnotes
Address reprint requests to G. Johan A. Offerhaus, Academic Medical Center, Department of Pathology, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands. E-mail: g.j.offerhaus@amc.uva.nl.
This work was supported by the Netherlands Organization for Scientific Research (grant 950-10-643).
References
- 1.Stadtländer CT, Waterbor JW: Molecular epidemiology, pathogenesis and prevention of gastric cancer. Carcinogenesis 1999, 20:2195-2208 [DOI] [PubMed] [Google Scholar]
- 2.Laurén P: The two main types of gastric carcinoma: diffuse and so-called intestinal-type carcinomas: an attempt at a histo-clinical classification. Acta Pathol Microbiol Scand 1965, 64:31-49 [DOI] [PubMed] [Google Scholar]
- 3.Schlemper RJ, Riddell RH, Kato Y, Borchard F, Cooper HS, Dawsey SM, Dixon MF, Fenoglio-Preiser CM, Flejou JF, Geboes K, Hattori T, Hirota T, Itabashi M, Iwafuchi M, Iwashita A, Kim YI, Kirchner T, Klimpfinger M, Koike M, Lauwers GY, Lewin KJ, Oberhuber G, Offner F, Price AB, Rubio CA, Shimizu M, Shimoda T, Sipponen P, Solcia E, Stolte M, Watanabe H, Yamabe H: The Vienna classification of gastrointestinal epithelial neoplasia. Gut 2000, 47:251-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solcia E, Fiocca R, Luinetti O, Villani L, Padovan L, Calistri D, Ranzani GN, Chiaravalli A, Capella C: Intestinal and diffuse gastric cancers arise in a different background of Helicobacter pylori gastritis through different gene involvement. Am J Surg Pathol 1996, 20:S8-S22 [DOI] [PubMed] [Google Scholar]
- 5.Fearon ER, Vogelstein B: A genetic model for colorectal tumorigenesis. Cell 1990, 61:759-767 [DOI] [PubMed] [Google Scholar]
- 6.Taylor DN, Blaser MJ: The epidemiology of Helicobacter pylori infection. Epidemiol Rev 1991, 13:42-59 [DOI] [PubMed] [Google Scholar]
- 7.: International Agency for Research on Cancer: Infection with H Pylori. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, vol. 61, Schistosomes, Liver Flukes and Helicobacter pylori. 1994:pp 177-240 IARC; Lyon [PMC free article] [PubMed]
- 8.Shibata D, Weiss LM: Epstein-Barr virus-associated gastric adenocarcinoma. Am J Pathol 1992, 140:769-774 [PMC free article] [PubMed] [Google Scholar]
- 9.Sugiura M, Imai S, Tokunaga M, Koizumi S, Uchizawa M, Okamoto K, Osato T: Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: unique viral latency in the tumour cells. Br J Cancer 1996, 74:625-631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.zur Hausen A, Brink AA, Craanen ME, Middeldorp JM, Meijer CJ, van den Brule AJ: Unique transcription pattern of Epstein-Barr virus (EBV) in EBV-carrying gastric adenocarcinomas: expression of the transforming BARF1 gene. Cancer Res 2000, 60:2745-2748 [PubMed] [Google Scholar]
- 11.Subramanian C, Cotter MA, II, Robertson ES: Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23–H1: a molecular link to cancer metastasis. Nat Med 2001, 7:350-355 [DOI] [PubMed] [Google Scholar]
- 12.Kang GH, Lee S, Kim WH, Lee HW, Kim JC, Rhyu MG, Ro JY: Epstein-Barr virus-positive gastric carcinoma demonstrates frequent aberrant methylation of multiple genes and constitutes CpG island methylator phenotype-positive gastric carcinoma. Am J Pathol 2002, 160:787-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spender LC, Cornish GH, Sullivan A, Farrell PJ: Expression of transcription factor AML-2 (RUNX3, CBFalpha-3) is induced by Epstein-Barr virus EBNA-2 and correlates with the B-cell activation phenotype. J Virol 2002, 76:4919-4927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, Kim HM, Kim WJ, Yamamoto H, Yamashita N, Yano T, Ikeda T, Itohara S, Inazawa J, Abe T, Hagiwara A, Yamagishi H, Ooe A, Kaneda A, Sugimura T, Ushijima T, Bae SC, Ito Y: Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 2002, 109:113-124 [DOI] [PubMed] [Google Scholar]
- 15.Tamura G, Sakata K, Nishizuka S, Maesawa C, Suzuki Y, Terashima M, Eda Y, Satodate R: Allelotype of adenoma and differentiated adenocarcinoma of the stomach. J Pathol 1996, 180:371-377 [DOI] [PubMed] [Google Scholar]
- 16.Choi SW, Park SW, Lee KY, Kim KM, Chung YJ, Rhyu MG: Fractional allelic loss in gastric carcinoma correlates with growth patterns. Oncogene 1998, 17:2655-2659 [DOI] [PubMed] [Google Scholar]
- 17.Yustein AS, Harper JC, Petroni GR, Cummings OW, Moskaluk CA, Powell SM: Allelotype of gastric adenocarcinoma. Cancer Res 1999, 59:1437-1441 [PubMed] [Google Scholar]
- 18.Offerhaus GJ: Gastric stump cancer: lessons from old specimens. Lancet 1994, 343:66-67 [DOI] [PubMed] [Google Scholar]
- 19.Baas IO, van Rees BP, Musler A, Craanen ME, Tytgat GN, van den Berg FM, Offerhaus GJ: Helicobacter pylori and Epstein-Barr virus infection and the p53 tumour suppressor pathway in gastric stump cancer compared with carcinoma in the non-operated stomach. J Clin Pathol 1998, 51:662-666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cawkwell L, Bell SM, Lewis FA, Dixon MF, Taylor GR, Quirke P: Rapid detection of allele loss in colorectal tumours using microsatellites and fluorescent DNA technology. Br J Cancer 1993, 67:1262-1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S: A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998, 58:5248-5257 [PubMed] [Google Scholar]
- 22.Blok P, Craanen ME, Offerhaus GJ, Dekker W, Kuipers EJ, Meuwissen SG, Tytgat GN: Molecular alterations in early gastric carcinomas. No apparent correlation with Helicobacter pylori status. Am J Clin Pathol 1999, 111:241-247 [DOI] [PubMed] [Google Scholar]
- 23.Wu MS, Shun CT, Wang HP, Sheu JC, Lee WJ, Wang TH, Lin JT: Genetic alterations in gastric cancer: relation to histological subtypes, tumor stage, and Helicobacter pylori infection. Gastroenterology 1997, 112:1457-1465 [DOI] [PubMed] [Google Scholar]
- 24.Wu MS, Shun CT, Wu CC, Hsu TY, Lin MT, Chang MC, Wang HP, Lin JT: Epstein-Barr virus-associated gastric carcinomas: relation to H. pylori infection and genetic alterations. Gastroenterology 2000, 118:1031-1038 [DOI] [PubMed] [Google Scholar]
- 25.zur Hausen A, van Grieken NC, Meijer GA, Hermsen MA, Bloemena E, Meuwissen SG, Baak JP, Meijer CJ, Kuipers EJ, van den Brule AJ: Distinct chromosomal aberrations in Epstein-Barr virus-carrying gastric carcinomas tested by comparative genomic hybridization. Gastroenterology 2001, 121:612-618 [DOI] [PubMed] [Google Scholar]
- 26.Baylin SB, Herman JG: DNA hypermethylation in tumorigenesis. Epigenetics joins genetics. Trends Genet 2000, 16:168-174 [DOI] [PubMed] [Google Scholar]
- 27.Szekely L, Selivanova G, Magnusson KP, Klein G, Wiman KG: EBNA-5, an Epstein-Barr virus-encoded nuclear antigen, binds to the retinoblastoma and p53 proteins. Proc Natl Acad Sci USA 1993, 90:5455-5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilentz RE, Su GH, Dai JL, Sparks AB, Argani P, Sohn TA, Yeo CJ, Kern SE, Hruban RH: Immunohistochemical labeling for dpc4 mirrors genetic status in pancreatic adenocarcinomas: a new marker of DPC4 inactivation. Am J Pathol 2000, 156:37-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell SM, Harper JC, Hamilton SR, Robinson CR, Cummings OW: Inactivation of Smad4 in gastric carcinomas. Cancer Res 1997, 57:4221-4224 [PubMed] [Google Scholar]