Abstract
The regenerative capacity of mammalian adult liver reflects the ability of a number of cell populations within the hepatic lineage to take action. Limited information is available regarding factors and mechanisms that determine the specific lineage level at which liver cells contribute to liver repair as well as the fate of their progeny in the hostile environment created by liver injury. In the present study, we attempted to identify novel molecules preferentially involved in liver regeneration by recruitment of transit-amplifying, ductular (oval) cell populations. With a subtractive cDNA library screening approach, we identified 48 enriched, nonredundant gene products associated with liver injury and oval cell proliferation in the adult rat liver. Of these, only two, namely α-fetoprotein and a novel transcript with high homology to human DMBT1 (deleted in malignant brain tumor 1), were specifically associated with the emergence of ductular (oval) cell populations in injured liver. Subsequent cloning and characterization of the rat DMBT1 homologue revealed a highly inducible expression in ductular reactions composed of transit-amplifying ductular (oval) cells, but not in ductular reactions after ligation of the common bile duct. In human liver diseases, DMBT1 was expressed in ductular reactions after infection with hepatitis B and acetaminophen intoxication, but not in primary biliary cirrhosis, primary sclerosing cholangitis, and obstruction of the large bile duct. The expression heterogeneity in ductular reactions and multiple functions of DMBT1 homologues point to intriguing roles in regulating not only tissue repair but also fate decision and differentiation paths of specific cell populations in the hepatic lineage.
The proficiency of the adult mammalian liver to regenerate under pathophysiological conditions has long been recognized. However, only recently has it been firmly established that this regenerative capacity reflects the ability of several recognized populations of cells with stem-like characteristics to respond to damage of liver tissue. Because hepatic cells with stem-like properties are ideal therapeutic targets in patients suffering chronic liver failure, further knowledge of the mechanisms regulating the commitment of a particular cell type to participate in liver repair is important both for identification of therapeutic potentials and in understanding developmental processes and tissue homeostasis.
The hallmarks of stem cells include competency to renew repeatedly, to renew the stem cell population, and to generate sufficient differentiated progeny to maintain or regenerate the functional capacity of a tissue. 1 In rodent models, reconstruction of liver mass lost to surgical resection is achieved through proliferation of fully differentiated, normally quiescent hepatocytes and bile duct cells in the residual tissue. These mature cells in the hepatic lineage are numerous, can respond rapidly, and give rise to a large number of progeny while maintaining their differentiated phenotype. 2,3 However, in certain types of toxic hepatic injury impairing the replication of hepatocytes, a large population of ductular epithelial cells, possibly originating from endogenous stem cells in the canal of Hering, is produced. The resulting intricate network of ductular structures invade the parenchyma, where the ductular epithelial cells may differentiate to hepatocytes or bile duct cells to reconstitute the architecture and function of the damaged liver tissue. 4-6 The ductular epithelial cells are, therefore, at least bipotential progenitor cells and have, because of their cytological appearance with an oval-shaped nucleus and a high nuclear to cytoplasmic ratio, been named oval cells. Nevertheless, regeneration in response to other classes of hepatotoxins impairing hepatocyte replication seems to be accomplished by vigorously proliferating small hepatocyte-like progenitor cells expressing phenotypic characteristics of fetal hepatoblasts and adult mature hepatocytes. 7,8 It is not established if these small incompletely differentiated hepatocyte-like cells resemble the population of mature hepatocytes in adult liver giving rise only to new hepatocytes or the bipotential fetal hepatoblasts capable of differentiation into mature hepatocytes and bile duct cells during fetal liver development. Finally, there may be two origins of stem cells in the liver: endogenous stem cells located in the canal of Hering and exogenous stem cells derived from the bone marrow and capable of differentiation into hepatocytes and bile duct cells after homing to the injured liver. 9,10
The existence of hepatic cells with stem-like properties in humans is less well established. However, the ability of hepatocytes and bile epithelial cells to reconstitute liver mass and function after surgical resection is well known. Furthermore, ductular reactions recognized in a variety of genetic, acute, and chronic liver diseases display cells with phenotypic characteristics of the bipotential ductular (oval) cells in rodent models. 11-15 As in rodent liver, the intrahepatic origin of these stem-like cells is likely to be the canals of Hering. 16 Finally, it has recently been shown that in human transplant recipients, hepatocytes and bile duct cells can be derived from extrahepatic circulating multipotent stem cells, probably of bone marrow origin, and that these cells are involved in replenishment of hepatic epithelial cells. 17,18 Thus, the tremendous regenerative capacity of the mammalian liver seems to be reflected by the ability to call forth a cellular response at different levels in the hepatic lineage. This has led to the hypothesis that similar to other organ systems, the cellular lineage of the liver consists of true stem cells located in the canal of Hering, progenitor cells [transit-amplifying bipotential ductular (oval) cells], and mature hepatocytes and bile duct cells. 19
Limited information is available regarding factors and mechanisms that determine the contribution of a liver cell population at a specific lineage level to liver repair as well as the fate of its progeny in the hostile environment created by liver injury. However, the ability of a particular cell population in the hepatic lineage to respond and participate in liver regeneration seems to be associated with the type of injury inflicted. 2,7,20 These differences are reflected in the hepatic expression patterns of several growth regulatory molecules. 21-24 Hence, it is tempting to hypothesize that the specific microenvironment created in response to a particular liver injury may determine fate decision and differentiation paths of the recruited cell populations, allowing the progeny to survive and proliferate in a hostile environment. Therefore, if a more targeted approach to decreasing tissue injury and enhancing repair/regeneration in humans is to be achieved a better understanding of the cellular and molecular mechanisms allowing stem cells to repopulate, proliferate, and differentiate in a hostile environment is required.
In the present study, we have focused our efforts on identifying novel molecules and mechanisms defining the microenvironment necessary for proliferation of transit-amplifying ductular (oval) cells in injured liver. We have used a selective subtractive cDNA library screening approach applying a polymerase chain reaction (PCR)-based cDNA library construction and cDNA array analysis, extensive Northern blot analysis of identified molecules in representative models of rat liver regeneration, PCR-based cDNA cloning, and immunostaining of formalin-fixed tissue from experimental rat models as well as from a number of human liver diseases displaying ductular reactions. Here we report the identification and characterization of DMBT1 and its rat homologue dmbt1 4.7kb as novel molecules in liver regeneration. The molecules are rapidly induced after liver injury, and show strong heterogeneity of expression in ductular reactions of adult human and rat liver dependent on the injury induced. These findings point to intriguing roles of the molecules as factors in the microenvironment regulating not only tissue repair but also fate decision and differentiation paths of transit-amplifying ductular (oval) cells.
Materials and Methods
Experimental Animal Models
Male Wistar rats, 8 weeks of age, were purchased from M&B A/S (Ry, Denmark) and kept under standardized conditions with access to food and water ad libitum. Liver regeneration through replication of mature hepatocytes and bile duct cells was achieved by surgical resection of the median and left lateral liver lobes removing ∼70% of the liver mass (PHx protocol). 2 Liver regeneration by transit-amplifying ductular (oval) cells was achieved using the AAF/PHx protocol in which treatment with 2-acetylaminofluorene (2-AAF) (9 days, 20 mg/kg/day by gavage) was interrupted at day 5 to perform a 70% hepatectomy. 6,25 Liver regeneration through proliferation of small hepatocyte-like progenitor cells was achieved by inhibiting replication of mature hepatocytes by two intraperitoneal injections of retrorsine (30 mg/kg) 2 weeks apart, followed by a 70% hepatectomy 5 weeks after the last treatment with retrorsine (retrorsine/PHx protocol). 7 Control groups included: 1) a sham operation with laparotomy only; 2) treatment with 2-AAF (9 days, 20 mg/kg/day by gavage) combined with a sham operation at day 5; and 3) treatment with retrorsine (30 mg/kg i.p. 2 weeks apart) followed by a sham operation 5 weeks after the last treatment with toxin. Finally, proliferation of mature bile duct cells was induced by ligation of the common bile duct for 5 days. 26 Groups of three animals were sacrificed by cervical dislocation at the time points indicated in the figures, and parts of the livers were snap-frozen in liquid nitrogen for RNA extraction, or fixed for histological examination. The Danish Council for Supervision with Experimental Animals had approved the studies.
Human Tissue Samples
Normal liver tissue from liver transplant donors or taken for diagnostic purposes (n = 7) served as control samples. Diseased tissue samples were obtained from transplant recipients with primary biliary cirrhosis (n = 5, cirrhotic stage with active inflammation), primary sclerosing cholangitis (n = 3, cirrhotic stage with active inflammation), obstruction of the large bile duct (n = 2, cirrhotic stage), acetaminophen intoxication (n = 5, submassive liver cell necrosis), and hepatitis B infection (n = 3, submassive liver cell necrosis). A few tissue samples were received fresh allowing one part to be snap-frozen in liquid N2 for later extraction of total RNA and one part to be fixed in formalin. The remaining samples were only fixed in formalin. All fixed tissues were embedded in paraffin for routine histology and immunohistochemistry.
Differential Expression Cloning and Sequencing
The suppression subtractive hybridization cDNA library was constructed with the PCR-Select cDNA subtraction kit (Clontech Laboratories Inc., Palo Alto, CA) according to the manufacturer’s instructions. The rat livers chosen for library construction were selected according to histological evaluations of liver morphology and cellular composition. A rat liver from the AAF/PHx protocol at day 10 after partial hepatectomy harboring a very large population of transit-amplifying ductular (oval) cells was chosen as the tester sample. As the driver sample a normal liver from an untreated rat was used. Polyadenylated RNA [poly(A)+ RNA] was prepared from snap-frozen liver tissue by ultracentrifugation through a cesium chloride cushion and enrichment by oligo(dT)-cellulose chromatography. Samples of 2 μg of poly(A)+ RNA were reverse-transcribed to cDNA. Subsequently, tester and driver cDNA were hybridized, the remaining unhybridized sequences amplified by PCR, and cloned into pCRII plasmid vectors (TA cloning kit; Invitrogen, Carlsbad, CA). Differentially expressed cDNA sequences were identified by cDNA array analysis as described in the PCR-Select cDNA subtraction kit protocol (Clontech). In the present study, 600 Escherichia coli transformants carrying the pCRII plasmid vector were analyzed for cDNA inserts by PCR. Of these, 196 transformants containing cDNA inserts larger than 250 bp were chosen for further array analysis. The cDNA products were amplified by PCR and arrayed on nylon membranes. Subsequently, the membranes were hybridized to cDNA probes produced by forward and reverse subtraction and labeled with [32P]dCTP (Rediprime; Amersham Pharmacia Biotech, Uppsala, Sweden). Clones containing cDNA inserts that exclusively hybridized to the forward-subtracted probe [representing cDNAs expressed highly in liver regenerating by recruitment of transit-amplifying ductular (oval) cells] were sequenced using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit with an automated sequencer (ABI373; PE Applied Biosystems, Foster City, CA). DNA sequences and conceptual translations were compared with known nucleotide and protein sequences using the BLAST algorithm (www.nci.nlm.nih.gov/BLAST/). Four publicly accessible databases were searched: SwissProt, GenBank nr protein, GenBank nr nucleotide, and dbEST-expressed sequence tags.
Cloning of Rat dmbt1 4.7kb
The full-length cDNA for rat dmbt1 4.7kb was cloned by reverse transcribing 1 μg of total RNA (AAF/PHx protocol 10 days after PHx) into cDNA with the Smart Race cDNA amplification kit (Clontech). The resulting cDNA was then amplified in a 50-μl 5′ Race PCR reaction composed of 2.5 μl of cDNA template (diluted 1:100), 5 μl of Universal primer mix, 5 μl of Buffer A, and 5 μl of Buffer B (Elongase Enzyme Mix System; Life Technologies, Palo Alto, CA), 200 μmol/L each of dNTP, 1 μl Elongase Enzyme Mix, and 400 nmol/L gene-specific primer (Eb-3-UTR rev1, 5′-CTAGCTAGAGAAAGGATGGTGATGCCA-3′). The cDNA was amplified by an initial denaturing step for 2 minutes at 94°C followed by 35 cycles of 30 seconds at 94°C, 30 seconds at 60°C, and 12 minutes at 68°C. The PCR products were cloned into the pCR-XL-TOPO plasmid vector (TOPO XL PCR cloning kit, Invitrogen) and sequenced.
Northern Blot and Semiquantitative Reverse Transcriptase-PCR Analyses
In experiments on liver regeneration, animals were sacrificed as indicated and total RNA extracted from ∼50 mg of snap-frozen liver tissue (RNeasy kit; Qiagen Inc., Santa Clarita, CA). Populations of viable nonparenchymal cells were isolated by perfusion of the liver in situ as described. 22 The nonparenchymal cell populations were isolated 7 days after partial hepatectomy in the AAF/PHx protocol combining treatment with 2-AAF and a 70% hepatectomy, or 0 (untreated), 3, 6, 24, 48, and 96 hours after initiation of treatment with 2-AAF alone (20 mg/kg/day). Hepatocytes were isolated 0 (untreated), 3, 6, 24, 48, and 96 hours after treatment with 2-AAF alone (20 mg/kg/day) by a two-step in situ perfusion procedure. 27 All cell preparations were snap-frozen in liquid nitrogen and stored at −70°C until total RNA was isolated (RNAstat Reagent; Tel-Test Inc., Friendswood, TX).
Northern blot analysis was performed by electrophoresis of 10 μg of total RNA in 1% agarose/0.2 formaldehyde gels and transfer onto nylon membranes. A rat Multiple Tissue Northern (MTN) blot was purchased from Clontech. For quantification, a slot blot analysis was performed by immobilizing 10 μg of total RNA onto nylon membranes. Membranes were hybridized to cDNA probes labeled with [32P]dCTP. The probe for rat dmbt1 4.7kb encompassed nucleotides 4260 to 4623 of the 3′-untranslated sequence (GenBank accession number to be deposited), and for rat α-fetoprotein (AFP) nucleotides 101 to 329 (GenBank accession no. X02361). Both cDNA fragments were isolated in the subtraction library analysis.
For semiquantitative reverse transcriptase-PCR analysis of DMBT1 in human liver, samples of 1 μg of total RNA were reverse-transcribed and amplified using the Advantage 2 PCR enzyme system (Clontech) and the following forward and reverse primers: hDMBT1-ZP-fwd3 (5′-CCTGCTCTGTCTGCCAAATCACATGCAAGCC-3′) and hDMBT-3-UTR rev1 (5′-GCATGGATTCTGGGACTGCAGGTCTATGGGCCA-3′). As internal standard of the RNA amount used in the PCR, a primer pair of human β-actin (5′-TCTGGCCGTACCACTGGCAT-3′) and (5′-cactgtgttggcgtacaggt-3′) were used for amplification. Amplification conditions were 35 cycles of 30 seconds at 94°C, 30 seconds at 65°C, and 2 minutes at 72°C.
Immunohistochemistry
Tissue specimens were fixed in 4% neutral formalin or paraformaldehyde for 24 hours and processed for routine histology. Immunostaining of liver tissue was performed on 5-μm paraffin sections that were cleared with xylene and rehydrated through a graded series of alcohols (5 minutes each) ending with 10 minutes of incubation in phosphate-buffered saline. After quenching of endogenous peroxidase activity (15 minutes of incubation in methanol containing 2% H2O2), antigenic unmasking was accomplished by either heating in 10 mmol/L of sodium citrate (pH 6.0) for 15 minutes (DMBT1 and cytokeratin 7) or treatment with 0.1% trypsin for 15 minutes at 37°C (AFP). Nonspecific activity was blocked by a 60-minute incubation in blocking solution composed of 100 mmol/L Tris (pH 7.6), 550 mmol/L NaCl, 10 mmol/L KCl, 0.05% Triton X-100, 1% bovine serum albumin, and 1.5% serum of the secondary antibody species in a humidified chamber at room temperature. The sections were subsequently incubated overnight with the primary antibodies diluted in the above solution at 4°C. Specific binding of antibody was revealed by streptavidin/biotin immunoperoxidase techniques (Vectastain ABC Elite kit; Vector Laboratories, Burlingame, CA) with diaminobenzidine as substrate followed by light staining with Mayer’s hematoxylin. The primary antibodies used on rat tissue were rabbit polyclonal against gp-340/DMBT1 (rabbit anti-human 421-gp-340, 28,29 diluted 1:400), and AFP (rabbit anti-human AFP, diluted 1:800; DAKO, Glostrup, Denmark). Immunostaining of human tissues was achieved with mouse monoclonal antibodies against human gp-340/DMBT1 (mouse monoclonal Hyb213-6, 28,29 diluted 1:100) and human cytokeratin 7 (mouse monoclonal anti-human cytokeratin 7, diluted 1:100; BioGenex, San Ramon, CA). The specificities of the gp-340/DMBT1 antibodies were verified by Western blot analysis of DMBT1 protein purified from rat liver and human bronchi alveolar lung washings (data not shown).
Results
Identification of Genes Preferentially Expressed in Rat Liver Harboring Transit-Amplifying Ductular (Oval) Cells
We approached the isolation of genes preferentially expressed in liver regenerating by recruitment of transit-amplifying ductular (oval) cells by identifying uniquely expressed transcripts in the AAF/PHx protocol as compared to the normal liver. Our overall screening approach, encompassing PCR-based construction of a subtractive cDNA library and cDNA array analysis combined with sequence acquisition and bioinformatics of resulting library clones, identified 48 enriched, nonredundant gene products (Table 1) ▶ .
Table 1.
Subtractive cDNA Library of Genes Highly Expressed in Rat Liver Regeneration by Transit-Amplifying Ductular (Oval) Cells
| No. | Insert identity | Accession no. | No. | Insert identity | Accession no. | No. | Insert identity | Accession no. |
|---|---|---|---|---|---|---|---|---|
| 1 | Endomembrane protein emp70* | AF160213 | 17 | 6.2 kda, protein | NM_019059 | 33 | MtN3-like protein | AF151726 |
| 2 | α-Fetoprotein* | V01254 | 18 | Ebnerin/dmbt1* | U32681 | 34 | hnRNP H | Y14196 |
| 3 | Carbamyl phosphate synthetase 1* | M12322 | 19 | 14-kda, ubiquitin conjugating enzyme | U04308 | 35 | Lysosomal-associated transmembrane protein 4A | NM_008640 |
| 4 | α-1-acid glycoprotein* | V01216 | 20 | Receptor for activated kinase C | AJ132860 | 36 | Voltage-dependent anion channel 3 (Vdac3)* | NM_011696 |
| 5 | Na-, K-ATPase β subunit* | M14137 | 21 | Lysine-ketoglutarate reductase/saccharo-pine dehydrogenase* | AJ224761 | 37 | Guanine nucleotide binding protein, α-inhibiting polypeptide (Gnai3) | NM_013106 |
| 6 | FGF-4 induced serine/threonine type 2C phosphatase homologue (FIN13)* | U42383 | 22 | Cytochrome P450 IVB1 (Cyp4b1)* | NM_016999 | 38 | Unknown* | |
| 7 | Amino acid transporter system A (ATA2)* | AF249673 | 23 | RNA binding motif protein (Rbm3)* | NM_016809 | 39 | Phenylalanine hydroxylase* | K02599 |
| 8 | Corticoid steroid-binding globulin* | X70533 | 24 | Translation elongation factor 2 (EF-2)* | NM_017245 | 40 | Elongation factor 1 α (EF-1)* | L19339 |
| 9 | Ribosomal protein L24* | X78443 | 25 | Stem-loop binding protein (Slbp)* | NM_009193 | 41 | Anthracycline-associated resistance protein (Arx)* | U35833 |
| 10 | Tyrosine amino transferase* | X02741 | 26 | p68 RNA helicase* | X65627 | 42 | CDC23* | NM_004661 |
| 11 | Fibronectin* | L29191 | 27 | Urate oxidase* | M24396 | 43 | mdr1b* | M81855 |
| 12 | Interferon-related putative protein (TIS7)* | X17400 | 28 | Proliferation-associated protein 1 (Plfap)* | NM_011119 | 44 | MHC class II RT1.D α* | AJ001998 |
| 13 | Aspartate amino transferase (cAspAT)* | D00252 | 29 | Guanine nucleotide-releasing protein (mss4) | L10336 | 45 | Histone H1.2 | Y12291 |
| 14 | Thioredoxin interacting factor | U30789 | 30 | Peroxisomal phytanoyl-CoA hydroxylase (PHYH) | AF121345 | 46 | Chaperonin containing TCP-1 epsilon (Ccte) | Z31555 |
| 15 | Stromal cell derived factor receptor 2 (Sdfr2, SDR2)* | NM_009146 | 31 | Leucine amino peptidase | NM_015907 | 47 | Chaperonin subunit 7 (eta) (Cct7) | NM_007638 |
| 16 | Glvr-1 | M73696 | 32 | Proton/phosphate symporter | M23984 | 48 | γ-Actin* | X52815 |
*cDNA inserts used in the comparative Northern blot analysis.
To confirm a preferential expression in liver regenerating from transit-amplifying ductular (oval) cells, a comparative Northern blot analysis of 30 gene products listed in Table 1 ▶ was performed using a number of experimental models to induce regeneration from cells at different stages in the hepatic lineage. Regeneration was achieved by: 1) mature hepatocytes and bile duct cells using the PHx protocol; 2) transit-amplifying ductular (oval) cells using the AAF/PHx protocol; and 3) small hepatocyte-like progenitor cells using the retrorsine/PHx protocol. Control groups included a sham operation, treatment with 2-AAF combined with a sham operation, and treatment with retrorsine combined with a sham operation. Animals were sacrificed 1, 3, 5, 7, and 9 days after partial hepatectomy and gene expression patterns analyzed. All products studied were expressed at higher steady-state levels in the liver harboring a large population of transit-amplifying ductular (oval) cells (AAF/PHx protocol day 9 after PHx) as compared to sham-treated animals, confirming the efficacy of the subtraction analysis. Interestingly, of the 30 transcripts studied only transcripts encoded by AFP and a gene sequence with high identity to rat Ebnerin, 30 mouse CRP-ductin, 31 and human gp-340/DMBT1 29,32 showed preferential expression in the AAF/PHx protocol (Figure 1A) ▶ . The steady-state levels of the remaining 28 transcripts were clearly elevated in the AAF/PHx protocol but were also increased to variable degrees in other protocols of injury (data not shown).
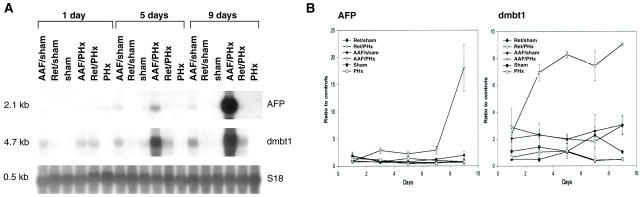
Figure 1.
Gene expression analysis of AFP and the DMBTt1 homologue in rat liver. A: Northern blot analysis. Each lane contains 10 μg of total RNA isolated from one rat liver. Membranes were hybridized to cDNA probes labeled with [32P]dCTP and visualized by autoradiography. Hybridization with S18 was used to assess integrity and equal loading of RNA samples. B: Quantification of gene expression by slot blot and phosporimager analysis. Each point represents the mean of hepatic gene expression in three animals normalized to the expression in a control group of three untreated rats. AAF/sham, treatment with 2-AAF and laparotomy only; Ret/sham, treatment with retrorsine and laparotomy only; sham, laparotomy only; AAF/PHx, treatment with 2-AAF and a 70% hepatectomy; Ret/PHx, treatment with retrorsine and a 70% hepatectomy; PHx, 70% hepatectomy only. Animals were sacrificed 1, 3, 5, 7, and 9 days after the surgical procedure.
Expression of AFP in the rat is normally restricted to the hepatoblasts in the fetal liver. However, during development of the bipotent transit-amplifying ductular (oval) cell phenotype in adult liver AFP expression is acquired. 4,5,23,25,33 In the AAF/PHx model of liver regeneration, transit-amplifying ductular (oval) cells proliferate to form an intricate network of ductular structures. The number of transit-amplifying ductular (oval) cells reaches a maximum between 8 and 11 days after the partial hepatectomy 33 in accordance with the increasing expression of AFP from day 7 to day 9 after hepatectomy in the present study (Figure 1B ▶ , AAF/PHx protocol). In contrast, increased levels of AFP transcripts were not detectable by Northern blot analysis in liver regeneration from small hepatocyte-like progenitor cells (Figure 1B ▶ , Ret/PHx protocol) or from mature hepatocytes and bile duct epithelial cells (Figure 1B ▶ , PHx protocol). Therefore, expression of AFP seems to be a highly specific marker for transit-amplifying ductular (oval) cells in the regenerating adult rat liver. Expression of the DMBT1 rat homologue also appeared to be strongly associated with transit-amplifying ductular (oval) cells (Figure 1B ▶ , AAF/PHx model). However, increased expression of the DMBT1 homologue was detected already at day 3 after hepatectomy (Figure 1B ▶ , AAF/PHx protocol). Furthermore, expression of the DMBT1 homologue was detectable in the AAF/sham and Ret/PHx protocols in accordance with the previously described emergence of small populations of transit-amplifying ductular (oval) cells in portal areas. 7,25
Immunostaining with antibodies against AFP and DMBT1 detected both proteins in structures of transit-amplifying ductular (oval) cells at day 9 after hepatectomy in the AAF/PHx protocol (Figure 2, a and b) ▶ . Expression of dmbt1 in newly formed bile ducts (Figure 2b) ▶ and in structures resembling intestinal epithelium (Figure 2d) ▶ was in contrast to the lack of AFP expression in the same structures (Figure 2, a and c) ▶ . In the large foci of newly formed, small basophilic hepatocytes only entrapped structures of transit-amplifying ductular (oval) cells expressed both proteins (Figure 2, e and f) ▶ .
Figure 2.
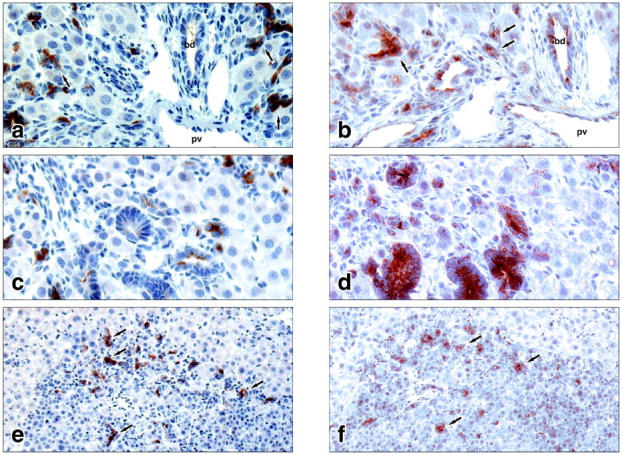
Cellular localization of AFP (a, c, e) and the rat homologue of DMBT1 (b, d, f) at day 9 after a 70% hepatectomy in the AAF/PHx protocol. a and b feature a portal area with an intricate network of transit-amplifying ductular (oval) cells penetrating the hepatic parenchyma. c and d feature structures of intestinal-type tissue staining positive for dmbt1 but not for AFP. e and f feature a focus of small basophilic hepatocytes entrapping structures of ductular (oval) cells staining positive for both AFP and dmbt1. pv, portal vein; bd, bile duct; arrows point to structures of ductular (oval) cells. Original magnifications: ×400 (c and d); ×200 (e and f).
We have previously described heterogeneity in AFP expression among the bile ductules and the larger bile ducts after activation of transit-amplifying ductular (oval) cells. 25,33 In the AAF/PHx protocol, only a small population of the total number of bile ductules expressed AFP at early time points (up to 5 days after PHx) 25,33 whereas the larger bile ducts consistently were negative. At later time points, from 5 days after PHx and beyond, all cells in ductular structures attained the ability to express AFP. 33 Therefore, we investigated if the expression of the DMBT1 rat homologue would follow a similar induction pattern. In control liver, a few cells of the larger hepatic bile ducts occasionally showed a positive reaction with the antibody against DMBT1. Cells in the bile ductules, endothelial cells, and hepatocytes were negative (Figure 3A, a) ▶ . Interestingly, induction of DMBT1 expression was observed in all ductular epithelial cells in models in which transit-amplifying ductular (oval) cells have been identified [ie, AAF treatment alone (Figure 3A, c) ▶ , AAF/PHx model (Figure 3A, d) ▶ , and retrorsine/PHx model (Figure 3A, f) ▶ ]. After PHx, DMBT1 expression was also detected in ductular epithelial cells at the time when they proliferate to reconstitute the bile epithelial structures 2 (Figure 3A, e) ▶ . Hepatocytes proliferating after PHx (Figure 3A, e) ▶ , small hepatocyte-like progenitor cells proliferating in response to treatment with retrorsine/PHx (Figure 3A, f) ▶ , and ductular cells proliferating after ligation of the common bile duct (Figure 3A, b) ▶ did not react with the antibodies. In the AAF/PHx protocol 5 days after PHx, some hepatic endothelial cells also stained positive for dmbt1 protein (Figure 3A, d) ▶ . A summary of AFP and dmbt1 protein expression in epithelial cell populations under various conditions of liver injury and regeneration in the adult rat liver is presented in Table 2 ▶ . Northern blot analysis of separated hepatocyte and nonparenchymal cell populations after AAF treatment alone confirmed that induction of dmbt1 expression was exclusive to the nonparenchymal cell populations. The induction was observed in cell isolates as early as 24 hours after initiation of the AAF treatment (Figure 3B) ▶ .
Figure 3.
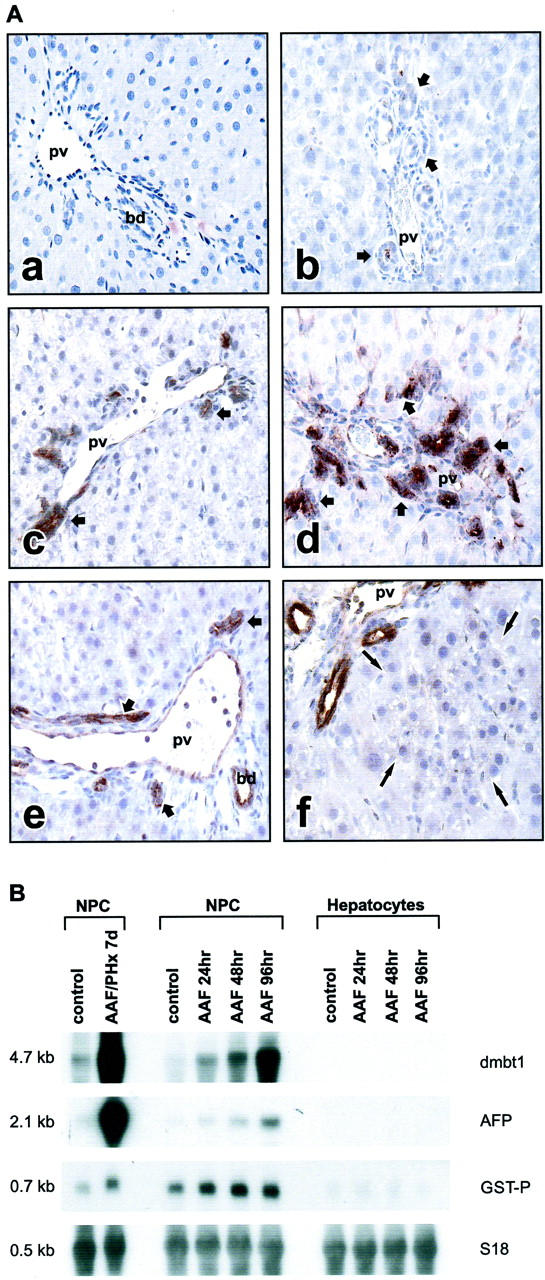
A: Localization of rat dmbt1 in normal rat liver (a), 5 days after ligation of the common bile duct (b), 5 days after initiation of 2-AAF treatment (c), 5 days after partial hepatectomy in the AAF/PHx protocol (d), 5 days after partial hepatectomy (e), and 5 days after partial hepatectomy in the Ret/PHx model (f). Short arrows point to ductular structures; long arrows point to a focus of small hepatocyte-like cells. pv, portal vein; bd, bile duct. B: Analysis of dmbt1 and AFP transcripts in isolated liver cell populations by Northern blotting of 10 μg of total RNA. Hybridization to the placental form of glutathione S-transferase (GST-P) was used as a common marker for bile duct and ductular (oval) cells to verify the purity of nonparenchymal (NPC) versus parenchymal (hepatocytes) cell populations. Hybridization with S18 was used to assess integrity and equal loading of RNA. Original magnifications, ×200 (a to f).
Table 2.
Summary of dmbt1 and α-Fetoprotein Protein Expression in Epithelial Cell Compartments Under Various Conditions of Liver Injury and Regeneration in the Adult Rat Liver
| Hepatocytes | Epithelial cells in bile ducts and ductules | Epithelial cells in ductular reactions | Epithelial cells in foci of small hepatocytes | Epithelial cells in structures of intestinal-type tissue | |
|---|---|---|---|---|---|
| Control | −/−* | −/− | NP§ | NP | NP |
| Sham, 5 days | −/− | −/− | NP | NP | NP |
| BDL, 5 days | −/− | −/− | −/− | NP | NP |
| PHx, 5 days | −/− | +/−† | NP | NP | NP |
| AAF/Sham, 5 days | −/− | +/− | +/(+)‡ | NP | NP |
| AAF/PHx, 5 days | −/− | +/− | +/+ | NP | NP |
| AAF/PHx, 9 days | −/− | +/− | +/+ | (+)/(+) | +/− |
| Ret/Sham, 5 days | −/− | −/− | NP | NP | NP |
| Ret/PHx, 5 days | −/− | +/− | +/(+) | −/− | NP |
*−/−, Immunostaining of dmbt1/AFP not detectable.
†+, Immunostaining detectable.
‡(+), Immunostaining not detectable in all epithelial cells in the structure.
§Not present by histological examination.
The DMBT1 Homologue Expressed in Regenerating Rat Liver Is a Specific Splice Variant
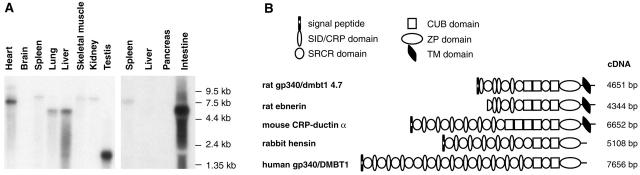
The transcript size of the DMBT1 rat homologue expressed in regeneration from transit-amplifying ductular (oval) cells was calculated from the Northern blot analysis in Figure 1A ▶ to be ∼4700 bp (hereafter referred to as rat dmbt1 4.7kb). Northern blot analysis of normal, adult rat tissues revealed that the 4.7-kb splice variant appeared specifically in the injured liver. With the exception of the intestine, expression levels in adult tissues were generally very low (Figure 4A) ▶ . A number of different splice variants were detected by the cDNA probe specific to the 3′-untranslated region of rat dmbt1 4.7kb, including a 6.0-kb transcript in normal liver, lung, and intestine, a 8.4-kb variant in spleen, skeletal muscle, and kidney, a 7.7-kb variant in heart, and a 1.7-kb variant in testis (Figure 4A) ▶ . Long-range PCR with a primer designed from the subtraction clone used in the Northern blot analysis with identity to the 3′ end of human DMBT1 cDNA, resulted in amplification of a single product from rat liver harboring a large population of transit-amplifying ductular (oval) cells. The 4651-bp cDNA product contained a 5′-untranslated region of 17 bp, followed by an ORF of 4273 bp, and a 3′-untranslated region of 378 bp. The 3′-untranslated region was included in the cDNA sequence isolated in the subtraction library screen. The nucleotide sequence of rat dmbt1 4.7kb cDNA has been deposited at the European Molecular Biology Laboratory nucleotide sequence database.
Figure 4.
A: Northern blot analysis of DMBT1 homologues in normal adult rat tissues. Left: Samples of 2 μg of poly (A+) RNA are loaded in each lane and the membrane exposed to X-ray film for 7 days. Right: Samples of 10 μg of total RNA are loaded in each lane and the membrane exposed to X-ray film for 2 days. B: Domain organization of rat dmbt1 4.7kb and members of the SRCR superfamily group B with high homology to dmbt1 4.7kb.
The ORF encoded a polypeptide chain of 1380 amino acids, including a putative signal peptide of 20 residues (Figure 4B) ▶ . When the signal peptide was omitted, the calculated molecular mass of the polypeptide chain was ∼149 kd. The domain organization of rat dmbt1 4.7kb was similar to the organization of the human, mouse, and rabbit homologues 28-31,34 and features the putative signal peptide followed by the first SID/CRP domain, and three SRCR (scavenger receptor cysteine-rich) domains separated by fours SID/CRPs. The third SRCR domain was followed by a short Thr-rich region, a CUB (C1r/C1 seconds, sea urchin epidermal growth factor, bone morphogenetic protein-1) domain, a short Ser-Thr-Pro-rich region, a second CUB domain, a second short Ser-Thr-Pro-rich region, a fourth SRCR domain, a third short Ser-Thr-Pro-rich region, a third CUB domain, a ZP (zona pellucida) domain, and finally an amino acid sequence containing a transmembrane domain and a short cytosolic tail of 23 and18 amino acids (Figure 4B) ▶ . Rat dmbt1 4.7kb represented a longer cDNA sequence of rat Ebnerin originally cloned from von Ebner’s glands 30 and reported to be expressed during transit-amplifying ductular (oval) cell proliferation in a previous study. 23
Heterogeneity of DMBT1 Expression Revealed in Ductular Reactions of Human Liver
By reverse transcriptase-PCR analysis, specific transcripts encoded by DMBT1 were amplified from clinical tissue samples displaying ductular reactions in livers with submassive necrosis caused by acetaminophen intoxication (n = 1) and hepatitis B infection (n = 2) but not from normal liver (n = 1), primary biliary cirrhosis (n = 1), and primary sclerosing cholangitis (n = 1) (data not shown). These preliminary results encouraged us to investigate the expression of DMBT1 protein in ductular reactions of a larger number of clinical tissue samples using immunohistochemical staining against cytokeratin 7 (CK7) as a marker for duct or ductular-derived epithelial cells. 35 In liver showing a normal morphology (n = 7) no reaction with the antibody against DMBT1 was observed in CK7-positive ductular structures [Figure 5, a (DMBT1) and b (CK7) ▶ ]. In liver regenerating after parenchymal destruction because of hepatitis B infection (n = 3) or acetaminophen intoxication (n = 5), DMBT1 staining was observed in association with reactive ductular structures formed by CK7-positive cells [Figure 5c (DMBT1) and d (CK7); e (DMBT1) and f (CK7) ▶ ]. DMBT1 protein appeared to be located in the apical membrane of ductular epithelial cells (Figure 5, c and e) ▶ . Consistent with previous observations 35 both reactive bile ductules and intermediate (transitional) hepatocyte-like cells stained for CK7 in livers with end-stage primary biliary cirrhosis, primary sclerosing cholangitis, and obstruction of the large bile duct (Figure 5; h, j, l) ▶ . However, neither the ductules nor the intermediate hepatocyte-like cells stained positive for DMBT1 in these three diseases (Figure 5; g, i, k) ▶ .
Figure 5.
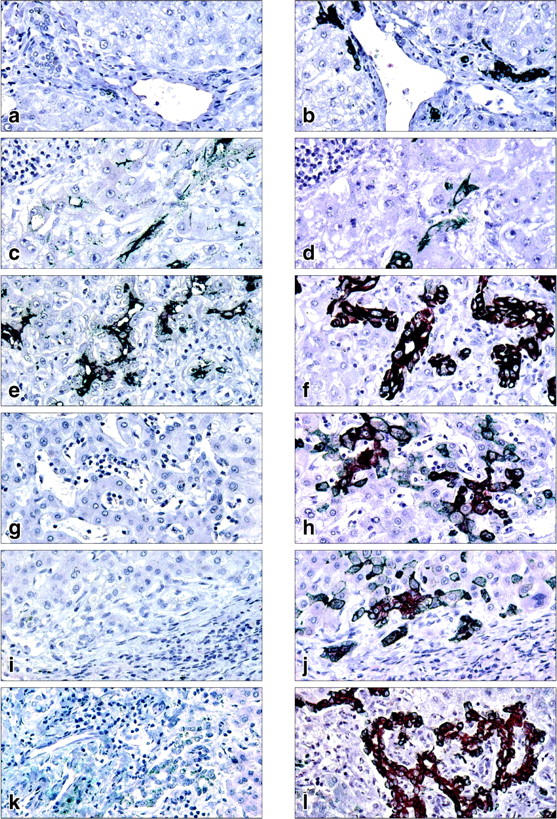
Representative immunostaining of DMBT1 (a, c, e, g, i, k) or cytokeratin 7 (CK7) (b, d, f, h, j, l) in human liver disease with ductular reactions. Pairs of serial sections were used in the analysis. Normal liver (a, b), hepatitis B infection (c, d), acetaminophen intoxication (e, f), primary biliary cirrhosis (g, h), primary sclerosing cholangitis (i, j), and large bile duct obstruction (k, l). Original magnifications, ×400.
Discussion
Throughout the past several years, it has become increasingly clear that liver regeneration is fundamentally regulated by several peptides that act in a highly coordinated manner to ensure rapid restoration of tissue function and mass. These include members of several distinct growth factor and cytokine families operating through cognate cell surface receptors on responsive epithelial cell populations in both autocrine and paracrine modes of action. 2,21,22 In a previous study, we used a similar, although less stringent subtractive cDNA library approach, to demonstrate that modulation of molecules involved in interferon-γ-mediated events was associated with emergence of transit-amplifying ductular (oval) cells in regenerating rat liver. 23 This first study was designed to identify modulation of molecules associated with the nonparenchymal cell population and was performed on isolated, nonparenchymal cell populations enriched in transit-amplifying ductular (oval) cells. The aim of the present study was to define molecules associated with changes in the microenvironment of the liver regenerating from transit-amplifying ductular (oval) cells and, therefore, homogenates from regenerating liver harboring a large population of transit-amplifying ductular (oval) cells were used. Furthermore, a stringent cDNA array and expression analysis were added in the present study to identify molecules that were strongly associated with regeneration from transit-amplifying ductular (oval) cells. Therefore, it is not surprising that the molecules reported previously as being modulated during regeneration from transit-amplifying ductular (oval) cells 23 are not identical to the molecules presented in Table 1 ▶ . Interestingly, two molecules, namely AFP and DMBT1 (Ebnerin), have been identified as strongly associated with liver regeneration from transit-amplifying ductular (oval) cells in both studies indicating that these molecules in combination with the events controlling their modulation may be particularly important as factors in fate decision and differentiation of transit-amplifying ductular (oval) cells within the hepatic lineage.
Considering the possible importance of DMBT1 as a novel molecule in liver regeneration from transit-amplifying ductular (oval) cells, we decided to clone the complete cDNA sequence of the dmbt1 splice variant expressed in regenerating rat liver. The cloned cDNA sequence, rat dmbt1 4.7kb, has a size of 4651 bp and encodes a protein composed of a putative signal peptide sequence, four group B SRCR domains, three CUB domains, one ZP domain, a transmembrane region, and a short cytoplasmic tail with potential phosphorylation sites. The protein has a domain organization similar to the SRCR group B proteins Ebnerin, 30 CRP-ductin, 31 hensin, 34 and gp-340/DMBT1 29,32 in the rat, mouse, rabbit, and human, respectively. SRCR domains are capable of mediating protein-protein interactions and the majority of proteins containing SRCR domains have been implicated in functions within the immune defense system. 36 The CUB domain was first identified in tolloid, a protein that activates decapentaplegic (dpp, the Drosophila transforming growth factor-β homologue). The domain has been recognized as a common motif in proteins involved in embryo- and organogenesis including bone morphogenetic protein-1, 37 but are also found in plasma proteins such as serine protease molecules including complement subcomponents Cls/Clr- and MBL-associated serine proteases. The ZP domain, present in ZP sperm receptor proteins, bears some similarity to another transforming growth factor-β-binding protein, the type III receptor betaglycan. 38 Consequently, the domain structure of rat dmbt1 4.7kb implies a role for the protein as a putative signaling receptor in pathways involved in liver regeneration from transit-amplifying ductular (oval) cells.
At least two distinct functions of DMBT1 homologues have been identified. Clearly, the proteins have one function in the mucosal defense system and a second in epithelial differentiation. A splice variant of human DMBT1 also called gp-340 binds the collectin protein surfactant protein D (SP-D). 28,29 SP-D is generally present on the apical side of all mucosal surfaces including the intrahepatic bile ducts, 39 and the protein has been shown to play an important role in opsonization of bacteria, viruses, and allergens, in modulating the immune response by suppressing macrophage activation and oxidant production as well as inhibiting T-lymphocyte proliferation, and in maintaining surfactant phospholipid homeostasis. 40-42 Furthermore, DMBT1 homologues have recently been identified as putative trefoil factor receptors. 43 Trefoil factors, a family comprised of three small proteins designated TFF1 (pS2), TFF2 (SP), and TFF3 (ITF), are secreted onto the apical side of mucosal surfaces including the intrahepatic bile ducts. Although their mechanisms of action are unknown, trefoil factors can protect epithelial surfaces from a variety of deleterious agents, including bacterial toxins, chemicals, and drugs, and seem to play a key role in promoting epithelial regeneration after injury. 44,45 Induced expression of dmbt1 in transit-amplifying ductular (oval) cells in regenerating rat liver and of DMBT1 in ductular reactions of human liver after submassive parenchymal necrosis caused by acetaminophen intoxication or hepatitis B infection may, therefore, comprise an important protective mechanism in the hostile environment created by liver injury and facilitate functional restoration of the damaged liver. Interestingly, neither ductular structures nor intermediate (transitional) hepatocyte-like cells expressed DMBT1 in diseases such as primary biliary cirrhosis, primary sclerosing cholangitis, and obstruction of the large bile duct in which the progression is from initial primary damage of the biliary tree to secondary injury of the lobule in the end stage. This fundamental difference between ductular reactions in human liver disease may suggest that the direct damage of the biliary tree and/or a different signaling from the extracellular matrix could affect the expression of DMBT1 in these diseases and point to the use of DMBT1 as an interesting marker protein for diagnostic purposes.
Several lines of evidence also support an intriguing role of DMBT1 homologues in epithelial differentiation. 31,46,47 In the small intestine, mouse CRP-ductin is expressed in the crypt epithelium from the stem cells in the crypt base to the terminally differentiating cells in the crypt top. However, as the cells move from the crypt to the villus, the gene seems to turn off. 31 Therefore, it was reasoned that CRP-ductin might be involved in differentiation of intestinal epithelium. Recent biochemical studies have established that rabbit hensin is able to switch the polarity and induce processes of terminal differentiation in kidney epithelial cells. Hensin seems to induce a reorganization of the apical membrane and its associated cytoskeleton with the expression of cytokeratin 19 and Villin. The result is an apical-basal cell polarization and epithelial columnarization similar to what occurs in terminal differentiation of other epithelia. 34,46 It has been proposed that a population of nonpolarized, undifferentiated cells located in the biliary ductules and with a blast-like phenotype, replicate and differentiate to transitional cells exhibiting a similar apical-basal polarity and phenotype as transit-amplifying ductular (oval) cells. 48 Therefore, it is tempting to speculate that expression of DMBT1 may be a critical determinant of fate decision and differentiation of transit-amplifying ductular (oval) cells in adult liver injury and repair.
Acknowledgments
We thank Gerda Demant Olesen, Bjørg Krog, and Kirsten Priisholm for excellent technical assistance.
Footnotes
Address reprint requests to Hanne Cathrine Bisgaard, Ph.D., Department of Medical Biochemistry and Genetics, The Panum Institute, University of Copenhagen, Blegdamsvej 3, DK-2200 Copenhagen, Denmark. E-mail: hcb@imbg.ku.dk.
Supported by grants from the Danish Natural Science and Medical Research Councils (to H. C. B.), the Novo Nordisk Foundation (to H. C. B. and N. T.), the Fonden til Lægevidenskabens Fremme (to N. T.), the Savværksejer Jeppe Juhl og Hustru Ovita Juhl’s Mindefond (to H. C. B.), and the Benzon Foundation (to U. H.).
References
- 1.Blau HM, Brazelton TR, Weimann JM: The evolving concept of a stem cell: entity or function. Cell 2001, 105:829-841 [DOI] [PubMed] [Google Scholar]
- 2.Michalopoulos GK, De Frances MC: Liver regeneration. Science 1997, 276:60-66 [DOI] [PubMed] [Google Scholar]
- 3.Overturf K, Al-Dhalimy M, Ou C, Grompe M: Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. Am J Pathol 1997, 51:1273-1280 [PMC free article] [PubMed] [Google Scholar]
- 4.Lemire JM, Shiojiri N, Fausto N: Oval cell proliferation and the origin of small hepatocytes in liver injury induced by D-galactosamine. Am J Pathol 1991, 139:535-552 [PMC free article] [PubMed] [Google Scholar]
- 5.Dabeva MD, Shafritz D: Activation, proliferation, and differentiation of progenitor cells into hepatocytes in the D-galactosamine model of liver regeneration. Am J Pathol 1993, 43:1606-1620 [PMC free article] [PubMed] [Google Scholar]
- 6.Evarts RP, Hu Z, Omori N, Omori M, Marsden ER, Thorgeirsson SS: Precursor-product relationship between oval cells and hepatocytes: comparison between tritiated thymidine and bromodeoxyuridine as tracers. Carcinogenesis 1996, 17:2143-2151 [DOI] [PubMed] [Google Scholar]
- 7.Gordon GJ, Coleman WB, Hixson DC, Grisham JW: Liver regeneration in rats with retrorsine-induced hepatocellular injury proceeds through a novel cellular response. Am J Pathol 2000, 156:607-619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon GJ, Coleman WB, Grisham JW: Temporal analysis of hepatocyte differentiation by small hepatocyte-like progenitor cells during liver regeneration in retrorsine-exposed rats. Am J Pathol 2000, 157:771-786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen BE, Bowen WC, Patrene KD, Mars WM, Sullivan AK, Murase N, Boggs SS, Greenberger JS, Goff JP: Bone marrow as a potential source of hepatic oval cells. Science 1999, 284:1168-1170 [DOI] [PubMed] [Google Scholar]
- 10.Theise ND, Badve S, Saxena R, Henegariu O, Sell S, Crawford JM, Krause DS: Derivation of hepatocytes from bone marrow cells in mice after radiation-induced myeloablation. Hepatology 2000, 31:235-240 [DOI] [PubMed] [Google Scholar]
- 11.Hsia CC, Evarts RP, Nakatsukasa H, Marsden ER, Thorgeirsson SS: Occurrence of oval-type cells in hepatitis B virus-associated human hepatocarcinogenesis. Hepatology 1992, 16:1327-1333 [DOI] [PubMed] [Google Scholar]
- 12.De Vos R, Desmet V: Ultrastructural characteristics of novel epithelial cell types identified in human pathological liver specimens with chronic ductular reactions. Am J Pathol 1992, 140:1441-1450 [PMC free article] [PubMed] [Google Scholar]
- 13.Sell S: Comparison of liver progenitor cells in human atypical ductular reactions with those seen in experimental models of liver injury. Hepatology 1998, 27:317-331 [DOI] [PubMed] [Google Scholar]
- 14.Lowes KN, Brennan BA, Yeoh GC, Olynyk JK: Oval cell numbers in human chronic liver diseases are directly related to disease severity. Am J Pathol 1999, 154:537-541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crosby HA, Hubscher S, Fabris L, Joplin R, Sell S, Kelly D, Strain AJ: Immunolocalization of putative human liver progenitor cells in livers from patients with end-stage primary biliary cirrhosis and sclerosing cholangitis using the monoclonal antibody OV-6. Am J Pathol 1998, 152:771-779 [PMC free article] [PubMed] [Google Scholar]
- 16.Theise ND, Saxena R, Portmann BC, Thung SN, Yee H, Chiriboga L, Kumar A, Crawford JM: The canals of Hering and hepatic stem cells in humans. Hepatology 1999, 30:1425-1433 [DOI] [PubMed] [Google Scholar]
- 17.Theise ND, Nimmakayalu M, Gardner R, Illei PB, Morgan G, Teperman L, Henegariu O, Krause DS: Liver from bone marrow in humans. Hepatology 2000, 32:11-16 [DOI] [PubMed] [Google Scholar]
- 18.Alison MR, Poulsom R, Jeffery R, Dhillon AP, Quaglia A, Jacob J, Novelli M, Prentice G, Williamson J, Wright NA: Hepatocytes from non-hepatic adult stem cells. Nature 2000, 406:257. [DOI] [PubMed] [Google Scholar]
- 19.Sell S: Heterogeneity and plasticity of hepatocyte lineage cells. Hepatology 2001, 33:738-750 [DOI] [PubMed] [Google Scholar]
- 20.Alison M, Golding M, Lalani E, Sarraf C: Wound healing in the liver with particular reference to stem cells. Phil Trans R Soc Lond B 1998, 353:877-894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy P, Bisgaard HC, Schnur J, Thorgeirsson SS: Studies on hepatic gene expression in different liver regenerative models. Biochem Biophys Res Commun 2000, 272:591-595 [DOI] [PubMed] [Google Scholar]
- 22.Bisgaard HC, Santoni-Rugiu E, Nagy P, Thorgeirsson SS: Modulation of the plasminogen activator/plasmin system in rat liver regenerating by recruitment of oval cells. Lab Invest 1998, 78:237-246 [PubMed] [Google Scholar]
- 23.Bisgaard HC, Müller S, Nagy P, Rasmussen LJ, Thorgeirsson SS: Modulation of the gene network connected to interferon-γ in liver regeneration from oval cells. Am J Pathol 1999, 155:1075-1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tygstrup N, Bangert K, Ott P, Bisgaard HC: Messenger RNA profiles in liver injury and stress: a comparison of lethal and nonlethal rat models. Biochem Biophys Res Commun 2002, 290:518-525 [DOI] [PubMed] [Google Scholar]
- 25.Bisgaard HC, Nagy P, Santoni-Rugiu E, Thorgeirsson SS: Proliferation, apoptosis, and induction of hepatic transcription factors are characteristics of the early response of biliary epithelial (oval) cells to carcinogens. Hepatology 1996, 23:62-70 [DOI] [PubMed] [Google Scholar]
- 26.Polimeno L, Azzarone A, Zeng QH, Panella C, Subbotin V, Carr B, Bouzahzah B, Francavilla A, Starzl TE: Cell proliferation and oncogene expression after bile duct ligation in the rat: evidence of a specific growth effect on bile duct cells. Hepatology 1995, 21:1070-1078 [PMC free article] [PubMed] [Google Scholar]
- 27.Gant TW, Silverman JA, Bisgaard HC, Burt RK, Marino PA, Thorgeirsson SS: Regulation of 2-acetylaminofluorene- and 3-methylcholanthrene-mediated induction of multidrug resistance and cytochrome P450IA gene family expression in primary hepatocyte cultures and rat liver. Mol Carcinog 1991, 4:499-509 [DOI] [PubMed] [Google Scholar]
- 28.Holmskov U, Lawson P, Teisner B, Tornøe I, Willis AC, Morgan C, Koch C, Reid KBM: Isolation and characterization of a new member of the scavenger receptor subfamily, glycoprotein-340 (gp-340), as a lung surfactant protein-D binding molecule. J Biol Chem 1997, 272:13743-13749 [DOI] [PubMed] [Google Scholar]
- 29.Holmskov U, Mollenhauer J, Madsen J, Vitved L, Grønlund J, Tornøe I, Kliem A, Reid KBM, Poustka A, Skjødt K: Cloning of gp-340, a putative opsonin receptor for lung surfactant protein D. Proc Natl Acad Sci USA 1999, 96:10794-10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Snyder SH: Molecular cloning of ebnerin, a von Ebner’s gland protein associated with taste buds. J Biol Chem 1995, 270:17674-17679 [DOI] [PubMed] [Google Scholar]
- 31.Cheng H, Bjerknes M, Chen H: CRP-ductin: a gene expressed in intestinal crypts and in pancreatic and hepatic ducts. Anat Rec 1996, 244:327-343 [DOI] [PubMed] [Google Scholar]
- 32.Mollenhauer J, Wiemann S, Scheurlen W, Korn B, Hayashi Y, Wilgenbus KK, von Deimling A, Poustka A: DMBT1, a new member of the SRCR family, on chromosome 10q25.3-26.1 is deleted in malignant brain tumours. Nat Genet 1997, 17:32-39 [DOI] [PubMed] [Google Scholar]
- 33.Bisgaard HC, Nagy P, Ton PT, Hu Z, Thorgeirsson SS: Modulation of cytokeratin 14 and α-fetoprotein expression during hepatic oval cell proliferation and liver regeneration. J Cell Physiol 1994, 159:475-484 [DOI] [PubMed] [Google Scholar]
- 34.Takito J, Yan L, Ma J, Hikita C, Vijayakumar S, Warburton D, Al-Awqati Q: Hensin: the polarity reversal protein, is encoded by DMBT1, a gene frequently deleted in malignant gliomas. Am J Physiol 1999, 277:F277-F289 [DOI] [PubMed] [Google Scholar]
- 35.Roskams T, De Vos R, Van Eyken P, Mayzaki H, Van Damme B, Desmet V: Hepatic OV-6 expression in human liver disease and rat experiments: evidence for hepatic progenitor cells in man. J Hepatol 1998, 29:454-462 [DOI] [PubMed] [Google Scholar]
- 36.Aruffo A, Bowen MA, Patel DD, Haynes BF, Starling GC, Gebe JA, Bajorath J: CD-6 ligand interactions: a paradigm for SRCR domain function. Immunol Today 1997, 18:498-504 [DOI] [PubMed] [Google Scholar]
- 37.Bork P, Beckmann G: The CUB domain. A widespread module in developmentally regulated proteins. J Mol Biol 1993, 231:539-545 [DOI] [PubMed] [Google Scholar]
- 38.Bork P, Sander C: A large domain common to sperm receptors (ZP2 and ZP3) and TGF-beta type III receptor. FEBS Lett 1992, 300:237-240 [DOI] [PubMed] [Google Scholar]
- 39.Madsen J, Kliem A, Tornøe I, Skjødt K, Koch K, Holmskov U: Localization of lung surfactant protein D on mucosal surfaces in human tissues. J Immunol 2000, 164:5866-5870 [DOI] [PubMed] [Google Scholar]
- 40.Wert SE, Yoshida M, LeVine AM, Ikegami M, Jones T, Ross GF, Fisher JH, Korfhagen TR, Whitsett JA: Increased metalloproteinase activity, oxidant production, and emphysema in surfactant protein D gene-inactivated mice. Proc Natl Acad Sci USA 2000, 97:5972-5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borron PJ, Crouch EC, Lewis JF, Wright JR, Possmayer F, Fraher LJ: Recombinant rat surfactant-associated protein D inhibits human T lymphocyte proliferation and IL-2 production. J Immunol 1998, 161:4599-4603 [PubMed] [Google Scholar]
- 42.Korfhagen TR, Sheftelyevich V, Burhans MS, Bruno MD, Ross GF, Wert SE, Stahlman MT, Jobe AH, Ikegami M, Whitsett JA, Fisher JH: Surfactant protein-D regulates surfactant phospholipid homeostasis in vivo. J Biol Chem 1998, 273:28438-28443 [DOI] [PubMed] [Google Scholar]
- 43.Thim L, Mørtz E: Isolation and characterization of putative trefoil peptide receptors. Regul Pept 2000, 90:61-68 [DOI] [PubMed] [Google Scholar]
- 44.Playford RJ, Marchbank T, Goodlad RA, Chinery RA, Poulsom R, Hanby AM, Wright NA: Transgenic mice that over express the human trefoil peptide pS2 have an increased resistance to intestinal damage. Proc Natl Acad Sci USA 1996, 93:2137-2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mashimo H, Wu D, Podolsky DK, Fishman MC: Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science 1996, 274:262-265 [DOI] [PubMed] [Google Scholar]
- 46.Vijayakumar S, Takito J, Hikata C, Al-Awqati Q: Hensin remodels the apical cytoskeleton and induces columnarization of intercalated epithelial cells: processes that resemble terminal differentiation. J Cell Biol 1999, 144:1057-1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mollenhauer J, Herbertz S, Holmskov U, Tolnay M, Krebs I, Merlo A, Schrøder HD, Maier D, Breiling F, Wiemann S, Gröne H, Poustka A: DMBT1 encodes a protein involved in the immune defense and in epithelial differentiation and is highly unstable in cancer. Cancer Res 2000, 6:1704-1710 [PubMed] [Google Scholar]
- 48.Novikoff PM, Yam A, Oikawa I: Blast-like cell compartment in carcinogen-induced proliferating bile ductules. Am J Pathol 1996, 148:1473-1492 [PMC free article] [PubMed] [Google Scholar]