Abstract
The replicative spread of retrotransposons in the genome creates new insertional polymorphisms, increasing retrotransposon numbers and potentially both their share of the genome and genome size. The BARE-1 retrotransposon constitutes a major, dispersed, active component of Hordeum genomes, and BARE-1 number is positively correlated with genome size. We have examined genome size and BARE-1 insertion patterns and number in wild barley, Hordeum spontaneum, in Evolution Canyon, Lower Nahal Oren, Mount Carmel, Israel, along a transect presenting sharply differing microclimates. BARE-1 has been sufficiently active for its insertional pattern to resolve individuals in a way consonant with their ecogeographical distribution in the canyon and to distinguish them from provenances outside the canyon. On both slopes, but especially on the drier south-facing slope, a simultaneous increase in the BARE-1 copy number and a decrease in the relative number lost through recombination, as measured by the abundance of solo long terminal repeats, appear to have driven the BARE-1 share of the genome upward with the height and dryness of the slope. The lower recombinational loss would favor maintenance of more full-length copies, enhancing the ability of the BARE-1 family to contribute to genome size growth. These local data are consistent with regional trends for BARE-1 in H. spontaneum across Israel and therefore may reflect adaptive selection for increasing genome size through retrotransposon activity.
Retrotransposons resemble retroviruses in their structure and life cycle (1, 2). They are ubiquitous (3–5) and contribute a large proportion of the total repetitive DNA of some plant genomes (6). Retrotransposons are mobilized by a replicative mechanism that has the capacity to generate and insert many new daughter copies into the genome, thereby increasing genome size (7). The error-prone nature of their replication by reverse transcriptase (8), the mutagenic potential of their transpositional integration (9), and the effects of their accumulation and recombination (10) together suggest that active retrotransposons may be major contributors to genome diversification in the plants. Genomic changes induced by retrotransposons can be tracked by the joints between the flanking DNA and the conserved retrotransposon termini created upon integration. Marker techniques based on PCR amplification between retrotransposons and flanking DNA recently have been developed (11–13).
Accumulated data indicate that retrotransposons in plants (14–16), animals, and fungi respond to various forms of stress. When stress factors in the environment vary ecogeographically, retrotransposon prevalence and insertion patterns may vary accordingly. The immediate wild ancestor of cultivated barley (Hordeum vulgare), Hordeum spontaneum, is ideal for analyzing retrotransposon insertions and their role in the genome because of the presence of a large and active retrotransposon family and the availability of well-studied wild populations distributed in diverse habitats (17–21). The BARE-1 family of retrotransposons comprises on average 14 × 103 copies in the genomes of Hordeum species (10). Members of this family are transcriptionally (22) and translationally (23) active, encoding both a polyprotein (24, 25) and processing signals (26), which are functionally conserved. The BARE-1 copy number is positively correlated with both genome size and habitat aridity (10), factors that are themselves correlated (27) regionally in H. spontaneum.
We have examined the role of the BARE-1 retrotransposon in genome diversification in individuals at the Evolution Canyon microsite, Lower Nahal Oren, Mount Carmel, Israel (28–30). This 400-m-wide erosion gorge (see Fig. 4, which is published as supplementary data on the PNAS web site, www.pnas.org), dating from the Plio-Pleistocene era, presents north- and south-facing slopes (NFS and SFS, respectively) with common geologies and macroclimates but microclimates sharply differing in solar irradiation and aridity. Biotically, the NFS is Eurasian and the SFS is Afro-Asian within the Mediterranean context (28, 30). We have examined H. spontaneum along a north-south transect across the canyon slopes to test whether regional patterns (10) can be detected locally. The BARE-1 copy number and patterns of insertional polymorphism, as well as total genome size, were determined for accessions from the canyon.
Materials and Methods
Plant Materials.
Spikes from individual H. spontaneum plants were collected at six stations located along a 300-m north-south transect across the NFS and SFS of Evolution Canyon (28). The stations previously described (29, 31) as NFS (stations 5–7) and SFS (stations 1–3) are referred to here as: NH (north high), NM (north middle), NL (north low), SL (south low), SM (south middle), and SH (south high). From each station, seeds of 10 individual plants, separated by at least 1 m from each other, were used as the samples. The seeds were grown to seedlings for preparation of DNA and nuclei.
Retrotransposen-Microsatellite Amplified Polymorphism (REMAP) Amplification.
DNAs were prepared by the cetyltrimethylammonium bromide method (32). For REMAP PCR amplification, primers facing outward from the long terminal repeats (LTRs) were combined with anchored simple sequence repeat (SSR) primers. The PCR conditions and the SSR primers and annealing temperatures are as specified (see Table 1, which is published as supplementary data). The cycling program was: 94°C, 2 min; 30 cycles of 94°C for 30 sec, 54–58°C, depending on the primer pair, for 30 sec, a ramp of +0.5°C (sec)−1 to 72°C, and 72°C for 2 min, 3 sec at 72°C being added with each cycle; 72°C for 10 min; maintenance at 4°C. Primers to the BARE-1 LTR were LTR-Z, 5′-ctc gct cgc cca CTA CAT CAA CCG CGT TTA TT-3′, a forward primer matching bases 1993–2012 of BARE-1a (GenBank accession no. Z17327), the lowercase bases indicating a cloning tail, and LTR-A, 5′-gga att cat aGC ATG GAT AAT AAA CGA TTA TC-3′, a reverse primer matching 369–393 of BARE-1a. The products were resolved on 2% NuSieve 3:1 agarose (FMC) and detected by ethidium bromide staining.
REMAP Data Analysis.
Gel banding patterns were scored, and tables of band presence and absence were created. Principal component analyses were run with genstat 5, release 4.1 (NAG, Oxford, U.K.), and the plots were generated with the group average agglomerative clustering function as used previously (33). Statistical tests were carried out with sigmaplot 5.0 (SPSS, Chicago). Nonparametric correlation analyses were made by Spearman Rank Order (generating the correlation coefficient rS) as implemented in sigmastat 2.01 (SPSS). Values for P in the text represent the likelihood of falsely rejecting the null hypothesis that the variables are not correlated. Data values in the text are expressed as means and SE.
BARE-1 Copy Number Estimation.
Copy number was determined by reconstruction from dot blot hybridizations. To control for differences in loading, 139.4 ng of lambda DNA was added to each μg of plant DNA, giving 1.2 × 104 copies of lambda per barley genome equivalent. Dot blots were prepared with multiple replicates by using 1 ng or 10 ng of genomic DNA per sample and cross-linked under UV light. Isolated plasmids (0.1–10 ng) containing the fragments for hybridization probes served as controls on each filter. LTR (NheI–BsteII, 743 bp) and integrase (in, HpaI–BsmI, 589 bp) probes were subcloned from BARE-1a. Probes were random-primed (Rediprime or Megaprime, Amersham Pharmacia) and 32P-labeled. Filters were hybridized in 50% formamide, 1.25 × standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), 5 × Denhardt's reagent, 0.5% SDS, and 20 μg(ml)−1 herring sperm DNA overnight at 42°C.
Hybridized filters were washed successively with 2 × SSC, 0.1% SDS (10 min, 25°C), twice in 2 × SSC, 0.1% SDS (10 min, 65°C), and once in 0.2 × SSC (20 min, 65°C). Bound radiation was quantified by exposure of a PhosphorImager screen for 45 min followed by scanning on a Fuji PhosphorImager. The same filter was probed in series with in, LTR, and lambda probes. Hybridization response to the in and LTR probes was corrected to the average value for the lambda hybridization response and relative copy number calculated. Absolute copy number was calculated from the hybridization response of the genomic DNA compared with the control plasmids: copies(ng)−1 = (genomic cpm)(ng)−1 × (plasmid copies)(plasmid cpm)−1. Copies(ng)−1 were converted to copies(genome)−1 by using the Hordeum genome size data determined here.
Preparation of Nuclei and Flow Cytometry.
All leaf samples (≈50 mg each) were collected from the three-leaf stage. Protoplasts were isolated as before (34), then resuspended in 1 ml of nuclear buffer {30 mM sodium citrate, pH 7.0/45 mM MgCl2/20 mM Mops [3(N-morpholino)propanesulfonic acid]/1% (wt/vol) Triton X-100/5 μl(ml)−1 β-mercaptoethanol} (35, 36), to which 20 μm(ml)−1 propidium iodide then was added for staining. The samples were stained for 15 min and then centrifuged for 10 sec at 14,000 rpm. The supernatant was discarded and the pellet was suspended in 200 μl of nuclear buffer supplemented with 2.4 μl(ml)−1 RNase and 1 μl(ml)−1 of an internal standard solution (37) containing chicken red blood cells (2C = 2.33 pg; ref. 36). The tubes were incubated at 37°C for 15 min and then chilled on ice before analysis. An average of three separate determinations per individual were made on a 1,023-channel flow cytometer (FACSort, Becton Dickinson) having an argon-ion laser of 488-nm excitation wavelength. The output data were processed with the cellquest program supplied with the cytometer. The estimates for the nuclear DNA amount for the samples were calculated by using the median position of the plant nuclear peak.
Results
Genome size was measured by flow cytometry (see Fig. 5, which is published as supplementary material) for accessions collected at each of six stations along a north-south transect across Evolution Canyon (Fig. 4). Taking all stations together, a diploid genome size of 9.037 ± 0.027 (SE) pg was observed and is within 1.1% of the average of previous observations for a set of Israeli H. spontaneum accessions (38). The transect through the canyon presents two position variables, relative height (lower, middle, or upper) and orientation (NFS or SFS). With the limited precision of flow cytometry, the observed genome sizes were not distinct by sampling site within the canyon (Kruskal–Wallis ANOVA on Ranks). However, linear regression analyses indicate that genome size is weakly associated with slope orientation (R = 0.167, t = 1.684, P = 0.095), the SFS having larger genomes than the NFS.
Two regions of BARE-1, the enzyme-encoding in of the internal domain and the terminal LTRs, served as probes for copy number determinations. The in and LTR regions both are conserved (25, 26) and were used in earlier BARE-1 copy number determinations for Hordeum (10). The in probe is used to estimate the number of full-length BARE-1 elements. Copy number and genome size were estimated on the same accessions, and together gave an average of 1.40 ± 0.04 × 104 (range, 0.83 to 2.21 × 104) BARE-1 copies, equivalent to 2.98 ± 0.08% (range, 1.77% to 4.70%), of the haploid genome. This was in the range seen earlier for more broadly distributed H. spontaneum (10).
Because BARE-1 and other LTR-retrotransposons contain an LTR at each end, two are expected for each internal domain. However, the LTR copy number greatly exceeds that of the internal domain in barley, H. spontaneum, and throughout the Hordeum genus (10), because of the presence of large numbers of solo LTRs, hypothesized to result primarily from intraelement recombination between the LTRs and consequent loss of the internal domain. Here, we detected an average of 7.5 ± 0.2 × 104 LTRs per genome, 5.4 ± 0.1-fold more LTRs than internal domains. This finding indicates that the average genome measured contains 4.7 × 104 LTRs not attributable to full-length BARE-1 elements. These solo LTRs contribute an additional 8.4 × 107 bp or 2.03 ± 0.07% to the genome.
Each solo LTR represents a minimum of one integration event followed by recombinational loss of the internal domain and an LTR. Assuming no other changes in the repetitive DNA complement, BARE-1 therefore would comprise at least 11.7 ± 0.3% of the genome if none were lost through recombination. Taking all accessions together, the number of full-length BARE-1 elements (measured by in response) is positively and highly significantly correlated (Fig. 1A, rP = 0.432, P = 0.001) with the number of both total LTRs and solo LTRs (Pearson Product Moment, rP = 0.714, P = 1.35 × 10−9). The slope for the regression plotting the growth in LTR numbers (Fig. 1A) is 3.7 ± 0.5 (P < 0.001), whereas as a slope of 2 would be expected if the increase in LTR number were to come only from those remaining in full-length elements. These calculations imply that those genomes that have more BARE-1 elements at present generally have also lost more through recombination.
Figure 1.
BARE-1 copy number in H. spontaneum accessions at transect stations in Evolution Canyon. (A) LTR number as a function of in number for all accessions. (B) The in number as a function of transect station. (C) LTR number as a function of transect station. (D) Ratio of LTR number to in number as a function of station. For all plots, ▵ are samples from the NH station; □, NM; ▿, NL; ▾, SL; ▪, SM; ▴, SH. For plots B–D, means and SE are shown.
Given that the number of LTRs, in particular the solo LTRs, rises more rapidly than the number of BARE-1 elements, one would expect a positive correlation between the BARE-1 genome share and the ratio of LTRs to full-length elements. However, in the canyon, the LTR/in ratio is negatively and highly significantly correlated (rP = −0.351, P = 0.009) with the contribution of BARE-1 to genome size, as indeed was earlier seen for the genus Hordeum as a whole (10). Therefore, the higher the abundance of LTRs relative to full-length BARE-1 elements, the smaller the fraction of the genome occupied by BARE-1. Hence, variation in recombinational loss of BARE-1 may be linked to the genome share of the family both throughout the genus and for one species at a single geographical microsite.
In view of these correlations and the copy number and genome share variations among the H. spontaneum accessions, we examined whether measures of BARE-1 prevalence might vary with the height and orientation of the transect positions (Fig. 1B). The BARE-1 number is positively and significantly correlated (rP = 0.386, P = 0.004) with the height of the accession site. Furthermore, the accessions from the top of the canyon (NH, SH), when considered together, have a distinctly and significantly greater (Student's t test, t = 2.657, P = 0.01) number of BARE-1 copies than found in the bottom- and mid-slope accessions. The SH station, the most stressed site in the canyon, alone is distinct from all others (t = 3.107, P = 0.003). The number of LTRs in the genome is positively but not significantly associated with height in the canyon, particularly on the SFS (Fig. 1C). If only the SFS is considered, the correlation between LTR number and height on the slope becomes very strong and nearly significant (rP = 0.991, P = 0.084).
Variation in recombinational loss of BARE-1 with respect to insertional activity, reflected in the LTR/in ratio, is significantly and negatively correlated (rP = −0.351, P = 0.009) with height of the accession on the canyon slopes (Fig. 1D). Furthermore, the accessions from NH and SH, taken together, have a distinctly smaller proportion of solo LTRs than all other accessions (t = −2.985, P = 0.004). These data suggest that the higher in the canyon, the more full-length BARE-1 elements are maintained relative to the number lost through recombination. Consistent with this, BARE-1 comprises a significantly increasing proportion of the genome (rP = 0.402, P = 0.0025) with increasing height in the canyon. The SH individuals display a distinctly greater proportion of the genome than all of the rest of accessions (t = 3.082, P = 0.003), the distinction by height also being maintained when SH and NH are considered together (t = 2.769, P = 0.008) and when the top- and mid-slope accessions are compared with the two lowest stations (t = 2.594, P = 0.012). By multiple regression analyses, height in the canyon on both slopes is a good predictor of the BARE-1 share of the genome whether (P = 0.01) or not (P = 0.008) the SH station is included in the data set.
Using the REMAP method (12), integration joints between BARE-1 copies and flanking genomic sequences were detected by PCR amplification in reactions containing LTR primers in combination with primers to SSRs anchored at their 3′ end. Because BARE-1 elements have a tendency to insert into regions containing SSRs (12, 26, 39), many of the BARE-1 insertions thereby can be detected. Marker bands generated by this system are, in principal, insensitive to intraelement LTR recombination; the remaining, recombinant LTR would still serve as a priming site for PCR oriented in either direction.
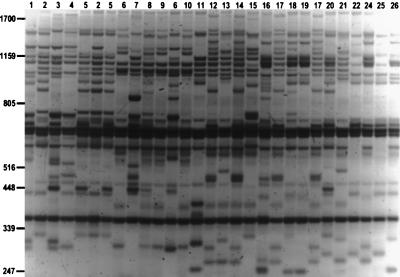
Ten individuals from each of the six transect stations were typed with REMAP. Earlier results (12) showed that the REMAP products generally do not derive from amplification between pairs of SSR domains, but rather from element insertions. The inter-SSR products were generally longer than the REMAP products, indicating that the SSRs pairs are more widely interspersed than LTR/SSR pairs. In control experiments containing the SSR primer but not the LTR primer, none of the bands produced by amplification between SSR loci in the genome had mobilities identical to the REMAP bands (see Fig. 6, which is published as supplementary material). In the gel of Fig. 2, the generally high degree of polymorphism detected between individuals from the canyon is evident. Seven sets of REMAP primer combinations were used to generate 316 bands from the accessions (Table 1). Of these, 277 or 88% were polymorphic.
Figure 2.
Banding patterns generated by REMAP amplification. The reaction was carried out with primers LTR-A and (CAC)7T. Lanes are labeled by the genotype of the sample (Table 2); two different accessions are shown for genotypes 2, 5, and 17. The products have been stained with ethidium bromide after agarose gel electrophoresis; the gel is shown as a negative image. Size markers in bp derive from a bacteriophage λ PstI digest.
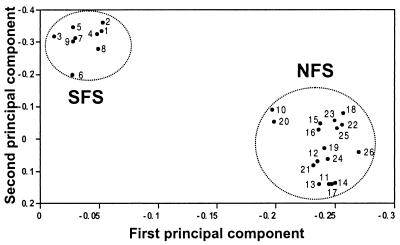
A total of 26 distinct banding genotypes were detected among the accessions (see Table 2, which is published as supplementary material). The particular primer combination (LTR-A, (CAC)7T) used in Fig. 2 does not distinguish genotypes 22 and 23, although other combinations do. The genotypes distinguished all cases but one (genotype 6) within collecting stations and represented three or fewer of the individuals per genotype, excepting NFS genotype 17 with four individuals and SFS genotype 6 with 16 individuals. Genotype 6 also contained both SL and SM individuals. The slopes were clearly distinct in the number of genotypes represented, the SFS having only nine genotypes whereas the NFS had 17. A mean of 114 ± 1 bands were detected in total for each of the stations except SM, for which only 65 bands were scored (see Table 3, which is published as supplementary material). This yielded average frequencies from 0.1 to 0.7 for a given genotype and an intrastation similarity index of 0.47 ± 0.05, a value of 1 indicating all bands are shared. The banding patterns generated for each individual were used to estimate genetic distances between them. The REMAP banding data were examined by principal component analysis (Fig. 3), which allows comparison of overall genotypic similarities in the absence of phylogenetic considerations. The analyses completely separated the individuals from the NFS from those of the SFS.
Figure 3.
Principle component analysis of Evolution Canyon H. spontaneum derived from variation in REMAP banding patterns. Numerals refer to the corresponding genotypes; those from the same slope have been circled.
Discussion
The number of full-length BARE-1 copies in individuals of H. spontaneum in Evolution Canyon, at the Lower Nahal Oren microsite, was found to range from 8.3 to 22.1 × 103 per haploid genome equivalent. This almost 3-fold variation in the number of full-length BARE-1 elements among individuals at a single microsite, within the range seen earlier for the genus Hordeum as a whole (10), indicates that this retrotransposon family has been highly and recently insertionally active.
An excess of LTRs, likely solo, were detected in all accessions. These are abundant across the genus Hordeum (10) and appear to result from intrachromosomal recombination between the LTR pairs within full-length elements. The excess in LTRs increases with the number of BARE-1 elements, consistent with earlier evidence that recombination is additive between elements in the genome (40). The results here furthermore confirm what was earlier shown (10) for the genus as a whole: the greater the number of solo LTRs relative to full-length BARE-1 elements, the smaller the part of the genome comprised by BARE-1. This indicates that variations in the relative rates of recombination and integration affect the success of a retrotransposon family in spreading within the genome, and that these variations may act within a single species at a single locale.
The data, moreover, suggest a linkage between BARE-1 numbers and the ecogeography of the Evolution Canyon microsite. More BARE-1 copies and proportionally fewer solo LTRs are found in the upper, drier sites within the canyon, particularly at the top of the SFS, than at lower sites. Earlier studies indicated a decrease in angiosperm species diversity (29, 31) and an increase in allozymic (30) and randomly amplified polymorphic DNA (28) diversity upward in the canyon, all correlated with increasing stress upward on both slopes, with the most stressful slope being the SFS. The upper stations on each slope are, furthermore, generally drier even during the wet season because of the movement of runoff down-slope. The local data at this single microsite mirror regional observations across Israel (10) that BARE-1 copy number was correlated with aridity across the range of the H. spontaneum, both sets of data being consistent with the presence within the BARE-1 promoter of abscisic acid-response elements typical for water stress-induced genes (24). The data therefore suggest that expression and propagation of BARE-1 may be stress induced and also that, the higher in the canyon, the lower the rate of loss of integrated copies through recombination.
Polymorphism detectable with REMAP markers yields a complete distinction between individuals growing on the NFS and SFS of Evolution Canyon, which are separated from each other by a maximum of 300 m. Because the REMAP pattern derives from short-range amplifications (hundreds of bases), the differences observed by REMAP are likely to have been generated by retrotransposon BARE-1 insertion, independent of other genetic changes among the individuals. Given the small percentage of the total BARE-1 copies visualized by the seven primer combinations, the data imply that BARE-1 integrational activity in the canyon has been greater than genomic homogenization driven by gene flow through pollen dispersal among the largely selfing (average 98.4%, ref. 41) H. spontaneum or by seed dispersal. The REMAP marker data show more genotypes in the NFS than in the SFS individuals, which we interpret as being caused by the patchiness of the NFS, having open areas suitable for H. spontaneum interrupted by shaded, tree-growing areas, in contrast to the SFS. Under sufficiently high rates of BARE-1 integrational activity, the patches appear to have become genotypically distinct with regard to the BARE-1 insertion pattern.
Classically “selfish” self-replicating units such as retrotransposons might be expected, independent of the genome as a whole, to undergo selection for increasingly efficient propagation. However, the observed combination of decreased recombinational loss together with increases in the number of full-length copies suggests that plant-level selection is operating to increase BARE-1 copy number. Increasing numbers of transposon copies have been thought to be associated with decreased fitness through increasing lethality (42, 43). However, the tendency of retrotransposons to insert into repetitive DNA in barley and other cereals (6, 12, 26, 39) mitigates their deleterious potential.
The insertion and maintenance of full-length BARE-1 copies would marginally increase genome size, albeit against the background of fluctuations in the content of other retrotransposons and repetitive DNA. Selection for large genomes has been hypothesized to occur in the Mediterranean basin (44), a region where growth takes place primarily in the cool, wet winters and not in the dry summers. Growth is more efficient under cool conditions by increase in cell volume rather than by increase in cell number because cell division rates are decreased by low temperatures (44, 45). The potential for large cell volumes has been directly correlated with genome size and associated nuclear volume in a wide range of organisms (46, 47). Retrotransposon integrational activity, by increasing genome size, may be adaptive.
Supplementary Material
Acknowledgments
We thank Anne-Mari Narvanto for her always excellent technical assistance. This research was supported by the Academy of Finland Genome Research Program and the European Union Directorate for Biotechnology research program on Molecular Tools for Biodiversity. E.N. thanks the Israel Discount Bank Chair of Evolutionary Biology, the Ancell-Teicher Research Foundation for Genetics and Molecular Evolution, and the Israel Science Foundation (Grant 157/98) for financial support.
Abbreviations
- in
integrase
- LTR
long terminal repeat
- NFS
north-facing slope
- NH
north high
- NM
north middle
- NL
north low
- REMAP
retrotransposen-microsatellite amplified polymorphism
- SFS
south-facing slope
- SH
south high
- SL
south low
- SM
south middle
- SSR
simple sequence repeat
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110587497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110587497
References
- 1.Boeke J D, Corces V G. Annu Rev Microbiol. 1989;43:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- 2.Doolittle R F, Feng D-F, Johnson M S, McClure M A. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 3.Voytas D F, Cummings M P, Konieczny A K, Ausubel F M, Rodermel S R. Proc Natl Acad Sci USA. 1992;89:7124–7128. doi: 10.1073/pnas.89.15.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flavell A J, Dunbar E, Anderson R, Pearce S R, Hartley R, Kumar A. Nucleic Acids Res. 1992;20:3639–3644. doi: 10.1093/nar/20.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suoniemi A, Tanskanen J, Schulman A H. Plant J. 1998;13:699–705. doi: 10.1046/j.1365-313x.1998.00071.x. [DOI] [PubMed] [Google Scholar]
- 6.SanMiguel P, Tikhonov A, Jin Y K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 7.SanMiguel P, Gaut B S, Tikhoniv A, Nakajima Y, Bennetzen J L. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 8.Gabriel A, Willems M, Mules E H, Boeke J D. Proc Natl Acad Sci USA. 1996;93:7767–7771. doi: 10.1073/pnas.93.15.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wessler S R, Bureau T E, White S E. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 10.Vicient C M, Suoniemi A, Anamthawat-Jónsson K, Tanskanen J, Beharav A, Nevo E, Schulman A H. Plant Cell. 1999;11:1769–1784. doi: 10.1105/tpc.11.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flavell A J, Knox M R, Pearce S R, Ellis T H N. Plant J. 1998;16:643–650. doi: 10.1046/j.1365-313x.1998.00334.x. [DOI] [PubMed] [Google Scholar]
- 12.Kalendar R, Grob T, Regina M, Suoniemi A, Schulman A H. Theor Appl Genet. 1999;98:704–711. [Google Scholar]
- 13.Waugh R, McLean K, Flavell A J, Pearce S R, Kumar A, Thomas B B T, Powell W. Mol Gen Genet. 1997;253:687–694. doi: 10.1007/s004380050372. [DOI] [PubMed] [Google Scholar]
- 14.Takeda S, Sugimoto K, Otsuki H, Hirochika H. Plant Mol Biol. 1998;36:365–376. doi: 10.1023/a:1005911413528. [DOI] [PubMed] [Google Scholar]
- 15.Mhiri C, Morel J-B, Vernhettes S, Casacuberta J M, Lucas H, Grandbastien M A. Plant Mol Biol. 1997;33:257–266. doi: 10.1023/a:1005727132202. [DOI] [PubMed] [Google Scholar]
- 16.Wessler S R. Curr Biol. 1996;6:959–961. doi: 10.1016/s0960-9822(02)00638-3. [DOI] [PubMed] [Google Scholar]
- 17.Nevo E, Beiles A, Zohary D. Biol J Linn Soc. 1986;27:335–380. [Google Scholar]
- 18.Nevo E, Zohary D, Brown A H D, Haber M. Evolution. 1979;33:815–833. doi: 10.1111/j.1558-5646.1979.tb04737.x. [DOI] [PubMed] [Google Scholar]
- 19.Nevo E, Baum B, Beiles A, Johnson D A. Genet Resour Crop Evol. 1998;45:151–159. [Google Scholar]
- 20.Pakniyat H, Powell W, Baird E, Handley L L, Robinson D, Scrimgeour C M, Nevo E, Hackett C A, Caligari P D S, Forster B P. Genome. 1997;40:332–341. doi: 10.1139/g97-046. [DOI] [PubMed] [Google Scholar]
- 21.Forster B P, Russell J R, Ellis R P, Handley L L, Robinson D, Hackett C A, Nevo E, Waugh R, Gordon D C, Keith R, Powell W. New Phytol. 1997;137:141–147. [Google Scholar]
- 22.Suoniemi A, Narvanto A, Schulman A. Plant Mol Biol. 1996;31:295–306. doi: 10.1007/BF00021791. [DOI] [PubMed] [Google Scholar]
- 23.Jääskeläinen M, Mykkänen A-H, Arna T, Vicient C, Suoniemi A, Kalendar R, Savilahti H, Schulman A H. Plant J. 1999;20:413–422. doi: 10.1046/j.1365-313x.1999.00616.x. [DOI] [PubMed] [Google Scholar]
- 24.Manninen I, Schulman A H. Plant Mol Biol. 1993;22:829–846. doi: 10.1007/BF00027369. [DOI] [PubMed] [Google Scholar]
- 25.Suoniemi A, Tanskanen J, Pentikäinen O, Johnson M S, Schulman A H. Mol Biol Evol. 1998;15:1135–1144. doi: 10.1093/oxfordjournals.molbev.a026021. [DOI] [PubMed] [Google Scholar]
- 26.Suoniemi A, Schmidt D, Schulman A H. Genetica. 1997;100:219–230. [PubMed] [Google Scholar]
- 27.Turpeinen T, Kulmula J, Nevo E. Genome. 1999;42:1094–1099. doi: 10.1139/g99-066. [DOI] [PubMed] [Google Scholar]
- 28.Nevo E. Proc R Soc London Ser B. 1995;262:149–155. [Google Scholar]
- 29.Nevo E. Theor Popul Biol. 1997;52:231–243. doi: 10.1006/tpbi.1997.1330. [DOI] [PubMed] [Google Scholar]
- 30.Nevo E, Apelbaum-Elkaher I, Garty J, Beiles A. Heredity. 1997;78:373–382. [Google Scholar]
- 31.Nevo E, Fragman O, Dafni A, Beiles A. Isr J Plant Sci. 1999;47:49–59. [Google Scholar]
- 32.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, Albright L M, Coen D M, Varki A. In: Current Protocols in Molecular Biology. Janssen K, editor. 1, Suppl. 7. New York: Wiley; 1995. pp. 4.7.1–4.7.8. [Google Scholar]
- 33.Russell J, Fuller J, Young G, Thomas B, Taramino G, Macaulay M, Waugh R, Powell W. Genome. 1997;40:442–450. doi: 10.1139/g97-059. [DOI] [PubMed] [Google Scholar]
- 34.Valkonen J P T, Nygren M, Ylönen A, Mannonen L. Genetica. 1994;92:203–207. [Google Scholar]
- 35.Villemont E, Dubois F, Sangwan R S, Vasseur G, Bourgois Y, Sangwan-Norreel B S. Planta. 1997;201:160–172. [Google Scholar]
- 36.Galbraith D W, Harkins K R, Maddox J M, Ayres N M, Sharma D P, Firoozabady E. Science. 1983;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- 37.Arumuganathan K, Earle E D. Plant Mol Biol Rep. 1991;9:229–241. [Google Scholar]
- 38.Kankaanpää J, Mannonen L, Schulman A H. Genome. 1996;39:730–735. doi: 10.1139/g96-092. [DOI] [PubMed] [Google Scholar]
- 39.Ramsay L, Macaulay M, Cardle L, Morgante M, degli Ivanissevich S, Maestri E, Powell W, Waugh R. Plant J. 1999;17:415–426. doi: 10.1046/j.1365-313x.1999.00392.x. [DOI] [PubMed] [Google Scholar]
- 40.Puchta H, Hohn B. Trends Plant Sci. 1996;1:340–348. doi: 10.1016/S1360-1385(03)00004-9. [DOI] [PubMed] [Google Scholar]
- 41.Brown A H D, Zohary D, Nevo E. Heredity. 1978;41:49–62. [Google Scholar]
- 42.Charlesworth B, Sniegowski P, Stephan W. Nature (London) 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 43.Wilke C M, Maimer E, Adams J. Genetica. 1992;86:155–173. doi: 10.1007/BF00133718. [DOI] [PubMed] [Google Scholar]
- 44.Grime J P, Mowforth M A. Nature (London) 1982;299:151–153. [Google Scholar]
- 45.Francis D, Barlow P W. Symp Soc Exp Biol. 1988;42:181–201. [PubMed] [Google Scholar]
- 46.Bennett M D. New Phytol. 1987;106,Suppl.:177–200. [Google Scholar]
- 47.Cavelier-Smith T. Annu Rev Biophys Bioeng. 1982;11:273–302. doi: 10.1146/annurev.bb.11.060182.001421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.