Abstract
Thymoma is the most frequent tumor arising in human thymus. In this study, we performed a detailed mapping of deleted regions on chromosome 6 shown previously to harbor the most frequent genetic aberrations in this cancer. We analyzed 40 thymomas using 41 microsatellites. Two hundred ninety-four (23.5%) of 1253 informative genotypes showed loss of heterozygosity (LOH), only 39 (2.4%) were positive for microsatellite instability (MSI). Genetic aberrations on chromosome 6 were found in 31 of 40 cases (77.5%) in five hot spots. The most frequent LOHs (48.6%) occurred in region 6q25.2 within a 0.7-Mb interval flanked by markers D6S441 and D6S290. Another hot spot showing LOH in 32.4% of tumors was located between markers D6S442 and D6S1708 (0.4 Mb apart) on 6q25.2-25.3, just 1.1 Mb from the D6S441-D6S290 deletions. The third hot spot (30%) showing LOH appeared in region 6p21.31 including the MHC locus (markers D6S1666-D6S1560, 1 Mb apart). The fourth hot spot (26.3%) was detected on 6q14.1-14.3 (D6S1596-D6S284, 5.2 Mb apart). Some tumors (21.6%) showed LOHs within a fifth hot spot on 6q21 (D6S447-D6S1592, 0.3 Mb apart). Thus, several tumor suppressor genes on chromosome 6 seem to be involved in the pathogenesis of thymoma.
Human thymoma is a mediastinal neoplasm derived from thymic epithelial cells. It is well known for its association with autoimmune diseases, especially myasthenia gravis. 1,2 However, genetic abnormalities common in this neoplasm are less well characterized. No distinct primary genetic abnormality has been established to date. Cytogenetic studies describing karyotypes of thymomas are mostly case studies showing no recurrent aberrations. 3-5 The only exception is translocation t(15;19), found in altogether three undifferentiated thymic carcinomas (type C thymoma) so far. 6-8
Using comparative genomic hybridization (CGH) and microsatellite analysis, we have characterized some of the common genetic abnormalities found in World Health Organization (WHO) types A, B3, and C thymoma in previous studies. 9,10 The most frequent genetic abnormality detected was loss of genetic material or LOH on the long arm of chromosome 6. Other consistent LOHs were detected in regions 3p22-24.2, 3p14.2 (FHIT gene locus), 5q21 (APC), 6p21, 6q21-22.1, 7p21-22, 8q11.21-23, 13q14 (RB), and 17p13.1 (p53). Comparing the allelotypes of the analyzed thymomas, we were able to identify two pathogenetic pathways these tumors develop along, characterized by the 6q23.3-25.3 and 5q21 LOHs, respectively. The APC aberration on 5q21 showed significant associations with LOH in the 3p22-24.2, 13q14, and 17p13.1 regions. Interestingly, type A thymomas presented with consistent LOH in the region 6q23.3-25.5 only, they did not reveal any aberrations in the APC, RB, and p53 gene loci, or regions 3p22-24.2 and 8q11.21-23.
Deletions and rearrangements involving chromosome 6q have been reported in a number of human malignancies, including breast carcinoma, 11 malignant melanoma, 12 renal cell carcinoma, 13 salivary gland adenocarcinoma, 14 ovarian carcinoma, 15 acute lymphoblastic leukemia, and nodal non-Hodgkin’s lymphomas. 16 In the latter, three regions (6q21, 6q23, and 6q25-27) were determined to show frequent deletions by cytogenetic analysis. 17 We described two hot spots of deletions on chromosome 6 in extranodal gastric high-grade large B-cell lymphoma in a previous study. 18
Because chromosome 6 abnormalities are so frequent in thymomas and to gain more insight into the contribution of chromosome 6 aberrations to thymomagenesis, we performed a detailed search for loss of heterozygosity (LOH) on chromosome 6 in thymoma to identify chromosomal regions containing putative and known tumor suppressor genes playing a role in its development. A reason why so few genetic alterations in thymoma have been characterized so far might also be the high number of nonneoplastic lymphocytes infiltrating the tumors. Analyzing the lymphocyte-rich thymoma types, we therefore used laser-assisted microdissection and primary cell culture to isolate the neoplastic cells. This approach helped to minimize contamination of the analyzed material by DNA from normal lymphocytes ubiquitously present especially in mixed and cortical types of thymoma. We performed an extensive mapping of chromosome 6 aberrations using a screening panel of 41 repeats to analyze 40 thymomas of various types. We were able to identify five chromosomal regions on chromosome 6 showing frequent aberrations in these tumors.
Materials and Methods
Patients
Forty thymomas were chosen for this study from the files of the Institute of Pathology at the Würzburg University. The tumors were classified according to the WHO Classification of Thymic Epithelial Tumors described recently. 19,20 Analyzed were 7 cases of type A, 9 of type AB, 5 of type B2, 15 of type B3, and 4 of type C tumors. According to Masaoka’s 21 clinical staging system, 9 cases were classified as stage I, 14 cases as stage II, 10 cases as stage III, and 7 cases as stage IV. The patients’ ages ranged from 29 to 84 years with a mean of 58.4 years. There were 20 females and 20 males included in the study.
Microdissection
Laser-assisted microdissection was performed to collect epithelial tumor cells in type AB thymoma, which contains both neoplastic epithelium-rich and normal lymphocyte-rich areas. Frozen sections (7-μm thick) were stained using an α-CD3 antibody (Immunotech, Marseille, France) and DAKO ChemMate DAB kit K5001 (DAKO, Glostrup, Denmark) followed by hematoxylin staining to discriminate between epithelium-rich and lymphocyte-rich areas. Tumor cells in the lymphocyte-poor areas were isolated by laser-assisted microdissection using an IX 70 microscope (Olympus, Tokyo, Japan) with Robot-MicroBeam program (PALM, Bernried, Germany). Dissected tumor cells were then catapulted into microfuge caps coated with DNA digestion buffer. Manual microdissection under a microscope was performed on type B3 thymomas having relatively large epithelium-rich areas. Twenty serial 10-μm-thick tissue sections were cut. The first and last cuts were stained with hematoxylin and eosin to assure high tumor content and as guidance for the following dissection. Fresh frozen tissue sections were visualized under the microscope and tumor-rich areas were scraped. Paraffin-embedded tissue sections were additionally stained with Nuclear-Fast Red to precisely delineate tumor areas.
Primary Culture of Neoplastic Thymic Epithelial Cells
Neoplastic thymic epithelial cells derived from primary cultures were used as a source of tumor DNA in type B2 thymomas. This type of thymoma by definition contains a heavy load of nonneoplastic normal lymphocytes. Fresh surgical specimens of thymoma were minced and cultured for 5 days in the presence of 1.35 mmol/L of 2-deoxyguanosine (2-dGuo; Sigma, Steinheim, Germany) in Minimum Essential Medium-D-Val (Life Technologies Ltd., Paisley, UK) supplemented with 10% fetal calf serum. 2-dGuo-treated tissue chunks were incubated in 0.25% trypsin-0.02% ethylenediaminetetraacetic acid for 30 minutes at 37°C and vortexed vigorously. Dispersed cells were cultured for 2 days and only adherent tumor cells were then collected. The purity of the isolated tumor cells was >95% as confirmed by immunohistochemistry using a mixture (CAM 5.2) of anti-cytokeratin-8 and -18 antibodies (Becton-Dickinson, San Jose, CA). Only tumor cells harvested from the first passage of primary culture within 1 week after surgical resection were used for microsatellite analysis. To check whether cultured cells were an appropriate source of tumor DNA, we established primary cultures from lymphocyte-poor type A (n = 1) and B3 (n = 3) thymomas and compared the genotypes of cultured cells to genotypes of cells derived directly from a tumor block. This control study showed identical allelic imbalances in cultured tumor cells and the tumor block (data not shown). In addition, the genotypes of nonneoplastic thymic epithelial cells from a primary culture of a hyperplastic thymus were compared to genotypes of peripheral blood lymphocytes of the same patient. In vitro acquisition of LOH was not found in these cultured epithelial cells (data not shown).
DNA Extraction and Microsatellite Analysis
DNA extraction was performed according to conventional protocols. 22 Tumor tissue (types A, AB, B3, and C thymoma) or pellet of tumor cells (type B2) were digested using proteinase K. DNA was extracted using phenol-chloroform, followed by ethanol precipitation. Control DNA was obtained from peripheral blood lymphocytes or adjacent normal thymus tissue not involved by the tumor. For microsatellite analysis, 41 repeats spanning chromosome 6 were used (Table 1) ▶ . First, chromosome 6 was screened for genetic aberrations using the Chromosome 6 Linkage Mapping Set (LMS-MD10) from Applied Biosystems (Weiterstadt, Germany). Median distance between repeats contained in this panel is ∼10 cM. These markers were then supplemented by additional repeats to narrow the hot spots of aberrations to meaningful sizes. Primer sequences for amplification of the latter microsatellites were retrieved from the Genome Data Base (GDB; http://gdbwww.gdb.org/). Chromosomal location of each marker was derived from the gene map on the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov) and the Ensembl database at the Sanger Institute (http://www.ensembl.org/Homo_sapiens/). Polymerase chain reaction primers were synthesized at MWG-Biotech (Munich, Germany) or Applied Biosystems (Weiterstadt, Germany) and one oligonucleotide of each primer pair was labeled with fluorescent dye phosphoramidites FAM, HEX, NED, or TAMRA. Paired normal and tumor DNA samples from each thymoma patient were amplified with AmpliTaq Gold enzyme (Applied Biosystems, Foster City, CA) under conditions specified by GDB. Thirty polymerase chain reaction cycles were performed in a MWG Primus Gold thermal cycler (MWG Biotech, Munich, Germany) in a total volume of 20 μl. Aliquots of polymerase chain reaction products were mixed with Genescan 350-ROX size standard (Applied Biosystems, Warrington, UK) and formamide, denatured, and subjected to electrophoresis on a 4.5% polyacrylamide gel using a 377 DNA Sequencer (Applied Biosystems). Automatically collected data were analyzed using Genescan and Genotyper software as described in the manufacturer’s manual. Only patients heterozygous for a given locus were informative; homozygosity and microsatellite instability (MSI) rendered the particular locus unevaluable for LOH. In heterozygous cases, ratios of both alleles in normal and tumor samples were calculated. If the ratios showed a difference of >20%, the locus was further evaluated for possible allelic imbalance. LOH was defined as at least 40% signal reduction of one allele in the tumor sample when compared to the control. All genetic aberrations were confirmed minimally twice.
Table 1.
Microsatellite Markers on Chromosome 6 Used in the Analysis of Thymoma
| Markers | Location |
|---|---|
| D6S1574 | 6p25.1 |
| D6S309 | 6p24.3 |
| D6S470 | 6p24.2 |
| D6S289 | 6p22.3 |
| D6S422, D6S276 | 6p22.1 |
| D6S105 | 6p21.3 |
| D6S1666, D6S1560 | 6p21.31 |
| D6S1610 | 6p21.2 |
| D6S294, D6S257 | 6p12.1 |
| D6S402, D6S430 | 6q12 |
| D6S1596 | 6q14.1 |
| D6S284 | 6q14.3 |
| D6S460 | 6q14-15 |
| D6S445 | 6q15 |
| D6S462, D6S1716 | 6q16.3 |
| D6S434, D6S447, D6S1592 | 6q21 |
| D6S287 | 6q22.31 |
| D6S262 | 6q23.1 |
| D6S292 | 6q23.2 |
| D6S308, D6S310, D6S1704 | 6q24.1 |
| D6S440 | 6q25.1 |
| D6S441, D6S290, D6S473, D6S442 | 6q25.2 |
| D6S1708, D6S415, D6S1612, D6S1581 | 6q25.3 |
| D6S264, D6S281, D6S446 | 6q27 |
CGH
To confirm the microsatellite analysis data and distinguish amplification from LOH, CGH was performed as described previously. 9
Immunohistochemistry for hMLH1 and hMSH2
In two cases (no. 24 and no. 28) showing increased levels of MSI, immunohistochemical staining for mismatch repair gene products hMLH1 and hMSH2 [α-hMLH1 antibody was from hybridoma clone G168-15 (Pharmingen, San Diego, CA); α-hMSH2 antibody was from hybridoma clone FE11 (Calbiochem, San Diego, CA)] was performed on formalin-fixed/paraffin-embedded tissue sections using a standard immunoperoxidase technique. Normal cells on the same slide served as a control.
Statistical Analysis
Statistical differences were analyzed by chi-square test or Fisher’s exact test using the commercially available Stat View statistical program (Abacus Concepts, Inc., Berkeley, CA).
Results
Frequent Allelic Imbalances on Chromosome 6 in Human Thymoma
Forty thymomas were screened for LOH using 41 microsatellites spanning chromosome 6 (Table 1) ▶ . LOH was found in 294 (23.5%) of 1253 informative genotypes in 30 of 40 tumors (75%). Two cases of type B3 thymoma showed LOH in all informative loci analyzed corresponding to monosomy 6. To verify the microsatellite analysis data and to distinguish amplification from LOH, the results were compared with CGH analyses performed on 25 thymomas included in our study. An example of CGH data with the results of a corresponding microsatellite analysis is shown in Figure 1 ▶ . CGH results on 18 thymomas (types A, B3, and C) had already been published previously. 9 Eight tumors were analyzed additionally (types AB and B2). Ten thymomas showed complete or partial losses of chromosome 6. No gain was found by CGH. Losses of the whole chromosome 6 occurred in one type B2, four B3, and two type C thymomas. Partial losses were found in three thymomas: loss of 6q in one type B3, 6pter-p21 loss in one type A, and loss of 6q24-qter in one type AB thymoma. All losses revealed by CGH were also detected by microsatellite analysis.
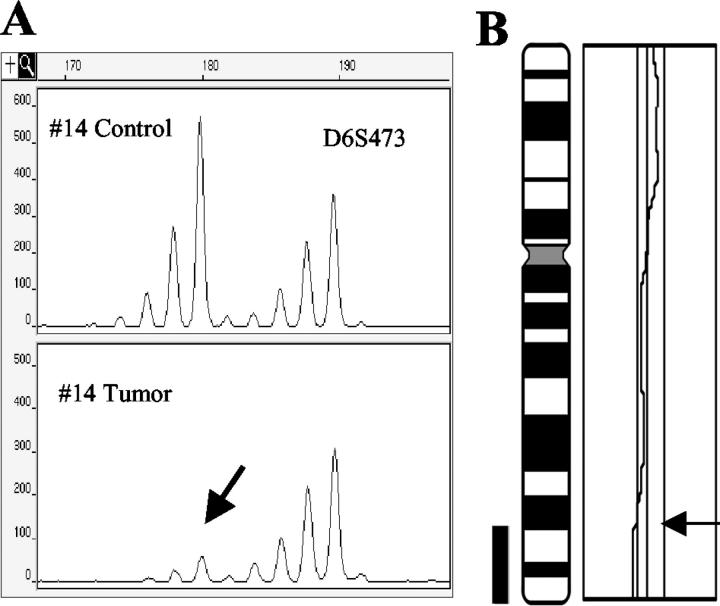
Figure 1.
Example of microsatellite analysis and CGH results. A: LOH. Control and tumor DNAs of case no. 14 were amplified with marker D6S473. The first allele signal intensity (180 bp, arrow) decreased by 90% as compared to the control. B: Corresponding partial loss of 6q24-qter detected by CGH.
Hot Spots of Frequent LOHs on Chromosome 6
We identified five hot spots showing frequent LOHs on chromosome 6 in thymoma. The hot spot showing the most frequent deletion was located in region 6q25.2 flanked by markers D6S441 and D6S290 (0.7 Mb apart, Figure 2A ▶ ). Eighteen of 37 cases (48.6%) displayed a LOH in this region. All thymoma types analyzed suffered this deletion, the percentages of affected tumors within individual WHO thymoma types ranged from 33.3 to 100%. There was no increased prevalence of this LOH in higher stages or more aggressive thymoma types. A separate hot spot of deletions appeared just 1.1-Mb telomeric from the former deletion locus, in the region 6q25.2-25.3, flanked by markers D6S442 and D6S1708 (0.4 Mb apart, Figure 2A ▶ ). LOH in this region could be detected in 12 of 37 informative cases (32.4%). The third hot spot was located in the MHC locus on 6p21.31, within a 1-Mb region flanked by markers D6S1666 and D6S1560 (Figure 2B) ▶ . We found LOHs in this hot spot in 12 of 40 informative cases (30%). Although 8 (66.7%) of these 12 cases manifested a concomitant myasthenia gravis (MG), the association between the 6p21.31 LOH and MG was not statistically significant. However, a type A thymoma (case no. 1) with LOH in the MHC locus manifested myasthenia gravis, which is a somewhat unusual association for this thymoma type. The fourth hot spot was detected within a 5.2-Mb interval on 6q14.1-14.3 defined by markers D6S1596 and D6S284 (Figure 2C) ▶ showing LOHs in 10 of 38 informative cases (26.3%). This LOH occurred more frequently in the more aggressive B3 and C types of thymoma (8 of 17, 47.1%) as compared to types A, AB, and B2 (2 of 21, 9.5%) (Fisher’s exact test, P = 0.0232). The last hot spot appeared within a 0.3-Mb region flanked by markers D6S447 and D6S1592 on 6q21 (Figure 2D) ▶ . Eight of 37 cases (21.6%) suffered LOH in this region.
Figure 2.

A: 6q25.2 and 6q25.2-25.3 LOH hot spots as defined by markers D6S441-D6S290 and D6S442-D6S1708, respectively. Status of each locus and its position (according to the Ensembl database) is indicated: filled boxes, LOH; vertically striped boxes, MSI; boxes with right hexagonal lines, retention of heterozygosity; open boxes, homozygosity; NA, no amplificate. B: 6p21.31 LOH hot spot as defined by markers D6S1666 and D6S1560. C: 6q14.1-14.3 LOH hot spot as defined by markers D6S1596 and D6S284. D: 6q21 LOH hot spot as defined by markers D6S447 and D6S1592.
MSI on Chromosome 6 in Thymoma
MSI was found in 39 (2.4%) of 1624 genotypes in 4 of 40 cases (10%). An example of MSI is shown in Figure 3 ▶ . Tumors positive for MSI were type B2 (one case) and B3 (three cases) thymomas. One of the latter cases (case no. 24) revealed high-level MSI (61% MSI-positive genotypes). The other three cases (no. 28, no. 27, and no. 19) showed low-level MSI (26.8%, 4.9%, and 2.4% MSI-positive genotypes, respectively). Immunohistochemical staining for the mismatch repair gene proteins hMLH1 and hMSH2 was performed to further investigate the cause of MSI in cases no. 24, no. 28, and no. 27. However, the tumor cells showed positive nuclear staining, confirming the presence of these mismatch repair proteins (data not shown).
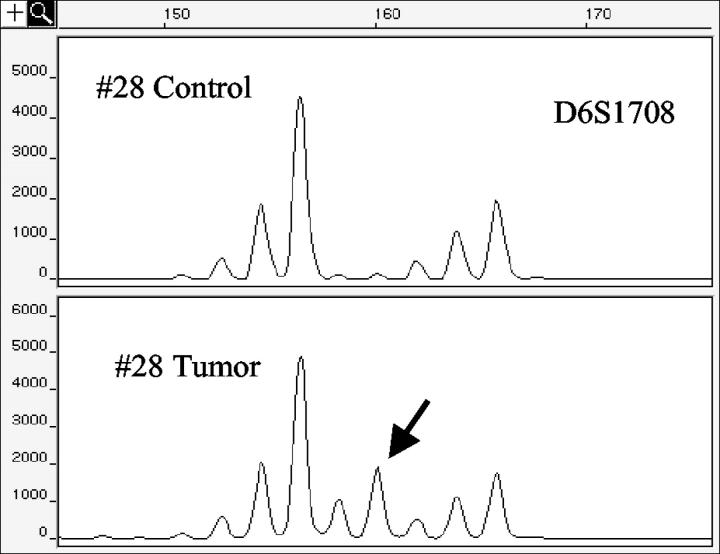
Figure 3.
Example of MSI. Control and tumor DNAs of case no. 28 were amplified with marker D6S1708. Tumor DNA shows a new additional allele (160 bp, arrow).
Frequency of Genetic Aberrations Correlates with Clinical Characteristics of Thymoma
When the analyzed thymomas were grouped according to their histological type as described in the new WHO thymoma classification, an association appeared between the thymoma type and the frequency of genetic aberrations expressed as percentage of microsatellite repeats showing either allelic imbalance or MSI. On chromosome 6 genetic aberrations were found in 10.8% of genotypes of type A thymoma, 12.6% of type AB, 20% of type B2, 26% of type B3, and 34.8% of type C thymoma (Table 2) ▶ .
Table 2.
Frequency of Genetic Aberrations on Chromosome 6 According to WHO Type
| Thymoma type | Genetic aberration frequency (%) |
|---|---|
| A | 30/279 (10.8%) |
| AB | 46/364 (12.6%) |
| B2 | 41/205 (20.0%) |
| B3 | 160/615 (26.0%) |
| C | 56/161 (34.8%) |
Significant differences (chi-square-test) were found in type A versus B2 (P = 0.0045), A versus B3 (P < 0.0001), A versus C (P < 0.0001), AB versus B2 (P = 0.0191), AB versus B3 (P < 0.0001), AB versus C (P < 0.0001), B2 versus C (P = 0.0015), and B3 versus C (P = 0.0271).
Another association was found between the thymoma stage as determined according to the Masaoka’s staging system 21 and the overall frequency of LOH on chromosome 6 in a particular stage. LOH was found in 7% of genotypes of stage I thymomas, 26.5% of stage II, 21.4% of stage III, and 24% of stage IV tumors (Table 3) ▶ . Frequency of LOH in stage I noninvasive thymomas was significantly lower than that in stage II to IV invasive thymomas (P < 0.0001).
Table 3.
Frequency of LOH on Chromosome 6 According to Masaoka’s Stage
| Stage | Genetic aberration frequency (%) |
|---|---|
| I | 25/357 (7.0%) |
| II | 152/573 (26.5%) |
| III | 87/407 (21.4%) |
| IV | 69/287 (24.0%) |
A significant difference (chi-square-test) was found between stages I (noninvasive thymoma) and II to IV (invasive thymoma) (P < 0.0001).
Discussion
Multiple malignancies display genetic aberrations on chromosome 6. Colorectal, breast, prostate, and ovarian cancer, leukemias, and lymphomas all have been reported to suffer frequent deletions especially on the long arm of this chromosome, although the location of the hot spots of deletions varies among different types of cancer. Such reports suggest the presence of several putative tumor suppressor genes on chromosome 6 contributing to carcinogenesis. Previously, we screened several types of thymoma, the most frequent type of neoplasm arising in thymus, for genetic abnormalities. We analyzed these tumors first using CGH followed by a focused microsatellite analysis, and could show that LOH on chromosome 6 is the most frequent genetic abnormality in thymoma. 9,10 To determine the exact location of LOH hot spots on chromosome 6 we now performed a detailed survey of chromosome 6 aberrations in thymoma (including for the first time WHO types AB and B2) using a panel of 41 microsatellite markers on a group of 40 tumors.
The most frequent LOH we detected occurred on 6q25.2; 48.6% of thymomas suffered a deletion in this locus. We narrowed down the region showing deletions to 0.7 Mb flanked by markers D6S441 and D6S290. Peculiarly, in the region 6q25.2-25.3, just 1.1 Mb from this hot spot toward the telomere, a second hot spot appeared flanked by markers D6S442 and D6S1708. Some of the thymomas (32.4%) showed LOH in this region. The 6q25.1-25.3 segment was reported to be a LOH hot spot harboring putative tumor suppressor genes in breast, 23 ovarian cancer, 24 and non-Hodgkin’s lymphoma. 18 The 6q25.2 region contains besides tens of expressed sequence tag (ESTs) also several already cloned or predicted genes such as the ribosomal protein L27A, F-box protein, or regulator of G-protein signaling 17 (RGS17). The last one is probably the most suitable candidate from the described genes for a putative tumor suppressor gene involved in thymomagenesis. Namely, proteins containing the RGS domain constitute a family of molecules that seem to function as negative regulators of heterotrimeric G protein signaling. 25
The region showing the third most frequent deletions (30% of informative cases) was 6p21.31. Deletions in this region were rather large spanning over a 1-Mb interval between microsatellites D6S1666 and D6S1560. This region contains among others the genes for the HLA class II major histocompability complex. Loss of the MHC genes has been reported in numerous tumors: in colorectal cancer, 26 head and neck squamous cell carcinoma, 27 cervical cancer, 28 leukemia, 29 and astrocytoma. 30 The MHC class I and II genes encode highly polymorphic cell surface glycoproteins that bind antigenic peptides for presentation to CD4- and CD8-bearing T lymphocytes, respectively. Because of the role of the MHC molecules in presenting immunogenic peptides to cytotoxic T cells, defects in the MHC gene expression allow the tumor cells to evade the immune response that might be expected to be generated after multiple mutations of genes, whose abnormal products are potential targets for the immune system. Allelic loss of the MHC genes may thus confer a clonal selective advantage providing tumor cells with a mechanism to escape recognition by the immune defense system. This selection pressure would explain why the MHC gene loss was present in the analyzed thymomas with such a high frequency. The finding of LOH at 6p21.31 might be also important for thymoma-associated autoimmunity. Specifically, taking into account the affinity-avidity model of T-cell development in normal thymus, 31-35 reduced level of MHC expression on neoplastic thymic epithelial cells may contribute to the establishment of a nontolerant T-cell repertoire in thymoma. 36-39 Although we found only a trend, not a statistically significant relationship between the LOH at the MHC locus in thymoma and MG in the respective patients, further investigation with a larger number of patients is necessary to clarify the significance of LOH at MHC locus for the pathogenesis of thymoma-associated MG.
Some of the patients (26.3%) showed LOHs within a 5.2-Mb interval flanked by markers D6S1596 and D6S284 on 6q14.1-14.3. Interestingly, a deletion of this region detected by cytogenetic analysis was described in a case of thymoma previously. 3 Similar deletions were reported also in malignant mesothelioma, 40 non-Hodgkin’s lymphoma, and acute lymphoblastic leukemia. 41 Although the overall frequency of allelic losses in this region was in general not so high in the analyzed thymomas, there were definitely more 6q14.1-14.3 LOHs in malignant thymoma types B3-C than in the more benign types A-B2 (47.1% and 9.5%, respectively; chi-square-test, P = 0.0232). These results suggest that 6q14.1-14.3 might harbor a potential tumor suppressor gene involved in the malignant progression or metastasis of thymoma. So far, no known tumor suppressor gene has been identified among tens of known and predicted genes residing in this region.
The fifth LOH hot spot was detected on 6q21 flanked by markers D6S447 and D6S1592 (0.3 Mb apart, 21.6% of informative cases). Losses in this region were reported in other malignancies such as breast cancer, 11,42 lymphoid leukemia, 43 non-Hodgkin’s lymphoma, 18 and ovarian cancer 24 as well. However, no known tumor suppressor gene has been identified in this region so far. The invasiveness of thymoma in general increases from type A, B, to type C of the WHO classification and seems to correlate with the degree of genetic instability displayed by the individual thymoma types. In this study, we found a significant increase in the frequency of genetic aberrations in more malignant thymoma types (Table 2) ▶ . Thus, the WHO classification of thymic epithelial tumors, which is reported to reflect both clinical features and immunological functions of tumor cells, 44 may also reflect the genetic characteristics of thymomas, although our study was limited to chromosome 6 only. However, staging of the thymomas according to the Masaoka’s 21 staging system did not show such a strong correlation with genetic aberration frequency. Only stage I noninvasive thymomas showed significantly lower frequency of such aberrations than stage II-IV invasive thymomas (Table 3) ▶ .
What role MSI plays in thymomagenesis is currently unknown. Also among the thymomas, some tumors display an increased level of MSI. We found four (10%) MSI-positive thymomas among the cases analyzed. The percentage of MSI-positive genotypes ranged from 4.9% to 61%. Interestingly, all of these MSI-positive tumors were type B thymomas (one B2 type and three B3 type cases). Thus, this finding might delineate another pathway of pathogenesis especially in cortical thymomas. However, the percentage of MSI-positive genotypes depends on the repeat panel used for the analysis. Using only repeats on chromosome 6 showing the most frequent abnormalities in thymoma implies probably an automatic bias toward more abnormalities. Moreover, those three cases showing >4% MSI-positive genotypes were stained for the mismatch repair proteins hMLH1 and hMSH2 and turned out to be positive for these enzymes. MSI in these cases was thus not related to the deficiency of these mismatch repair gene products.
In summary, we determined that chromosome 6 is a target of frequent chromosomal aberrations in thymoma. We identified five hot spots of frequent deletions on chromosome 6 flanked by markers D6S441-D6S290 (6q25.2), D6S442-D6S1708 (6q25.2-25.3), D6S1666-D6S1560 (6p21.31), D6S1596-D6S284 (6q14.1-14.3), and D6S447-D6S1592 (6q21). These results suggest the presence of several putative tumor suppressor genes on chromosome 6 that may contribute to the pathogenesis of thymoma.
Acknowledgments
We thank Dr. S. Schwarz for help with laser-assisted microdissection, E. Werder and C. Kohout for expert technical assistance, and E. Oswald for advice on cell culture methods.
Footnotes
Address reprint requests to Petr Starostik, M.D., Roswell Park Cancer Institute, Department of Pathology and Laboratory Medicine, Elm & Carlton Sts., Buffalo, NY 14263. E-mail: petr.starostik@roswellpark.org.
Supported by a Humboldt Research Fellowship from the Alexander von Humboldt Foundation (to M. I.) and a grant from the Deutsche Krebshilfe (grant no.10-1740St to P. S.).
References
- 1.Willcox N: Myasthenia gravis. Curr Opin Immunol 1993, 5:910-917 [DOI] [PubMed] [Google Scholar]
- 2.Müller-Hermelink HK, Marx A: Thymoma. Curr Opin Oncol 2000, 12:426-433 [DOI] [PubMed] [Google Scholar]
- 3.Kristoffersson U, Heim S, Mandahl N, Akerman M, Mitelman F: Multiple clonal chromosome aberrations in two thymomas. Cancer Genet Cytogenet 1989, 41:93-98 [DOI] [PubMed] [Google Scholar]
- 4.Sonobe H, Takeuchi T, Ohtsuki Y, Taguchi T, Shimizu K: A thymoma with clonal complex chromosome abnormalities. Cancer Genet Cytogenet 1999, 110:72-74 [PubMed] [Google Scholar]
- 5.Mirza I, Kazimi SN, Ligi R, Burns J, Braza F: Cytogenetic profile of a thymoma. A case report and review of the literature. Arch Pathol Lab Med 2000, 124:1714-1716 [DOI] [PubMed] [Google Scholar]
- 6.Kees UR, Mulcahy MT, Willoughby MLN: Intrathoracic carcinoma in an 11-year-old girl showing a translocation t(15;19). Am J Pediatr Hematol Oncol 1991, 13:459-464 [DOI] [PubMed] [Google Scholar]
- 7.Kubonishi I, Takehara N, Iwata J, Sonobe H, Ohtsuki Y, Abe T, Miyoshi I: Novel t(15;19)(q15;p13) chromosome abnormality in a thymic carcinoma. Cancer Res 1991, 51:3327-3328 [PubMed] [Google Scholar]
- 8.Lee ACW, Kwong Y-L, Fu KH, Chan GCF, Ma L, Lau Y-L: Disseminated mediastinal carcinoma with chromosomal translocation (15;19). A distinctive clinicopathologic syndrome. Cancer 1993, 72:2273-2276 [DOI] [PubMed] [Google Scholar]
- 9.Zettl A, Ströbel P, Wagner K, Katzenberger T, Ott G, Rosenwald A, Peters K, Krein A, Semik M, Müller-Hermelink HK, Marx A: Recurrent genetic aberrations in thymoma and thymic carcinoma. Am J Pathol 2000, 157:257-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou R, Zettl A, Ströbel P, Wagner K, Müller-Hermelink HK, Zhang S, Marx A, Starostik P: Thymic epithelial tumors can develop along two different pathogenetic pathways. Am J Pathol 2001, 159:1853-1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theile M, Seitz S, Arnold W, Jandrig B, Frege R, Schlag PM, Haensch W, Guski H, Winzer KJ, Barrett JC, Scherneck S: A defined chromosome 6q fragment (at D6S310) harbors a putative tumor suppressor gene for breast cancer. Oncogene 1996, 13:677-685 [PubMed] [Google Scholar]
- 12.Bastian BC, LeBoit PE, Hamm H, Brocker EB, Pinkel D: Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res 1998, 58:2170-2175 [PubMed] [Google Scholar]
- 13.Reutzel D, Mende M, Naumann S, Störkel S, Brenner W, Zabel B, Decker J: Genomic imbalances in 61 renal cancers from the proximal tubulus detected by comparative genomic hybridization. Cytogenet Cell Genet 2001, 93:221-227 [DOI] [PubMed] [Google Scholar]
- 14.Jin C, Martins C, Jin Y, Wiegant J, Wennerberg J, Dictor M, Gisselsson D, Strömbeck B, Fonseca I, Mitelman F, Tanke HJ, Hoglund M, Mertens F: Characterization of chromosome aberrations in salivary gland tumors by FISH, including multicolor COBRA-FISH. Genes Chromosom Cancer 2001, 30:161-167 [PubMed] [Google Scholar]
- 15.Wan M, Sun T, Vyas R, Zheng J, Granada E, Dubeau L: Suppression of tumorigenicity in human ovarian cancer cell lines is controlled by a 2 cM fragment in chromosomal region 6q24–q25. Oncogene 1999, 18:1545-1551 [DOI] [PubMed] [Google Scholar]
- 16.Jackson A, Carrara P, Duke V, Sinclair P, Papaioannou M, Harrison CJ, Foroni L: Deletion of 6q16–q21 in human lymphoid malignancies: a mapping and deletion analysis. Cancer Res 2000, 60:2775-2779 [PubMed] [Google Scholar]
- 17.Offit K, Parsa NZ, Gaidano G, Filippa DA, Louie D, Pan D, Jhanwar SC, Dalla-Favera R, Chaganti RSK: 6q deletions define distinct clinico-pathologic subsets of non-Hodgkin’s lymphoma. Blood 1993, 82:2157-2162 [PubMed] [Google Scholar]
- 18.Starostik P, Greiner A, Schultz A, Zettl A, Peters K, Rosenwald A, Kolve M, Müller-Hermelink HK: Genetic aberrations common in gastric high-grade large B-cell lymphoma. Blood 2000, 95:1180-1187 [PubMed] [Google Scholar]
- 19.Rosai J, Sobin LH: Histological typing of tumors of the thymus. Rosai J Sobin LH eds. WHO International Histological Classification of Tumors. 1999:pp 1-65 SpringerVerlag, Berlin
- 20.Müller-Hermelink HK, Marx A: Pathological aspects of malignant and benign thymic disorders. Ann Med 1999, 31(Suppl 2):5-14 [PubMed] [Google Scholar]
- 21.Masaoka A, Monden Y, Nakahara K, Tanioka T: Follow-up study of thymomas with special reference to their clinical stages. Cancer 1981, 48:2485-2492 [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Frisch EF, Maniatis T: Molecular Cloning: A Laboratory Manual. 1989. Cold Spring Harbor Press, Cold Spring Harbor
- 23.Utada Y, Haga S, Kajiwara T, Kasumi F, Sakamoto G, Nakamura Y, Emi M: Mapping of target regions of allelic loss in primary breast cancers to 1-cM intervals on genomic contigs at 6q21 and 6q25.3. J Cancer Res 2000, 91:293-300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang VW, Bell DA, Berkowitz RS, Mok SC: Whole genome amplification and high-throughput allelotyping identified five distinct deletion regions on chromosome 5 and 6 in microdissected early-stage ovarian tumors. Cancer Res 2001, 61:4169-4174 [PubMed] [Google Scholar]
- 25.Druey KM: Bridging with GAPs: receptor communication through RGS proteins. Sci STKE 2001, 104:RE14. [DOI] [PubMed] [Google Scholar]
- 26.Honchel R, McDonnell S, Schaid DJ, Thibodeau SN: Tumor necrosis factor-α allelic frequency and chromosome 6 allelic imbalance in patients with colorectal cancer. Cancer Res 1996, 56:145-149 [PubMed] [Google Scholar]
- 27.Feenstra M, Veltkamp M, van Kuik J, Wiertsema S, Slootweg P, van den Tweel J, de Weger R, Tilanus M: HLA class I expression and chromosomal deletions at 6p and 15q in head and neck squamous cell carcinomas. Tissue Antigens 1999, 54:235-245 [DOI] [PubMed] [Google Scholar]
- 28.Koopman LA, Corver WE, van der Slik AR, Giphart MJ, Fleuren GJ: Multiple genetic alterations cause frequent and heterogeneous human histocompatibility leukocyte antigen class I loss in cervical cancer. J Exp Med 2000, 191:961-975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bevan S, Catovsky D, Matutes E, Antunovic P, Auger MJ, Ben-Bassat I, Bell A, Berrebi A, Gaminara EJ, Júnior ME, Mauro FR, Quabeck K, Rassam SMB, Reid C, Ribeiro I, Stark P, van Dongen JJM, Wimperis J, Wright S, Marossy A, Yuille MR, Houlston RS: Linkage analysis for major histocompatibility complex-related genetic susceptibility in familial chronic lymphocytic leukemia. Blood 2000, 96:3982-3984 [PubMed] [Google Scholar]
- 30.Saitoh Y, Bruner JM, Levin VA, Kyritsis AP: Identification of allelic loss on chromosome arm 6p in human astrocytomas by arbitrarily primed polymerase chain reaction. Genes Chromosom Cancer 1998, 22:165-170 [DOI] [PubMed] [Google Scholar]
- 31.Willcox N, Schluep M, Ritter MA, Schuurman HJ, Newsom-Davis J, Christensson B: Myasthenic and nonmyasthenic thymoma. An expansion of a minor cortical epithelial cell subset? Am J Pathol 1987, 127:447-460 [PMC free article] [PubMed] [Google Scholar]
- 32.Ashton-Rickardt PG, Tonegawa S: A differential-avidity model for T-cell selection. Immunol Today 1994, 15:362-366 [DOI] [PubMed] [Google Scholar]
- 33.Jameson SC, Hogquist KA, Bevan MJ: Positive selection of thymocytes. Annu Rev Immunol 1995, 13:93-126 [DOI] [PubMed] [Google Scholar]
- 34.Fukui Y, Ishimoto T, Utsuyama M, Gyotoku T, Koga T, Nakao K, Hirokawa K, Katsuki M, Sasazuki T: Positive and negative CD4+ thymocyte selection by a single MHC class II/peptide ligand affected by its expression level in the thymus. Immunity 1997, 6:401-410 [DOI] [PubMed] [Google Scholar]
- 35.Barton GM, Rudensky AY: Requirement for diverse, low-abundance peptides in positive selection of T cells. Science 1999, 283:67-70 [DOI] [PubMed] [Google Scholar]
- 36.Inoue M, Fujii Y, Okumura M, Takeuchi Y, Shiono H, Miyoshi S, Matsuda H, Shirakura R: Neoplastic thymic epithelial cells of human thymoma support T cell development from CD4−CD8− cells to CD4+CD8+ cells in vitro. Clin Exp Immunol 1998, 112:419-426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue M, Okumura M, Miyoshi S, Shiono H, Fukuhara K, Kadota Y, Shirakura R, Matsuda H: Impaired expression of MHC class II molecules in response to interferon-gamma (IFN-γ) on human thymoma neoplastic epithelial cells. Clin Exp Immunol 1999, 117:1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller-Hermelink HK, Wilisch A, Schultz A, Marx A: Characterization of the human thymic microenvironment: lymphoepithelial interaction in normal thymus and thymoma. Arch Histol Cytol 1997, 60:9-28 [DOI] [PubMed] [Google Scholar]
- 39.Kadota Y, Okumura M, Miyoshi S, Kitagawa-Sakakida S, Inoue M, Shiono H, Maeda Y, Kinoshita T, Shirakura R, Matsuda H: Altered T cell development in human thymoma is related to impairment of MHC class II transactivator expression induced by interferon-gamma (IFN-γ). Clin Exp Immunol 2000, 121:59-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell DW, Jhanwar SC, Testa JR: Multiple regions of allelic loss from chromosome arm 6q in malignant mesothelioma. Cancer Res 1997, 57:4057-4062 [PubMed] [Google Scholar]
- 41.Menasce LP, Orphanos V, Santibanez-Koref M, Boyle JM, Harrison CJ: Common region of deletion on the long arm of chromosome 6 in non-Hodgkin’s lymphoma and acute lymphoblastic leukaemia. Genes Chromosom Cancer 1994, 10:286-288 [DOI] [PubMed] [Google Scholar]
- 42.Noviello C, Courjal F, Theillet C: Loss of heterozygosity on the long arm of chromosome 6 in breast cancer: possibly four regions of deletion. Clin Cancer Res 1996, 2:1601-1606 [PubMed] [Google Scholar]
- 43.Jackson A, Panayiotidis P, Foroni L: The human homologue of the Drosophila tailless gene (TLX): characterization and mapping to a region of common deletion in human lymphoid leukemia on chromosome 6q21. Genomics 1998, 50:34-43 [DOI] [PubMed] [Google Scholar]
- 44.Okumura M, Miyoshi S, Fujii Y, Takeuchi Y, Shiono H, Inoue M, Fukuhara K, Kadota Y, Tateyama H, Eimoto T, Matsuda H: Clinical and functional significance of WHO classification on human thymic epithelial neoplasms. A study of 146 consecutive tumors. Am J Surg Pathol 2001, 25:103-110 [DOI] [PubMed] [Google Scholar]