Abstract
Seed dormancy is defined as the failure of a viable seed to germinate under favorable conditions. Besides playing an adaptive role in nature by optimizing germination to the most suitable time, a tight control of dormancy is important in crop plants. Extensive genetic and physiological studies have identified the involvement of several factors, but the molecular mechanisms underlying this process are still largely unknown. We cloned the HISTONE MONOUBIQUITINATION1 (HUB1) gene, of which the mutant (previously identified as reduced dormancy4) has reduced seed dormancy and several pleiotropic phenotypes. HUB1 encodes a C3HC4 RING finger protein. The Arabidopsis thaliana genome contains one HUB1 homolog, which we named HUB2. The hub2 mutant also has reduced seed dormancy and is not redundant with hub1. Homologs of HUB1 and HUB2 in other species are required for histone H2B monoubiquitination. In agreement with this, the ubiquitinated form of histone H2B could not be detected in the hub1 and hub2 mutants. In yeast and human cells, histone H2B monoubiquitination is associated with actively transcribed genes. The hub1 mutant showed altered expression levels for several dormancy-related genes. We propose a role for chromatin remodeling in seed dormancy by H2B monoubiquitination through HUB1 and HUB2.
INTRODUCTION
The survival of a plant depends on the timing of transitions in its life cycle. The two main transitions are seed germination and the initiation of flowering. The molecular mechanisms underlying flowering initiation have been intensively studied and are increasingly well known (Putterill et al., 2004). However, the regulation of dormancy and germination is still poorly understood.
Seed dormancy is defined as the failure of an intact, viable seed to complete germination under favorable conditions (Bewley, 1997). A seed will germinate in appropriate environmental conditions only after it has lost dormancy. In Arabidopsis thaliana, dormancy is induced during seed maturation and released by aging (after-ripening) or imbibition of the seed at low temperatures (stratification). The transformation from dormancy to nondormancy is associated with changes in gene expression and protein patterns (Cadman et al., 2006; Chibani et al., 2006; Lee et al., 2006). An extensive interplay exists between environmental signals and endogenous developmental processes during the induction of dormancy, seed storage, and imbibition. This suggests the existence of a network of interactions between different genes in the control of seed germination.
The analysis of seed maturation mutants in Arabidopsis indicated that dormancy is induced during the later stages of development because mutants that are defective in seed maturation also lost dormancy (Bentsink et al., 2007). A dormant Arabidopsis seed is prevented from germinating because the embryo is unable to overcome the constraints of the surrounding tissues. When the seed coat of a dormant seed is removed, the embryo can usually germinate. The importance of the strength of the seed coat was confirmed by the observation of reduced dormancy in testa mutants of Arabidopsis (Debeaujon et al., 2000).
Physiological and genetic studies in Arabidopsis and tomato (Solanum lycopersicum) revealed a critical role for abscisic acid (ABA) in seed dormancy (Koornneef et al., 2002; Gubler et al., 2005; Finch-Savage and Leubner-Metzger, 2006). Mutants with defects in ABA biosynthesis or its mode of action show reduced seed dormancy (Koornneef et al., 1982, 1984; Giraudat et al., 1992; Léon-Kloosterziel et al., 1996a). Conversely, the ABA-supersensitive mutant era1 confers enhanced seed dormancy (Cutler et al., 1996). Recent studies also showed that ABA catabolism plays a role in dormancy release (Kushiro et al., 2004; Saito et al., 2004; Millar et al., 2006). Additional roles in the regulation of dormancy have been found for gibberellins, ethylene, sugars, phytochrome, brassinosteroids, and nitrate (Bentsink et al., 2007). The underlying molecular mechanisms and the events upstream and downstream of these factors are unknown, but genes that are not obviously related to the hormone pathways could play a role. Examples of such genes are the DOF transcription factors DAG1 and DAG2 (Papi et al., 2000, 2002; Gualberti et al., 2002) and the heterotrimeric G protein GPA1 (Ullah et al., 2002). However, these genes do not have strong effects on dormancy.
Quantitative trait loci (QTL) analysis for seed dormancy in Arabidopsis identified several QTL, representing potentially novel factors in dormancy induction or release (Alonso-Blanco et al., 2003). One of these QTL, named DELAY OF GERMINATION1 (DOG1), has recently been cloned (Bentsink et al., 2006). A mutant in the DOG1 gene was also isolated and is characterized by absence of dormancy without any pleiotropic phenotypes. This suggests that DOG1 may play a key role in the onset of seed dormancy. However, due to the lack of protein domains with known functions, the identity of this gene did not reveal more about the molecular mechanism of dormancy (Bentsink et al., 2006).
Additional new factors might be found among four mutants with reduced dormancy (rdo) that were obtained in mutagenesis screens of the Landsberg erecta (Ler) accession (Léon-Kloosterziel et al., 1996b; Peeters et al., 2002). Mutants at these four loci did not have altered levels or changed sensitivity for ABA, and the rdo4 mutant did not show a change in GA requirement (Peeters et al., 2002). Furthermore, all four mutants showed pleiotropic phenotypes in the adult plant.
In this article, we describe the map-based cloning of RDO4, which was renamed HISTONE MONOUBIQUITINATION1 (HUB1). HUB1 encodes a C3HC4 RING finger protein, which probably functions as the E3 ligase responsible for monoubiquitination of histone H2B. We present evidence that HUB1 is necessary for histone H2B monoubiquitination in vivo and influences gene expression of dormancy-related genes. These results strongly suggest a role for chromatin remodeling in the regulation of seed dormancy.
RESULTS
The hub1-2 (rdo4) Mutant Shows Developmental Defects, Including a Reduction in Seed Dormancy
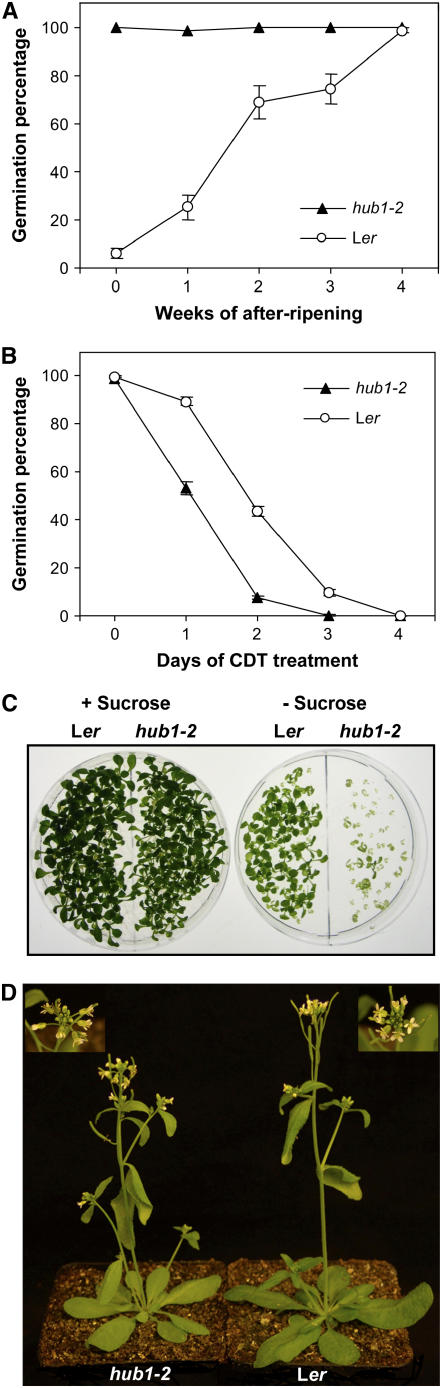
The hub1-2 (rdo4) mutant was originally isolated in a γ-irradiation mutagenesis screen on the basis of its reduced seed dormancy phenotype. In addition, the mutant showed pleiotropic phenotypes, such as pale-green leaf color, increased bushy appearance, and more open flower buds (Peeters et al., 2002). We analyzed the germination of hub1-2 mutants in more detail and confirmed its reduced dormancy (Figure 1A). In the dog1 mutant, reduced dormancy was shown to be correlated with reduced seed longevity (Bentsink et al., 2006). Analysis of hub1-2 seeds in a controlled deterioration test also showed a reduction in longevity (Figure 1B). Furthermore, the growth of hub1-2 seedlings on medium without sucrose is severely delayed compared with wild-type seedlings. Addition of 1.5% sucrose to the medium can rescue the delayed development of hub1-2, suggesting a defect in seedling establishment (Figure 1C). This could be caused by defective mobilization of reserves, which can be relieved by external sucrose (Eastmond and Graham, 2001). Examination of adult hub1-2 plants confirmed the pleiotropic phenotypes described by Peeters et al. (2002). The hub1-2 mutant plants are paler green and slightly more bushy than wild-type plants, and the flowers are positioned at increased angles to the main stem (Figure 1D). We could quantify one of the pleiotropic phenotypes by measurement of the chlorophyll content index, which was lower in hub1-2 than in the Ler wild-type (Figure 3C). These phenotypes suggest that HUB1 is part of a mechanism that plays a role in several processes in the plant.
Figure 1.
Seed Dormancy and Pleiotropic Phenotypes of the hub1-2 Mutant.
(A) Germination of Ler and hub1-2 seeds on water in the light after different periods of dry storage. Percentages are means (±se) of six seed bulks of each three plants.
(B) Germination of 6-month-old Ler and hub1-2 seeds after several days of controlled deterioration treatment (CDT) at 37°C and 85% relative humidity. Percentages are means (±se) of 12 plants.
(C) Seedling establishment of Ler and hub1-2 after 12 d of growth on Murashige and Skoog (MS) medium with or without 1.5% sucrose.
(D) Phenotype of 5-week-old Ler and hub1-2 plants grown in soil. The insets show the difference in inflorescence architecture between the mutant (left) and the wild type (right).
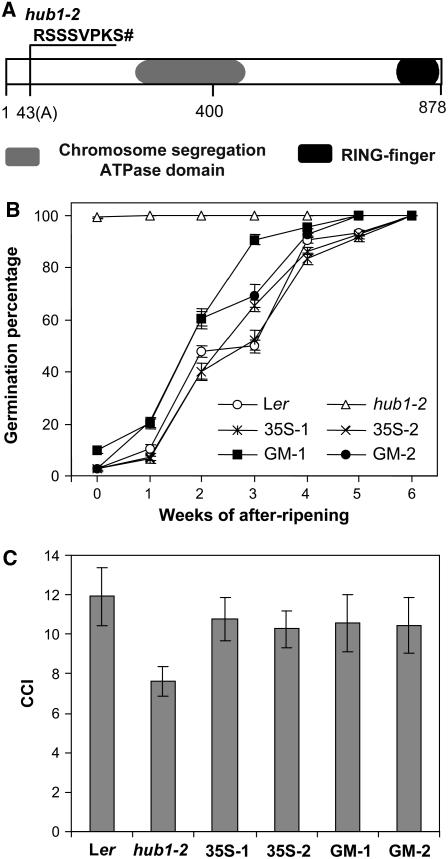
Figure 3.
Structure of HUB1 and Complementation of hub1-2.
(A) Schematic presentation of HUB1. The location of the hub1-2 mutation and the altered protein that it encodes are indicated, as well as the positions of the chromosome segregation ATPase domain and RING finger domain.
(B) Germination on water in the light after different periods of dry storage is shown for seeds of Ler, hub1-2, two homozygous transformants of hub1-2 with the HUB1 overexpressor (35S-1 and 35S-2), and two homozygous transformants of hub1-2 with the genomic fragment of HUB1 (GM-1 and GM-2). Percentages are means (±se) of six seed bulks of each three plants.
(C) Comparison of the chlorophyll content index (CCI) between Ler, hub1-2, two homozygous transformants of hub1-2 with the HUB1 overexpressor (35S-1 and 35S-2), and two homozygous transformants of hub1-2 with the genomic fragment of HUB1 (GM-1 and GM-2). Percentages are means (±sd) of 15 plants.
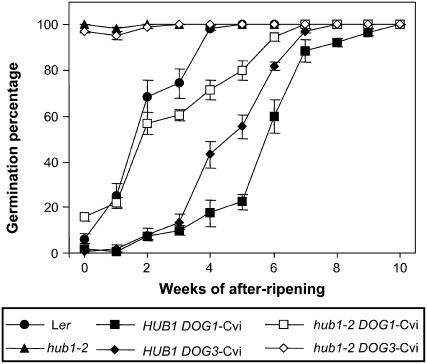
hub1-2 Is Epistatic to the DOG3 Locus
The hub1-2 dormancy phenotype was previously analyzed in the Ler accession, which has a relatively low level of dormancy. We were interested in the influence of the hub1-2 mutation on plants with higher dormancy levels. Therefore, the hub1-2 mutant was crossed into two near isogenic lines (NILs) that have a Cape Verde Islands (Cvi) introgression, containing the DOG1 or DOG3 QTL, in a Ler background (DOG1-Cvi and DOG3-Cvi; Alonso-Blanco et al., 2003). These introgressions substantially increased the dormancy level of Ler (Figure 2). Seeds from hub1-2 mutant plants with the DOG1-Cvi introgression had an intermediate dormancy level. This phenotype was confirmed by transformation of a 5.6-kb Cvi DOG1 genomic fragment into the hub1-2 mutant. Resulting homozygous transgenic lines showed a dormancy level between the hub1-2 mutant and DOG1-Cvi NIL (data not shown). We conclude from these results that HUB1-2 is not absolutely required for dormancy because seeds with a DOG1-Cvi allele, containing the hub1-2 mutation, showed dormancy levels similar to wild-type Ler. By contrast, seeds from the combination of hub1-2 with DOG3-Cvi were completely nondormant, indicating that hub1-2 is epistatic to DOG3 (Figure 2). This suggests that HUB1 regulates seed dormancy through the same pathway as DOG3.
Figure 2.
Seed Dormancy Levels of hub1-2 in DOG1 and DOG3 NILs.
Germination on water in the light after different periods of dry storage is shown for seeds of Ler, hub1-2, DOG1-Cvi, DOG3-Cvi, and the genotypes hub1-2 DOG1-Cvi and hub1-2 DOG3-Cvi. Percentages are means (±se) of six seed bulks of each three plants.
HUB1 Encodes a C3HC4 RING Finger Protein
HUB1 was previously mapped to the bottom of chromosome two, using a segregating population from the second backcross of hub1-2 (rdo4) with Columbia (Col) (Peeters et al., 2002). This backcross was necessary because natural variation in dormancy between Ler and Col accessions (van Der Schaar et al., 1997) causes modifications of the mutant phenotype in the progeny of the mapping cross. To facilitate fine-mapping, which requires accurate scoring of the mutant phenotype, we used an available NIL that contained an 11-Mb Cvi introgression encompassing the hub1-2 region, in a Ler background (LCN2-17; Keurentjes et al., 2006). This line was selected based on the known approximate map position of hub1-2 (Peeters et al., 2002). In the progeny of a cross between hub1-2 and this NIL, the dormancy phenotype of hub1-2 could be followed without difficulty. In addition, the pleiotropic phenotypes of hub1-2 were used to preselect homozygous mutants. For those recombinant plants that were essential for the mapping of the mutation, the mutant phenotype was confirmed by seed dormancy analysis. In a population of 2500 F2 plants, the location of hub1-2 could be confined to a region of 30 kb, containing five genes, in the overlap of BACs T13E15 and T14P1 between the markers T13E15-R5 and T14P1-R1. Based on the structure of these genes, analyzed in The Arabidopsis Information Resource (http://www.arabidopsis.org), and their expression pattern, analyzed with Genevestigator (https://www.genevestigator.ethz.ch; Zimmermann et al., 2004), three candidate genes were selected for sequence comparison of the mutant and Ler wild type. In the sequence of the hub1-2 mutant in one of these genes, At2g44950, a T-to-AC substitution in the second exon was detected, causing a frameshift and an early stop codon eight amino acids after the mutation (Figure 3A). The same gene was also identified by the angusta4-1 (ang4-1/hub1-1) mutation (Fleury et al., 2007), which was isolated on the basis of altered leaf shape (Berná et al., 1999). We renamed the RDO4/ANG4 gene HUB1 on the basis of its function (see below). This gene consists of 19 exons and encodes a C3HC4 RING finger protein of 878 amino acids with an ATPase domain, belonging to the RING-type ubiquitin E3 ligase family of Arabidopsis (Stone et al., 2005). RING domains are characteristic for a large class of ubiquitin ligases and are likely to be diagnostic for this activity (Kraft et al., 2005; Stone et al., 2005). In the hub1-2 mutant, translation is terminated after 50 amino acids, and the resulting protein lacks both the ATPase and RING domains. Therefore, hub1-2 is expected to be functionally a null mutant.
The identity of At2g44950 as the HUB1 gene was confirmed in a complementation experiment. A 9-kb Ler genomic fragment, containing the coding sequence of At2g44950 and 2.9-kb 5′ and 0.6-kb 3′ sequences, was transformed into hub1-2 mutant plants. T3 seeds from two independent transformants (GM-1 and GM-2), segregating for a single insertion event of the transgene, showed similar dormancy levels as wild-type Ler plants (Figure 3B). The pleiotropic phenotypes of hub1-2 were also complemented in these transformants, as shown for the chlorophyll content index in Figure 3C.
We also created transgenic Arabidopsis plants that overexpress HUB1 by placing the cDNA under control of the 35S promoter, followed by transformation into the hub1-2 mutant. Seeds from homozygous T2 transformants, segregating for a single insertion event, have dormancy levels that are slightly higher than that of the transformants of the genomic complementation (Figure 3B). The chlorophyll content index of the HUB1 overexpression transformants is similar to that of the genomic transformants (Figure 3C).
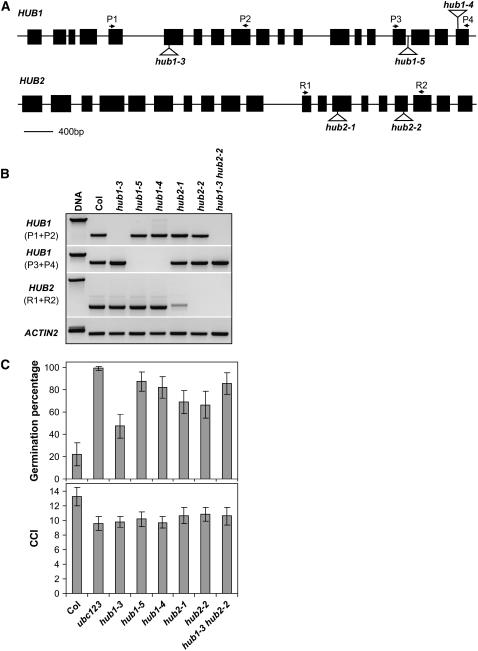
Additional mutant alleles in the Col background (hub1-3, hub1-4, and hub1-5) were obtained from the Salk insertion mutant collection (http://signal.salk.edu) and from the GABI-Kat collection (http://www.gabi-kat.de; Rosso et al., 2003). The location of the insertions is shown in Figure 4A. None of these T-DNA insertion mutants was a complete knockout, as indicated by RT-PCR analysis showing that the 3′ region of HUB1 was still transcribed in hub1-3 and the 5′ region was still transcribed in hub1-4 and hub1-5 (Figure 4B). The hub1-3 mutant seeds had slightly reduced dormancy, compared with Col wild-type seeds, but hub1-4 and hub1-5 mutant seeds showed stronger phenotypes (Figure 4C). Therefore, hub1-3 might still encode a partially functional HUB1 protein. However, the chlorophyll content index of all three alleles was lower than that of wild-type Col. Altogether, the sequencing, complementation, and T-DNA insertion mutant analysis confirm the identification of the gene At2g44950 as HUB1.
Figure 4.
Characterization of HUB1 and HUB2 T-DNA Insertion Lines.
(A) Schematic illustration of the gene structure of HUB1 and HUB2 with the positions of the T-DNA insertions. The positions of the primers, used for the RT-PCR analysis in (B), are indicated on top of the structures. Exons are shown as black boxes and introns as lines.
(B) RT-PCR analysis of the HUB1 and HUB2 transcripts in leaves of wild-type and T-DNA insertion mutants. Two different primer pairs were used for HUB1. The ACTIN2 gene was used as a loading control.
(C) Germination on water in the light of freshly harvested seeds and the chlorophyll content index (CCI) are shown for wild-type Col, the ubc1 ubc2 ubc3 triple mutant, and single and double hub1 and hub2 mutants. For the germination experiment, percentages are means (±sd) of six seed bulks of each three plants. For the chlorophyll content index experiment, percentages are means (±sd) of 15 plants.
HUB1 Is Expressed Ubiquitously, and Its Protein Is Confined to the Nucleus
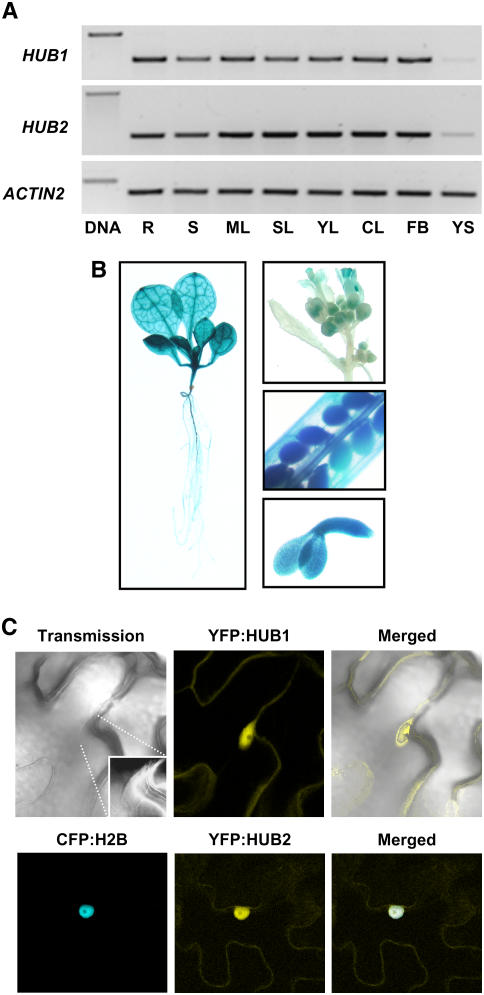
RT-PCR analysis showed the presence of HUB1 transcript in all tissues, with the lowest amount in young siliques (Figure 5A). This expression pattern is in agreement with data reported by Genevestigator (https://www.genevestigator.ethz.ch/at/; Zimmermann et al., 2004). In addition, we studied the expression pattern of HUB1 by fusing the 2.3-kb region 5′ of the HUB1 gene to the β-glucuronidase (GUS) reporter gene. This construct was transformed into wild-type Ler plants, and homozygous T2 transformants with a single insertion event were selected. GUS detection in these plants showed staining in all tissues, including embryos in developing seeds (Figure 5B), confirming the RT-PCR results. This suggests that HUB1 could function in all plant organs, which is in accordance with the observed pleiotropic phenotypes in the mutant.
Figure 5.
Expression and Localization Studies of HUB1 and HUB2.
(A) RT-PCR analysis of the HUB1 and HUB2 transcripts in organs of 6-week-old Arabidopsis plants. R, roots; S, stem, ML, mature leaves; SL, senescent leaves; YL, young leaves; CL, cauline leaves; FB, flowers and buds; YS, young siliques. The ACTIN2 gene was used as a loading control.
(B) Expression analysis by an HUB1 promoter:GUS construct in transgenic Arabidopsis plants reveals GUS signals throughout the entire plant. The left panel shows an 8-d-old plant; the right top panel shows an inflorescence; the right middle panel shows part of a silique 12 d after pollination; and the right bottom panel shows an isolated embryo 12 d after pollination.
(C) HUB1 and HUB2 proteins are located in the nucleus. The top panels show YFP signals in confocal microscopic images of cells from transgenic Arabidopsis plants stably transformed with the P35S:YFP:HUB1 construct. The inset (magnification with a higher contrast) shows the position of the nucleus. The bottom panels show CFP and YFP signals in confocal microscopic images of N. benthamiana cells transiently expressing P35S:CFP:H2B (to detect the nucleus) and P35S:YFP:HUB2.
To investigate the cellular localization of the HUB1 protein, the yellow fluorescent protein (YFP) was fused in frame to the N-terminal end of HUB1, behind the 35S promoter. This construct was transformed into hub1-2 mutant plants, and T2 transformants with a single insertion event were selected. These transgenic plants showed complementation of the hub1-2 phenotype. YFP signal was detected in the nucleus (Figure 5C), which indicated that the HUB1 protein functions in the nucleus.
Mutations in HUB2, the Arabidopsis Homolog of HUB1, Cause Reduced Seed Dormancy
Analysis of the Arabidopsis genomic sequence revealed that the gene F7A10.17 (At1g55250 + At1g55255) has high homology with HUB1. We named this gene HUB2. HUB2 is 57% identical with HUB1 at the nucleic acid level and 30% at the amino acid level. HUB2 has the same number of exons as HUB1, and the protein contains the same two domains. We studied one T-DNA insertion mutant for HUB2 from the GABI-Kat collection (hub2-1) and one from the Salk insertion mutant collection (hub2-2; Figure 4A). RT-PCR analysis with HUB2-specific primers did not show any amplification in hub2-2 and only a weak band in hub2-1 (Figure 4B). Sequencing revealed that this band contains HUB2 transcript with a 34-bp insertion that creates a stop codon. Both insertion mutants showed a similar pale-green leaf color as the hub1 mutants and also had a reduced chlorophyll content index (Figure 4C). Analysis of freshly harvested seeds showed a higher germination (∼60%) of hub2-1 and -2 compared with wild-type Col (20%) and a slightly lower germination than hub1-4 and -5 (Figure 4C). Therefore, hub2 mutants also have reduced seed dormancy. The expression pattern of HUB2, as determined by RT-PCR, is very similar to that of HUB1 (Figure 5A). Transient expression of a P35S:YFP:HUB2 fusion protein in Nicotiana benthamiana showed that the location of the HUB2 protein is confined to the nucleus (Figure 5C). A double mutant between hub1-3 and hub2-2 was made to analyze whether these two genes are redundant (Figure 4B). The chlorophyll content index of the double mutant did not differ from the single mutants, and the germination percentage of freshly harvested seeds was slightly higher than that of the single mutants (Figure 4C). Moreover, a double mutant between hub1-5 and hub2-2 showed the same phenotypes as the single mutants (data not shown). These data show that HUB1 and HUB2 influence the same processes in the plant and are weakly or not redundant.
HUB1 Is Homologous to Bre1, an Evolutionary Conserved Gene That Is Required for Histone H2B Monoubiquitination
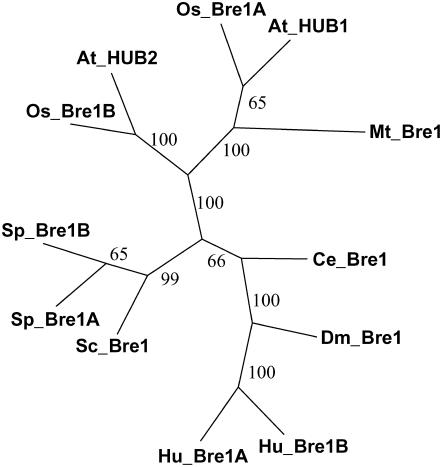
A database search revealed sequence homology of HUB1 and HUB2 to Bre1 genes in different organisms, including Medicago truncatula (Mt_Bre1), rice (Oryza sativa; Os_Bre1A and Os_Bre1B), yeast (Sc_Bre1), and human (Hu_Bre1A and Hu_Bre1B). Phylogenetic analysis of Bre1 homologs from different organisms showed that plant Bre1 genes form a separate cluster (Figure 6). The Bre1 protein functions as an E3 ligase, necessary for monoubiquitination of histone H2B, in yeast (Hwang et al., 2003; Wood et al., 2003), human (Kim et al., 2005; Zhu et al., 2005), and Drosophila melanogaster (Bray et al., 2005). Monoubiquitination of histone H2B is a prerequisite for histone H3 methylation at Lys-4 and -79 and is associated with actively transcribed genes (Sun and Allis, 2002; Wood et al., 2003; Zhu et al., 2005). In human cells, the two Bre1 homologs probably act as a tetramer consisting of two copies of each protein (Zhu et al., 2005). The two homologs in Arabidopsis, HUB1 and HUB2, could function in a similar way. This would explain the lack of a strong additive effect in the double mutant hub1-3 hub2-2 (Figure 4C). In yeast, ubiquitination of H2B by Bre1 is dependent on the ubiquitin-conjugating enzyme Sc_UBC2 (Robzyk et al., 2000). Phylogenetic analysis showed that Arabidopsis has three UBC2 homologs, At_UBC1, At_UBC2, and At_UBC3 (Kraft et al., 2005). One or more of these three genes could function as the E2 enzyme supplying ubiquitin, which can be transferred to H2B by HUB1. This hypothesis was supported by analysis of the ubc1 ubc2 ubc3 triple mutant. Similar to hub1 and hub2, this triple mutant has reduced dormancy, a pale-green leaf color, and a lower chlorophyll content index (Figure 4C).
Figure 6.
Phylogenetic Tree of HUB1 Homologs.
The unrooted phylogram was generated with PHYLIP3.66 using the alignment shown in Supplemental Figure 1 online. Bootstrap values (×10) from 1000 replications for each branch are shown.
hub1-2 Has Reduced Levels of H2B Monoubiquitination
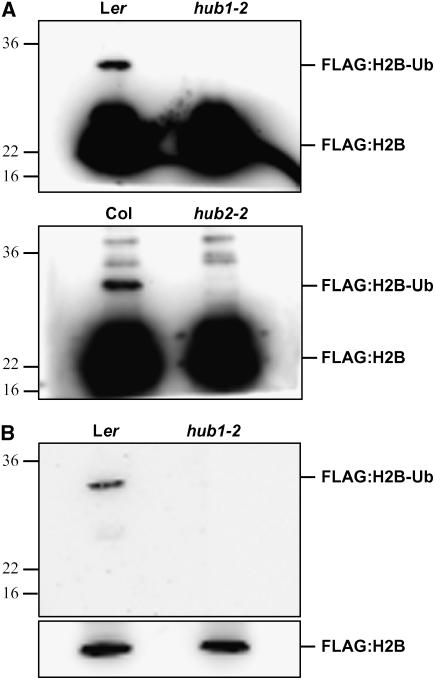
If HUB1 and HUB2 function as E3 ligases, necessary for monoubiquitination of H2B, reduced levels of H2B ubiquitination can be expected in the hub1 and hub2 mutants. To test this hypothesis, a construct containing FLAG-tagged H2B, driven by the 35S promoter, was transformed into the hub1-2 and hub2-2 mutants and their corresponding wild-types, Ler and Col. Histone-enriched protein was isolated from young seedlings, followed by immunoprecipitation with M2 agarose. Histone H2B was detected after SDS-PAGE and immunoblotting with FLAG antibodies. In yeast and human, monoubiquitination of H2B can be detected as a slower migrating form (Hwang et al., 2003; Kim et al., 2005). We observed this slower migrating form of H2B in wild-type Ler and Col but could not detect it in the hub1-2 and hub2-2 mutants (Figure 7A), indicating that HUB1 and HUB2 are both necessary for H2B monoubiquitination. Ubiquitination of this slower migrating band was confirmed by its detection with ubiquitin antibodies (Figure 7B). The Arabidopsis genome contains at least 10 histone H2B-like genes, and the absence of ubiquitinated FLAG-tagged H2B might be specific for the H2B gene that we used for our assay (At5g22880). To exclude this possibility, we also transformed another FLAG-tagged H2B (At3g45980) into the wild type and mutants and obtained the same results as for At5g22880 (data not shown). Therefore, we assume that monoubiquitination of H2B is absent in hub1 and hub2 mutants.
Figure 7.
In Vivo Analysis of Histone H2B Monoubiquitination.
(A) The slower migrating ubiquitinated form of FLAG:H2B can only be detected in the wild type and not in the hub1-2 and hub2-2 mutants. Histone-enriched protein was isolated from seedlings of P35S:H2B:FLAG transformants and immunoprecipitated with M2 agarose. Protein gel blots were probed with FLAG antibodies. On the hub2-2 blot, a few nonspecific bands were detected (see Supplemental Figure 2 online).
(B) A similar blot as in (A) was probed with ubiquitin antibodies to confirm ubiquitination of the slower migrating band. The bottom panel shows the same blot after probing with FLAG antibodies as a loading control.
Transcription Levels of Dormancy Related Genes Are Altered in the hub1-2 Mutant
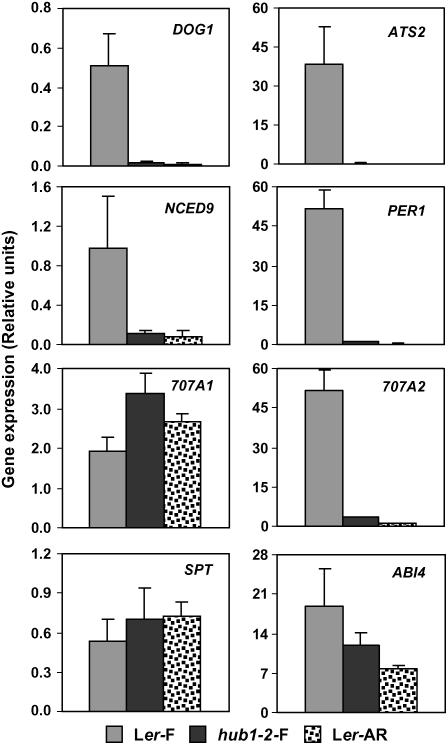
Based on its molecular function and the seed dormancy phenotypes of the mutant, we assume that HUB1 and HUB2 probably influence seed dormancy through ubiquitination of H2B, leading to changes in histone H3 methylation. These modifications likely result in changes in the expression of genes that influence seed dormancy. We analyzed the expression of several seed dormancy-related genes in seeds, imbibed for 24 h, by quantitative RT-PCR and compared freshly harvested wild-type Ler and hub1-2 mutant seeds with after-ripened nondormant Ler seeds. The genes DOG1, ATS2, NCED9, PER1, CYP707A1, CYP707A2, SPT, and ABI4 were selected for this purpose. DOG1 encodes a protein with unknown function that is essential for dormancy (Bentsink et al., 2006); ATS2 encodes a caleosin-like protein (Toorop et al., 2005); NCED9 is required for ABA biosynthesis in seeds (Lefebvre et al., 2006); PER1 has similarity to the peroxiredoxin family of antioxidants (Haslekås et al., 1998); CYP707A1 and CYP707A2 encode ABA 8′-hydroxylases, responsible for the regulation of ABA levels (Okamoto et al., 2006); SPT is a basic helix-loop-helix transcription factor that represses seed germination and mediates the germination response to temperature (Penfield et al., 2005); and ABI4 encodes an APETALA2 domain protein (Finkelstein et al., 1998). The expression of DOG1, ATS2, NCED9, PER1, and CYP707A2 in freshly harvested nondormant hub1-2 seeds is much lower than in freshly harvested dormant wild-type Ler seeds and comparable to after-ripened nondormant Ler (Figure 8). The same pattern was observed for ABI4, although its expression in hub1-2 fresh seeds and after-ripened wild-type seeds was only slightly reduced compared with fresh wild-type seeds. The genes CYP707A1 and SPT did not show clear expression differences between the different samples, apart from a slightly higher CYP707A1 expression in hub1-2 fresh seeds (Figure 8). Overall, our data show a strong similarity in gene expression of freshly harvested hub1-2 mutant seeds with after-ripened nondormant wild-type seeds, indicating a role for HUB1 in mediating aspects of seed dormancy that can be overcome in the wild type by after-ripening.
Figure 8.
hub1-2 Influences the Expression of Seed Dormancy-Related Genes.
Transcript levels of DOG1, ATS2, NCED9, PER1, CYP707A1, CYP707A2, SPT, and ABI4 were determined by quantitative RT-PCR. cDNA was generated from 24-h-imbibed freshly harvested seeds from wild-type Ler (Ler-F) and hub1-2 (hub1-2-F) or from after-ripened seeds from Ler (Ler-AR). The expression values of the individual genes were normalized using the expression level of ACTIN2 as an internal standard. The mean expression values and se values were calculated from the results of three independent experiments.
DISCUSSION
The importance of chromatin remodeling in a wide range of processes has become increasingly clear during the last decade. In seed development, for instance, chromatin remodeling is involved in the imprinting process (Gehring et al., 2004). A well-studied chromatin remodeling factor that is expressed in seeds is PICKLE (PKL). PKL represses the expression of embryonic traits during germination (Li et al., 2005) but does not influence seed dormancy (W.J.J. Soppe, unpublished data). Despite the lack of a dormancy phenotype for pkl, chromatin remodeling is likely to play a role in dormancy because changes in DNA methylation and histone H3 and H4 acetylation were observed during release of dormancy in potato tubers (Law and Suttle, 2002, 2004). HUB1 is an example of a gene influencing dormancy levels that is directly involved in chromatin remodeling. The hub1-2 (rdo4) mutant, which was originally isolated on the basis of reduced seed dormancy, has a mutation in a gene that encodes a C3HC4 RING finger protein, which is required for monoubiquitination of H2B.
The Function of HUB1 Is Not Restricted to Seed Dormancy
In addition to its dormancy phenotype, the hub1-2 mutant showed several pleiotropic phenotypes in the adult plant, specifically alterations in leaf color, plant architecture, and flower morphology, as well as a defect in seedling establishment (Figure 1). This list of pleiotropic phenotypes is probably not saturated, and careful examination is likely to yield additional phenotypes, such as the leaf shape modifications described by Fleury et al. (2007). The identification of HUB1 as an E3 ligase, required for H2B monoubiquitination explains these pleiotropic phenotypes. Ubiquitination of H2B is a fundamental process that is very likely to affect the expression of many different genes involved in various cellular processes.
HUB1 and HUB2 Are Both Required for Monoubiquitination of H2B
Our results strongly support a role for HUB1 and HUB2 as E3 ligases responsible for monoubiquitination of H2B. The phenotype of the double mutant hub1-3 hub2-2 is very similar to that of the single mutants, apart from a slightly higher germination of fresh seeds (Figure 4C). Although this could be caused by weak redundancy, a likely cause is leakiness of the hub2-2 mutant because a part of the gene was still expressed in the mutant (Figure 4B). In addition, hub1-3 has a higher dormancy level than all the other hub1 mutant alleles, and the double mutant of hub2-2 with hub1-5 does not show a stronger dormancy phenotype than the single mutants. An explanation for the absence of an enhanced phenotype in the double mutant compared with the single mutants could be that HUB1 and HUB2 function in a similar way as their human homologs, RNF20 and RNF40 (Hu_Bre1A and Hu_Bre1B). There are indications that these proteins function as a tetramer, with two copies of each polypeptide (Zhu et al., 2005). If HUB1 and HUB2 would also form such a complex, the absence of a single protein would destroy the tetramer and result in a similar phenotype as absence of both proteins. This would also explain our failure to detect ubiquitination activity of HUB1 in an in vitro assay (data not shown). Analysis of the HUB1 protein complex should confirm this hypothesis.
We could not detect any H2B ubiquitination in the hub1-2 and hub2-2 mutants with our FLAG-tagged H2B assay. Despite this dramatic reduction in H2B monoubiquitination, the mutant plants only showed mild phenotypic defects, suggesting a relatively minor influence on gene regulation. The Bre1 mutant in yeast is also viable, with its main defect being an increase in cell size (Hwang et al., 2003). However, Drosophila Bre1 mutants are lethal (Bray et al., 2005). In general, plants are more tolerant and can often survive mutations in chromatin modifying factors that are embryo lethal for animals (Li et al., 2002).
How Does HUB1 Affect Seed Dormancy?
Among the various pleiotropic phenotypes of the hub1-2 mutant, the effect on seed dormancy is relatively strong. This implies that histone H2B monoubiquitination plays an important role in the induction and/or maintenance of dormancy levels. In yeast, histone H2B monoubiquitination is correlated with gene transcription (Sun and Allis, 2002; Henry et al., 2003), and in human cells, the ubiquitination complex colocalizes with transcriptionally active genes (Kim et al., 2005; Zhu et al., 2005). H2B monoubiquitination probably influences gene transcription indirectly through an increase in methylation levels at histone H3 Lys-4 and -79 (Sun and Allis, 2002; Wood et al., 2003). A reduction in expression of specific genes can be expected in the hub1-2 mutant, causing reduced seed dormancy. In agreement with this, we found reduced expression of several dormancy-related genes, including DOG1, ATS2, NCED9, PER1, and CYP707A2 (Figure 8). However, these genes could also be indirectly regulated and act downstream of the primary targets of histone H2B monoubiquitination.
Although we could not detect DOG1 expression in freshly harvested seeds of the hub1-2 mutant, our genetic analysis showed that HUB1 is not epistatic to DOG1 but has an additive effect. The main function of DOG1 probably occurs during seed maturation (Bentsink et al., 2006), and it is possible that the gene is still expressed at this moment in the hub1-2 mutant. The dog1 mutant is completely nondormant, independent from the genetic background, whereas hub1-2 in combination with the DOG1-Cvi allele still exhibits low levels of dormancy (Figure 2). These data suggest that, although HUB1 acts upstream of DOG1, it is probably not the only factor controlling DOG1 expression. Genetic analysis indicated that HUB1 functions in the same pathway as the DOG3 locus. The gene, responsible for the DOG3 QTL, has not yet been identified. Future work should reveal at which stage during seed maturation, storage, and imbibition HUB1 acts on dormancy.
Many endogenous and environmental factors, such as hormones, seed coat, temperature, and light, take part in dormancy induction and release. With the identification of HUB1, we demonstrated the involvement of chromatin remodeling in the seed dormancy mechanism. The relation between histone ubiquitination and the factors that influence seed dormancy is still unclear but could be clarified by the identification of genes that are direct targets of HUB1. Two other important objectives are to identify additional chromatin remodeling factors that influence seed dormancy and to determine whether chromatin remodeling has a supportive or major role in the induction and release of seed dormancy.
METHODS
Plant Material and Growth Conditions
Identification of the hub1-2 (rdo4) mutant in the Ler background was described by Peeters et al. (2002). For mapping, a NIL, LCN2-17, containing a Cvi introgression fragment in the bottom of chromosome 2 (Keurentjes et al., 2006) was used. Seeds were sown on soil and grown in the greenhouse under photoperiodic cycles of 16 h of light and 8 h of dark at 20°C (day temperature) and 18°C (night temperature). Seeds grown on half-strength MS medium were first sterilized with 30% (v/v) bleach and 0.01% (v/v) SDS. Double mutants were generated using standard procedures, and the double mutant hub1-2 DOG1-Cvi was provided by L. Bentsink. Germination tests were done as described (Alonso-Blanco et al., 2003). Genotypes that were compared were grown together, harvested, and stored in identical ways, and germination assays were done in the same conditions at the same time. Chlorophyll content index was measured using a handheld chlorophyll meter (CCM-200; OPTI-Sciences).
Seed Longevity Measurement
Seed longevity was determined as germination viability after a controlled deterioration test. The controlled deterioration test was performed as follows: half-year-old seeds were equilibrated at 85% relative humidity and 37°C in the dark for 0, 3, 6, 9, and 12 d and then dried back at 32% relative humidity and 20°C for 3 d. The germination was tested on moist filter paper at 25°C and a 12-h-dark/12-h-light cycle by visually inspecting root tip emergence after 7 d.
Fine Mapping and Identification of HUB1
The genomic DNA of F2 plants was isolated using the Qiagen MagAttract 96 DNA plant core kit. For fine mapping, plants were genotyped with simple sequence length polymorphism, cleaved-amplified polymorphic sequence, and single-strand conformation polymorphism (SSCP) markers. The new markers were designed based on the Monsanto Arabidopsis Polymorphism database. The primer sequences of the markers adjacent to HUB1 are as follows: T13E15-R5 (SSCP), forward 5′-CAAGAAACCACGAAAAGGTTTCAC-3′ and reverse 5′-ATTAGGATGTACCCGGGGAGTG-3′; T14p1-R1 (SSCP), forward 5′-AGATATTGATATTGCGGCTCGTG-3′ and reverse 5′-CCAAATTTGCTCATGTCTTCCAC-3′.
For sequencing, 1-kb DNA fragments were amplified by PCR using 200 ng of genomic DNA isolated from wild-type plants and the hub1-2 mutant as templates. The primers were designed by Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3) based on the Col-0 sequence. DNA sequences were determined by the MPIZ DNA core facility on Applied Biosystems Abi Prism 377, 3100, and 3730 sequencers using BigDye terminator v3.1 chemistry. Premixed reagents were from Applied Biosystems. Oligonucleotides were purchased from Invitrogen and Operon.
Sequence similarity/homology analysis was performed using Blast2-WU and Blast2-NCBI (www.ebi.ac.uk). Multiple sequence alignment was performed by ClustalW (see Supplemental Figure 1 online). Phylogenetic analysis was performed by PHYLIP3.66.
Constructs and Plant Transformation
Total RNA was isolated from Ler young leaves using the Qiagen RNeasy kit. cDNAs were generated by SuperScript II reverse transcriptase (Invitrogen). 35S:HUB1 was constructed by inserting HUB1 cDNA into the pLEELA vector, which is a derivative of pJawohl3-RNAi (GenBank accession number AF404854) containing a GATEWAY cassette introduced into the HpaI site. For the genomic complementation, a 9-kb Ler genomic DNA fragment containing HUB1 was amplified by the expand long-template PCR system (Roche) and cloned into the XmaI site of the pBAR-A vector (GenBank accession number AJ251013). hub1-2 plants were transformed by Agrobacterium tumefaciens strain GV3101 pm90RK or GV3101 using the floral dip method (Clough and Bent, 1998). Transformants were selected based on their ability to survive after being sprayed twice with 150 mg/L BASTA. The 3:1 segregating transformant lines were selected on MS medium with 5 μg/mL dl-phosphinothricin. T3 homozygous transgenic plants were used for phenotypic analyses. All of the constructs used in this study were confirmed by sequencing.
Expression and Localization Studies
Expression was analyzed by RT-PCR with total RNA isolated from various tissues from 6-week-old plants. RT-PCR was performed with 25 amplification cycles for ACTIN2 and 30 cycles for HUB1 and HUB2 with the following gene-specific primers: ACTIN2, forward 5′-GTATGGTGAAGGCTGGATTTGC-3′ and reverse 5′-TGAGGTAATCAGTAAGGTCACGTCC-3′; HUB1, forward 5′-GGGATCTGCAAGACATGGAAAC-3′ and reverse 5′-TAGAACCCCAGAGAGGGACTGG-3′; and HUB2, forward 5′-GGTTTTGGAACTTAAGGAGGG-3′ and reverse 5′-AACTTCTTCCTGGAGCCTCAC-3′.
The PHUB1:GUS construct was made by insertion of the HUB1 promoter (−2431 to −202 relative to ATG of HUB1) into the pGWB3 vector (a gift from T. Nakagawa) via Gateway technology (Invitrogen). Transgenic plants were selected on MS medium with 50 μg/mL kanamycin and 25 μg/mL hygromycin. Homozygous T3 populations from 3:1 segregating T2 lines were selected for GUS assay (Kroj et al., 2003). The HUB1 full-length cDNA was cloned into pENSG-YFP (Jakoby et al., 2006) to generate an N-terminal YFP:HUB1 fusion protein for the localization study. Confocal microscopy (Leica TCS SP2) was used to detect YFP signal in T2 plants grown on MS medium.
HUB2 sublocalization was studied by coexpressing pENSG-YFP:RDL4 and pENSG-CFP:H2B in Nicotiana benthamiana as described (Wu et al., 2004).
For the expression analysis of dormancy-related genes, total RNA was isolated from 24-h imbibed seeds using the RNAqueous kit with plant RNA isolation aid (Ambion) and purified with the Qiagen RNeasy mini kit. cDNA was synthesized using a QuantiTect reverse transcription kit (Qiagen). Quantitative PCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen) and run on the Mastercycler ep realplex (Eppendorf) according to the manufacturer's instructions. ACTIN2 was used as an internal standard to normalize the data. We used the following primers: ACTIN2, forward 5′-CTCTCCTTGTACGCCAGTGGTC-3′ and reverse 5′-TAAGGTCACGTCCAGCAAGGTC-3′; DOG1, forward 5′-TAGGCTCGTTTATGCTTTGTGTGG-3′ and reverse 5′-CGCACTTAAGTCGCTAAGTGATGC-3′; ABI4, forward 5′-GCTTCCCAACATCAACACAACC-3′ and reverse 5′-TTGAGCGGAGGAAGTTGATGAG-3′; CYP707A1, forward 5′-TCCATCGCTCAAGACTCTCTCC-3′ and reverse 5′-ACCTCGTCTTTTCCGAAGATCG-3′; CYP707A2, forward 5′-CAATTCCTTCTTCGCCACTCG-3′ and reverse 5′-GCCTCTGGTCCAATCATACGC-3′; SPT, forward 5′-GGAGCTAGTGGCAACGAGACAG-3′ and reverse 5′-TGAACTTCAGCAGCTCTGCATC-3′; AtNCED9, forward 5′-ATCGACCGGAGAGATTCGAAAG-3′ and reverse 5′-TCACCTTCTCCTCGTCGTGAAC-3′; PER1, forward 5′-ACGGTGCCGAACCTAGAAGTG-3′ and reverse 5′-GTATTTGGCCATCGCACCAAG-3′; and ATS2, forward 5′-TTACTCGCGTCGCTTATCTTGG-3′ and reverse 5′-TTAGAGTCGCTTCCGTGCTTTG-3′. The specificity of the amplifications was verified by analysis of the PCR products on agarose gels and by melting curve analysis. The efficiency of the amplifications was confirmed by the analysis of standard curves and ranged from 0.95 to 1.08.
Screening of T-DNA Insertion Lines and Isolation of Additional hub1 and hub2 Alleles
T-DNA insertions lines for HUB1 (At2g44950) and HUB2 (At1g55250 + At1g55255 = F7A10.17) were obtained from the SALK collection (hub1-4, hub1-5, and hub2-2) or GABI-Kat collection (hub1-3 and hub2-1; generated in the context of the GABI-Kat program and provided by B. Weisshaar) with the following seed stock numbers: hub1-3, 276D08 GABI; hub1-4, salk_122512; hub1-5, salk_044415; hub2-1, 634H04 GABI; and hub2-2, salk_071289. PCR-based screening was used to identify individuals homozygous for T-DNA insertions in the HUB1 and HUB2 genes. The gene-specific primers designed by the SIGnAL T-DNA verification primer design program were used in combination with T-DNA left border primers. RT-PCR with RNA isolated from leaves was performed to confirm the homozygous knockout lines. PCR was performed with 25 cycles for ACTIN2 and 40 cycles for HUB1 and HUB2 with the following gene-specific primers: for HUB1, P1 5′-GCGGTCAGCTAGCTCTGAGTG-3′, P2 5′-CGTCTTTCGAGAAACATCACC-3′, P3 5′-TCAGCTTTTCTTGGAAGGCATAAC-3′, and P4 5′-TTGGTGCGGTCATATGTAGATAGG-3′; and for HUB2, R1 5′-ATGCTAACAAAGGCAGACGAACAG-3′ and R2 5′-TTCGAGGCTGATAACGAGGTGACG-3′.
In Vivo Ubiquitination Assay
FLAG:H2B (At5g22880; Boisnard-Lorig et al., 2001) was transformed into wild-type Ler and Col and the hub1-2 and hub2-2 mutants. Histone protein was extracted from 10-d-old T2 seedlings as described (Tariq et al., 2003). The pellet was washed twice with acetone and resuspended in 1 mL of immunoprecipitation lysis buffer (FLAG-tagged protein immunoprecipitation kit; Sigma-Aldrich). The histone solution was supplemented with 20 μL (bead volume) of M2 agarose. After a 3-h incubation at 4°C, beads were washed three times with wash buffer, and precipitated proteins were recovered by boiling with SDS sample loading buffer and analyzed by immunoblots using anti-FLAG-HRP (Sigma-Aldrich). For detection of monoubiquitinated histone H2B, the blot was autoclaved for 30 min (Swerdlow et al., 1986), followed by analysis with ubiquitin antibody (Abcam). Protein gel blots were developed by the chemiluminescent SuperSignal system (Pierce), and results were visualized and quantified in a Lumi-Imager detector (Boehringer Manheim).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AAL91211 (At_HUB1), AAG51572 (At_HUB2), ABE92765 (Mt_Bre1), XP_473416 (Os_Bre1A), ABB47997 (Os_Bre1B), CAA98640 (Sc_Bre1), NP_587845 (Sp_Bre1A), CAA22646 (Sp_Bre1B), AAK21443 (Ce_Bre1), AAF50744 (Dm_Bre1), BAB14005 (Hu_Bre1A), and AAH18647 (Hu_Bre1B).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment Used for PHYLIP3.66 Phylogenetic Analysis.
Supplemental Figure 2. Detection of Nonspecific Bands in Col Using FLAG Antibodies.
Supplementary Material
Acknowledgments
We thank Andreas Bachmair for providing the triple mutant ubc1 ubc2 ubc3, Joost Keurentjes for the NIL LCN2-17, Leónie Bentsink for the double mutant hub1-2 DOG1-Cvi, and Tsuyoshi Nakagawa for the pGWB3 vector. We also thank Elmon Schmelzer, Amir Sattarzadeh, and Rainer Franzen for help with confocal microscopy and Sigi Effgen, Christina Philipp, Regina Gentges, Mariana Harperscheidt, and Alexandra Kalde for the great technical support. Finally, we thank Kazumi Nakabayashi for discussions and Rebecca Silady for critical reading of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Wim J.J. Soppe (soppe@mpiz-koeln.mpg.de).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso-Blanco, C., Bentsink, L., Hanhart, C.J., Blankestijn-de Vries, H., and Koornneef, M. (2003). Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164 711–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink, L., Jowett, J., Hanhart, C.J., and Koornneef, M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink, L., Soppe, W., and Koornneef, M. (2007). Genetic aspects of seed dormancy. In Seed Development, Dormancy and Germination. K. Bradford and H. Nonogaki, eds (Oxford, UK: Blackwell Publishing), pp 113–132.
- Berná, G., Robles, P., and Micol, J.L. (1999). A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley, J.D. (1997). Seed germination and dormancy. Plant Cell 9 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisnard-Lorig, C., Colon-Carmona, A., Bauch, M., Hodge, S., Doerner, P., Bancharel, E., Dumas, C., Haseloff, J., and Berger, F. (2001). Dynamic analyses of the expression of the HISTONE∷YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, S., Musisi, H., and Bienz, M. (2005). Bre1 is required for notch signaling and histone modification. Dev. Cell 8 279–286. [DOI] [PubMed] [Google Scholar]
- Cadman, C.S.C., Toorop, P.E., Hilhorst, H.W.M., and Finch-Savage, W.E. (2006). Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 46 805–822. [DOI] [PubMed] [Google Scholar]
- Chibani, K., Ali-Rachedi, S., Job, C., Job, D., Jullien, M., and Grappin, P. (2006). Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiol. 142 1493–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cutler, S., Ghassemian, M., Bonetta, D., Cooney, S., and McCourt, P. (1996). A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273 1239–1241. [DOI] [PubMed] [Google Scholar]
- Debeaujon, I., Léon-Kloosterziel, K.M., and Koornneef, M. (2000). Influence of the testa on seed dormancy, germination and longevity in Arabidopsis thaliana. Plant Physiol. 122 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond, P.J., and Graham, I.A. (2001). Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6 72–77. [DOI] [PubMed] [Google Scholar]
- Finch-Savage, W.E., and Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytol. 171 501–523. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S., and Goodman, H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes and APETALA2 domain protein. Plant Cell 10 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury, D., et al. (2007). The Arabidopsis thaliana ortholog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, M., Choi, Y., and Fischer, R.L. (2004). Imprinting and seed development. Plant Cell 16 (suppl.): S203–S213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberti, G., Papi, M., Bellucci, L., Ricci, L., Bouchez, D., Camilleri, C., Costantino, P., and Vittorioso, P. (2002). Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Millar, A., and Jacobsen, J.V. (2005). Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 8 183–187. [DOI] [PubMed] [Google Scholar]
- Haslekås, C., Stacy, R.A.P., Nygaard, V., Culiáñez-Macià, F.A., and Aalen, R.B. (1998). The expression of a peroxiredoxin antioxidant gene, AtPer1, in Arabidopsis thaliana is seed-specific and related to dormancy. Plant Mol. Biol. 36 833–845. [DOI] [PubMed] [Google Scholar]
- Henry, K.W., Wyce, A., Lo, W.-S., Duggan, L.J., Emre, N.C.T., Kao, C.-F., Pillus, L., Shilatifard, A., Osley, M.A., and Berger, S.L. (2003). Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17 2648–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, W.W., Venkatasubrahmanyam, S., Ianculescu, A.G., Tong, A., Boone, C., and Madhani, H.D. (2003). A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11 261–266. [DOI] [PubMed] [Google Scholar]
- Jakoby, M.J., Weinl, C., Pusch, S., Kuijt, S.J.H., Merkle, T., Dissmeyer, N., and Schnittger, A. (2006). Analysis of the subcellular localization, function, and proteolytic control of the Arabidopsis cyclin-dependent kinase inhibitor ICK1/KRP1. Plant Physiol. 141 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes, J.J., Bentsink, L., Alonso-Blanco, C., Hanhart, C.J., Blankestijn-De Vries, H., Effgen, S., Vreugdenhil, D., and Koornneef, M. (December 18, 2006). Development of a Near Isogenic Line population of Arabidopsis thaliana and comparison of mapping power with a Recombinant Inbred Line population. Genetics http://dx.doi.org/10.1534/genetics.106.066423. [DOI] [PMC free article] [PubMed]
- Kim, J., Hake, S.B., and Roeder, R.G. (2005). The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell 20 759–770. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Jorna, M.L., Brinkhorst-van der Swan, D.L.C., and Karssen, C.M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in nongerminating gibberellins sensitive lines of Arabidopsis thaliana (L.). Heynh. Theor. Appl. Genet. 61 385–393. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Bentsink, L., and Hilhorst, H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5 33–36. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Reuling, G., and Karssen, C. (1984). The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant 61 377–383. [Google Scholar]
- Kraft, E., Stone, S.L., Ma, L., Su, N., Gao, Y., Lau, O.-S., Deng, X.-W., and Callis, J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139 1597–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroj, T., Savino, G., Valon, C., Giraudat, J., and Parcy, F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130 6065–6073. [DOI] [PubMed] [Google Scholar]
- Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S., Asami, T., Hirai, N., Koshiba, T., Kamiya, Y., and Nambara, E. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 23 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, R.D., and Suttle, J.C. (2002). Transient decreases in methylation at 5′-CCGG-3′ sequences in potato (Solanum tuberosum L.) meristem during progression of tubers through dormancy precede the resumption of sprout growth. Plant Mol. Biol. 51 437–447. [DOI] [PubMed] [Google Scholar]
- Law, R.D., and Suttle, J.C. (2004). Changes in histone H3 and H4 multi-acetylation during natural and forced dormancy break in potato tubers. Physiol. Plant 120 642–649. [DOI] [PubMed] [Google Scholar]
- Lee, C.S., Chien, C.T., Lin, C.H., Chiu, Y.Y., and Yang, Y.S. (2006). Protein changes between dormant and dormancy-broken seeds of Prunus campanulata Maxim. Proteomics 6 4147–4154. [DOI] [PubMed] [Google Scholar]
- Lefebvre, V., North, H., Frey, A., Sotta, B., Seo, M., Okamoto, M., Nambara, E., and Marion-Poll, A. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45 309–319. [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel, K.M., Gil, M.A., Ruijs, G.J., Jacobsen, S.E., Olszewski, N.E., Schwartz, S.H., Zeevaart, J.A., and Koornneef, M. (1996. a). Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 10 655–661. [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel, K.M., van de Bunt, G.A., Zeevaart, J.A.D., and Koornneef, M. (1996. b). Arabidopsis mutants with a reduced seed dormancy. Plant Physiol. 110 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., Hall, T.C., and Holmes-Davis, R. (2002). Plant chromatin: Development and gene control. Bioessays 24 234–243. [DOI] [PubMed] [Google Scholar]
- Li, H.-C., Chuang, K., Henderson, J.T., Rider, S.D., Jr., Bai, Y., Zhang, H., Fountain, M., Gerber, J., and Ogas, J. (2005). PICKLE acts during germination to repress expression of embryonic traits. Plant J. 44 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A., Jacobsen, J.V., Ross, J.J., Helliwell, C.A., Poole, A.T., Scofield, G., Reid, J.B., and Gubler, F. (2006). Seed dormancy and ABA metabolism in Arabidopsis and barley: The role of ABA 8′-hydroxylase. Plant J. 45 942–954. [DOI] [PubMed] [Google Scholar]
- Okamoto, M., Kuwahara, A., Seo, M., Kushiro, T., Asami, T., Hirai, N., Kamiya, Y., Koshiba, T., and Nambara, E. (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi, M., Sabatini, S., Altamura, M.M., Hennig, L., Schäfer, E., Costantino, P., and Vittorioso, P. (2002). Inactivation of the phloem-specific Dof zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiol. 128 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi, M., Sabatini, S., Bouchez, D., Camilleri, C., Costantino, P., and Vittorioso, P. (2000). Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev. 14 28–33. [PMC free article] [PubMed] [Google Scholar]
- Peeters, A.J.M., Blankestijn de Vries, H., Hanhart, C.J., Léon-Kloosterziel, K.M., Zeevaart, J.A.D., and Koornneef, M. (2002). Characterization of mutants with reduced seed dormancy at two novel rdo loci and a further characterization of rdo1 and rdo2 in Arabidopsis. Physiol. Plant 115 604–612. [DOI] [PubMed] [Google Scholar]
- Penfield, S., Josse, E.-M., Kannangara, R., Gilday, A.D., Halliday, K.J., and Graham, I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15 1998–2006. [DOI] [PubMed] [Google Scholar]
- Putterill, J., Laurie, R., and Macknight, R. (2004). It's time to flower: The genetic control of flowering time. Bioessays 26 363–373. [DOI] [PubMed] [Google Scholar]
- Robzyk, K., Recht, J., and Osley, M.A. (2000). Rad6-dependent ubiquitination of histone H2B in yeast. Science 287 501–504. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Saito, S., Hirai, N., Matsumoto, C., Ohigashi, H., Ohta, D., Sakata, K., and Mizutani, M. (2004). Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134 1439–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L., Hauksdóttir, H., Troy, A., Herschleb, J., Kraft, E., and Callis, J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z.-W., and Allis, C.D. (2002). Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418 104–108. [DOI] [PubMed] [Google Scholar]
- Swerdlow, P.S., Finley, D., and Varshavsky, A. (1986). Enhancement of immunoblot sensitivity by heating of hydrated filters. Anal. Biochem. 156 147–153. [DOI] [PubMed] [Google Scholar]
- Tariq, M., Saze, H., Probst, A.V., Lichota, J., Habu, Y., and Paszkowski, J. (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100 8823–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toorop, P.E., Barroco, R.M., Engler, G., Groot, S.P.C., and Hilhorst, H.W.M. (2005). Differentially expressed genes associated with dormancy or germination of Arabidopsis thaliana seeds. Planta 221 637–647. [DOI] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.G., Wang, S.C., and Jones, A.M. (2002). Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol. 129 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Der Schaar, W., Alonso-Blanco, C., Léon-Kloosterziel, K., Jansen, R.C., van Ooijen, J.W., and Koornneef, M. (1997). QTL analysis of seed dormancy in Arabidopsis using recombinant inbred lines and MQM mapping. Heredity 79 190–200. [DOI] [PubMed] [Google Scholar]
- Wood, A., Krogan, N.J., Dover, J., Schneider, J., Heidt, J., Boateng, M.A., Dean, K., Golshani, A., Zhang, Y., Greenblatt, J.F., Johnston, M., and Shilatifard, A. (2003). Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11 267–274. [DOI] [PubMed] [Google Scholar]
- Wu, A.-J., Andriotis, V.M.E., Durrant, M.C., and Rathjen, J.P. (2004). A patch of surface-exposed residues mediates negative regulation of immune signaling by tomato Pto kinase. Plant Cell 16 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, B., Zheng, Y., Pham, A.D., Mandal, S.S., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. (2005). Monoubiquitination of human histone H2B: T1he factors involved and their roles in HOX gene regulation. Mol. Cell 20 601–611. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.