Abstract
AvrBsT is a type III effector from Xanthomonas campestris pv vesicatoria that is translocated into plant cells during infection. AvrBsT is predicted to encode a Cys protease that targets intracellular host proteins. To dissect AvrBsT function and recognition in Arabidopsis thaliana, 71 ecotypes were screened to identify lines that elicit an AvrBsT-dependent hypersensitive response (HR) after Xanthomonas campestris pv campestris (Xcc) infection. The HR was observed only in the Pi-0 ecotype infected with Xcc strain 8004 expressing AvrBsT. To create a robust pathosystem to study AvrBsT immunity in Arabidopsis, the foliar pathogen Pseudomonas syringae pv tomato (Pst) strain DC3000 was engineered to translocate AvrBsT into Arabidopsis by the Pseudomonas type III secretion (T3S) system. Pi-0 leaves infected with Pst DC3000 expressing a Pst T3S signal fused to AvrBsT-HA (AvrBsTHYB-HA) elicited HR and limited pathogen growth, confirming that the HR leads to defense. Resistance in Pi-0 is caused by a recessive mutation predicted to inactivate a carboxylesterase known to hydrolyze lysophospholipids and acylated proteins in eukaryotes. Transgenic Pi-0 plants expressing the wild-type Columbia allele are susceptible to Pst DC3000 AvrBsTHYB-HA infection. Furthermore, wild-type recombinant protein cleaves synthetic p-nitrophenyl ester substrates in vitro. These data indicate that the carboxylesterase inhibits AvrBsT-triggered phenotypes in Arabidopsis. Here, we present the cloning and characterization of the SUPPRESSOR OF AVRBST-ELICITED RESISTANCE1.
INTRODUCTION
Xanthomonas campestris pv vesicatoria (Xcv) is a foliar pathogen that causes bacterial spot on tomato (Solanum lycopersicum) and pepper (Capsicum annuum) plants (Jones et al., 1998). Xcv enters the leaf via wounds or natural openings created by guard cells and hydrathodes. Once in the apoplast, Xcv becomes infectious and uses the type III secretion (T3S) apparatus to secrete and translocate effector proteins into plant cells (Gurlebeck et al., 2006). Mutant bacteria deficient in T3S or lacking T3S proteins are less pathogenic or nonpathogenic. Thus, T3S and its effectors are essential virulence determinants required for Xcv colonization and persistent in host plants.
T3S effectors were initially named avirulence (Avr) proteins because bacterial pathogens that inject such proteins into resistant plant cells were unable to grow in that host and thus were classified as avirulent. However in other genetic backgrounds (i.e., susceptible hosts), the same bacteria were able to grow, indicating that the strains were fully virulent. Subsequent detailed molecular studies in resistant hosts have shown that Avr T3S effectors are recognized either directly or indirectly inside plant cells by resistance (R) proteins of the nucleotide binding site, leucine-rich repeat class (NB-LRR) (Ellis et al., 2000; Dangl and Jones, 2001). Such recognition launches a defense-signaling cascade that halts pathogen growth at early stages of pathogenesis (Nimchuk et al., 2003). Plants that lack cognate R proteins fail to recognize pathogens expressing Avr proteins. Thus, the bacteria are able to escape host detection and grow within the apoplast, eventually causing disease.
Bioinformatics and functional genetic screens have been used to identify and characterize the T3S effector proteome in the genome of Xcv 85-10, a model strain used to study bacterial spot (Roden et al., 2004a; Thieme et al., 2005; Gurlebeck et al., 2006). Although numerous effectors (>30) have been identified (Gurlebeck et al., 2006), the biochemical function of these proteins in promoting bacterial colonization within the host, resulting in either disease or resistance signaling, is poorly understood. Yet, a central theme emerging from the study of different phytopathogenic strains indicates that bacteria deliberately suppress plant immune responses to interfere with the activation of disease resistance protein–mediated pathways (reviewed in Mudgett, 2005).
We are interested in elucidating the biochemical function of the YopJ family of T3S effectors that are prevalent in Xcv strains. YopJ is a T3S effector that was originally identified in Yersinia pestis, the causal agent of bubonic plague. YopJ inhibits mitogen-activated protein kinase (MAPK) and nuclear factor κB signaling pathways involved in animal innate immunity by preventing the activation of the family of MAPK kinases (MAPKK) (Orth et al., 1999, 2000). YopJ contains a catalytic domain similar to clan CE of Cys proteases, which includes the adenoviral protease family and the ubiquitin-like protease family (Orth et al., 2000). This fact prompted the hypothesis that YopJ and possibly other members of this family may function as proteases to disrupt the posttranslational conjugation of small ubiquitin modifiers to host proteins during infection. Only indirect evidence supports this hypothesis (Orth et al., 2000). Intriguingly, recent in vitro and transfection studies have shown that YopJ functions as an acetyltransferase that blocks the activation of MAPKK6 by acetylating critical Ser and Thr residues, thereby inhibiting their phosphorylation (Mukherjee et al., 2006).
To date, four YopJ-like proteins have been characterized in different Xcv strains: AvrBsT, AvrXv4, AvrRxv, and XopJ (Whalen et al., 1988; Ciesiolka et al., 1999; Astua-Monge et al., 2000; Noel et al., 2001). Each protein contains the catalytic triad (His, Glu, and Cys) conserved in clan CE Cys proteases. In AvrBsT (Orth et al., 2000), AvrXv4 (Roden et al., 2004b), and AvrRxv (Bonshtien et al., 2005), these residues are required to induce the hypersensitive response (HR) inside resistant plant cells, suggesting that proteolysis or modification of host proteins is the initial signal that triggers immune responses. The host target(s) for these proteins is unknown, apart from AvrXv4, which displays weak small ubiquitin modifier isopeptidase activity in planta (Roden et al., 2004b).
YopJ-like effectors also exist in several other bacterial plant pathogens, including Erwinia amylovora (Oh et al., 2005), Pseudomonas syringae (Alfano et al., 2000; Arnold et al., 2001), and Ralstonia solanacearum (Deslandes et al., 2003; Lavie et al., 2004), as well as one plant symbiont (Ciesiolka et al., 1999). The maintenance of this class of effectors in diverse pathogens suggests that each may target a similar substrate or use a conserved catalytic mechanism to alter eukaryotic signal transduction. Despite such conservation, some plant hosts can recognize individual YopJ-like effectors and activate robust immune responses (e.g., Lycopersicon pennellii LA716 Xv4 plants recognize Xcv AvrXv4 [Astua-Monge et al., 2000]; tomato cv Hawaii 7998 and bean [Phaseolus vulgaris] cv Sprite Rxv plants recognize AvrRxv [Whalen et al., 1988, 1993]; and Arabidopsis thaliana ecotype Nd-1 RRS1-R plants recognize R. solanacearum PopP2 [Deslandes et al., 2002]).
Interestingly, RRS1-R and PopP2 interact physically in yeast and colocalize to the plant nucleus in transient expression assays (Deslandes et al., 2003). Furthermore, RRS1-R nuclear import is dependent on the presence of PopP2. RRS1-R is an unusual recessive R gene encoding the typical N-terminal TIR-NB-LRR domains fused to a C-terminal WRKY transcription factor domain (Deslandes et al., 2002). The mechanism by which PopP2 alters host signaling and RRS1-R abrogates its action is unclear. However, the functional domains of RRS1-R suggest that this R protein can recognize PopP2 and then directly activate defense gene induction in the nucleus.
A long-term goal of our work is to elucidate the virulence function of AvrBsT in Xcv's natural hosts, tomato and pepper. We set out to use plants that differentially respond to Xcv AvrBsT infection to reveal AvrBsT-specific resistance pathways. Unfortunately, all of the commonly used tomato cultivars we tested are susceptible, whereas all of the pepper cultivars are resistant. This precluded us from performing detailed molecular analyses using one plant species. Therefore, we used the Arabidopsis model system to initiate the molecular dissection of AvrBsT function and recognition in a nonhost plant. We examined the natural variation that exists among Arabidopsis ecotypes to identify lines resistant and susceptible to AvrBsT. In our screen, we discovered only one ecotype (Pi-0) that is resistant to bacteria expressing AvrBsT. Positional cloning was then used to identify the gene involved in AvrBsT-triggered defense responses.
Here, we report the cloning and characterization of a carboxylesterase that acts as a SUPPRESSOR OF AVRBST-ELICITED RESISTANCE1 (SOBER1) in Arabidopsis.
RESULTS
HR Screen for Arabidopsis Resistance to Xanthomonas campestris pv campestris Strain 8004 Expressing AvrBsT
We exploited the natural variation that exists among Arabidopsis ecotypes to identify a locus governing resistance to the AvrBsT type III effector from Xcv (Ciesiolka et al., 1999). The host range of Xcv is exclusive and extensive in the Solanaceae, primarily infecting tomato and pepper plants (Jones et al., 1998). Therefore, we used the Brassica bacterial pathogen Xanthomonas campestris pv campestris (Xcc) strain 8004 to infect Arabidopsis and deliver the AvrBsT protein into plant cells by the Xcc T3S system. Xcc 8004 causes mild disease symptoms on Arabidopsis plants, similar to those induced on its natural Brassica host turnip (Brassica rapa) (Parker et al., 1993). The use of Xcc 8004 enabled us to screen for Arabidopsis ecotypes that induce the HR (i.e., localized, rapid programmed cell death [Greenberg and Yao, 2004]) specifically in response to AvrBsT activity in plant cells.
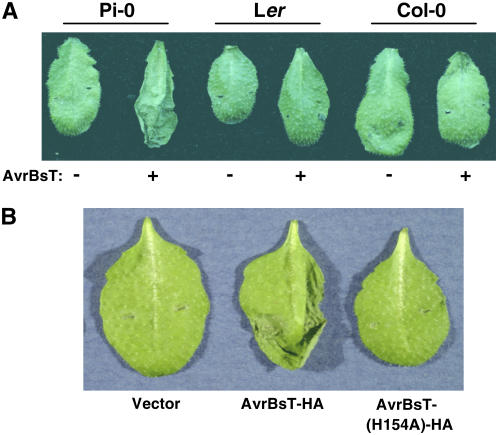
We inoculated 71 Arabidopsis ecotypes with Xcc 8004 carrying pDD62(avrBsT-HA) and a pDD62 empty vector control. Only the Pi-0 ecotype infected with Xcc 8004 expressing AvrBsT-HA elicited a strong HR at 24 h after inoculation (Figure 1A). Pi-0 plants infected with Xcc 8004 alone did not produce the HR. The Landsberg erecta (Ler) and Columbia (Col-0) ecotypes did not elicit HR symptoms in response to either strain (Figure 1A). This screen suggested that Pi-0 is resistant to Xcc expressing AvrBsT, whereas most Arabidopsis ecotypes are susceptible.
Figure 1.
AvrBsT-Dependent HR Phenotypes in Xcc 8004–Infected Arabidopsis Leaves.
(A) Xcc 8004 expressing AvrBsT-HA elicits the HR in Arabidopsis Pi-0 plants. Pi-0, Ler, and Col-0 leaves were hand-inoculated with a 3 × 108 colony-forming units (cfu)/mL suspension of Xcc 8004 carrying pDD62 and pDD62(avrBsT-HA), designated as − and + AvrBsT-HA, respectively. Symptoms were photographed at 24 h after inoculation.
(B) Pi-0 plants do not elicit a HR when infected with Xcc 8004 expressing AvrBsT(H154A)-HA. Pi-0 leaves were hand-inoculated with a 3 × 108 cfu/mL suspension of Xcc 8004 carrying pDD62, pDD62(avrBsT-HA), or pDD62(avrBsT-H154A-HA), designated as vector, AvrBsT-HA, and AvrBsT(H154A)-HA, respectively. Symptoms were photographed at 24 h after inoculation.
To determine whether the putative Cys protease activity of AvrBsT is required to elicit a HR in Pi-0 plants, we infected plants with Xcc 8004 pDD62(avrBsT-H154A-HA) expressing a mutant AvrBsT protein in which the conserved catalytic His residue was substituted with Ala. Wild-type AvrBsT, but not catalytic core mutants, elicits a HR in Nicotiana benthamiana leaves when expressed by Xcc 8004 or when transiently expressed in planta using Agrobacterium tumefaciens (Orth et al., 2000). Similarly, Pi-0 plants infected with Xcc 8004 expressing AvrBsT(H154A)-HA did not elicit a HR (Figure 1B). This finding indicates that resistant Pi-0 plants recognize either a structural component of the AvrBsT protein or the product(s) generated by AvrBsT proteolysis or modification within plant cells.
Pseudomonas–Arabidopsis Pathosystem to Study AvrBsT-Elicited Disease Resistance
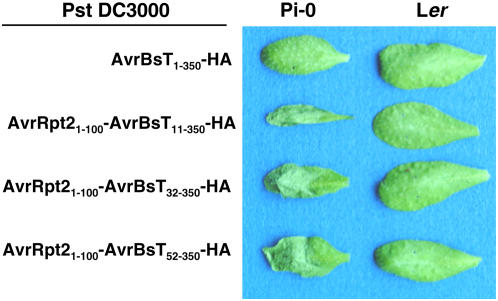
We next verified that the HR response observed in Pi-0 results in reduced bacterial growth during plant infection. Xcc 8004, however, is a vascular bacterial pathogen and grows poorly in leaves under the conditions normally used to study foliar bacterial pathogens. Thus, we used Pseudomonas syringae pv tomato (Pst) strain DC3000, a robust foliar bacterial pathogen of Arabidopsis, to study the impact of the AvrBsT-dependent HR on bacterial growth in Pi-0 plants. We engineered Pst DC3000 to express the Xanthomonas AvrBsT effector and translocate it into plant cells through the Pst DC3000 T3S system. We first tested whether or not Pst DC3000 can recognize AvrBsT and its native nonconserved T3S signal sequence. Pst DC3000 expressing AvrBsT1-350-HA did not elicit a HR in Pi-0 plants (Figure 2), indicating that the AvrBsT T3S signal sequence is not recognized by the Pst T3S apparatus. To circumvent this delivery problem, we constructed N-terminal deletions of AvrBsT to remove its Xanthomonas T3S signal sequence (generally contained within the first 50 amino acids) and replaced it with a functional Pseudomonas T3S signal sequence (contained within amino acids 1 to 100) from the AvrRpt2 effector protein (Guttman and Greenberg, 2001). Agrobacterium-mediated transient expression of the AvrBsT-HA mutant proteins (AvrBsT11-350-HA, AvrBsT32-350-HA, and AvrBsT52-350-HA) in N. benthamiana did not impair HR induction (data not shown). This finding indicated that AvrBsT amino acids 52 to 350 are sufficient to trigger HR activity in planta.
Figure 2.
AvrBsT-Dependent HR Phenotypes in Pst DC3000–Infected Arabidopsis Leaves.
Pi-0 and Ler leaves were hand-inoculated with a 3 × 108 cfu/mL suspension of Pst DC3000 carrying pDD62(avrBsT-HA), pVSP61(avrRpt21-100-avrBsT11-350-HA), pVSP61(avrRpt21-100-avrBsT32-350-HA), or pVSP61(avrRpt21-100-avrBsT52-350-HA), designated as AvrBsT1-350-HA, AvrRpt21-100-AvrBsT11-350-HA, AvrRpt21-100-AvrBsT32-350-HA, and AvrRpt21-100-AvrBsT52-350-HA, respectively. Labeling refers to amino acid number in the respective protein. Symptoms were photographed at 12 h after inoculation.
We next determined whether Pst DC3000 expressing AvrBsT mutant proteins (AvrBsT11-350-HA, AvrBsT32-350-HA, and AvrBsT52-350-HA) or the corresponding AvrRpt2-AvrBsT fusion proteins (AvrRpt21-100-AvrBsT11-350-HA, AvrRpt21-100-AvrBsT32-350-HA, and AvrRpt21-100-AvrBsT52-350-HA) could elicit the HR in Arabidopsis Pi-0 leaves. As expected, none of the Pst DC3000 strains expressing the N-terminal AvrBsT-HA deletion mutants elicited a HR in Pi-0 leaves (data not shown). However, all of the Pst DC3000 strains expressing the AvrRpt2-AvrBsT-HA hybrid proteins induced a strong AvrBsT-dependent HR in Pi-0 leaves and no response in Ler leaves (Figure 2). Pi-0 leaves infected with Pst DC3000 expressing AvrRpt21-100-AvrBsT32-350-(H154A)-HA, a hybrid protein containing a mutation in the catalytic core, did not trigger HR (see Supplemental Figure 1 online), indicating that the Pi-0 HR is triggered by the putative enzymatic activity of AvrBsT and not by an artificial activity elicited by the AvrRpt21-100 fusion protein. Overall, these data demonstrate that the AvrRpt2 T3S signal sequence can target AvrBsT's C-terminal effector domain into Arabidopsis cells via the Pst DC3000 T3S apparatus and that the hybrid proteins maintain AvrBsT-dependent HR specificity.
Arabidopsis Pi-0 Plants Reduce Pathogen Growth and Express PR1 in Response to AvrBsT
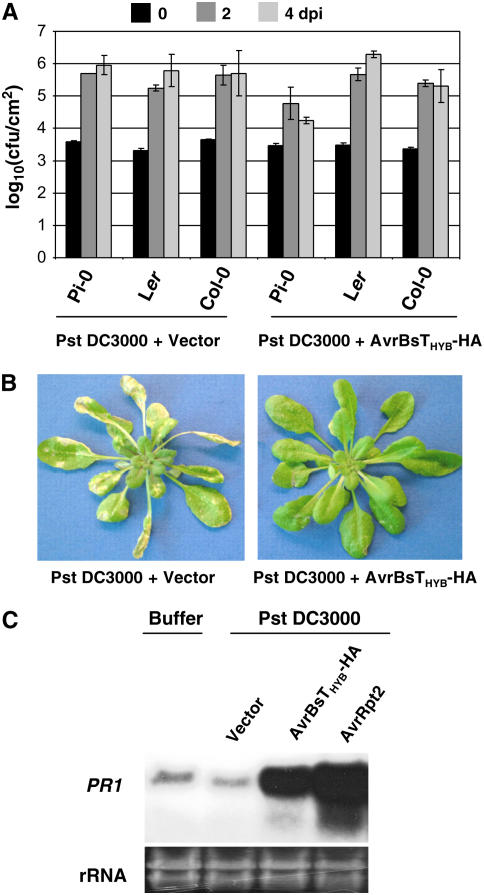
We next measured the growth of Pst DC3000 expressing AvrRpt2-AvrBsT11-350-HA (hereafter referred to as AvrBsTHyb-HA) in infected Pi-0, Ler, and Col-0 plants. Arabidopsis leaves were hand-inoculated with Pst DC3000 strains, and bacterial growth in planta was quantified by constructing a bacterial growth curve (Mudgett and Staskawicz, 1999). Pst DC3000 was able to grow to high levels (∼105 to 106 cfu/cm2) in Col-0, Pi-0, and Ler plants (Figure 3A). However, the growth of Pst DC3000 expressing AvrBsTHyb-HA was restricted in Pi-0 plants (∼104 cfu/cm2) and unaffected in Ler and Col-0 plants (Figure 3A). Pi-0 leaves infected with Pst DC3000 carrying the empty vector for 4 to 5 d exhibited severe chlorosis and necrotic lesions, whereas leaves infected with Pst DC3000 expressing AvrBsTHyb-HA for 4 to 5 d were nearly free of disease symptoms (Figure 3B).
Figure 3.
Pi-0 Plants Recognize AvrBsT and Restrict Pst DC3000 Growth.
(A) Growth of Pst DC3000 strains in Pi-0, Ler, and Col-0. Leaves were hand-inoculated with a 105 cfu/mL suspension of bacteria carrying pVSP61 or pVSP61(avrRpt21-100-avrBsT11-350-HA), designated as Pst DC3000 + Vector and Pst DC3000 + AvrRpt2-AvrBsT11-350-HA, respectively. Bacteria present in leaves were monitored at 0 d (black bars), 2 d (dark gray bars), and 4 d (light gray bars) after inoculation (dpi). Data points represent the mean log10 (cfu/cm) ± sample sd for four replicates.
(B) Pi-0 plants are susceptible to Pst DC3000 and resistant to Pst DC3000 expressing AvrRpt2-AvrBsT11-350-HA. Pi-0 plants were dipped in a 2 × 108 cfu/mL suspension of Pst DC3000 carrying pVSP61 (left panel) or pVSP61(avrRpt21-100-avrBsT11-350-HA) (right panel). Symptoms were photographed at 5 d after inoculation.
(C) RNA gel blot showing PR1 mRNA expression in Pst DC3000–infected Pi-0 plants. Leaves were hand-inoculated with 1 mM MgCl2 (buffer) or a 5 × 107 cfu/mL suspension of Pst DC3000 carrying pVSP61, pVSP61(avrRpt21-100-avrBsT11-350-HA), or pVSP61(avrRpt2), designated as Vector, AvrRpt2-AvrBsT11-350-HA, and AvrRpt2, respectively. Total RNA was isolated at 8 h after inoculation. The blot was hybridized to a probe specific for the PR1 gene. Twenty micrograms of total RNA was loaded in each lane. Ethidium bromide–stained rRNA present in each lane served as a loading control.
We also verified that expression of the PR1 defense gene was induced in Pi-0 in response to Pst DC3000 AvrBsTHYB-HA infection. Pi-0 leaves were infiltrated with MgCl2 buffer or Pst DC3000–expressing empty vector, AvrBsTHyb-HA or AvrRpt2, for 8 h and then total RNA was extracted. AvrRpt2 activates RPS2 disease resistance (Bent et al., 1994; Mindrinos et al., 1994), a pathway that is operational in Pi-0 (data not shown). As expected, PR1 mRNA was highly expressed in the Pst DC3000 AvrRpt2-infected leaves relative to the basal level present in the buffer and Pst DC3000 vector controls (Figure 3C). Pst DC3000 AvrBsTHyb-HA infection also induced high levels of PR1 gene expression (Figure 3C). Collectively, these data reveal that AvrBsT recognition in planta results in defense gene expression that limits pathogen growth.
AvrBsT-Elicited Resistance in Pi-0 Is Monogenic and Recessive
To determine the genetic basis of immunity, resistant Pi-0 was crossed to susceptible Ler and the segregation of AvrBsT-triggered HR was scored in the F1 progeny. None of the F1 progeny produced HR, indicating that disease resistance in Pi-0 was not dominant. F1 plants were allowed to self-pollinate, and then the segregation of HR was monitored in the F2 generation: 151 of 625 plants showed HR. This closely approximates a 3:1 ratio (χ2 = 0.235; P > 0.5), indicating that the HR phenotype was caused by a recessive mutation at a single locus. We initially expected that Pi-0 might express a novel R protein (which we refer to as BST, following conventional Avr-R protein nomenclature) that recognizes AvrBsT. However, the recessive nature of the phenotype indicated that the genetic basis of resistance might be a mutation in the AvrBsT target or in a negative regulator of AvrBsT-dependent resistance. Both scenarios could result in the activation of defense responses that are normally silent in unchallenged plants. Hence, we designated the locus SOBER1 and the corresponding Pi-0 allele sober1-1.
Genetic Components of AvrBsT-Elicited Disease Resistance
We took advantage of the available defense mutants defining nodes in the plant innate immune network (Hammond-Kosack and Parker, 2003) to determine the genetic components, if any, required for AvrBsT-triggered defense responses in Arabidopsis. Tests of epistasis were performed by crossing resistant Pi-0 sober1-1 plants with the following mutants: Col-0 ndr1-1 (Century et al., 1997), Ler eds1-2 (Falk et al., 1999), Col-0 pad4- 1 (Jirage et al., 1999), Col-0 jar1-1 (Staswick et al., 2002), Col-0 sid2-1 (Dewdney et al., 2000; Wildermuth et al., 2001), and Col-0 npr1-1 (Cao et al., 1997). In planta growth curves were determined with F3 families established from independent F2 double mutant individuals to quantify the growth of Pst DC3000 AvrBsTHyb-HA.
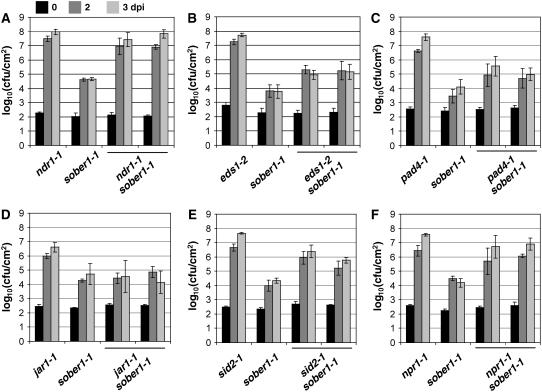
Defense pathways obeying classical gene-for-gene relationships generally rely on either NDR1 or the EDS1/PAD4 complex, depending on the structural domains of R gene products (Aarts et al., 1998; Feys et al., 2001). Interestingly, sober1-1 immunity is completely blocked in the ndr1-1 background (Figure 4A), whereas sober1-1 resistance is only partially impaired in eds1-2 sober1-1 (Figure 4B) and pad4-1 sober1-1 (Figure 4C). Pst DC3000 AvrBsTHyb-HA was able to grow ∼10-fold higher in these double mutants compared with the sober1-1 mutant. These phenotypes are consistent with the cooperative action of EDS1 and PAD4 in defense signal transduction (Feys et al., 2001).
Figure 4.
Pathogen Growth in Known Arabidopsis Defense Mutants, sober1-1, and Respective Double Mutants.
sober1-1 epistasis analysis in the ndr1-1 (A), eds1-2 (B), pad4-1 (C), jar1-1 (D), sid2-1 (E), and npr1-1 (F) genetic backgrounds. Leaves were hand-infiltrated with a 105 cfu/mL suspension of Pst DC3000 carrying pVSP61(avrRpt21-100-avrBsT11-350-HA). Bacteria present in leaves were monitored at 0 d (black bars), 2 d (dark gray bars), and 3 d (light gray bars) after inoculation (dpi). Data points represent the mean log10 (cfu/cm) ± sample sd for four replicates of the single mutant parents and two independent double mutant F3 families. The data represent trends observed in at least two experiments using several (two to six) independent mutant F3 families.
We also determined whether small defense signaling molecules played a role in sober1-1 resistance. Jasmonic acid, a lipid-derived hormone and a central regulator of plant responses to insect feeding and necrotrophic pathogens (Staswick et al., 2002), is probably not involved in sober1-1 defense signaling, because the pathogen grew equally well in jar1-1 sober1-1 and sober1-1 plants (Figure 4D). By contrast, the sid2-1 mutation in isochorismate synthase, an enzyme required for salicylic acid (SA) biosynthesis, significantly compromised sober1-1 immunity, as shown by the ∼100-fold increase in pathogen growth in sid2-1 sober1-1 compared with sober1-1 plants (Figure 4E). Similarly, the npr1-1 mutant markedly impaired sober1-1 resistance (Figure 4F), indicating that both SA and this downstream defense regulator are required for defense against AvrBsT-containing pathogens.
Positional Cloning of SOBER1
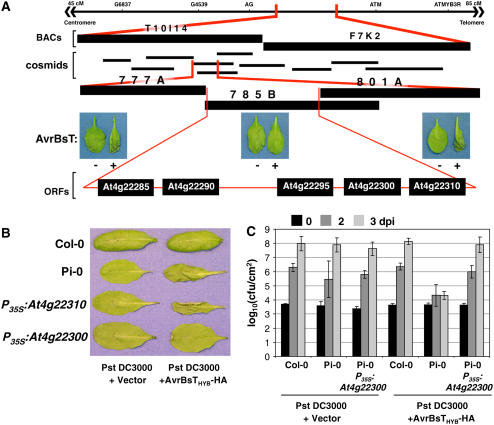
To map SOBER1, we genotyped 34 resistant F2 progeny obtained from a cross between Pi-0 (resistant parent) and Ler (susceptible parent). SOBER1 was mapped to chromosome IV between the genetic markers AG (63 centimorgans [cM]) and ATMYB3R (83 cM) (Figure 5A). We then took advantage of the tools available for the susceptible Col-0 ecotype to obtain two overlapping BACs that spanned the interval. Col-0 BAC T10I14 and BAC F7K2 covered the left and right ends, respectively (Figure 5A). A Col-0 genomic library was screened using BAC-specific probes to generate a cosmid contig spanning T10I14 and F7K2. To fill in gaps in the contig, the BACs were used to make a cosmid library in the binary vector pCLD04541. DNA gel blot hybridization and sequencing of the cosmid ends were used to assemble 10 cosmids (Figure 5A). Cosmid clones mobilized into A. tumefaciens GV3101 pMP90 were then used to independently transform Pi-0. We predicted that the dominant Col-0 SOBER1-susceptible allele would suppress an AvrBsT-dependent HR in transgenic Pi-0 plants. As expected, two Pi-0 T1 transgenic lines independently expressing cosmid clones 785B (Figure 5A) and 795A (see Supplemental Figure 2 online) failed to induce HR after Pst DC3000 AvrBsTHyb-HA infection. By contrast, Pi-0 T1 transgenic lines expressing cosmid clones 777A and 801A still elicited HR (Figure 5A). The minimal HR-suppressing genomic region shared by cosmid clones 785B and 795A spanned ∼14 kb and contained five predicted genes, according to the Munich Information Center for Protein Sequences (MIPS) database annotation (Schoof et al., 2002).
Figure 5.
Cloning of SOBER1.
(A) Physical mapping of the SOBER1 locus. The top line represents the genomic region on chromosome IV between markers AG and ATM, to which the SOBER1 locus was first mapped. Two Col-0 BACs (T10I14 and F7K2) and 10 cosmid clones spanning this region were then isolated. Col-0 cosmid clones 777A, 785B, and 801A were independently transformed into Pi-0 for complementation analysis. T1 transgenic lines were infected with a 3 × 108 cfu/mL suspension of Pst DC3000 pVSP61 and Pst DC3000 pVSP61(avrRpt21-100-avrBsT11-350-HA), designated as AvrBsT − and +, respectively, and scored for HR at 10 to 12 h after inoculation. The complementing Col-0 cosmid clone 785B contains five genes: At4g22285, At4g22290, At4g22295, At4g22300, and At4g22310.
(B) Functional complementation of sober1-1 with At4g22300. Col-0, Pi-0, and T1 Pi-0 transgenic lines expressing P35S-At4g22310 and P35S-At4g22300 were infected with a 3 × 108 cfu/mL suspension of Pst DC3000 pVSP61 and Pst DC3000 pVSP61(avrRpt21-100-avrBsT11-350-HA), designated as Pst DC3000 and Pst DC3000 + AvrBsTHyb-HA, respectively, and scored for HR at 10 to 12 h after inoculation.
(C) In planta bacterial growth in a T1 Pi-0 transgenic line expressing P35S-At4g22300. Leaves were hand-inoculated with the 105 cfu/mL suspension of bacteria described for (B). Bacteria present in leaves were monitored at 0 d (black bars), 2 d (dark gray bars), and 3 d (light gray bars) after inoculation (dpi). Data points represent the mean log10 (cfu/cm) ± sample sd. The data represent phenotypes observed in at least three independent T1 Pi-0 transgenic lines.
To identify SOBER1, the Col-0 allele for each candidate gene was transformed into Pi-0 and the respective T1 lines were scored for an AvrBsT-elicited HR. Pi-0 transgenic plants constitutively expressing the Col-0 At4g22300 cDNA (P35S-At4g22300) did not induce HR in response to Pst DC3000 AvrBsTHyb-HA infection (Figure 5B), whereas Pi-0 P35S-At4g22310 transgenic plants, for example, and the Pi-0–resistant control triggered massive tissue collapse (Figure 5B). Moreover, Pi-0 P35S-At4g22300 T1 lines did not induce HR in response to Xcc 8004 AvrBsT infection, whereas Pi-0 P35S-At4g22310 T1 leaves fully collapsed as a result of HR (see Supplemental Figure 3 online). These data indicate that Col-0 At4g22300 can suppress HR when AvrBsTHyb-HA is delivered by Pst DC3000 or when native AvrBsT is delivered by Xcc 8004.
To verify that the Col-0 At4g22300 allele also restored disease susceptibility, several independent Pi-0 P35S-At4g22300 transgenic lines were challenged with Pst DC3000 and Pst DC3000 AvrBsTHyb-HA. In planta growth curves show that Pi-0 P35S-At4g22300 plants were fully susceptible to Pst DC3000 AvrBsTHyb-HA (Figure 5C). These complementation data demonstrate that the Col-0 At4g22300 cDNA is sufficient to suppress HR and disease resistance in the Pi-0 background. Thus, the At4g22300 gene locus corresponds to SOBER1.
Molecular Nature of sober1-1 Resistance
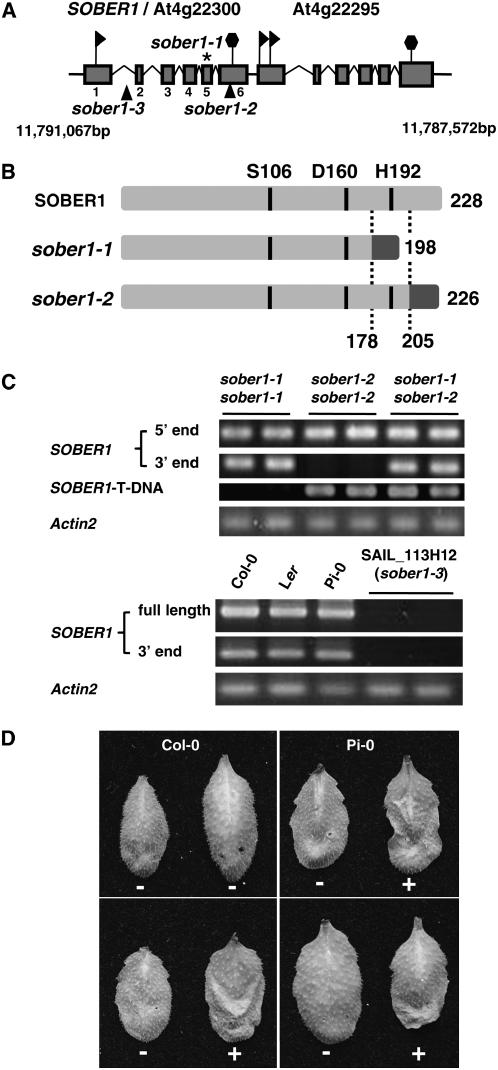
The SOBER1/At4g22300 coding sequence is composed of six exons spanning 1510 bp on the reverse strand of chromosome IV between positions 11,791,067 and 11,789,558 bp (Figure 6A). At4g22295 resides just downstream of SOBER1, possessing an undefined promoter region. Originally, this region was annotated as a single gene; however, cDNAs exist for each gene, providing evidence that the region encodes two discrete proteins that are very similar (75% identity). This architecture is suggestive of tandem duplication of an ancestral gene.
Figure 6.
Mutations in SOBER1 Confer Resistance to AvrBsT in Arabidopsis.
(A) Scheme of the genomic locus for SOBER1/At4g22300 and At4g22295. Exons are represented with shaded boxes and introns with broken lines. The asterisk denotes the Pi-0 sober1-1 mutation in exon 6, and the triangles denote the T-DNA insertion in Col-0 sober1-2 (SALK_036632) and Col-0 sober1-3 (SAIL_113H12) lines. Flags and hexagons indicate coding sequence start and stop codons, respectively. Chromosome IV coordinates are designated in base pairs.
(B) Scheme of the predicted peptides encoded by the SOBER1, sober1-1, and sober1-2 alleles. The conserved catalytic residues (Ser [S], Asp [D], and His [H]) in the predicted Ser hydrolase are indicated. Domains in light gray are identical to wild-type SOBER1. Domains in dark gray are unrelated to SOBER1.
(C) SOBER1 mRNA expression in sober1-1, sober1-2, and sober1-3 plants. The 5′ end (327 bp of exon 1) and 3′ end (309 bp of exon 6) of SOBER1 mRNA were amplified by RT-PCR using total RNA extracted from unchallenged leaves from Pi-0 sober1-1, Col-0 sober1-2, and F2 individuals from a Pi-0 × SALK_036632 cross (sober1-1 sober1-2). The SOBER1 T-DNA chimeric cDNA fragment (377 bp) was amplified with an exon 4 primer and a T-DNA left border primer. In all cases, the PCR products matched the expected cDNA fragment size. To analyze sober1-3 lines, the full-length (802 bp) and 3′ end of SOBER1 mRNA were amplified by RT-PCR using total RNA extracted from unchallenged leaves from Col-0, Ler, Pi-0, and two sober1-3 homozygous individuals (SAIL_113H12 line). All PCR products were sequenced. Actin2 (At3g18780) was used as a reference control.
(D) HR phenotype of F1 individuals from a Pi-0 × SALK_036632 cross (sober1-1 sober1-2). In each panel, the right and left leaves were infiltrated with a 3 × 108 cfu/mL suspension of Pst DC3000 pVSP61 and Pst DC3000 pVSP61(avrRpt21-100-avrBsT11-350-HA), respectively. + = HR and − = no visible response at 10 to 12 h after inoculation.
To determine the molecular nature of sober1-1 resistance, the genomic regions of At4g22300 from Pi-0, Col-0, and Ler were sequenced. In addition, we analyzed the sequences of full-length cDNAs and the 3′ ends generated by RT-PCR as well as the Col-0 cDNA U22920 obtained from the ABRC (Yamada et al., 2003). A full-length cDNA was amplified for each ecotype (Figure 6B), indicating that the gene is expressed in Arabidopsis. The transcript from Col-0 is 1036 bp long with a 136-bp 5′ untranslated region, an open reading frame of 684 bp, and a 3′ untranslated region of 216 bp. The deduced polypeptide contains 228 amino acids with a theoretical molecular mass of 24.8 kD, consistent with the MIPS annotated sequence (http://mips.gsf.de/). SOBER1 is predicted to encode a Ser hydrolase containing a highly conserved catalytic core containing the three residues Ser-106, Asp-160, and His-192 (Figure 6B).
Examination of the resistant Pi-0 allele compared with the susceptible Col-0 and Ler alleles revealed that sober1-1 contains a Phe-to-Leu substitution at codon 174 identical to the Ler allele and a distinctive single nucleotide deletion in codon 179 within exon 5. The deletion shifts the downstream reading frame of sober1-1, resulting in a shorter polypeptide unrelated to SOBER1 (Figure 6B). Therefore, the Pi-0 sober1-1 allele encodes a polypeptide lacking His-192, suggesting that it is likely a nonfunctional enzyme.
Independent sober1 Mutants Are Resistant to AvrBsT
Two Col-0 T-DNA mutants in SOBER1 became available during the course of this study and were designated sober1-2 (SALK_036632) and sober1-3 (SAIL_113H12) (Alonso et al., 2003). In the sober1-2 mutant, the T-DNA was mapped downstream of codon 205 in exon 6. This generated a frameshift in the protein and a premature stop after codon 226 (Figure 6B). The T-DNA insertion in sober1-2 altered the 3′ end coding region but not the expression of the gene (Figure 6C). The predicted mutant protein has the conserved hydrolase catalytic core but contains different amino acids at positions 205 to 226. The substitution of these C-terminal amino acids is not expected to grossly alter the structure of the enzyme; however, it may interfere with substrate binding or protein–protein interactions, considering the close proximity to the active site (see Figures 7B and 7C below and Discussion). In the sober1-3 mutant, the T-DNA insertion was mapped 3′ of nucleotide 167 in the first intron, resulting in a 60-bp deletion (Figure 6A). Semiquantitative RT-PCR of homozygous mutants showed that no product (full-length open reading frame or 3′ end) could be amplified using total RNA isolated from sober1-3. Therefore, we concluded that the Col-0 sober1-3 mutation is a null allele.
Figure 7.
SOBER1 Belongs to a Ubiquitous Family of APT1/LysoPLA1-Related Proteins.
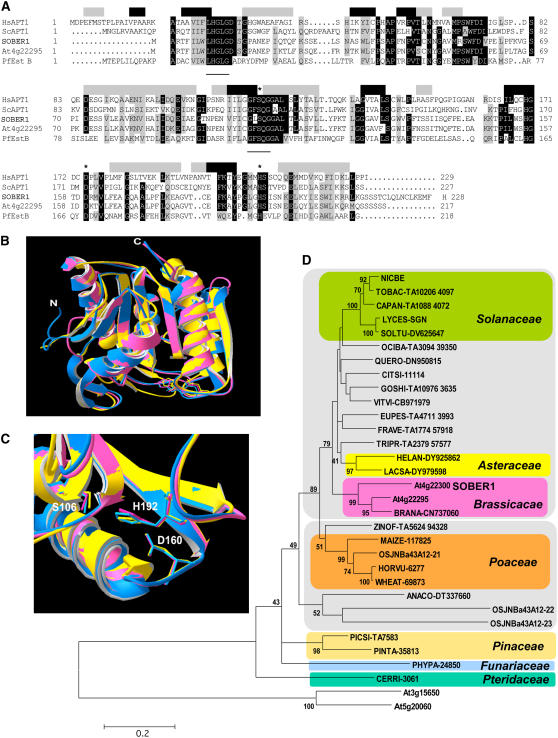
(A) Amino acid sequence alignment of SOBER1 with its closest Arabidopsis homolog At4g22295, the Pseudomonas fluorescens carboxylesterase EstB (Pf EstB), and two acyl-protein thioesterases from human (Hs APT1) and yeast (Sc APT1). Identical and highly conserved residues (four of five sequences) are shaded in black, and similar residues are shaded in gray. Shaded boxes above the alignment indicate Hs APT1 secondary structure (black, β-strand; gray, α-helices). Catalytic residues are labeled with asterisks. Two conserved motifs found in this class of Ser hydrolases are underlined: LHGLGD and GFSAG.
(B) Ribbon view of the predicted SOBER1 structure (pink) (amino acids 1 to 213) superposed onto its modeling templates Hs APT1 (blue) and Pf EstB (yellow).
(C) Closeup view of the catalytic core of the proteins shown in (B). Numbering of the catalytic residues refers to the SOBER1 sequence.
(D) Neighbor-joining phylogenic tree of plant SOBER1-related hypothetical peptides. Bootstrap consensus tree and support values >40 are reproduced on appropriate nodes. The tree was rooted on two Arabidopsis sequences belonging to a diverging APT1-like family. Sequence designations refers to gene name or the UniProt Knowledgebase (http://www.ebi.ac.uk/uniprot/ or http://www.ebi.ac.uk/newt/display for direct links to the taxonomy browser) identification code of the organism from which they originate followed by a unique code corresponding to their accession number in the source database. Sequences in colored boxes belong to the same taxonomic family. The two gray boxes delineate dicotyledons (top) from monocotyledons (bottom). The scale bar represents 0.2 JTT distance matrix units.
We next investigated whether or not the sober1-2 and sober1-3 plants are resistant to Pst DC3000 expressing AvrBsTHYB-HA. None of the homozygous sober1-2 or sober1-3 leaves infected with Pst DC3000 AvrBsTHYB-HA elicited HR (data not shown). This finding was not unexpected considering that resistance did not segregate as a monogenic, recessive trait in the F2 progeny from a Col-0 × Pi-0 cross, as observed in F2 progeny from the Ler × Pi-0 mapping cross. Thus, we speculated that the sober1 mutant phenotype might require one or more factors from the Pi-0 ecotype that is absent or different in the Col-0 background. To test this notion, the Col-0 sober1-2 (SALK_036632) and Col-0 sober1-3 (SAIL_113H12) lines were each crossed to Pi-0 sober1-1 and the resulting F1 progeny were scored for HR. Both F1 progeny expressing the heterozygous mutant alleles (i.e., sober1-1 sober1-2 [Figure 6C] and sober1-1 sober1-3) elicited HR in response to Pst DC3000 AvrBsTHYB-HA and no response to Pst DC3000 alone (Figure 6D). HR segregated 3:1 in the resulting F2 progeny, indicating that a single recessive gene in Col-0 abrogates the HR phenotype mediated by sober1 (A. Wilson and M.B. Mudgett, unpublished results). Moreover, we identified resistant homozygous sober1-3 sober1-3 F2 individuals, indicating that AvrBsT resistance in Arabidopsis results from a loss of SOBER1 function. We are currently cloning the gene from Pi-0 required for AvrBsT-dependent HR to identify a second player involved in the AvrBsT/SOBER1 pathway.
SOBER1 Encodes an Arabidopsis Ortholog of Acyl Protein Thioesterase/Lysophospholipase
SOBER1 belongs to the α/β hydrolase-2 protein family (Pfam02230) that is structurally related to the α/β hydrolase superfamily (Pfam00561) (Finn et al., 2006). Members of this family (currently 413) possess phospholipase and carboxylesterase activity with broad substrate specificity. Figure 7A shows the amino acid alignment of Arabidopsis SOBER1 and At4g2295 along with the most related enzymes characterized at the biochemical level. Pseudomonas fluorescens esterase EstB (Pf EstB) belongs to a conserved cluster of family VI bacterial lipolytic enzymes (Hong et al., 1991; Prim et al., 2006) and shares 30% identity and 49% similarity with SOBER1. The human acyl protein thioesterase (Hs APT1), which shares 33% identity and 49% similarity with SOBER1, was initially characterized as a lysophospholipase (LysoPLA1) capable of hydrolyzing esters on a broad range of lysophospholipids (LysoPLs) producing free fatty acids and glycerolphosphate derivatives (Zhang and Dennis, 1988; Sugimoto et al., 1996; Wang et al., 1997a, 1999, 2000; Portilla et al., 1998). It was later discovered that rat APT1 (Duncan and Gilman, 1998), yeast Sc APT1 (Duncan and Gilman, 2002), and Hs APT1 (Wang et al., 2000) catalyze the removal of thioacyl groups from modified Gα subunits of heterotrimeric G protein complexes (a process also known as depalmitoylation), underscoring the diverse nature of this class of enzymes.
SOBER1 contains three regions that are highly conserved in Ser hydrolases: (1) the N-terminal LHGLGD motif; (2) the GXSXG motif containing the Ser nucleophile; and (3) the conserved catalytic core containing the nucleophile-acid-His–ordered sequence (S, D, H) (Figure 7A). Analysis of SOBER1 using fold recognition and threading programs (PHYRE, http://www.sbg.bio.ic.ac.uk/∼phyre/; LOOPP, http://cbsuapps.tc.cornell.edu/loopp.aspx) predicted with high confidence (Phyre E-value = 5.2e−22) extensive secondary structure conservation with Hs APT1 and Pf EstB. The three-dimensional structures of Hs APT1 and Pf EstB (Protein Data Bank identifiers IFJ2 and 1AUO, respectively) were shown to fit the canonical α/β hydrolase fold and to exhibit pronounced relatedness (Kim et al., 1997; Devedjiev et al., 2000). Such conservation allowed us to model SOBER1's structure using the archetypal hydrolase structures. Figure 7B shows the first approximation model of SOBER1's ribbon structure obtained with the SWISS-MODEL server (http://swissmodel.expasy.org//SWISS-MODEL.html; Schwede et al., 2003). The putative backbone conformation of SOBER1 displays good similarity with the modeling templates, with major differences or modeling conflicts restricted to loops of solvent-exposed regions (Figure 7B). In addition, SOBER1's conserved catalytic triad aligns with the functional triad of the Hs APT1 and Pf EstB enzymes. The Asp-His dyad in Ser hydrolases is predicted to function as a proton sink that activates the nucleophilic Ser (Devedjiev et al., 2000).
Phylogenic Analysis of SOBER1-Like Proteins in Plants
In the Cluster of Orthologous Groups (COG) database (Tatusov et al., 2003), SOBER1 and APT1 enzymes are assigned to the COG0400 cluster that contains 200 BLAST hits in 149 unique species belonging to most major phylogenetic lineages with the exception of Archaea and viruses, emphasizing the wide distribution of this gene family in nature. We BLASTed plant genome and EST databases and retrieved sequences similar to that of SOBER1 (E value < 10−10) in the kingdom Viridiplantae. In the latest annotation of the Arabidopsis genome, besides At4g22295, at least two other genes are predicted to encode proteins related to SOBER1 with the Pfam α/β hydrolase-2 protein family signature. Similarity mining in The Arabidopsis Information Resource and Arabidopsis thaliana Plant Genome Database genomic and EST databases identified At3g15650 and At5g20060 proteins, supported by experimental cDNA, which display lower similarity, with 31 and 28% identity and 43 and 44% similarity, respectively. Although they possess the three putative catalytic residues embedded in conserved blocks, alignment with genuine acyl protein thioesterase/lysophospholipase (APT1/LysoPLA1) enzyme sequences necessitated the inclusion of large gaps (see Supplemental Figure 4 online). Thus, these proteins likely define a distinct hydrolase subgroup that includes other related eukaryotic proteins from rice (Oryza sativa) and mammals. Moreover, in preliminary phylogenetic analysis, At3g15650 and At5g20060 consistently clustered away from the APT1/LysoPLA1 clade (Figure 7D).
Evolutionary relationships between Arabidopsis sequences and the additional 28 plant SOBER1-related sequences (ranging between 39 and 75% identity and 51 and 80% similarity to SOBER1) were inferred from phylogenic analysis (Figure 7D). The predicted catalytic residues were invariable in all of the sequences except OSJNBa43A12-23. The overall topology of the resulting tree is consistent with current plant phylogeny. SOBER1 family members were found in a wide spectrum of evolutionarily divergent species. SOBER1-like sequences from species belonging to the same taxonomic family clustered together. SOBER1 and At4g22295 fall into the Brassicaceae monophyletic group, suggesting that a gene duplication event occurred relatively recently in evolution. Interestingly, the rice genome contains three genes highly related to SOBER1 (Figure 7D). These genes exist in a tandem array on chromosome IV, reminiscent of the genomic organization in Arabidopsis. Arabidopsis and rice were the only species possessing multiple SOBER1 paralogs, but this situation is probably attributable to the incomplete coverage of the genomes.
Recombinant SOBER1 Displays Carboxylesterase Activity in Vitro
To test whether SOBER1 encodes an active carboxylesterase, we purified recombinant His6-tagged proteins from Escherichia coli and assayed a variety of acylated colorimetric substrates. We expressed Col-0 His6-SOBER1, Pi-0 His6-SOBER1-1, His6-ScAPT1, and a catalytic core mutant, Col-0 His6-SOBER1(H192A), in which the His was substituted with Ala. This type of mutation abrogated the biochemical activity of the mouse APT1/LysoPLA1 enzyme with minimal impact on global protein conformation (Wang et al., 1997b). Preliminary experiments testing protein expression and solubility indicated that although all recombinant proteins were produced at high levels in E. coli strain BL21(DE3)pLysS, Pi-0 His6-SOBER1-1 protein was not soluble in the buffers and conditions tested. By contrast, Col-0 His6-SOBER1, His6-SOBER1(H192A), and His6-ScAPT1 were purified under native conditions (see Supplemental Figure 5 online).
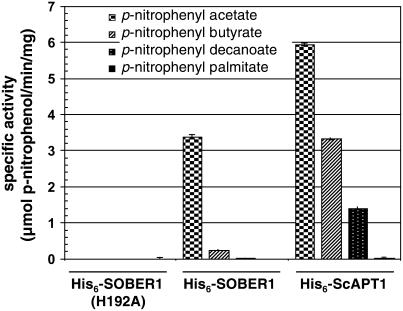
We used standard spectrophotometric assays to monitor bacterial lipase/esterase activity to study the SOBER1 enzyme. Such assays monitor the formation of p-nitrophenol resulting from the hydrolysis of synthetic p-nitrophenyl ester substrates (Kuznetsova et al., 2005). Both His6-SOBER1 and His6-ScAPT1 recombinant proteins were able to cleave p-nitrophenyl acetate (C2 acyl chain); however, SOBER1 specific activity was approximately twofold lower (Figure 8). By contrast, His6-SOBER1(H192A) activity was indistinguishable from that of buffer controls lacking recombinant protein (Figure 8), confirming that His-192 is essential for catalysis.
Figure 8.
SOBER1 Has in Vitro Carboxylesterase Activity on Short Acyl-Chain p-Nitrophenyl Esters.
Enzyme assays were performed in triplicate at 23°C with 0.45 mM substrate and 0.1 μg of each recombinant protein (∼18.5 nM). Proteins assayed were wild-type Col-0 His6-SOBER1, the catalytic core mutant His6-SOBER1(H192A), and the yeast ortholog His6-ScAPT1. Values represent the average carboxylesterase activity (micromoles of p-nitrophenol per minute per milligram of protein) ± sd. Reactions were performed in triplicate. Similar results were obtained in two independent experiments.
We also studied the effect of acyl chain length on enzyme activity to determine whether the enzymes favored substrates containing long carbon chains and hence exhibited true lipase activity. We found that the ability of the control His6-ScAPT1 enzyme to hydrolyze ester bonds gradually decreased with long acyl chain substrates (i.e., the specific activities for p-nitrophenyl butyrate and p-nitrophenyl decanoate were 50 and 25%, respectively, that of p-nitrophenyl acetate) (Figure 8). Cleavage of p-nitrophenyl palmitate by His6-ScAPT1 was observed only with a 10-fold higher concentration of protein and prolonged incubations. His6-SOBER1 specific activity was 10-fold lower for p-nitrophenyl butyrate, whereas little or no activity was detected for p-nitrophenyl decanoate and p-nitrophenyl palmitate. Thus, SOBER1 has carboxylesterase activity (EC 3.1.1.1) with a preference for short acyl chain substrates.
SOBER1 Enzyme Activity Is Required for the Suppression of AvrBsT-Elicited Defense in Arabidopsis
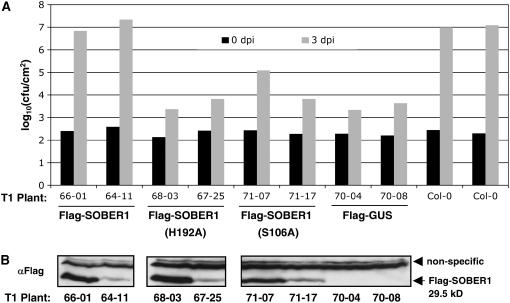
Our genetic and biochemical analyses support the hypothesis that SOBER1 enzyme activity may be required to inhibit AvrBsT-elicited immunity in Arabidopsis. To test this hypothesis, we asked whether or not the SOBER1(H192) mutant protein could suppress Pst DC3000 AvrBsTHYB-HA growth in Pi-0–like wild-type SOBER1 (Figure 5C). We generated Pi-0 transgenic plants expressing Flag-tagged SOBER1, Flag-SOBER1(H192A), Flag-SOBER1(S106A), a nucleophile mutant, and Flag-GUS (for β-glucuronidase) as a control. Proteins were constitutively expressed under the control of the cauliflower mosaic virus 35S promoter. Independent T1 Pi-0 lines were monitored for Pst DC3000 AvrBsTHYB-HA multiplication and protein expression by immunoblot analysis.
Flag-SOBER1 Pi-0 transgenic plants allowed the proliferation of Pst DC3000 AvrBSTHYB-HA at 3 d after inoculation to similar titers as the Col-0 untransformed susceptible control plants (Figure 9A). By contrast, both Flag-SOBER1(H192A) and Flag-SOBER1(S106A) failed to restore susceptibility in the Pi-0 background, although protein expression levels were comparable to those of Flag-SOBER1 (Figures 9A and 9B). Bacterial populations in Flag-SOBER1(H192A) and Flag-SOBER1(S106A) transgenic lines were not significantly different from those of the Flag-GUS resistant control plants (Figure 9A). These data strongly support the hypothesis that SOBER1 enzyme activity is required for the suppression of AvrBsT-elicited defense responses in Arabidopsis.
Figure 9.
SOBER1 Enzyme Activity Is Required for Suppression of AvrBsT Resistance in Pi-0.
(A) In planta bacterial growth in Pi-0 T1 transgenic lines constitutively expressing Flag-SOBER1, Flag-SOBER1-H192A, Flag-SOBER1-S106A, or Flag-GUS. Leaves infected with a 105 cfu/mL suspension of Pst DC3000 pVSP61(avrRpt21-100-avrBsT11-350-HA) were sampled from individual plants at 0 d (black bars) and 3 d (gray bars) after inoculation (dpi). Flag-GUS Pi-0 T1 lines and untransformed Col-0 plants served as resistant and susceptible controls, respectively. Data for two independent T1 lines are presented. Similar results were obtained with four independent transformants per test construct.
(B) Immunoblot analysis of the uninfected, independent Pi-0 T1 transgenic plants used for (A). Total protein (∼50 μg) was analyzed by gel blot analysis using Flag antisera. Only SOBER1 protein expression is shown. Flag-GUS protein expression was detected in the appropriate lines (data not shown). Data presented were obtained from a series of immunoblots from a single day. The top bands represent nonspecific proteins, and the bottom band represents the Flag-tagged SOBER1 protein.
Arabidopsis SOBER1 Compromises AvrBsT-Dependent HR in N. benthamiana
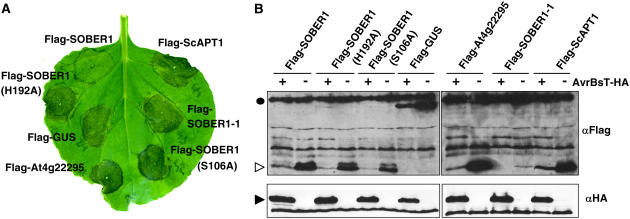
AvrBsT triggers a rapid HR when transiently expressed in N. benthamiana (Orth et al., 2000). We next asked whether or not ectopic expression of SOBER1 in N. benthamiana could suppress AvrBsT-elicited HR. To test this notion, we transiently coexpressed AvrBsT-HA with wild-type Flag-SOBER1 or mutant protein [Flag-SOBER1(H192A) and Flag-SOBER1(S106A)] or a Flag-GUS control. N. benthamiana leaves transiently infected with A. tumefaciens strains containing AvrBsT-HA and Flag-GUS underwent confluent cell death and tissue necrosis (i.e., the HR) at 3 d after infection (Figure 10A), consistent with AvrBsT's previously reported phenotype (Orth et al., 2000). HR was dependent on AvrBsT, because strains carrying vector controls produced no visible reactions (data not shown). Coexpression of AvrBsT and Flag-SOBER1, however, did not result in HR at the same time point (Figure 10A), even though both proteins were expressed (Figure 10B). Suppression of HR was dependent on a wild-type hydrolase catalytic core, because neither Flag-SOBER1(H192A) nor Flag-SOBER1(S106A) prevented AvrBsT-dependent tissue collapse (Figure 10A). Similarly, expression of mutant Flag-SOBER1-1 (the Pi-0 allele) did not block HR. This mutant protein accumulated at low levels in N. benthamiana, suggesting that this truncated enzyme may be inherently unstable or poorly expressed in this system. We also note that coexpression of AvrBsT-HA with the test Flag proteins led to lower expression of the respective test protein. The significance of this observation is not yet clear.
Figure 10.
Arabidopsis SOBER1 Enzyme Activity Suppresses AvrBsT-Elicited HR in N. benthamiana.
(A) A. tumefaciens transient coexpression of AvrBsT-HA with individual N-terminal Flag-tagged proteins in N. benthamiana: Flag-SOBER1 (Col-0 wild-type allele); Flag-SOBER1-H192 and Flag-SOBER1-S106A (two Col-0 catalytic core mutant alleles); Flag-GUS; Flag-At4g22295 (Col-0 SOBER1 homolog); Flag-SOBER1-1 (Pi-0 mutant allele); and Flag-ScAPT1 (yeast homolog). The AvrBsT-elicited HR phenotype was photographed at 4 d after inoculation.
(B) Immunoblot analysis of proteins transiently expressed in N. benthamiana leaves as shown in (A). A. tumefaciens expressing AvrBsT-HA (+) or vector (−) was coexpressed with each Flag-tagged protein described for (A). Total protein (∼50 μg) was extracted from tissue at 43 h after inoculation (before HR) and then examined by gel blot analysis using FLAG (top panels) and HA (bottom panels) antisera. The black circle, white arrowhead, and black arrowhead refer to the molecular mass: 99, 30, and 42 kD, respectively. These experiments were repeated three times with similar results.
Finally, we took advantage of this convenient transient assay to test whether proteins highly related to SOBER1 could compromise AvrBsT-elicited HR. Interestingly, neither Flag-At4g22295 nor Flag-ScAPT1 coexpression with AvrBsT-HA blocked HR (Figure 10A). The fact that Sc APT1 is an active hydrolase suggests that enzyme activity per se is not sufficient for biological function. Rather, it is likely that differences in substrate specificity and/or the regulation of the enzyme in the host are important for the downregulation of AvrBsT phenotypes in N. benthamiana and possibly Arabidopsis.
DISCUSSION
Our results reveal that AvrBsT-specific disease resistance in the Arabidopsis Pi-0 ecotype is attributable to a single nucleotide deletion in a Ser hydrolase in the α/β hydrolase-2 family. The mutation is predicted to result in a truncated polypeptide missing an invariant His residue in its predicted catalytic core (Figure 6C). The recessive nature of the Pi-0 sober1-1 allele and the fact that homozygous sober1-3 sober1-3 F2 plants from a Col-0 sober1-3 × Pi-0 sober1-1 cross are HR-positive support the hypothesis that plant immunity to AvrBsT results from a loss of SOBER1 enzymatic activity. Consistent with this hypothesis, the recombinant wild-type Col-0 His6-SOBER1 protein is an active carboxylesterase that cleaves acylated substrates in vitro. Recombinant protein lacking the conserved His residue in the hydrolase active site, His6-SOBER1(H192A), was inactive, demonstrating that the His residue is required for enzyme activity (Figure 8). Furthermore, T1 Pi-0 transgenic lines expressing the wild-type Col-0 allele (Flag-SOBER1) are susceptible to Pst DC3000 AvrBsTHYB-HA infection, whereas T1 lines expressing catalytic core mutants [Flag-SOBER1(H192A) and Flag-SOBER1(S106A)] are resistant (Figure 9).
Additional evidence indicating that the Pi-0 allele lacking His-192 is likely an inactive enzyme comes from the unique conservation of protein structure for enzymes containing the α/β hydrolase fold. The enzyme core in this superfamily consists of an α/β sheet formed by eight β-sheets connected by α-helices. The catalytic residues that make up the nucleophile-acid-His active site have been highly preserved, but not the substrate binding site. These residues are dispersed on loops that are the highly conserved structural features of the fold. Only the His residue is invariant in this superfamily; the nucleophile and acid loops can contain other amino acid residues (Holmquist, 2000). Substitution of His-208 to Ala in the murine ortholog abolished lysophospholipase activity without causing global structural changes, demonstrating that the His residue is required for substrate hydrolysis (Wang et al., 1997b). Interestingly, the sober1-2 mutant contains a functional catalytic core, indicating that enzyme activity alone may not be sufficient to alter AvrBsT-dependent phenotypes in planta. Rather, both enzyme activity and a functional C-terminal protein domain may be required to suppress resistance.
Small (∼25 kD) eukaryotic α/β hydrolases with the most similarity to SOBER1 are in the acyl protein thioesterase/lysophospholipase (APT1/LysoPLA1) subfamily. Hydrolases in this family differ in their substrate specificity and contribution to cellular signaling. Nonetheless, the common denominator is their structure that facilitates the cleavage of ester bonds. For instance, these enzymes hydrolyze thioester bonds to remove palmitate from G protein α subunits and RAS. Thioacylation and deacylation of Gα and RAS dynamically control their membrane localization and thus their role in signal transduction (Smotrys and Linder, 2004). These enzymes show remarkable in vitro esterase versatility, in that they can also deacylate ghrelin growth hormone (Shanado et al., 2004) and viral membrane glycoproteins (Veit and Schmidt, 2001).
This subfamily also cleaves the acylester bond in LysoPLs with lower affinity (Wang et al., 1997a, 1997b). LysoPLA1s strictly control the level of LysoPL metabolites that serve many diverse roles within cells. Therefore, the loss or change in function of this conserved enzyme would likely affect cellular physiology. For example, in mammalian cells, at low nontoxic levels, LysoPLs act as lipid second messengers, transducing signals elicited from membrane receptors, whereas at high levels they disturb membrane conformation, affecting the activities of many membrane-bound enzymes, and lead to cell lysis. Moreover, LysoPLs can also potentiate immune responses in mammalian cells (Wang and Dennis, 1999). The situation is similar in plants, in which stress cues trigger the production of LysoPLs and the modulation of enzyme activity (Wang, 2001). In addition to our study, there is one report of a cellular phenotype associated with an APT1/LysoPLA1 mutation. Gene replacement of the Magnaporthe grisea LPL1 gene caused defects in apressoria formation and penetration into leaf cells (Kanamori et al., 2005).
Collectively, our data indicate that changes in APT1/LysoPLA1 enzyme activity inside plant cells may dramatically alter the host's ability to respond to pathogens expressing AvrBsT. Our working hypothesis is that SOBER1 enzyme activity inhibits or prevents AvrBsT-triggered defense responses in Arabidopsis. At this point, it is not clear whether SOBER1 is a unique component of AvrBsT defense signal transduction or a susceptibility factor (Chu et al., 2006; Yang et al., 2006). It is unlikely that SOBER1 plays a general role in bacterial resistance, considering that Pi-0, Col-0, and Ler plants are equally susceptible to Pst DC3000. Also, healthy Pi-0 plants do not display any abnormal morphological or molecular phenotypes, indicating that the sober1-1 mutation is not in the enhanced resistance/lesion-mimic class of defense mutants. Rather, the AvrBsT dependence of the sober1-1 phenotype implies that AvrBsT and SOBER1 may work on the same host substrate or that the two proteins may interact directly within the plant cell, potentially affecting each other's biological function(s).
Our search for plant SOBER1 homologs illustrates the wide distribution of this protein family in the plant kingdom. Interestingly, several SOBER1 homologs were identified in pepper and N. benthamiana. Considering that these two species are able to detect and respond to AvrBsT, we suspect that the corresponding genes surveyed are actually orthologous to the At4g22295 gene, whose product, in contrast with SOBER1, was unable to suppress HR after transient expression in N. benthamiana (Figure 10A). The structural/functional basis for the differential enzymatic activity of SOBER1 and At4g22295 in this bioassay is under investigation.
The biochemical role of AvrBsT is currently unknown; however, its Yersinia homolog, YopJ, is an acetyltransferase that inhibits MAPK signaling by acetylating MAPKK6 (Mukherjee et al., 2006). This raises the interesting possibility that AvrBsT and SOBER1 may work antagonistically within the cell by modulating posttranslational modification(s) on a common host substrate, perhaps by altering its acylation and/or sumoylation status. If AvrBsT and SOBER1 modify the same host target, then SOBER1 might be able to overcome or reverse the virulence action of AvrBsT. Thus, AvrBsT's action within the cell would essentially go unnoticed. Conversely, in the absence of SOBER1, AvrBsT would be able to modify its target. As a result, host physiology would be altered, which could in turn alarm the immune system and activate defense signaling and/or stress responses.
Epistasis tests revealed some of the defense components required for immunity in Pi-0 (Figure 4). NDR1 is a critical player in this pathway, as ndr1-1 sober1-1 double mutants were fully susceptible to Pst DC3000 AvrBsTHYB-HA infection. NDR1 is a plasma membrane glycophosphatidylinositol-anchored protein required for the activation of disease resistance signaling for a number of R proteins in Arabidopsis (Coppinger et al., 2004). Mutations in EDS1, PAD4, SID2, and NPR1 only partially compromised sober1-1 resistance, whereas a JAR1 mutant had no effect. Both EDS1 and PAD4 are homologous with acyl hydrolases, but no enzymatic activity has yet been confirmed. Still, it is clear that these proteins play a fundamental role in transducing redox signals in response to biotic and abiotic stress (Mateo et al., 2004). SID2 is an isochorismate synthase that synthesizes SA from chorismate (Wildermuth et al., 2001), and NPR1 is a novel ankyrin-repeat protein that functions downstream of SA to induce defense gene expression (Cao et al., 1997). JAR1 is an enzyme that adenylates jasmonic acid, an important modification required for hormone signaling (Staswick et al., 2002). Collectively, these genetic studies indicate that SA-dependent and SA-independent pathways both contribute to sober1-1–mediated immunity in Pi-0.
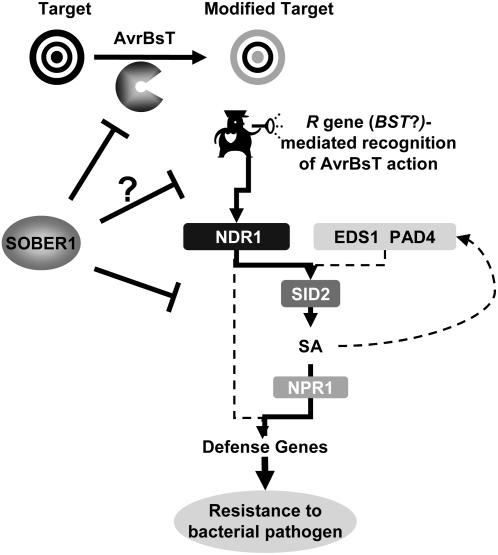
It remains to be determined whether or not an R gene mediates AvrBsT-dependent resistance in Pi-0. However, our genetic analyses indicate that a second gene in Pi-0 (and Ler) is required for sober1-1–mediated immunity. This gene may encode the elusive R gene. We refer to this putative R gene as BST to clearly indicate resistance against AvrBsT, following preexisting nomenclature used to describe resistance to Xanthomonas T3S effector proteins. Considering the NDR1 dependence of sober1-1 resistance, we speculate that BST may belong to the coiled-coil NB-LRR class of R proteins (Aarts et al., 1998). Alternatively, sober1-1 resistance could act in an NDR1-dependent, R gene–independent pathway. The working model proposed for the signaling network relaying AvrBsT-elicited immunity in Arabidopsis is shown in Figure 11.
Figure 11.
Working Model of a Signaling Network Relaying AvrBsT-Elicited Immunity in Arabidopsis.
Our genetic studies also revealed a second locus that controls AvrBsT defenses in Arabidopsis. The Col-0 sober1-2 T-DNA line infected with Pst DC3000 AvrBsTHYB-HA did not elicit HR, suggesting that another gene in Pi-0 is required for the phenotype of sober1. Indeed, F1 sober1-2 sober1-1 individuals were able to elicit HR in response to Pst DC3000 AvrBsTHYB-HA infection. Furthermore, HR segregated in three-fourths of the resulting F2 individuals, indicating that a single, recessive allele in Col-0 suppresses the phenotype of sober1-2. We are currently cloning this gene to further characterize the molecular basis of AvrBsT–SOBER1 interactions in Arabidopsis.
In summary, our work suggests that changes in SOBER1 enzymatic activity can dramatically affect the plant's ability to control bacterial growth. Whether or not SOBER1 functions as a negative regulator of AvrBsT-dependent responses or is in fact a susceptibility factor remains to be determined. The identification of host substrates and additional components operating in the AvrBsT/SOBER1 pathway will provide additional clues to the strategies used by bacterial pathogens to manipulate host physiology.
METHODS
Bacterial Growth and Plasmid Mobilization
Strains used in this study were as follows: Escherichia coli DH5a and Top10; Agrobacterium tumefaciens C58C1 pCH32 (Tai et al., 1999) and GV3101 pMP90 (Koncz and Schell, 1986); Pseudomonas syringae pv tomato strain DC3000 (originally obtained from Diane Cuppels); and Xanthomonas campestris pv campestris strain 8004 (originally obtained from Mike Daniels). E. coli and A. tumefaciens strains were grown on Luria agar medium (Sambrook et al., 1989) at 37 and 28°C, respectively. Pst DC3000 and Xcc 8004 strains were grown on nutrient yeast glycerol agar medium (Turner et al., 1984) at 28°C. Vectors were mobilized from E. coli into Pst, Xcc, and A. tumefaciens by standard triparental matings.
AvrBsT Plasmid Constructions
PCR was used to construct gene deletions and fusions. Primers and conditions used are available on request. All PCR-generated DNA fragments were sequenced. Numbering herein refers to the codon of the gene described.
To construct AvrRpt2-AvrBsT fusion proteins, a BglII-BamHI DNA fragment containing the promoter and amino acids 1 to 100 of AvrRpt2 was amplified by PCR using pDSK600(AvrRpt2) (Mudgett and Staskawicz, 1999) as template and then cloned into pCRII (Invitrogen), creating pCRII(avrRpt21-100). The BglII-BamHI fragment was subcloned into the BamHI site of the broad host range vector pVSP61, creating pVSP61(avrRpt21-100). This eliminated the 5′ BamHI site. Three N-terminal AvrBsT deletions (avrBsT113505-HA, avrBsT32-350-HA, and avrBsT52-350-HA) were constructed by PCR using pDD62(avrBsT-HA) (Orth et al., 2000) as DNA template. Each fragment was designed to have a BamHI site at the 5′ end and XhoI-BamHI sites at the 3′ end. The DNA fragments were cloned into pCRII (Invitrogen). For expression in Pst DC3000, the AvrBsT deletions were subcloned as BamHI fragments into the BamHI site of pVSP61(avrRpt21-100), creating pVSP61(avrRpt21-100-avrBsT11-350-HA), pVSP61(avrRpt21-100-avrBsT32-350-HA), and pVSP61(avrRpt21-100-avrBsT350-HA). For transient expression in planta, the AvrBsT deletions were subcloned as BamHI-XhoI fragments into the binary vector pMDD1 (Mudgett et al., 2000), creating pMDD1(avrBsT11-350-HA), pMDD1(avrBsT32-350-HA), and pMDD1(avrBsT52-350-HA).
Plant Growth and Pathogen HR Assays
Arabidopsis thaliana ecotypes were grown in pots in Metro-Mix 200 soil (Premier Horticulture) in growth chambers (22°C, 60% RH, 125 μE·m−2·s−1 fluorescent illumination) on an 8-h-light/16-h-dark cycle. Fully expanded leaves of 4- to 5-week-old plants were used for bacterial inoculations. Wild-type Arabidopsis Col-0, Ler, and Pi-0 ecotypes used for this study were obtained from B. Staskawicz (University of California, Berkeley). Mutant seeds used were as follows: Col-0 pad4-1 (J. Parker, Max Planck Institute); Ler eds1-2, Col-0 npr1-1, and Col-0 ndr1-1 (B. Staskawicz); and Col-0 jar1-1 and Col-0 sid2-1 (S. Somerville, Carnegie Institute).
Nicotiana benthamiana was grown in ProMix (Premier Horticulture) in pots under greenhouse conditions. Plants with two to four leaves were used for bacterial inoculations. Bacterial suspensions in 1 mM MgCl2 were hand-infiltrated into the extracellular spaces of a leaf through a small wound site using a 1-cc syringe. Xcc 8004 and Pst DC3000 strains were inoculated at 3 × 108 cells/mL. HR phenotypes were recorded 1 to 2 d after inoculation for Xcc 8004 and 10 to 12 h after inoculation for Pst DC3000.
Transient Protein Expression in N. benthamiana
Agrobacterium tumefaciens strain C58C1 pCH32 was used for transient protein expression in N. benthamiana. Strains were grown overnight at 28°C on Luria agar medium containing the appropriate antibiotics. Bacteria were collected and incubated in induction medium (10 mM MES, pH 5.6, 10 mM MgCl2, and 150 μM acetosyringone; Acros Organics) for 2 h before inoculation. Leaves were hand-inoculated with a 1.2 × 109 cells/mL suspension of bacteria in induction medium. Plants were incubated at room temperature under continuous low light for 2 to 4 d. Proteins were separated by SDS-PAGE and analyzed by immunoblot analysis as described (Mudgett et al., 2000). Proteins were visualized by chemiluminescence using monoclonal HA antiserum (Covance), monoclonal FLAG antiserum (Sigma-Aldrich), and peroxidase-conjugated secondary antibodies (Bio-Rad).
Pathogen Growth and Disease Analysis
To monitor Pst DC3000 growth in Arabidopsis leaves, plants were hand-infiltrated with bacterial suspensions of 1 × 105 cells/mL in 1 mM MgCl2. Plants were kept at high humidity (∼60 to 80%) in a growth chamber for 4 d. Tissue (four No. 1 cork-borer leaf discs) collected at 0, 2, and 3 d after inoculation was ground in 1 mM MgCl2, diluted, and then plated on nutrient yeast glycerol agar medium containing appropriate antibiotics and cycloheximide (50 μg/mL). Bacterial populations were determined for four replicate plants per treatment. To monitor disease phenotypes, plants grown in pots covered with plastic mesh were dipped into bacterial suspensions of 2 × 108 cells/mL in 1 mM MgCl2 and 0.02% Silwet L-77.
Genetic Analysis and Mapping SOBER1 Resistance
F2 progeny from a Pi-0 (resistant parent) × Ler (susceptible parent) cross were scored for HR after Pst DC3000 pVSP61(avrRpt21-100-avrBsT11-350-HA) infection. More than 4400 F2 progeny collected from three different F1 plants were scored for AvrBsT-dependent HR. Approximately 1050 F2 plants exhibited HR, ∼24.3%. Known molecular markers (cleaved-amplified polymorphic sequences, simple sequence length polymorphisms, and single nucleotide polymorphisms) that distinguish polymorphisms between Ler and Col-0 (http://Arabidopsis.org/) were used to design new cleaved-amplified polymorphic sequence markers to distinguish Pi-0 from Ler. These markers are available upon request. Thirty-four HR-positive F2 individuals were used to map SOBER1 to chromosome IV between AG (63 cM) and ATMYB3R (83 cM). For fine mapping, 1074 HR-positive F2 plants were genotyped with the left marker ARW11.1 and the right marker ARW334C to identify recombinants between these markers and the SOBER1 locus. Three recombinants narrowed the genetic interval to 130,400 bp.
Construction of a Cosmid Contig Spanning the SOBER1 Locus
A Col-0 genomic library constructed in the binary vector pBIC20 (a gift from C. Somerville, Carnegie Institute) was screened to generate a set of overlapping cosmid clones spanning SOBER1. The E. coli library was plated on Luria agar with tetracycline (10 μg/mL) and cycloheximide (50 μg/mL). Colonies were transferred to Hybond N+ nylon membranes (Amersham Biosciences) and the DNA was cross-linked. The AlkPhos direct labeling and detection system (GE Healthcare Bioscience) was used to label BAC-specific probes and to isolate clones. To fill contig gaps, we constructed and screened a binary cosmid library that was constructed using the two Col-0 BACs spanning the interval (ABRC, Ohio State University): T10l14 (left end) and F7K2 (right end). BAC DNA partially digested with Sau3AI was gel-purified and ligated to pCLD04541 (Bancroft et al., 1997) digested with BamHI. The cosmid clones were packaged into bacteriophage l particles with the Gigapack III packaging extract (Stratagene) and transduced into E. coli.
Complementation Analysis
The Col-0–derived binary cosmid clones spanning the SOBER1 locus were mobilized into A. tumefaciens strain GV3101 pMP90. The strains were used to transform Pi-0 plants by the floral dip method (Bent, 2000). Leaves of kanamycin-resistant T1 transgenic plants were infected with Pst DC3000 pVSP61(avrRpt21-100-avrBsT11-350-HA). Two Pi-0 T1 lines (11.785B and 11.795A) failed to induce an AvrBsT-dependent HR. The T-DNA region shared by 11.785B and 11.795A contains five genes annotated by the MIPS database: At4g22285, At4g22290, At4g2295, At4g22300, and At4g22310.
The Col-0 genomic region and/or cDNA for each candidate gene was independently transformed into Pi-0, and the respective T1 lines were scored for an AvrBsT-dependent HR. Specifically, Col-0 At4g22290 and At4922300 cDNAs (U24405 and U22920, respectively; ABRC) were cloned into binary vector pGWB2 (T. Nakagawa, Shimane University). At4g22285, At4g22295, and At4g22310 open reading frames were amplified by PCR using genomic DNA, sequenced, and then cloned into pGWB2. Binary plasmids were mobilized into A. tumefaciens strain GV3101 pMP90 to use for plant transformation.
RNA Isolation and RNA Gel Blot Analysis
Total RNA was isolated from adult leaves using Trizol reagent (Sigma-Aldrich) according to the manufacturer's instructions. For RNA gel blot hybridization, 20 μg of total RNA was denatured and separated on 1% agarose-formaldehyde gels (Sambrook et al., 1989). RNA was transferred onto a Hybond N+ membrane (GE Healthcare Bioscience) and cross-linked to the membrane using a Stratalinker UV cross-linking apparatus (Stratagene). The RNA gel blots were hybridized with a [32P]PR1 probe at 55°C overnight in 5× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), 0.5% SDS, 5× Denhardt's solution (1× Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), and 100 μg/mL salmon sperm DNA. Blots were washed in 1× SSC and 0.1% SDS buffer at 55°C and then exposed to film.
Sequencing of SOBER1 Alleles and RT-PCR
The genomic DNA sequence of At4g22300 from Pi-0, Ler, and Col-0 was sequenced using standard methods. To sequence the coding region, total RNA was isolated (as described above) to generate cDNA using 1 μg of RNA, oligo(dT) primers, and SuperScript III reverse transcriptase (Invitrogen). cDNA and gene-specific primers (available upon request) were used in semiquantitative RT-PCR and 3′ rapid amplification of cDNA ends. cDNA products were cloned into pCR8/TOPO/GW (Invitrogen).
Protein Purification and Enzyme Assays
Coding sequences of wild-type SOBER1, mutant SOBER1(H192A), and yeast (Saccharomyces cerevisiae) Sc APT1 (YRL118c) were amplified by PCR with primers incorporating restriction enzyme sites for cloning into the NdeI and BamHI sites of pET15b expression vector (Novagen/EMD Biosciences), generating His6-SOBER1, His6-SOBER1(H192A), and His6-ScAPT1, respectively. Plasmids were transformed into E. coli strain BL21(DE3) pLysS for protein expression. Bacteria induced with 0.5 mM isopropylthio-β-galactoside at 23°C for 4 h were lysed in buffer (50 mM phosphate buffer, pH 8, 300 mM NaCl, 0.5% Triton X-100, and 10 mM imidazole) and then sonicated. Insoluble material was removed by centrifugation at 17,000g for 20 min. The supernatant was incubated with nickel-nitrilotriacetic acid agarose Superflow agarose (Qiagen) for 30 min at 4°C and then poured into a column. After collecting the flow-through, the agarose was washed with 3 mL of lysis buffer and 10 mL of wash buffer (50 mM phosphate buffer, pH 7, 150 mM NaCl, 0.5% Triton X-100, 10 mM imidazole, and 20% glycerol). Recombinant protein was eluted in wash buffer containing a gradient of 10 to 200 mM imidazole. Protein fractions were pooled, desalted with a PD-10 column (Amersham Bioscience) to remove the imidazole, and then stored at −80°C.
The carboxylesterase assay was modified (Prim et al., 2003) to fit a 96-well format. Briefly, chromogenic p-nitrophenyl ester substrates (Sigma-Aldrich) prepared in DMSO were diluted in 50 mM phosphate buffer, pH 8, 0.1% gum arabic (Agros Organics), and 0.5% Triton X-100. This solution (230 μL) containing 0.45 mM substrate was incubated at 23°C with 3 μg of recombinant protein (20 μL) stored in buffer (50 mM phosphate buffer, pH 8, 150 mM NaCl, 0.5% Triton X-100, and 20% glycerol). p-Nitrophenol formation was recorded by monitoring absorbance at OD = 405 nm. The OD unit formation rates (OD/min) calculated by fitting a linear model to the data points (maximum rate over 10 min) were converted to micromoles of p-nitrophenol using a control calibration plot obtained in the same buffer conditions. Reactions were performed in triplicate. Carboxylesterase-specific activity is reported as micromoles of p-nitrophenol per minute per milligram of protein ± sd.
Modeling and Phylogenic Analyses
Protein structure modeling was performed with amino acids 1 to 213 of SOBER1 using two modeling templates, Hs APT1 (accession number O75608) and Pf EstB (accession number Q51758) according to the SWISS-MODEL (Schwede et al., 2003).
For neighbor-joining phylogenic tree reconstruction, SOBER1 amino acid sequence was queried against plant EST and cDNA sequences in the GenBank, The Institute for Genomic Research Plant Transcript Assemblies (http://plantta.tigr.org/), and PlantGDB-Assembled Unique Transcripts (http://www.plantgdb.org/) databases. Full-length hits with an E value < 10−10 were included in the analysis together with predicted Arabidopsis proteins At3g15650 and At5g20060 belonging to a diverging APT1-like family as an out group. Sequence alignments were initially performed using the ClustalW algorithm of the VectorNTI AlignX module with the BLOSUM62 matrix and default parameters; subsequently, they were manually refined in BioEdit (Tom Hall, Ibis Therapeutics). N- and C-terminal unrelated regions were deleted. Phylogenetic analyses and tree drawing were performed on the processed protein alignment (see Supplemental Figure 1 online) with MEGA version 3.1 software (Kumar et al., 2004) using the neighbor-joining method of inference with a JTT substitution model that assumed a uniform rate among sites and pairwise deletion of gaps. The inferred tree topology was tested by bootstrap analysis (1000 trials), and supporting values were included on the tree.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF100725 and EF100726.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Pst DC3000 Expressing the AvrBsTHyb Catalytic Core Mutant Protein Does Not Elicit a HR in Arabidopsis Pi-0 Leaves.
Supplemental Figure 2. Suppression of AvrBsT-Dependent HR in Pst DC3000–Infected Pi-0 Transgenic Lines.
Supplemental Figure 3. Xcc 8004 Expressing AvrBsT Does Not Elicit a HR on Pi-0 sober1-1 Transgenic Plants Complemented with the Col-0 At4g22300 cDNA.
Supplemental Figure 4. Protein Alignment Used for Phylogenetic Analyses and Tree Drawing for Figure 7D.
Supplemental Figure 5. Native Purification of the SOBER1 Protein.
Supplementary Material
Acknowledgments
We thank T. Nakagawa, J. Parker, C. Somerville, S. Somerville, B. Staskawicz, The Arabidopsis Information Resource, and the ABRC for reagents and seed stocks. We are also grateful to J. Monroe and S. Somerville for helpful discussions, S. Long and her laboratory for technical support, and J.-G. Kim for critical review. This work was funded by National Institutes of Health Grant 1RO1 GM-068886 and the Hellman's Scholar Fellowship awarded to M.B.M.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Mary Beth Mudgett (mudgett@stanford.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J.R., Charkowski, A.O., Deng, W.L., Badel, J.L., Petnicki-Ocwieja, T., van Dijk, K., and Collmer, A. (2000). The Pseudomonas syringae hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA 97 4856–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Arnold, D.L., Jackson, R.W., Fillingham, A.J., Goss, S.C., Taylor, J.D., Mansfield, J.W., and Vivian, A. (2001). Highly conserved sequences flank avirulence genes: Isolation of novel avirulence genes from Pseudomonas syringae pv. pisi. An. Microbiol. (Rio J.) 147 1171–1182. [DOI] [PubMed] [Google Scholar]
- Astua-Monge, G., Minsavage, G.V., Stall, R.E., Vallejos, C.E., Davis, M.J., and Jones, J.B. (2000). Xv4-AvrXv4: A new gene-for-gene interaction identified between Xanthomonas campestris pv. vesicatoria race T3 and the wild tomato relative Lycopersicon pennellii. Mol. Plant Microbe Interact. 13 1346–1355. [DOI] [PubMed] [Google Scholar]
- Bancroft, I., Love, K., Bent, E., Sherson, S., Lister, C., Cobett, C., Goodman, H.M., and Dean, C. (1997). A strategy involving the use of high redundancy YAC subclone libraries facilitates the contiguous representation in cosmid and BAC clones of 1.7 Mb of the genome of the plant Arabidopsis thaliana. Weeds World 4 1–9. [Google Scholar]
- Bent, A.F. (2000). Arabidopsis in planta transformation. Uses, mechanisms, and prospects for transformation of other species. Plant Physiol. 124 1540–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bonshtien, A., Lev, A., Gibly, A., Debbie, P., Avni, A., and Sessa, G. (2005). Molecular properties of the Xanthomonas AvrRxv effector and global transcriptional changes determined by its expression in resistant tomato plants. Mol. Plant Microbe Interact. 18 300–310. [DOI] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63. [DOI] [PubMed] [Google Scholar]
- Century, K.S., Shapiro, A.D., Repetti, P.P., Dahlbeck, D., Holub, E., and Staskawicz, B.J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278 1963–1965. [DOI] [PubMed] [Google Scholar]
- Chu, Z., Yuan, M., Yao, J., Ge, X., Yuan, B., Xu, C., Li, X., Fu, B., Li, Z., Bennetzen, J.L., Zhang, Q., and Wang, S. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20 1250–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesiolka, L.D., et al. (1999). Regulation of expression of avirulence gene avrRxv and identification of a family of host interaction factors by sequence analysis of avrBsT. Mol. Plant Microbe Interact. 12 35–44. [DOI] [PubMed] [Google Scholar]
- Coppinger, P., Repetti, P.P., Day, B., Dahlbeck, D., Mehlert, A., and Staskawicz, B.J. (2004). Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J. 40 225–237. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Deslandes, L., Olivier, J., Peeters, N., Feng, D.X., Khounlotham, M., Boucher, C., Somssich, I., Genin, S., and Marco, Y. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L., Olivier, J., Theulieres, F., Hirsch, J., Feng, D.X., Bittner-Eddy, P., Beynon, J., and Marco, Y. (2002). Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 99 2404–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devedjiev, Y., Dauter, Z., Kuznetsov, S.R., Jones, T.L., and Derewenda, Z.S. (2000). Crystal structure of the human acyl protein thioesterase I from a single X-ray data set to 1.5 A. Structure 8 1137–1146. [DOI] [PubMed] [Google Scholar]
- Dewdney, J., Reuber, T.L., Wildermuth, M.C., Devoto, A., Cui, J., Stutius, L.M., Drummond, E.P., and Ausubel, F.M. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24 205–218. [DOI] [PubMed] [Google Scholar]
- Duncan, J.A., and Gilman, A.G. (1998). A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J. Biol. Chem. 273 15830–15837. [DOI] [PubMed] [Google Scholar]
- Duncan, J.A., and Gilman, A.G. (2002). Characterization of Saccharomyces cerevisiae acyl-protein thioesterase 1, the enzyme responsible for G protein alpha subunit deacylation in vivo. J. Biol. Chem. 277 31740–31752. [DOI] [PubMed] [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. (2000). Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Biol. 3 278–284. [DOI] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R.D., et al. (2006). Pfam: Clans, web tools and services. Nucleic Acids Res. 34 D247–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J.T., and Yao, N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6 201–211. [DOI] [PubMed] [Google Scholar]
- Gurlebeck, D., Thieme, F., and Bonas, U. (2006). Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163 233–255. [DOI] [PubMed] [Google Scholar]
- Guttman, D.S., and Greenberg, J.T. (2001). Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol. Plant Microbe Interact. 14 145–155. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Parker, J.E. (2003). Deciphering plant-pathogen communication: Fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14 177–193. [DOI] [PubMed] [Google Scholar]
- Holmquist, M. (2000). Alpha/beta-hydrolase fold enzymes: Structures, functions and mechanisms. Curr. Protein Pept. Sci. 1 209–235. [DOI] [PubMed] [Google Scholar]
- Hong, K.H., Jang, W.H., Choi, K.D., and Yoo, O.J. (1991). Characterization of Pseudomonas fluorescens carboxylesterase: Cloning and expression of the esterase gene in Escherichia coli. Agric. Biol. Chem. 55 2839–2845. [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.B., Stall, R.E., and Baouzar, H. (1998). Diversity among xanthomonads pathogenic on pepper and tomato. Annu. Rev. Phytopathol. 36 41–58. [DOI] [PubMed] [Google Scholar]
- Kanamori, M., Saitoh, K.-I., Arie, T., Kmakura, T., and Teraoka, T. (2005). Possible roles and functions of LPL1 gene encoding lysophospholipase during early infection by Magnaporthe grisea. J. Gen. Plant Pathol. 71 253–262. [Google Scholar]
- Kim, K.K., Song, H.K., Shin, D.H., Hwang, K.Y., Choe, S., Yoo, O.J., and Suh, S.W. (1997). Crystal structure of carboxylesterase from Pseudomonas fluorescens, an alpha/beta hydrolase with broad substrate specificity. Structure 5 1571–1584. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Kuznetsova, E., Proudfoot, M., Sanders, S.A., Reinking, J., Savchenko, A., Arrowsmith, C.H., Edwards, A.M., and Yakunin, A.F. (2005). Enzyme genomics: Application of general enzymatic screens to discover new enzymes. FEMS Microbiol. Rev. 29 263–279. [DOI] [PubMed] [Google Scholar]
- Lavie, M., Seunes, B., Prior, P., and Boucher, C. (2004). Distribution and sequence analysis of a family of type III-dependent effectors correlate with the phylogeny of Ralstonia solanacearum strains. Mol. Plant Microbe Interact. 17 931–940. [DOI] [PubMed] [Google Scholar]
- Mateo, A., Muhlenbock, P., Rusterucci, C., Chang, C.C., Miszalski, Z., Karpinska, B., Parker, J.E., Mullineaux, P.M., and Karpinski, S. (2004). LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 136 2818–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos, M., Katagiri, F., Yu, G.-L., and Ausubel, F.M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78 1089–1099. [DOI] [PubMed] [Google Scholar]
- Mudgett, M.B. (2005). New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56 509–531. [DOI] [PubMed] [Google Scholar]
- Mudgett, M.B., Chesnokova, O., Dahlbeck, D., Clark, E.T., Rossier, O., Bonas, U., and Staskawicz, B.J. (2000). Molecular signals required for type III secretion and translocation of the Xanthomonas campestris AvrBs2 protein to pepper plants. Proc. Natl. Acad. Sci. USA 97 13324–13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett, M.B., and Staskawicz, B.J. (1999). Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: Demonstration of secretion and processing during bacterial pathogenesis. Mol. Microbiol. 32 927–941. [DOI] [PubMed] [Google Scholar]
- Mukherjee, S., Keitany, G., Li, Y., Wang, Y., Ball, H.L., Goldsmith, E.J., and Orth, K. (2006). Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312 1211–1214. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z., Eulgem, T., Holt, B.F., III, and Dangl, J.L. (2003). Recognition and response in the plant immune system. Annu. Rev. Genet. 37 579–609. [DOI] [PubMed] [Google Scholar]
- Noel, L., Thieme, F., Nennstiel, D., and Bonas, U. (2001). cDNA-AFLP analysis unravels a genome-wide hrpG-regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 41 1271–1281. [DOI] [PubMed] [Google Scholar]
- Oh, C.-S., Kim, J.F., and Beer, S.V. (2005). The Hrp atherogenicity island of Erwinia amylovora and identification of three novel genes required for systemic infection. Mol. Plant Pathol. 6 125–138. [DOI] [PubMed] [Google Scholar]
- Orth, K., Palmer, L.E., Bao, Z.Q., Stewart, S., Rudolph, A.E., Bliska, J.B., and Dixon, J.E. (1999). Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science 285 1920–1923. [DOI] [PubMed] [Google Scholar]
- Orth, K., Xu, Z.H., Mudgett, M.B., Bao, Z.Q., Palmer, L.E., Bliska, J.B., Mangel, W.F., Staskawicz, B., and Dixon, J.E. (2000). Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290 1594–1597. [DOI] [PubMed] [Google Scholar]
- Parker, J.E., Barber, C.E., Fan, M.J., and Daniels, M.J. (1993). Interaction of Xanthomonas campestris with Arabidopsis thaliana: Characterization of a gene from X. c. pv. raphani that confers avirulence to most A. thaliana accessions. Mol. Plant Microbe Interact. 6 216–224. [DOI] [PubMed] [Google Scholar]
- Portilla, D., Crew, M.D., Grant, D., Serrero, G., Bates, L.M., Dai, G., Sasner, M., Cheng, J., and Buonanno, A. (1998). cDNA cloning and expression of a novel family of enzymes with calcium-independent phospholipase A2 and lysophospholipase activities. J. Am. Soc. Nephrol. 9 1178–1186. [DOI] [PubMed] [Google Scholar]
- Prim, N., Bofill, C., Pastor, F.I., and Diaz, P. (2006). Esterase EstA6 from Pseudomonas sp. CR-611 is a novel member in the utmost conserved cluster of family VI bacterial lipolytic enzymes. Biochimie 88 859–867. [DOI] [PubMed] [Google Scholar]
- Prim, N., Sanchez, M., Ruiz, C., Javier Pastor, F.I., and Diaz, P. (2003). Use of methylumbelliferyl-derivative substrates for lipase activity characterization. J. Mol. Catal. B Enzym. 22 339–346. [Google Scholar]
- Roden, J., Belt, B., Ross, J., Tachibana, T., Vargas, J., and Mudgett, M.B. (2004. a). Genetic screen to isolate type III effectors translocated into plant cells during Xanthomonas campestris pathovar vesicatoria infection. Proc. Natl. Acad. Sci. USA 101 16624–16629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden, J., Eardley, L., Hotson, A., Cao, Y., and Mudgett, M.B. (2004. b). Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant Microbe Interact. 17 633–643. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schoof, H., Zaccaria, P., Gundlach, H., Lemcke, K., Rudd, S., Kolesov, G., Arnold, R., Mewes, H.W., and Mayer, K.F. (2002). MIPS Arabidopsis thaliana Database (MAtDB): An integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res. 30 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede, T., Kopp, J., Guex, N., and Peitsch, M.C. (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanado, Y., Kometani, M., Uchiyama, H., Koizumi, S., and Teno, N. (2004). Lysophospholipase I identified as a ghrelin deacylation enzyme in rat stomach. Biochem. Biophys. Res. Commun. 325 1487–1494. [DOI] [PubMed] [Google Scholar]
- Smotrys, J.E., and Linder, M.E. (2004). Palmitoylation of intracellular signaling proteins: Regulation and function. Annu. Rev. Biochem. 73 559–587. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E., Tiryaki, I., and Rowe, M.L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, H., Hayashi, H., and Yamashita, S. (1996). Purification, cDNA cloning, and regulation of lysophospholipase from rat liver. J. Biol. Chem. 271 7705–7711. [DOI] [PubMed] [Google Scholar]
- Tai, T., Dahlbeck, D., Stall, R.E., Peleman, J., and Staskawicz, B.J. (1999). High-resolution genetic and physical mapping of the region containing the Bs2 resistance gene of pepper. Theor. Appl. Genet. 99 1201–1206. [Google Scholar]
- Tatusov, R.L., et al. (2003). The COG database: An updated version includes eukaryotes. BMC Bioinformatics 4 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme, F., et al. (2005). Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187 7254–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, P., Barber, C., and Daniels, M. (1984). Behaviour of the transposons Tn5 and Tn7 in Xanthomonas campestris pv. campestris. Mol. Gen. Genet. 195 101–107. [Google Scholar]
- Veit, M., and Schmidt, M.F. (2001). Enzymatic depalmitoylation of viral glycoproteins with acyl-protein thioesterase 1 in vitro. Virology 288 89–95. [DOI] [PubMed] [Google Scholar]
- Wang, A., Deems, R.A., and Dennis, E.A. (1997. a). Cloning, expression, and catalytic mechanism of murine lysophospholipase I. J. Biol. Chem. 272 12723–12729. [DOI] [PubMed] [Google Scholar]
- Wang, A., and Dennis, E.A. (1999). Mammalian lysophospholipases. Biochim. Biophys. Acta 1439 1–16. [DOI] [PubMed] [Google Scholar]
- Wang, A., Johnson, C.A., Jones, Y., Ellisman, M.H., and Dennis, E.A. (2000). Subcellular localization and PKC-dependent regulation of the human lysophospholipase A/acyl-protein thioesterase in WISH cells. Biochim. Biophys. Acta 1484 207–214. [DOI] [PubMed] [Google Scholar]
- Wang, A., Loo, R., Chen, Z., and Dennis, E.A. (1997. b). Regiospecificity and catalytic triad of lysophospholipase I. J. Biol. Chem. 272 22030–22036. [DOI] [PubMed] [Google Scholar]
- Wang, A., Yang, H.C., Friedman, P., Johnson, C.A., and Dennis, E.A. (1999). A specific human lysophospholipase: cDNA cloning, tissue distribution and kinetic characterization. Biochim. Biophys. Acta 1437 157–169. [DOI] [PubMed] [Google Scholar]
- Wang, X. (2001). Plant phospholipases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 211–231. [DOI] [PubMed] [Google Scholar]
- Whalen, M.C., Stall, R.E., and Staskawicz, B.J. (1988). Characterization of a gene from a tomato pathogen determining hypersensitive resistance in non-host species and genetic analysis of this resistance in bean. Proc. Natl. Acad. Sci. USA 85 6743–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen, M.C., Wang, J.F., Carland, F.M., Heiskell, M.E., Dahlbeck, D., Minsavage, G.V., Jones, J.B., Scott, J.W., Stall, R.E., and Staskawicz, B.J. (1993). Avirulence gene avrRxv from Xanthomonas campestris pv. vesicatoria specifies resistance on tomato line Hawaii 7998. Mol. Plant Microbe Interact. 6 616–627. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565. [DOI] [PubMed] [Google Scholar]
- Yamada, K., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302 842–846. [DOI] [PubMed] [Google Scholar]
- Yang, B., Sugio, A., and White, F.F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 103 10503–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.Y., and Dennis, E.A. (1988). Purification and characterization of a lysophospholipase from a macrophage-like cell line P388D1. J. Biol. Chem. 263 9965–9972. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.