Abstract
The Arabidopsis thaliana MYB26/MALE STERILE35 (MS35) gene is critical for the development of secondary thickening in the anther endothecium and subsequent dehiscence. MYB26 is localized to the nucleus and regulates endothecial development and secondary thickening in a cell-specific manner in the anther. MYB26 expression is seen in anthers and also in the style and nectaries, although there is no effect on female fertility in the ms35 mutant. MYB26 expression in anthers occurs early during endothecial development, with maximal expression during pollen mitosis I and bicellular stages, indicating a regulatory role in specifying early endothecial cell development. Overexpression of MYB26 results in ectopic secondary thickening in both Arabidopsis and tobacco (Nicotiana tabacum) plants, predominantly within the epidermal tissues. MYB26 regulates a number of genes linked to secondary thickening, including IRREGULAR XYLEM1 (IRX1), IRX3, IRX8, and IRX12. Changes in expression were also detected in two NAC domain genes, NAC SECONDARY WALL–PROMOTING FACTOR1 (NST1) and NST2, which have been linked to secondary thickening in the anther endothecium. These data indicate that MYB26 regulates NST1 and NST2 expression and in turn controls the process of secondary thickening. Therefore, MYB26 appears to function in a regulatory role involved in determining endothecial cell development within the anther and acts upstream of the lignin biosynthesis pathway.
INTRODUCTION
In Arabidopsis thaliana, as in most flowering plant species, the anther wall comprises four cell layers: the epidermis, endothecium, middle layer, and tapetum (Dawson et al., 1993). Anther dehiscence is a multiple-step process that includes the lytic opening of a longitudinal line of weakness in the epidermis, known as the stomium, and retraction of the anther wall to widen the stomium and permit pollen release. During microspore maturation, cellulose and lignified thickenings are deposited in the endothecium (Dawson et al., 1999). The cells of the stomium and circular cell cluster (present in tobacco [Nicotiana tabacum]) are then enzymatically lysed (Goldberg et al., 1993). The endothecium is thought to be critical in generating the forces required for dehiscence. First, swelling of the endothecium cells provides an inwardly directed force, causing the weakened stomium to rupture. Then, desiccation of the endothecium causes differential shrinkage of thickened and unthickened parts of the cell wall, resulting in an outwardly bending force, leading to retraction of the anther wall and full opening of the stomium (Keijzer, 1987; Bonner and Dickinson, 1989).
A number of Arabidopsis dehiscence mutants have been characterized, such as dde1/opr3 (Sanders et al., 2000; Stintzi and Browse, 2000), coi1 (Feys et al., 1994; Xie et al., 1998), dad1 (Ishiguro et al., 2001), and aos/dde2-2 (Park et al., 2002; von Malek et al., 2002), which disrupt the jasmonic acid (JA) biosynthesis/signaling pathway. Analysis of these mutants suggests that water transport into the anthers is regulated by JA/ethylene (Ishiguro et al., 2001). The SUC1 protein, a plasma membrane H+-sucrose symporter, accumulates in the connective cells surrounding the vascular tissue during the final stages of anther development, and it has been suggested that this causes an accumulation of sucrose in these tissues that results in increased water uptake (Stadler et al., 1999). It is speculated that JA may be needed for the expression of SUC1 and other genes associated with water transport in anthers (Ishiguro et al., 2001).
The ms35/myb26 mutation appears to disrupt a pathway distinct from that defined by the JA dehiscence mutants. Anther and pollen development follows the same pattern as in wild-type plants, with lysis of the cells forming the stomium, leaving no bridge between the epidermal/endothecial cells on either side of the stomium. However, wall thickenings are not formed in the ms35 endothecium, resulting in the endothecial cells becoming flattened and distorted (Dawson et al., 1999). The lack of thickening means that the outward bending of the anther wall fails to occur, the endothecial cells collapse, and the anther fails to open. However, the pattern of lignification in the vascular tissue of the anthers, stems, and leaves is the same in wild-type and ms35/myb26 plants. Therefore, the ms35 mutation appears to specifically affect secondary wall deposition in the endothecium. We have shown that the failure of dehiscence in the ms35 mutant is attributable to a defect in the MYB26 gene that is homologous with the R2R3-type MYB transcription factors (Steiner-Lange et al., 2003). R2R3-type MYB transcription factors are thought to act either as transcriptional activators or repressors, controlling development and the determination of cell fate as well as the regulation of biosynthetic pathways (Stracke et al., 2001). They have also been specifically linked to a variety of metabolic pathways associated with the regulation of phenylpropanoid metabolism (Borevitz et al., 2000; Yang et al., 2001; Patzlaff et al., 2003; Newman et al., 2004; Rogers and Campbell, 2004; Goicoechea et al., 2005; Rogers et al., 2005; Deluc et al., 2006). Overexpression of many of these MYB factors results in the induction of genes associated with the phenylpropanoid biosynthesis pathway; for example, in Antirrhinum majus, Am MYB305 and Am MYB340 regulate flavanol biosynthesis (Moyano et al., 1996), whereas Am MYB308 (Tamagnone et al., 1998) and the Arabidopsis homolog At MYB4 (Jin et al., 2000) downregulate hydrocinnamic acid and monolignol biosynthesis.
The deposition of lignified secondary thickening is usually only seen once cell growth has stopped. Secondary cell walls are composed predominantly of cellulose, lignin, and xylan, and the biosynthesis and deposition of wall material involves the coordinated regulation of a number of complex metabolic pathways. Defects associated with the failure of secondary thickening exhibit a diversity of phenotypes, ranging from collapse of the xylem vessels (irregular xylem [irx]) (Turner and Somerville, 1997) to loss of dehiscence (Mitsuda et al., 2005). Two NAC domain transcription factors, NAC SECONDARY WALL–PROMOTING FACTOR1 (NST1) and NST2, have been proposed as regulators of secondary wall thickening and specifically endothecium thickening (Mitsuda et al., 2005). Mitsuda et al. (2005) proposed that NST1 and NST2 act redundantly to regulate secondary cell thickenings in anther walls and are required for anther dehiscence; they also implied that NST1 and NST2 may act by regulating MYB26 expression.
We have performed a functional analysis of MYB26 and demonstrated that MYB26 is localized to the nucleus and plays a critical role in the regulation of endothecial maturation and secondary thickening in a cell-specific manner in the anther. Overexpression of MYB26 results in the development of ectopic secondary thickening in both Arabidopsis and tobacco plants. MYB26 appears to regulate a number of genes linked to secondary thickening. Changes in expression were detected in two NAC domain genes, NST1 and NST2, and in IRX1, IRX3, IRX8, and IRX12, indicating that MYB26 may be involved in their regulation. These data suggest that MYB26 plays a regulatory role in determining cell development, or competence for secondary thickening, within the anther and acts upstream of the lignin biosynthesis pathway, possibly via NST1 and NST2.
RESULTS
Alterations in Secondary Thickening in the ms35 Mutant
The ms35 mutant is male-sterile as a result of a failure of anther dehiscence. In the wild type, secondary thickening occurs in the endothecium as bands of striated spring-like thickening that is composed of cellulose and lignin (Figures 1A, 1D, and 1N). Deposition commences during pollen mitosis I and is maximal by the end of pollen mitosis II (Figures 1G and 1I). Secondary thickening is absent from the surrounding cell layers of the anther and is necessary to create the shearing force required for anther dehiscence. In the ms35 mutant, no secondary thickening or lignification is seen in the anther endothecium (Figures 1B and 1E), although other tissues in the mutant plant undergo normal secondary thickening. This lack of strengthening of the endothecium means that as the anther dehydrates the endothecium layer distorts, shearing forces do not develop, and the anther fails to open. The appearance of the endothecium changes before the onset of secondary thickening (Figure 1G). In the wild type, as the endothecial cells develop they expand, particularly toward the center of the locule. However, in the ms35 mutant, the endothecial cell number does not appear changed, and these cells do not fully expand and do not have any secondary thickening (Figures 1H and 1J). Cell expansion is an essential prerequisite before laying down secondary thickening, because the rigidity of the secondary wall prevents subsequent expansion. This suggests that MYB26 acts at an early stage of endothecium development, governing cell expansion or competence, before the deposition of secondary thickening.
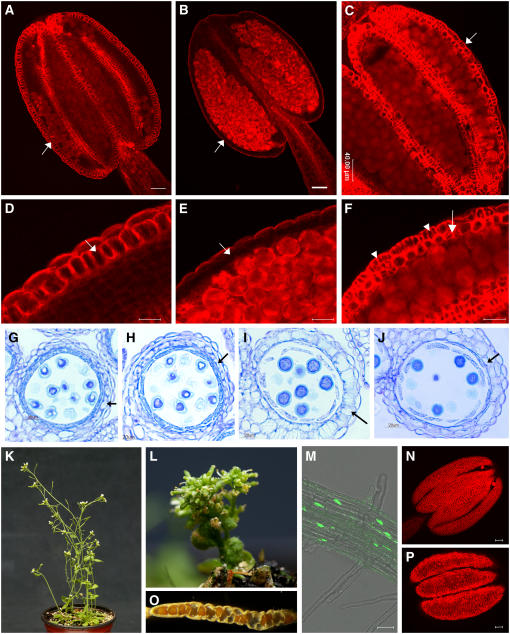
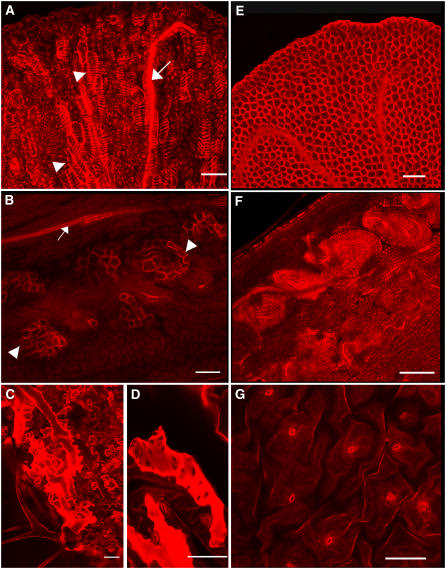
Figure 1.
Phenotypic Characterization of Arabidopsis ms35 Mutant and Arabidopsis Pro35S:MYB26 Overexpression Lines.
(A) to (F) Anthers were stained with acridine orange/ethidium bromide and visualized by confocal microscopy (excitation, 590 nm).
(A) Ler wild-type partially dehisced anther from open flowers. Secondary thickening is visible in the endothecium (arrow).
(B) ms35 mutant nondehiscent mature anther from open flowers. Secondary thickening is absent in the endothecium (arrow).
(C) Mature anther from open flowers of a MYB26 overexpression line.
(D) Close-up of (A).
(E) Close-up of (B).
(F) Close-up of (C). Secondary thickening is visible in the endothecium (arrow) and as ectopic islands in the epidermal tissues (arrowheads).
(G) to (J) Sections through wild-type anthers ([G] and [I]) and ms35 mutant anthers ([H] and [J]).
(G) During pollen mitosis I, endothecial cells begin to expand (arrow) in the wild type.
(H) At the corresponding stage in the ms35 mutant, minimal endothecium expansion is seen (arrow).
(I) During pollen mitosis II, endothecium expansion continues in the wild type and secondary thickening is clearly evident (arrow).
(J) At the corresponding stage in the mutant, the endothecium cells fail to fully expand and no secondary thickening occurs (arrow).
(K) to (M) and (O) MYB26 overexpression lines.
(K) The overexpression (OEx) line OEx4-8, showing less severe phenotypic changes.
(L) Line OEx8-1, showing an extreme dwarf phenotype and only partial fertility.
(M) Nuclear localization of Pro35S:MYB26:GFP OEx in root tissues.
(O) Mature silique from a reduced less extreme line, showing deformity attributable to increased secondary thickening.
(N) and (P) Overview of mature anthers from the wild type (N) and a MYB26 OEx line (P) showing increased lignification in the MYB26 OEx line.
Bars = 40 μm except in (G) to (J), where bars = 20 μm.
Regulation of MYB26 Expression
The ms35 mutation is attributable to a deletion and rearrangement 1288 bp upstream of the translational start of MYB26, which results in a severe downregulation of MYB26 expression and the failure of dehiscence (Steiner-Lange et al., 2003), indicating that the MYB26 regulatory region extends a considerable distance upstream from the predicted translational start site of the gene (Figure 2). There is a large region (8.124 kb) upstream of MYB26 before the next annotated gene that does not contain any predicted open reading frames, suggesting that this region may have some role in the regulation of MYB26 expression. We have identified and characterized two SALK knockout lines that contain insertions upstream of MYB26 (SALK_056264) and within the coding sequence (SALK_112372) (Figure 2). Homozygous knockout lines were generated and screened for the presence of insertions and phenotypic changes; both lines were male-sterile as a result of a lack of secondary thickening in the endothecium and the failure of anther dehiscence. These lines were crossed using pollen from heterozygous ms35MS35 plants; the resulting progeny from each cross segregated 1:1 for male sterility:fertility (data not shown), indicating that they were allelic to the ms35 mutation. These knockout lines gave the same phenotype of full sterility attributable to a lack of dehiscence; this was expected in the case of SALK_112372, which lies within the third exon of the MYB26 transcript. However, the flanking sequence from the SALK_056264 insertion maps to −1696 bp upstream of the translational start (Figure 2). Therefore, the MYB26 regulatory region may extend as far as −1696 bp upstream from the translational start site, and this region is required for functional expression of the MYB26 transcript. Bioinformatic analysis of this region has not indicated any definitive motifs that may be responsible for this regulation.
Figure 2.
Structure of the MYB26 Gene.
Map of the Arabidopsis At3g13890 region, showing ms35 deletion and positions of SALK knockout lines. Arrows indicate open reading frames.
A promoter:β-glucuronidase (GUS) fusion was constructed to localize the expression pattern of MYB26; the 3.1-kb region upstream of the transcriptional start site was fused to the GUS reporter gene and transformed into Landsberg erecta (Ler) wild-type plants. Expression of ProMYB26:GUS is seen only in floral tissues, with signal observed in the anthers and filaments of the flowers; strong signal is also detected in the style and nectaries of the flowers (Figure 3). Expression is observed in young postmeiotic buds through to open flowers (Figures 3A and 3C). Expression is seen at a low level throughout the anther in the tapetum, endothecium, and connective tissues (Figure 3B) and also in the filament tissues (Figures 3A and 3D). Strong GUS staining is also visible in the stylar tissue and nectaries after fertilization, during early seed development (Figure 3E). No signal is detected in vegetative tissues, and no vegetative expression is detected by RT-PCR analysis (data not shown). Although strong GUS staining is seen in the style and nectaries of the ProMYB26:GUS transgenic plants, no significant phenotypic differences associated with an alteration of lignification are observed in these tissues in the ms35 mutant. A slight reduction in lignification is apparent in ms35 mutant styles, although lignification is still present and no change in female fertility is detected, because full seed set can be obtained by crossing the male-sterile mutant with pollen from wild-type plants.
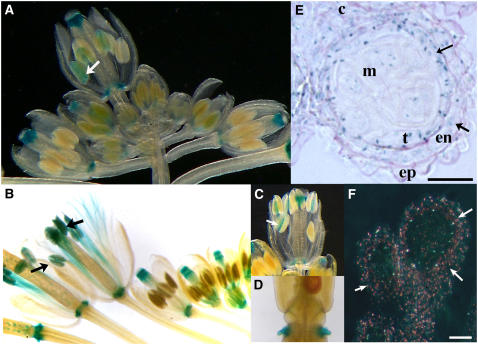
Figure 3.
ProMYB26:GUS Fusion Expression in Arabidopsis.
(A) and (B) Inflorescence showing expression of GUS in anthers, filaments, nectaries, and stylar tissues (arrows indicate expression in anther tissue). (A) is a dark-field image.
(C) Close-up dark-field image of buds before opening, showing expression in postmeiotic anthers (arrow).
(D) Immature silique showing GUS expression in nectaries.
(E) and (F) Sections through anther at pollen mitosis I stage. GUS expression can be seen throughout the anther, particularly in the endothecium and tapetal and connective tissues (arrows). (E) is a dark-field image. c, connective tissue; en, endothecium; ep, epidermis; m, microspores; t, tapetum. Bars = 40 μm.
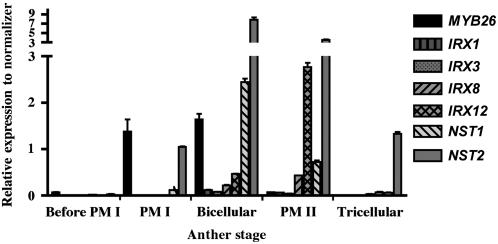
The ProMYB26:GUS fusion data were supported by quantitative PCR analysis of MYB26 expression in staged flowers and anthers (Figure 4). High levels of MYB26 expression were detected in excised anthers at pollen mitosis I and bicellular stages (Figure 4). Very low levels of expression were detected before pollen mitosis I and in pollen mitosis II, but at a significantly lower level than in pollen mitosis I and bicellular stages. Quantitative RT-PCR analysis of expression was performed in buds, which showed expression in older buds (tricellular pollen stage) and open flowers (see Supplemental Figure 1 online), indicating that this expression corresponds to that seen in ProMYB26:GUS fusion lines in the nectaries and stylar tissue and not to anther expression. Other genes associated with secondary thickening in the anther were also analyzed by quantitative RT-PCR (Figure 4); these show expression at stages after MYB26, indicating that they may be regulated by MYB26 or induced by changes in the cell competence linked to MYB26 expression. The timing of MYB26 expression, before secondary thickening formation, and the altered appearance of the endothecium cells implies that MYB26 may play a regulatory role at an earlier stage associated with the determination of cell expansion or maturation rather than by direct regulation of secondary cell wall thickening.
Figure 4.
Quantitative RT-PCR Expression in Wild-Type Arabidopsis Staged Excised Anthers.
Relative expression levels were determined compared with actin expression and expressed as fold changes relative to actin expression (Stratagene Mx3005P). The data pool consisted of two replicates repeated on at least two separate occasions; error bars show sd of expression changes. PM I, pollen mitosis I; PM II, pollen mitosis II.
Ectopic Lignification Attributable to Overexpression of MYB26
The MYB26 cDNA was cloned from closed buds into the pGWB5 vector (kindly provided by Tsuyoshi Nakagawa, Shimane University) under the control of the cauliflower mosaic virus (CaMV)35S promoter with a C-terminal green fluorescent protein (GFP) sequence to analyze the effect of ectopic overexpression. This construct was transferred into the wild-type Ler background and also into heterozygous ms35 gl1 plants. The Pro35S:MYB26 transcript partially complemented the ms35 mutation, causing some restoration of lignification within the endothecium that was sufficient to allow partial anther dehiscence and limited self-fertilization, although complete endothecial thickening did not occur. The fact that endothecial thickening was induced by the Pro35S:MYB26:GFP construct implies that the GFP tag does not affect the function of the MYB26 protein; rather, the CaMV35S promoter was unable to regulate expression in the anther to the same extent as seen with the native MYB26 promoter.
The pGWB5:MYB26 construct contains a C-terminal GFP tag, and expression of the GFP could be visualized by confocal microscopy within the nuclei, confirming that MYB26 is localized to the nucleus (Figure 1M), as predicted for its role as a putative transcription factor. Transgenic lines were tested by semiquantitative RT-PCR and quantitative RT-PCR to identify those showing overexpression of MYB26. Different levels of expression were seen between the different lines and in the different tissues (see Figures 7 and 8 below), and varying degrees of phenotypic changes were observed between different lines (Figures 1K and 1L). In all cases, the plants overexpressing MYB26 were reduced in size, which appeared to be attributable to a premature cessation of cell expansion rather than to altered cell number. This variation in size ranged from partially reduced to extremely stunted (Figures 1K and 1L). Changes in the thickening of tissues was evident in many organs of the plant, particularly leaves, petals, and ovules (Figure 5); however, no changes were detected in the root tissues, despite obvious expression in the roots (Figure 1M). Levels of fertility varied, with reduced fertility correlating with levels of ectopic thickening in the reproductive tissues (e.g., OEx8-1 and OEx8-3); very extreme lines were very stunted and completely sterile.
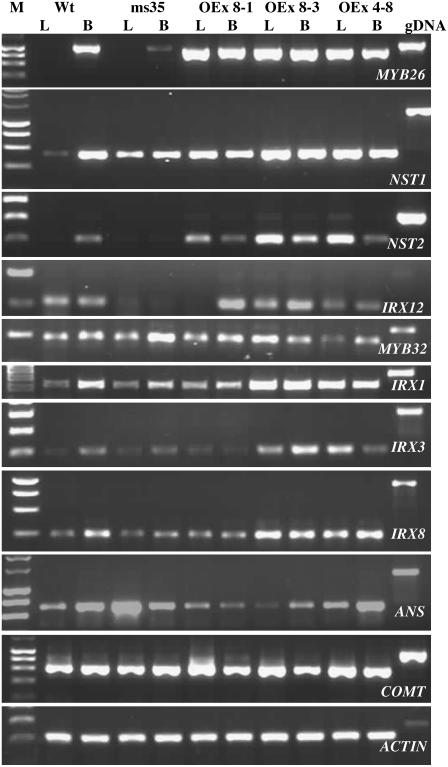
Figure 7.
RT-PCR Analysis of Genes Associated with Secondary Thickening in Arabidopsis Wild Type, ms35 Mutant, and MYB26 Overexpression Lines.
Overexpression lines were OEx8-1, OEx8-3, and OEx4-8 (Table 1; see Supplemental Table 1 online). M, 1-kb marker; Wt, wild type; L, leaf tissue; B, buds; gDNA, genomic DNA control.
Figure 8.
Quantitative RT-PCR of Selected Genes Showing Altered Regulation in the Arabidopsis MYB26 Overexpression Lines OEx8-1, OEx8-3, and OEx4-8.
Data are shown for bud tissues (A) and leaves (B). Relative expression levels were determined compared with actin leaf expression using the 2−ΔΔCT analysis method and are expressed as fold changes relative to actin. The data shown are from two replicates repeated on at least two separate occasions; error bars show sd of expression changes.
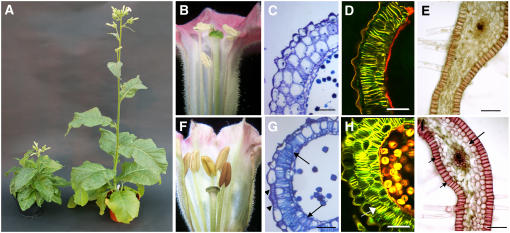
Figure 5.
Ectopic Secondary Thickening in Different Tissues in the Arabidopsis MYB26 Overexpression Lines and Wild-Type Ler.
Tissues ([A] to [D], At MYB OEx; [E] to [G], wild type) were stained with acridine orange/ethidium bromide and visualized by confocal microscopy (590 nm). Bars = 40 μm.
(A) Petal from a MYB26 OEx line, showing normal xylem development (arrow) and ectopic spring-like secondary thickening (arrowheads).
(B) Immature ovule from a MYB26 OEx line, showing normal xylem development (arrow) and ectopic secondary thickening in the epidermal tissues of the ovule (arrowheads).
(C) and (D) Leaf tissues from a MYB26 OEx line, showing net-like ectopic secondary thickening. A more detailed view is shown in (D).
(E) Wild-type petal, showing normal xylem development.
(F) Corresponding wild-type ovule.
(G) Wild-type leaf tissue.
In the floral and reproductive tissues, the ectopic thickening resembled the striated spring-like bands of thickening that are seen in the xylem tissues (Fukuda, 1997) and the anther endothecium (Dawson et al., 1999) (Figures 5A and 5B). However, in the leaf tissues, ectopic lignification of the epidermal cells appeared as a dense net-like pattern that covered the epidermal cell walls (Figures 5C and 5D). The leaves of the plants tended to be distorted and twisted as a consequence of the changes in the thickening of these tissues. In all cases, the thickening was caused by modifications in both secondary thickening and lignification, as indicated by phloroglucinol-HCl and also acridine orange/ethidium bromide staining; however, normal lignification of xylem tissues still occurred in these lines (Figures 5A and 5B).
Ectopic secondary thickening and lignification are frequently associated with the epidermal cells (Figures 1 and 5) and involve specific patches of cells within the different tissues rather then all cells within a tissue. This is particularly evident in the epidermal tissues of the anther. In the wild type, secondary thickening occurs in the anther endothecium as bands of striated spring-like thickening (Figures 1A, 1D, 1I, and 1N); this is not seen in the surrounding epidermal cell layers of the anther and is absent in the ms35 mutant (Figures 1B, 1E, and 1J). However, in the lines overexpressing MYB26, patches of epidermal cells within the anther also develop these spring-like secondary thickenings (Figures 1C, 1F, and 1P). These bands of thickening run along the longitudinal axis of the cells; in the resultant architecture, the thickening in the two cell layers, endothecium and epidermis, runs perpendicular to each other. This means that the bands of thickening run in opposing directions. The result of this is that as the anther dehydrates, opposing forces are generated, rather than the unidirectional force seen in wild-type anthers; therefore, the anther is unable to open fully and is effectively male sterile or of reduced fertility. When released manually, the pollen is viable; however, the extreme lines also show ectopic lignification of the style, ovaries, and ovules, with associated effects on female fertility, and therefore are completely sterile. In some less extreme cases, anther dehiscence and seed set occur, although siliques from these plants are frequently distorted as a consequence of increased thickening (Figure 1O).
Similar patches of ectopic secondary thickening were seen in most of the tissues analyzed except the roots (Figures 1 and 5). The selectivity in cells that developed ectopic thickening was also clearly evident within the ovary, where the ovule wall developed secondary thickening and the other cell layers did not (Figures 5B and 5F). The patchy development of secondary thickening in different cells may be a consequence of levels of MYB26 expression varying on a cell-to-cell basis, or more likely, given that regulation is by the CaMV35S promoter, a reflection of the competence of those cells to develop secondary thickening.
Ectopic Lignification in Tobacco by Overexpression of MYB26
The Arabidopsis Pro35S:MYB26:GFP constructs were also transferred into tobacco by Agrobacterium tumefaciens transformation of leaf explants and transgenic plants regenerated from transformed callus. The plants showed a similar phenotype to that seen in the Arabidopsis OEx lines; they were stunted, bushy, and smaller than wild-type equivalents (Figure 6A) and the leaves, flowers, and stems were much tougher and harder, with increased rigidity. The leaves showed abnormal epidermal cell shape and were contorted as a consequence of increased secondary thickening (Figure 6A). Increased amounts of lignin were associated with the secondary thickening, as indicated by increased levels of phloroglucinol-HCl staining in petals (Figures 6E and 6I). As seen in the Arabidopsis MYB26 OEx material, no changes in secondary thickening were seen in the root tissues and normal lignification was still observed in other parts of the plants. The most extreme MYB26 OEx plants were also both male- and female-sterile, with thickening occurring in the ovaries and a failure of dehiscence attributable to an increase in layers of opposing thickening in the anther (Figures 6B to 6D and 6F to 6H). The epidermal tissues of the anther were particularly affected, but the extent of thickening in the other cell layers was also increased greatly (Figures 6C, 6D, 6G, and 6H). Tobacco anthers undergo lignification of both the endothecium and middle cell layers (Sanders et al., 2005), resulting in a double layer of lignified cells surrounding the anther. However, in the wild type, these run in parallel, facilitating anther dehiscence, unlike the situation seen in the OEx lines, in which the epidermal thickening is in the opposite orientation and dehiscence cannot fully occur (Figure 6H).
Figure 6.
Phenotypic Characterization of Tobacco Pro35S:MYB26 Overexpression Lines.
(A) A MYB26 OEx line (left) and a wild-type tobacco plant (right).
(B) to (E) Wild-type tobacco.
(F) to (I) MYB26 OEx tobacco.
(B) and (F) Open flowers. Anthers in the At MYB35 OEx line (F) fail to fully dehisce, and petal pigmentation is reduced.
(C) and (G) Transverse toluidine blue–stained anther sections, showing ectopic secondary thickening in epidermal cells (arrowheads) and increased endothecium and middle cell layer thickenings (arrows) in the OEx lines (G).
(D) and (H) Acridine orange/ethidium bromide–stained anthers (merged images; excitation, 520 and 590 nm), showing increased lignification and secondary thickening in the epidermis, endothecium, and middle cell layer of the OEx lines (H).
(E) and (I) Phloroglucinol-stained petal sections from the central corolla region. Increased lignification, as indicated by stronger staining, is seen in the epidermis (small arrows) and the ground parenchyma cells of the OEx lines (large arrow).
Bars = 40 μm.
Petal and anther pigmentation varied in the tobacco OEx lines, with the most severe lines showing a significant reduction of pigmentation in the petals (Figure 6F). This finding suggests that in tobacco overexpression of the pathway regulating secondary thickening also affects flavonoid development; no such changes were evident in the Arabidopsis OEx lines.
Expression of Genes Associated with Secondary Thickening in Arabidopsis Overexpression Lines
Overexpression of MYB26 resulted in ectopic secondary thickening and lignification in various organs and cell types within the transgenic lines. The Arabidopsis lines carrying the Pro35S:MYB26:GFP construct and the ms35 mutant were analyzed by semiquantitative RT-PCR for alterations in the expression of selected genes linked to secondary thickening (Table 1). These genes fall into a number of distinct classes, including those associated with the regulation of secondary thickening, to key steps in the phenylalanine ammonia-lyase (PAL) pathway, or to cellulose synthesis and secondary thickening (Table 1; see Supplemental Table 1 online). Quantitative RT-PCR analysis was also conducted on a subset of these genes, and the data were analyzed by normalization and relative expression to actin controls using the 2−ΔΔCT method (Livak and Schmittgen, 2001) (Figure 8); the observed expression patterns mimic those seen by semiquantitative RT-PCR analysis.
Table 1.
Summary of Semiquantitative RT-PCR of Genes Associated with Lignin and Secondary Thickening
| Gene of Interest | Wild-Type Expression
|
ms35 Mutant
|
OEx Lines
|
Change in ms35 Mutant | Change in OEx Lines | Putative Role of MYB26 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| AGI Code | Leaves | Buds | Leaves | Buds | Leaves | Buds | ||||
| NST1 | At2g46770 | * | *** | ** | ** | *** | *** | Decrease in buds | Increase in leaves | Regulation |
| NST2 | At3g61910 | – | ** | * | * | ** | ** | Decrease in buds | Increase in leaves | Regulation |
| MYB32 | At4g34990 | ** | ** | ** | *** | ** | ** | Increase in buds | No change | Secondary effect |
| MYB61 | At1g09540 | * | ** | * | ** | * | * | No change | No change | No effect |
| IRX8 | At5g54690 | * | ** | * | * | *** | *** | Decrease in buds | Increase | Regulation |
| IRX1 | At4g18780 | * | ** | * | ** | ** | ** | Slight decrease | Increase in leaves | Regulation |
| IRX3 | At5g17420 | * | ** | * | ** | ** | ** | No change | Increase | Regulation |
| IRX5 | At5g44030 | ** | *** | ** | *** | ** | *** | No change | No change | No effect |
| IRX13 | At5g03170 | * | ** | * | ** | * | ** | No change | No change | No effect |
| IRX12 (laccase) | At2g38080 | ** | ** | * | * | ** | ** | Decrease | Increase | Regulation |
| XCIP | At4g35350 | ** | ** | ** | ** | ** | ** | No change | No change | No effect |
| ANS | At2g38240 | ** | ** | *** | ** | ** | ** | Increase in leaves | No change | No effect |
| C4H | At2g30490 | ** | ** | ** | ** | ** | ** | No change | No change | No effect |
| DFR | At1g08200 | *** | *** | *** | *** | *** | *** | No change | No change | No effect |
| COMT | At1g67980 | ** | ** | ** | ** | *** | ** | No change | Increase in leaves | Secondary effect |
| PAL2 | At3g53260 | ** | ** | ** | ** | ** | ** | No change | No change | No effect |
| IRX4 | At1g15950 | *** | *** | *** | *** | *** | *** | No change | No change | No effect |
| At3g62160 | At3g62160 | ** | ** | NT | ** | ** | ** | No change | No change | No effect |
| AtHB-8 | At4g32880 | ** | ** | NT | ** | ** | ** | No change | No change | No effect |
These data were determined using overexpression lines OEx8-1, OEx8-3, and OEx4-8 (Figure 7). ***, high level of expression; **, moderate level of expression; *, low level of expression; –, no expression detected. AGI, Arabidopsis Genome Initiative; NT, not tested.
As expected, MYB26 expression was decreased in the ms35 mutant buds and increased in the MYB26 OEx lines. The OEx lines showed varying levels of expression in the buds and in all cases showed ectopic expression in the leaf tissues, which normally do not express MYB26 (Figures 7 and 8). No changes in At MYB32 and At MYB61 expression were observed in the OEx lines, although the At MYB32 transcript increased slightly in the ms35 mutant buds. These MYB transcription factors have been linked to the regulation of phenylpropanoid metabolism and ectopic lignification in Arabidopsis (Newman et al., 2004; Preston et al., 2004) respectively. MYB32 has been shown to be expressed at high levels in the tapetum and to be important for stamen development (Preston et al., 2004; Mandaokar et al., 2006). Therefore, the expression change seen in the ms35 buds, and the associated lack of change in the OEx lines, are likely to be secondary consequences of changes in anther development and not of direct MYB26 regulation, indicating that MYB26 does not act via a pathway involving either of these transcription factors. We also observed a slight increase in expression of the ANTHOCYANIDIN SYNTHASE (ANS) gene in ms35 leaves; no phenotypic changes were seen in the ms35 mutant leaves, so this is likely to represent secondary changes in the regulation of pathways attributable to the lack of secondary thickening in the anther.
Two NAC domain genes, NST1 and NST2, have been linked to secondary thickening of the anther tissues (Mitsuda et al., 2005), with the overexpression of NST1 and NST2 reported to induce ectopic secondary thickening of the anther tissues. Mitsuda et al. (2005) suggested that they may be involved in regulating MYB26 expression; however, no data were presented to support this. By contrast, our analysis supports the reverse situation, with downregulation of NST1 and NST2 in the ms35 mutant and a corresponding increase of NST1 and NST2 expression in the Pro35S:MYB26 lines (Figures 7 and 8). Analysis of NST1 and NST2 expression in wild-type anthers showed that these genes are expressed after MYB26 expression at the bicellular and pollen mitosis II stages, although some expression of NST2 is also evident at the tricellular stage (Figure 4). No significant expression of NST1 and NST2 is seen in wild-type leaf tissues, although induction is observed in leaves when MYB26 is ectopically expressed. In general, the level of NST1 and NST2 induction in the different overexpression lines correlates with the level of ectopic MYB26 expression. In buds, line OEx4-8 shows fewer changes in expression of MYB26 compared with the wild type and the other overexpressing lines and a corresponding reduced enhancement of NST1 and NST2 expression; however, it also shows minimal changes in floral phenotype (Figure 1K). In other lines, such as OEx8-3, the induction of NST1 and NST2 is more pronounced, particularly for NST2 (Figure 8A). The same trends are observed in the leaf tissue, with greater expression of MYB26 resulting in more NST1 and NST2 expression (Figure 8B); this is particularly evident for NST2 expression. However, the level of change between the two genes, NST1 and NST2, is not always constant. NST1 and NST2 have already been shown to act redundantly (Mitsuda et al., 2005); therefore, the level of each is unlikely to be critical at inducing ectopic secondary thickening. This variability of induction indicates that other factors may influence NST1 and NST2 expression, particularly because some expression of NST1 and NST2 is still seen in the ms35 mutant, but it may also reflect variation in a cell-specific and temporal manner. NST1 and NST2 expression is reduced significantly in the ms35 mutant; however, a low level of NST1 and NST2 expression was still detectable by quantitative PCR, suggesting that there may be additional regulators of their expression. These data suggest that MYB26 acts upstream of these two NAC genes, NST1 and NST2, and in turn the formation of thickening in the endothecium. It is unclear at present whether this regulation occurs directly or indirectly; however, analysis of the promoter regions of NST1 and NST2 suggests that there are a number of putative MYB binding sites in the 5′ upstream regions (TRANSFAC; AthaMap).
A number of genes that have been linked to xylem tracheary element development and secondary cell wall thickening (Table 1; see Supplemental Table 1 online) were also analyzed in the ms35 mutant and OEx lines. Genes associated with secondary thickening (IRX1, IRX3, IRX12 [laccase], and IRX8) showed changes in expression correlated with levels of MYB26, with downregulation in the ms35 mutant buds and increased expression in the Pro35S:MYB26 lines (Figures 7 and 8). It has also been demonstrated previously that altered expression of NST1 results in the overexpression of many of these genes (e.g., IRX1, IRX3, IRX4, IRX5, IRX8, IRX13, and IRX12) (Mitsuda et al., 2005). This finding suggests that MYB26 directly, or indirectly possibly via NST1 and NST2, regulates their expression. However, no direct correlation with MYB26 expression could be seen for a number of key genes from the PAL pathway (Table 1), suggesting that lignification may not be directly regulated by MYB26 or may act via alternative sets of genes involved in lignification. Altered expression levels of ANS and caffeic acid O-methyltransferase (COMT) were seen in the mutant leaves and OEx leaves, respectively (Table 1, Figure 7); however, this did not correspond to changes in the OEx and mutant lines, respectively, suggesting that MYB26 does not directly regulate these genes and that the observed changes are the consequence of secondary changes in flux in the PAL pathway (Figure 9). These data suggest that although the absence of MYB26 results in a downregulation of many marker genes associated with secondary thickening, no direct changes in genes associated with the PAL pathway are seen. This may be a reflection of subtle changes in gene expression associated with only a limited number of cells within the tissues analyzed, a feature of temporal expression patterns indicating that increased expression of these genes only occurs for a limited period at the onset of the development of secondary thickening, or that the MYB26 does not directly regulate these genes but acts via an additional factor to cause increased secondary thickening.
Figure 9.
Scheme of the Branch Points in the Phenylpropanoid Pathway.
This figure is adapted from Deluc et al. (2006). ANS/LDOX, anthocyanidin synthase; CAD, cinnamyl alcohol dehydrogenase; CCR, cinnamyl-CoA reductase; C3H, 4-coumarate-3-hydroxylase; C4H, cinnamate 4-hydoxylase; CHI, chalcone isomerase; CHS, chalcone synthase; 4CL, 4-coumarate-CoA ligase; COMT, caffeic acid O-methyltransferase; DFR, dihydroflavonol 4-reductase; F3H, flavonoid 3-hydroxylase; F5H, ferulate 5-hydroxylase; 3GT, anthocyanidin 3-glucosyltransferase; PAL, phenylalanine ammonia-lyase; 3RT, anthocyanidin-3-glucoside rhamnosyl transferase.
DISCUSSION
Expression of the MYB26 Transcript
High-level expression of the MYB26 transcript appears to require a region extending beyond 1.6 kb upstream of the predicted translational start site, and the absence of putative enhancer sequences in this region results in the lack of dehiscence seen in the ms35 mutant and the SALK_056264 insertion line. Insertional SALK knockout lines within the MYB26 open reading frame and the upstream region of MYB26 have identical phenotypes to the ms35 mutant. This indicates that although low-level expression is detected in the ms35 mutant (Figures 7 and 8), the ms35 mutant serves as a null, with insufficient transcript being produced for endothecial lignification; however, the heterozygote is fully fertile, indicating that a single copy of the MYB26 transcript is sufficient for normal lignification of the endothecium.
Using a 3.1-kb upstream region from the predicted translational start site fused to a GUS reporter, expression was detected in a number of floral organs, but no vegetative expression was seen. High levels of expression were seen in the style and nectaries of the flowers. Female fertility appears unaffected in the ms35 mutant, indicating that the style is fully functional. Lignin development, although reduced, still occurs within the style, implying that there may be redundant genes regulating secondary thickening within the style, so that secondary thickening still occurs in the absence of MYB26, albeit at a slightly reduced level. Nectaries have a role in the production of nectar for insect attraction to facilitate pollination; however, because Arabidopsis is predominantly self-fertile, the nectaries may not have a functional role in pollination and fertilization. Expression of the MYB26 transcript is detected in these tissues, although no significant differences in secondary thickening were observed, which may either reflect a lack of functional MYB26 protein in these tissues or indicate a redundancy of gene regulation. Expression of a number of genes linked to PAL metabolism (Thoma et al., 1994) and dehiscence (Rajani and Sundaresan, 2001) has also been observed in Arabidopsis nectaries, although alterations in phenotype have not been linked to these expression patterns. Expression, as indicated by GUS promoter fusion data and quantitative RT-PCR, is seen in the anthers, from the start of pollen mitosis I through to bicellular pollen, although expression in the nectaries and style is maintained beyond these stages. This pattern of anther expression coincides with the differentiation of cell types within the anther and the expansion of the endothecial cells immediately before the initiation of secondary thickening in the endothecium and vascular tissues in the filament. Immediately after MYB26 expression, other genes directly linked to secondary thickening are expressed, implying that MYB26 acts upstream of secondary thickening via the regulation of genes such as the NAC domain genes NST1 and NST2 and may direct cell expansion and the change of cell competence toward secondary thickening, rather than directly inducing the expression of genes in the secondary thickening pathways.
Induction of Secondary Thickening
Ectopic secondary thickening was induced in both Arabidopsis and tobacco plants by overexpression of the MYB26 transcript. In general, the severity of the phenotype correlated with the level of MYB26 transcript, suggesting that the amount of MYB26 protein is linked to the extent of phenotypic changes. Increased secondary thickening and lignification were seen in these lines.
A number of MYB factors have been linked to PAL metabolism and secondary thickening. Eucalyptus grandis Eg MYB2, which has homology with Arabidopsis At MYB83 and Populus tremuloides Pt MYB4, has been shown to regulate expression by binding to MYB binding sites in the promoters of cinnamoyl-CoA reductase and cinnamyl alcohol dehydrogenase genes (Goicoechea et al., 2005). Tobacco plants overexpressing Eucalyptus MYB2 showed increased secondary cell wall thickness and altered lignification profiles, indicating that Eucalyptus MYB2 has a role in the coordinated control of genes involved in the monolignol-specific pathway; however, other core PAL genes remain unaffected (Goicoechea et al., 2005). Arabidopsis MYB61 has also been linked to the ectopic lignification and dark photomorphogenic phenotype of de-etiolated3 (Newman et al., 2004). Arabidopsis MYB32 has also been shown to regulate genes in the PAL pathway, with downregulation of the MYB32 gene increasing transcript levels of the COMT gene and reducing levels of the DIHYDROFLAVONOL 4-REDUCTASE (DFR) and ANS genes (Preston et al., 2004). Despite anther expression, the pattern of MYB32 expression is very different from that of MYB26; MYB32 is expressed in all major organs, but with a high level of expression in the tapetum and the stigma (Preston et al., 2004), whereas expression of MYB26 occurs specifically within the endothecium tissues, style, and nectaries. MYB32 does not appear to affect the dehiscence process but is thought to alter pollen wall formation by affecting wall development, resulting in collapsed inviable pollen (Preston et al., 2004). No change of expression was seen in either of these two MYB factors in the MYB26 overexpression lines, indicating that MYB26 is upstream of, or does not induce, secondary thickening via MYB32 or MYB61.
IRX1, IRX3, IRX8, and IRX12 expression appears to be correlated directly with levels of MYB26, suggesting direct or indirect regulation by MYB26. Similar increased expression of these genes was also observed in the Pro35S:NST1-overexpressing lines (Mitsuda et al., 2005). IRX1, IRX3, and IRX5 have been shown to be coexpressed in secondary cell walls and to encode essential components of the cellulose-synthesizing complex (Taylor et al., 1999, 2003). Transcriptomic analysis has shown that IRX1 and IRX3 exhibited similar expression patterns during hypocotyl and stem development, and their expression pattern has been linked by expression profiling to a number of genes associated with secondary thickening, including IRX8 (a glucosyl transferase involved in pectin synthesis) and IRX12 (laccase) (Brown et al., 2005). IRX3 and laccase (IRX12) have both been shown to be regulated by VND7 during transdifferentiation of xylem vessel elements (Kubo, 2005). The altered expression of these genes in the Pro35S:MYB26:GUS lines suggests that MYB26 directly or indirectly regulates a group of key genes associated with secondary wall formation, possibly via regulation of NST1 and NST2 expression.
Lignin is derived from the dehydrogenative polymerization of hydroxycinnamyl alcohols (monolignols) and other phenylpropanoids (Raes et al., 2003). Biosynthesis of monolignols requires enzymes of the phenylpropanoid (PAL) pathway (Figure 9) using derivatives of Phe. Lignin is a major component of secondary thickening of cell walls, resulting in the development of large amounts of biomass and providing physical strength for plants combined with the physical forces required for dehiscence, pod shatter, etc. Many mutants have been identified in biosynthetic pathways of lignin, but only recently have genes been identified associated with the regulation of this pathway (see Supplemental Table 1 online). Lignification is usually seen in sclerified cells and requires the coordinated expression of a number of genes from the PAL pathway and those associated with the polymerization of monolignols. Therefore, there is a need for a central regulatory system for the induction of lignin development, which then switches on the other components required for lignification. MYB proteins have been linked to such a regulatory role; two Antirrhinum MYB transcription factors have been shown to regulate the expression of genes in the PAL pathway (Tamagnone et al., 1998). MYB46 is a predicted ortholog of pine (Pinus) MYB4, a positive regulator of the lignification of xylem cells and phloem fibers in loblolly pine (Pinus taeda) (Patzlaff et al., 2003), and MYB52, a predicted ortholog of Populus tremula × P. tremuloides MYB21a, is a repressor of caffeoyl-CoA 3-O-methyltransferase expression (Karpinska et al., 2004). Expression of NAC and MYB factors has been associated with the transcriptome of xylem and phloem development in Arabidopsis root hypocotyls (Zhao et al., 2005). Conserved activator elements, which are critical for the regulation of expression, have been identified in promoters of PAL pathway genes (Neustaedter et al., 1999) and are similar to the motifs recognized by MYB proteins. In the case of the PAL promoter, these have been shown to bind and regulate GUS reporter expression (Sablowski et al., 1994). No changes were evident in pigment formation and flavonoid gene expression in the Arabidopsis OEx material, although changes in pigmentation in petal and anther were seen in the tobacco OEx lines, suggesting that there may be subtle alterations in the regulation of components of the PAL pathway in tobacco compared with Arabidopsis. Similar effects were observed in grapevine (Vitis vinifera) by overexpression of Vv MYB5a, which resulted in an increase in pigment formation in stamens, enhanced anthocyanin biosynthesis, and increased expression of genes in the flavonoid biosynthetic pathway, such as CHALCONE SYNTHASE, CHALCONE ISOMERASE, FLAVANONE 3-HYDROXYLASE, and DFR (del Rio et al., 2004).
The fact that ectopic production of lignin is seen in the MYB26 OEx Arabidopsis and tobacco lines suggests that MYB26 has a key role in the regulation of genes in the PAL and monolignol polymerization pathways and that the regulation of this pathway appears to be evolutionarily conserved between tobacco and Arabidopsis. However, no direct increase in gene expression of a number of specific genes from the PAL pathway was seen in OEx MYB26 lines, although changes in IRX12 (laccase) expression were seen. This finding suggests that the regulation of PAL pathway genes is either extremely transient and low-level or that the role of MYB26 in lignification may be indirect or via alternative members of these pathways, because there is a high level of redundancy in the PAL pathway. Our data indicate that MYB26 acts by inducing the expression of the NAC domain genes NST1 and NST2, which were previously linked to expression changes associated with secondary thickening and lignification (Mitsuda et al., 2005). The NAC transcription family has been linked to maintaining tissue boundaries, regulating the control of growth from cell division to expansion, and other developmental processes such as senescence (Zhao et al., 2005). NST1 and NST2 appear to act redundantly in the regulation of secondary cell walls in the anther (Mitsuda et al., 2005), and a similar phenotype to that seen in the Pro35S:MYB26 lines of ectopic secondary thickening was observed when NST1 or NST2 was expressed using the CaMV35S promoter. It was suggested previously that NST1 and/or NST2 may regulate the expression of MYB26 (Mitsuda et al., 2005); however, this contradicts the changes of NST1 and NST2 expression seen in our data. NST1 and NST2 exhibit different but overlapping expression patterns: NST1 is seen in many tissues of the plant, including inflorescence stems, mid ribs of leaves and anthers, filaments, stamens, and carpels, whereas NST2 shows a much more specific pattern, with expression in anther walls and pollen grains (Mitsuda et al., 2005). This is in contrast with the observations of MYB26 expression, which is seen only in the reproductive tissues and principally in the filament, anther, nectaries, and style. Therefore, MYB26 may play a role in establishing cell competence and determining which cells are capable of undergoing secondary thickening by regulating NST1 and NST2 expression in these tissues.
The induction of ectopic secondary thickening does not occur in all cells, but it is evident predominantly in epidermal cells of both Arabidopsis and tobacco MYB26 OEx lines. The epidermis has been shown to be a highly metabolically active cell type; for example, in Catharanthus roseus, concomitant expression of genes from at least four separate metabolic pathways, including those from the PAL pathway, was seen in the epidermal tissues (Mahroug et al., 2006). The development of thickening predominantly in the epidermis of the OEx lines may reflect the competence of the epidermal tissue for such metabolic activity. However, not all cells undergo thickening; some were visible as ectopic islands among cells showing normal development. This sporadic thickening has also been described in other reports of ectopic secondary thickening (Mitsuda et al., 2005). This may be a consequence of differences in the cell-specific expression of the transgene, but more likely it reflects a competence for secondary thickening in those cells that is associated either with the cessation of cell growth or with the ability to respond to signals required for secondary thickening.
This work has shown that the processes of secondary thickening are conserved between Arabidopsis and tobacco and that overexpression of MYB26 results in ectopic induction of secondary thickening and lignification. This is seen in selected cells within a variety of tissues, suggesting that some aspect of competence is required for the induction of thickening. These data suggest that MYB26 may either regulate genes associated with secondary thickening in the endothecium or else function in specifying cell competence and determining which cells undergo secondary thickening. MYB26 appears to act by regulating the expression of NST1 and NST2, which may directly or indirectly affect the levels of genes associated with the development of secondary thickening. The ability to regulate secondary thickening in a conserved manner has significant commercial applications for the wood and paper industries.
METHODS
Plant Growth Conditions
Seeds of Arabidopsis thaliana var Ler and the ms35gl mutant were sown onto a compost mix of Levington M3:vermiculite (3:1) and grown in a glasshouse at 21/17°C (day/night) with a 22/2-h photoperiod as described previously (Dawson et al., 1999). The T-DNA insertion lines SALK_056264 and SALK_112372 were generated by the Salk Institute Genomic Analysis Laboratory and obtained from the Nottingham Arabidopsis Stock Centre (NASC). Both T-DNA insertion lines and wild-type (ecotype Columbia) control plants were grown as described previously. Plants were genoptyped by PCR using SALK_LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′), SALK_112372_RP (5′-CATTGAGCTTCACAGCATTCTTGG-3′), and SALK_112372_LP (5′-GTCCACAAGAGATTGGCGACG-3′) primers for SALK_112372 and SALK_LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′), SALK_056264_LP (5′-CCGCGGGTTAAAATCTATTATGTGA-3′), and SALK_056264_RP (5′-TCGAACGTACGTATGTAGAGTCCT-3′) primers for SALK_056264.
Transformation of Tobacco
Tobacco (Nicotiana tabacum) seeds were surface-sterilized and sown onto MSR3 (Liu, 2005) medium under conditions of 16 h of daylight at 25°C followed by 8 h of dark at 18°C. Fully expanded, 3- to 4-week-old tobacco leaves were used as explants. Transgenic tobacco plants were generated by Agrobacterium tumefaciens–mediated leaf slice transformation (Liu, 2005) and selected for kanamycin resistance. Regenerated transgenic plants were moved onto compost and grown in the glasshouse as described for Arabidopsis.
MYB26 Promoter∷GUS Construct
A 3.127-kb region upstream of the MYB26 gene was amplified by PCR (MS35-Xho1-3540L, 5′-GCCTCGAGAAGGGAAGCCTCCGACAGAA-3′; MS35R-AVRII-14R, 5′-AGCCTAGGCTAGATCTCTATCGCTCTCTTAGTCT-3′), digested with XhoI/AvrII, and cloned upstream between the XhoI and XbaI sites of the uidA gene of the MOG402-based binary vector pMOGMS1:GUS (Wilson et al., 2001), replacing the MS1 promoter to create pMOGMYB26:GUS. The construct was then transferred into Agrobacterium (C58 pGV3850) by electroporation (Sambrook et al., 1989) and transformed into Arabidopsis (Ler) plants by floral dipping (Clough and Bent, 1998). β-Glucuronidase activity was visualized by staining the inflorescences overnight in 5-bromo-4-chloro-3-indolyl-β-glucuronic acid solution (Willemsen et al., 1998).
Overexpressing Lines
The 1135-bp MYB26 coding region was amplified by PCR (FGWMS35cDNA, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGGTCATCACTCATGCTG-3′; RGWMS35cDNA, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGTTATGACGTACTGTCCACAAGAGAT-3′) from Ler bud cDNA, cloned by recombination into Gateway pDONR201 entry vector (Invitrogen), and transferred into pGWB5 (Karimi et al., 2002) under the control of the CaMV35S promoter. The construct was then transformed into Arabidopsis and tobacco plants as described above. Plants were selected on Murashige and Skoog plates containing 50 mg/mL kanamycin and screened for the presence of the transgene by PCR.
Expression Analysis
RNA was isolated from buds and leaves (RNeasy; Qiagen), and cDNA was prepared using 5 μg of total RNA in a 20-μL reaction (SuperScript II reverse transcriptase; Invitrogen). This was used for RT-PCR analysis using 0.5 μL of cDNA template and the appropriate primer pairs (see Supplemental Table 1 online) for 30 cycles at 94°C for 30s, 58°C for 30s, and 72°C for 1 min. RT-PCR results were normalized using Arabidopsis actin-2 controls (Wilson et al., 2001).
Quantitative RT-PCR
Quantitative RT-PCR analyses were performed using the Mx3005P multiplex quantitative PCR system (Stratagene). Reactions were set up using the Brilliant SYBR Green QPCR Master Mix (Stratagene) in a final volume of 25 μL containing 0.5 μL of cDNA and 0.5 μL of the appropriate primers (see Supplemental Table 1 online). PCR cycling conditions for amplification were 95°C for 10 min followed by 40 cycles of 95°C for 30 s, 58°C for 1 min, and 72°C for 1 min. All samples were run at least in duplicate. Data acquisition and analyses were performed using Mx3005TM multiplex quantitative PCR system software. Relative expression levels were determined compared with actin leaf expression using the 2−ΔΔCT analysis method (Livak and Schmittgen, 2001).
Microscopy
For analysis of lignin, fresh samples or tissues were stained with phloroglucinol-HCl (Ruzin, 1999) and were observed with a light microscope (Nikon). For confocal microscopy (TCS SP2; Leica), a modified ethidium bromide/acridine orange stain was used (A.M. Patten, personal communication); the ethidium bromide stains lignified cells (red fluorescence; excitation, 590 nm) and the acridine orange stains nonlignified walls (green fluorescence; excitation, 520 nm). Fresh tissues were washed (1× PBS and 2% [v/v] Tween 20 for 10 min, then 1× PBS), stained with 0.01% (w/v) acridine orange (1 h, room temperature), washed (1× PBS), and then stained with 0.00005% (w/v) ethidium bromide (1 h, room temperature).
Plant material was also fixed and embedded in paraffin, and sections were stained with toluidine blue, as described previously (Vizcay-Barrena and Wilson, 2006). A minimum of 10 independent transformants were analyzed.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the key genes mentioned in this article are as follows: MYB26 (At3g13890), NST1 (At2g46770), NST2 (At3g61910), IRX1 (At4g18780), IRX3 (At5g17420), IRX5 (At5g44030), IRX8 (At5g54690), and IRX12 (At2g38080); others are listed in Table 1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Primer Sequences for Genes Associated with Lignin and Secondary Thickening.
Supplemental Figure 1. Quantitative RT-PCR Expression in Wild-Type Arabidopsis Flowers and Buds.
Supplementary Material
Acknowledgments
We thank Malcolm Bennett and Jerry Roberts for critical reading of the manuscript. Seed stocks and bioinformatics information were obtained from the NASC. This work was funded by the Biotechnology and Biological Science Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zoe A. Wilson (zoe.wilson@nottingham.ac.uk).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bonner, L., and Dickinson, H. (1989). Anther dehiscence in Lycopersicon esculentum Mill. New Phytol. 113 97–115. [DOI] [PubMed] [Google Scholar]
- Borevitz, J.O., Xia, Y., Blount, J., Dixon, R.A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.M., Zeef, L.A., Ellis, J., Goodacre, R., and Turner, S.R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Dawson, J., Sözen, E., Vizir, I., Van Waeyenberge, S., Wilson, Z.A., and Mulligan, B.J. (1999). Characterization and genetic mapping of a mutation (ms35) which prevents anther dehiscence in Arabidopsis thaliana by affecting secondary wall thickening in the endothecium. New Phytol. 144 213–222. [Google Scholar]
- Dawson, J., Wilson, Z.A., Briarty, L.G., and Mulligan, B.J. (1993). Development of anthers and pollen in male sterile mutants of Arabidopsis thaliana. In Arabidopsis: An Atlas of Morphology, J. Bowman, ed (Berlin: Springer-Verlag), pp. 282–295.
- del Rio, L.A., Corpas, F.J., and Barroso, J.B. (2004). Nitric oxide and nitric oxide synthase activity in plants. Phytochemistry 65 783–792. [DOI] [PubMed] [Google Scholar]
- Deluc, L., Barrieu, F., Marchive, C., Lauvergeat, V., Decendit, A., Richard, T., Carde, J.P., Merillon, J.M., and Hamdi, S. (2006). Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 140 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, H. (1997). Tracheary element differentiation. Plant Cell 9 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goicoechea, M., Lacombe, E., Legay, S., Mihaljevic, S., Rech, P., Jauneau, A., Lapierre, C., Pollet, B., Verhaegen, D., Chaubet-Gigot, N., and Grima-Pettenati, J. (2005). EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 43 553–567. [DOI] [PubMed] [Google Scholar]
- Goldberg, R., Beals, T., and Sanders, P. (1993). Anther development: Basic principles and practical applications. Plant Cell 5 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro, S., Kawai-Oda, A., Ueda, J., Nishida, I., and Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., Tonelli, C., Weisshaar, B., and Martin, C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Karpinska, B., Karlsson, M., Srivastava, M., Stenberg, A., Schrader, J., Sterky, F., Bhalerao, R., and Wingsle, G. (2004). MYB transcription factors are differentially expressed and regulated during secondary vascular tissue development in hybrid aspen. Plant Mol. Biol. 56 255–270. [DOI] [PubMed] [Google Scholar]
- Keijzer, C. (1987). The processes of anther dehiscence and pollen dispersal. I. The opening mechanism of longitudinally dehiscing anthers. New Phytol. 105 487–489. [DOI] [PubMed] [Google Scholar]
- Kubo, M. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. (2005). Manipulation of Plastid Morphology and Analysis of Plastid Gene Expression. PhD dissertation (Nottingham, UK: University of Nottingham).
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Mahroug, S., Courdavault, V., Thiersault, M., St-Pierre, B., and Burlat, V. (2006). Epidermis is a pivotal site of at least four secondary metabolic pathways in Catharanthus roseus aerial organs. Planta 223 1191–1200. [DOI] [PubMed] [Google Scholar]
- Mandaokar, A., Thines, B., Shin, B., Lange, B.M., Choi, G., Koo, Y.J., Choi, Y.D., and Browse, J. (2006). Transcriptional regulators of stamen development in Arabidopsis by transcriptional profiling. Plant J. 46 984–1008. [DOI] [PubMed] [Google Scholar]
- Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyano, E., Martinez-Garcia, J.F., and Martin, C. (1996). Apparent redundancy in myb gene function provides gearing for the control of flavonoid biosynthesis in Antirrhinum flowers. Plant Cell 8 1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neustaedter, D.A., Lee, S.P., and Douglas, C.J. (1999). A novel parsley 4CL1 cis element is required for developmentally regulated expression and protein-DNA complex formation. Plant J. 18 77–88. [DOI] [PubMed] [Google Scholar]
- Park, J.H., Halitschke, R., Kim, H.B., Baldwin, I.T., Feldmann, K.A., and Feyereisen, R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31 1–12. [DOI] [PubMed] [Google Scholar]
- Patzlaff, A., McInnis, S., Courtenay, A., Surman, C., Newman, L.J., Smith, C., Bevan, M.W., Mansfield, S., Whetten, R.W., Sederoff, R.R., and Campbell, M.M. (2003). Characterisation of a pine MYB that regulates lignification. Plant J. 36 743–754. [DOI] [PubMed] [Google Scholar]
- Preston, J., Wheeler, J., Heazlewood, J., Li, S.F., and Parish, R.W. (2004). AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 40 979–995. [DOI] [PubMed] [Google Scholar]
- Raes, J., Rohde, A., Christensen, J.H., Van de Peer, Y., and Boerjan, W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133 1051–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajani, S., and Sundaresan, V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11 1914–1922. [DOI] [PubMed] [Google Scholar]
- Rogers, L.A., and Campbell, M.M. (2004). The genetic control of lignin deposition during plant growth and development. New Phytol. 164 17–30. [DOI] [PubMed] [Google Scholar]
- Rogers, L.A., Dubos, C., Surman, C., Willment, J., Cullis, I.F., Mansfield, S.D., and Campbell, M.M. (2005). Comparison of lignin deposition in three ectopic lignification mutants. New Phytol. 168 123–140. [DOI] [PubMed] [Google Scholar]
- Ruzin, S.E. (1999). Plant Microtechnique and Microscopy. (New York: Oxford University Press).
- Sablowski, R.W., Moyano, E., Culianez-Macia, F.A., Schuch, W., Martin, C., and Bevan, M. (1994). A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 13 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sanders, P.M., Bui, A.Q., Le, B.H., and Goldberg, R.B. (2005). Differentiation and degeneration of cells that play a major role in tobacco anther dehiscence. Sex. Plant Reprod. 17 219–241. [Google Scholar]
- Sanders, P.M., Lee, P.Y., Biesgen, C., Boone, J.D., Beals, T.P., Weiler, E.W., and Goldberg, R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, R., Truernit, E., Gahrtz, M., and Sauer, N. (1999). The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J. 19 269–278. [DOI] [PubMed] [Google Scholar]
- Steiner-Lange, S., Unte, U.S., Eckstein, L., Yang, C., Wilson, Z.A., Schmelzer, E., Dekker, K., and Saedler, H. (2003). Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J. 34 519–528. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Welisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis. Curr. Biol. 4 447–456. [DOI] [PubMed] [Google Scholar]
- Tamagnone, L., Merida, A., Parr, A., Mackay, S., Culianez-Macia, F.A., Roberts, K., and Martin, C. (1998). The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Howells, R.M., Huttly, A.K., Vickers, K., and Turner, S.R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma, S., Hecht, U., Kippers, A., Botella, J., De Vries, S., and Somerville, C. (1994). Tissue-specific expression of a gene encoding a cell wall-localized lipid transfer protein from Arabidopsis. Plant Physiol. 105 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S.R., and Somerville, C. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcay-Barrena, G., and Wilson, Z.A. (2006). Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J. Exp. Bot. 57 2709–2717. [DOI] [PubMed] [Google Scholar]
- von Malek, B., van der Graaff, E., Schneitz, K., and Keller, B. (2002). The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216 187–192. [DOI] [PubMed] [Google Scholar]
- Willemsen, V., Wolkenfelt, H., de Vrieze, G., Weisbeek, P., and Sheres, B. (1998). The Hobbit gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125 521–531. [DOI] [PubMed] [Google Scholar]
- Wilson, Z.A., Morroll, S.M., Dawson, J., Swarup, R., and Tighe, P.J. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28 27–39. [DOI] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yang, S., Sweetman, J.P., Amirsadeghi, S., Barghchi, M., Huttly, A.K., Chung, W.I., and Twell, D. (2001). Novel anther-specific myb genes from tobacco as putative regulators of phenylalanine ammonia-lyase expression. Plant Physiol. 126 1738–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C., Craig, J.C., Petzold, H.E., Dickerman, A.W., and Beers, E.P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.