Abstract
Chromatin remodeling is emerging as a central mechanism for patterning and differentiation in multicellular eukaryotes. SWI/SNF chromatin remodeling ATPases are conserved in the animal and plant kingdom and regulate transcriptional programs in response to endogenous and exogenous cues. In contrast with their metazoan orthologs, null mutants in two Arabidopsis thaliana SWI/SNF ATPases, BRAHMA (BRM) and SPLAYED (SYD), are viable, facilitating investigation of their role in the organism. Previous analyses revealed that syd and brm null mutants exhibit both similar and distinct developmental defects, yet the functional relationship between the two closely related ATPases is not understood. Another central question is whether these proteins act as general or specific transcriptional regulators. Using global expression studies, double mutant analysis, and protein interaction assays, we find overlapping functions for the two SWI/SNF ATPases. This partial diversification may have allowed expansion of the SWI/SNF ATPase regulatory repertoire, while preserving essential ancestral functions. Moreover, only a small fraction of all genes depends on SYD or BRM for expression, indicating that these SWI/SNF ATPases exhibit remarkable regulatory specificity. Our studies provide a conceptual framework for understanding the role of SWI/SNF chromatin remodeling in regulation of Arabidopsis development.
INTRODUCTION
ATP-dependent chromatin remodeling is important for regulation of gene expression in eukaryotes, where the genomic template for transcription is chromatin. The cis-regulatory elements in the core DNA, which is wound around the histone octamer in the nucleosome, are not readily accessible to transcription factors. Chromatin remodeling ATPases use the energy derived from ATP hydrolysis to alter the accessibility of the core DNA by sliding the histone octamer to a new position by inducing conformational changes in the histone octamer/DNA interaction or by transiently displacing the histone octamer from the DNA (Kingston and Narlikar, 1999; Mohrmann and Verrijzer, 2005; Smith and Peterson, 2005; Saha et al., 2006).
SNF2 chromatin remodeling ATPases can be grouped into subfamilies that are conserved between the animal and plant kingdoms (Flaus et al., 2006). Three such subfamilies named SWI/SNF, ISWI, and CHD are implicated in regulation of transcription (de la Serna et al., 2006). Of these, the SWI/SNF subfamily is best characterized. SWI/SNF ATPases are central catalytic subunits of large (1 to 2 MD) chromatin remodeling complexes. The biochemically active chromatin remodeling core complex consists of one ATPase (hBRM or BRG1 in humans and Swi2/Snf2 or Sth1 in yeast), two SANT/SWIRM/Leu zipper–containing proteins termed SWI3 (BAF155 or BAF170 in humans and Swi3 or Rsc8 in yeast), and one protein with a repeat domain and a coiled-coil domain called SNF5 (hSNF5/INI1 in humans and Snf5 or Sfh1 in yeast) (Phelan et al., 1999; Mohrmann and Verrijzer, 2005). Holocomplexes can be distinguished by the presence of distinct accessory proteins. Accessory proteins assist in recruitment of the SWI/SNF complex to target DNA and may regulate the activity of the complex (Mohrmann and Verrijzer, 2005).
Chromatin remodeling complexes do not have DNA binding specificity on their own. Rather, they are targeted to promoter regions via interaction with transcription factors. Human BRG1 and hBRM have been shown to interact with different groups of transcription factors that bind to distinct motifs in the N-terminal domains of the two ATPases (Kadam and Emerson, 2003). Transcription factors also interact with other core or accessory complex components (Simone, 2006).
Most multicellular eukaryotes have multiple SWI/SNF ATPases (Flaus et al., 2006). Arabidopsis thaliana has four members of this family, while rice (Oryza sativa) has three and poplar (Populus spp) has six (http://www.chromdb.org/; Flaus et al., 2006; Su et al., 2006; this study). This raises the question of the functional overlap between individual members of this family. Plants also have multiple SWI3 proteins: Arabidopsis has four (ATSWI3A, ATSWI3B, ATSWI3C, and ATSWI3D), rice has six, and poplar has five (Sarnowski et al., 2002; Zhou et al., 2003; http://www.chromdb.org/). By contrast, only a single SNF5 ortholog is present in Arabidopsis, rice, and poplar (Brzeski et al., 1999; http://www.chromdb.org/).
The role of SWI/SNF chromatin remodeling in Arabidopsis development has recently been studied intensively (Wagner and Meyerowitz, 2002; Zhou et al., 2003; Farrona et al., 2004; Kwon et al., 2005, 2006; Sarnowski et al., 2005; Hurtado et al., 2006; Su et al., 2006). Null mutants have been described for two Arabidopsis SWI/SNF ATPases: SPLAYED (SYD) and BRAHMA (BRM) (Wagner and Meyerowitz, 2002; Hurtado et al., 2006; Kwon et al., 2006). Absence of either ATPase leads to pleiotropic developmental defects, but the plants are viable, allowing investigation of the role of these SWI/SNF ATPases throughout development. Morphological and molecular analyses suggest that syd and brm mutants exhibit both similar and distinct defects. Both mutants are slow growing and dwarfed, have defects in cotyledon separation, and exhibit reduced apical dominance (Wagner and Meyerowitz, 2002; Farrona et al., 2004; Hurtado et al., 2006; Kwon et al., 2006; Su et al., 2006). Null mutants in BRM also have unique root growth defects and are male sterile (Wagner and Meyerowitz, 2002; Hurtado et al., 2006; Kwon et al., 2006).
Similar phenotypes were described for mutants in other putative SWI/SNF core complex components. Antisense knockdown alleles of the SNF5 homolog BSH have pleiotropic phenotypes, including loss of apical dominance and sterility (Brzeski et al., 1999), but the bsh null mutant phenotype has not yet been described. atswi3c mutants closely resemble brm mutants (Farrona et al., 2004; Sarnowski et al., 2005; Hurtado et al., 2006; Kwon et al., 2006). However, atswi3d mutants have pleiotropic phenotypes that do not resemble that of any known SWI/SNF ATPase mutants (Sarnowski et al., 2005). In contrast with single brm and syd mutants, atswi3a and atswi3b mutants are embryonic lethal (Sarnowski et al., 2005). Furthermore, protein interaction studies revealed that the N-terminal domain of BRM (BRMN) interacts with ATSWI3C and more weakly with ATSWI3B (Farrona et al., 2004; Hurtado et al., 2006). In addition, homo- and heterodimers can form between several ATSWI3 proteins, and both ATSWI3A and ATSWI3B interact with BSH. It is currently not understood with which of these complex components SYD interacts.
Here, we investigate functional overlap, protein interactions, and specificity of the two SWI/SNF ATPases: BRM and SYD. Based on comparative genome analyses, it has been proposed that organismal complexity may arise from increasingly elaborate regulation of gene expression, including diversification of chromatin remodeling activities (Levine and Tjian, 2003). This raises the question of how function has evolved among individual members of a chromatin remodeling gene family. It is clear from the mutant phenotypes of BRM and SYD that these genes are not completely functionally redundant. However, the extent of their functional overlap remains to be determined. Our transcription profiling and protein interaction studies reveal that BRM and SYD have unique and shared targets and interaction partners. Consistent with these results, double mutants have more severe phenotypes than the single mutants. In addition, our data suggest occurrence of multiple distinct SWI/SNF core complexes in different Arabidopsis tissues. Finally, as previously suggested for the role of SYD in shoot apical meristem maintenance (Kwon et al., 2005), our studies show that the two SWI/SNF ATPases control expression of a very small number of genes and thus that they are specific transcriptional coregulators.
RESULTS
Expression Profiling of brm and syd Compared with the Wild Type Indicates That the ATPases Regulate Few Targets
To investigate the functional overlap between SYD and BRM, we performed genomic expression studies in plants homozygous null for SYD (syd-2), homozygous null for BRM (brm-101), and for an isogenic wild-type control (Landsberg erecta [Ler]). To minimize potential secondary effects of loss of ATPase activity, we performed the experiment at an early developmental stage, when few morphological differences between the mutants and the wild type are observed (Figure 1A). To minimize potential differences in gene expression due to precocious flowering in brm-101 or syd-2, we grew seedlings in long days, where subtle or no early flowering is observed (Hurtado et al., 2006; Su et al., 2006), and harvested 10-d-old seedlings after floral induction (Kobayashi et al., 1999; Blazquez and Weigel, 2000).
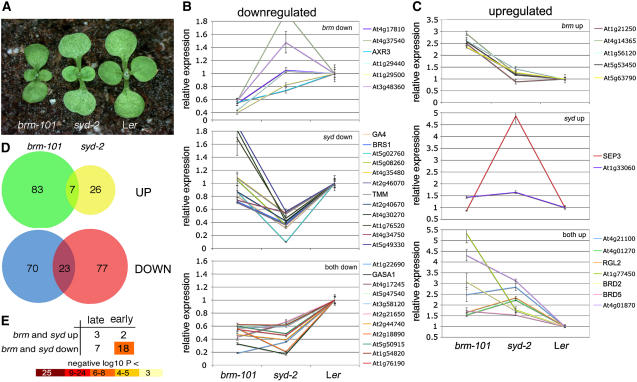
Figure 1.
Genomic Expression Studied in brm and syd Null Mutants.
(A) Ten-day-old long-day-grown brm-101, syd-2, and wild-type (Ler) seedlings were used for genomic expression studies. Both brm-101 and syd-2 are smaller than the wild type. The size of brm-101 is reduced compared with syd-2, and the two mutants exhibit unique subtle cotyledon and leaf shape abnormalities.
(B) and (C) Quantitative real-time PCR analysis of all predicted regulatory gene products (transcription factors and signaling molecules) identified using Rank Product (Breitling et al., 2004; FDR <10%; see also Table 1 and Supplemental Tables 1 to 4 online). Shown are 43 genes that were found to be misregulated by real-time PCR. The mean expression value for two biological replicates and three technical replicates per gene normalized by the value obtained for the ubiquitously expressed eukaryotic translation initiation factor EIF4A is indicated. Error bars denote the se of the mean. For ease of comparison, the value for the wild type (Ler) was set to 1. Genes downregulated in brm-101 and/or syd-2 are shown in (B), and those upregulated in brm-101 and/or syd-2 are shown in (C).
(D) Genes identified as misregulated in each mutant as described above were compared in a pairwise fashion. Genes upregulated or downregulated in both mutants are shown in the overlap of the Venn diagrams. Genes misregulated in one mutant only are indicated in the nonoverlapping segments of the Venn diagrams.
(E) Comparison of genes preferentially expressed during later stages of seedling development (late) and those preferentially expressed during early seedling development (early) identified from the developmental data set in AtGenExpress (Schmid et al., 2005; see Methods for details) versus those upregulated in both syd-2 and brm-101 and to those downregulated in both syd-2 and brm-101. The total number of late and early genes was 1919 and 2548, respectively. P values are based on a two-tailed Fisher's exact test.
Less than 1% of the genes present on the ATH1 Affymetrix array were differentially expressed in each mutant compared with the wild type based on our analyses (Rank Product; Breitling et al., 2004) using an estimated false discovery rate (FDR) of <10% (Storey and Tibshirani, 2003). For comparison, a commonly employed statistical approach (two-way mixed model analysis of variance [ANOVA]; P < 0.05; genes upregulated at least twofold) yielded gene lists of comparable size (Table 1). The genes identified in our analysis likely represent both direct and indirect targets of the chromatin remodeling ATPases. Consistent with this, we did not see strong enrichment in functional categories (http://www.arabidopsis.org/tools/bulk/go/index.jsp) among the genes with altered expression (data not shown). These data suggest that only a small subset of all genes requires SYD or BRM for proper expression in 10-d-old seedlings.
Table 1.
Number of Genes Misregulated in brm-101 Compared with the Wild Type and syd-2 Compared with the Wild Type
| Rank Product
|
Two-Way Mixed Model ANOVA
|
|
|---|---|---|
| Analysis | FDR <10% (P < 0.001) | P < 0.05 Twofold or More |
| Upregulated in brm-101 | 90 | 153 |
| Upregulated in syd-2 | 33 | 118 |
| Downregulated in brm-101 | 93 | 115 |
| Downregulated in syd-2 | 100 | 209 |
Number of genes identified as misregulated using global expression studies by implementation of different statistical tools. Rank Product was described by Breitling et al. (2004). Cutoffs were based on estimated FDRs (Storey and Tibshirani, 2003). The P values of the genes in these gene lists are all below 0.001. For comparison, a traditional two-way mixed model ANOVA analysis is shown.
To test the efficacy of the microarray-based identification of genes as dependent on BRM and/or SYD, we performed real-time PCR analyses on a subset of the genes misregulated in brm and in syd. We chose to focus first on genes encoding transcription factors or signaling molecules (based on database and manual annotation; see Supplemental Tables 1 to 4 online). Because SWI/SNF ATPases tend to control expression of regulatory genes (Tsukiyama, 2002; de la Serna et al., 2006), these categories of genes are perhaps more likely to be direct targets of BRM and SYD. Real-time PCR analyses were performed on 45 genes (Tables 2 and 3). Forty-three of these genes showed at least a 1.5-fold decrease or increase in expression in real-time PCR experiments compared with the wild type (Figures 1B and 1C), confirming the microarray results (Tables 2 and 3; observed FDR = 4.5%). Downregulated (Figure 1B) or upregulated (Figure 1C) genes were grouped based on their dependence on BRM, SYD, or both ATPases. Among the genes affected by these mutants are genes involved in auxin signaling (AXR3), gibberellic acid signaling (GA4, RGL2, and GASA1), and brassinosteroid signaling (BRS1) (Figures 1B and 1C, Tables 2 and 3). In addition, two genes implicated in chromatin regulation, BRD2 and BRD5 (http://www.chromdb.org/), are upregulated (Figure 1C, Table 3). These proteins contain a bromodomain, a motif that allows binding to acetylated Lys residues on histone tails. SEP3, a meristem identity regulation and flower patterning gene (Honma and Goto, 2001; Pelaz et al., 2001a, 2001b; Castillejo et al., 2005; Teper-Bamnolker and Samach, 2005; Sridhar et al., 2006), is strongly upregulated only in syd-2.
Table 2.
Regulatory Genes Downregulated in brm101 and/or syd-2 as Determined by Real-Time PCR
| FC brm | FC syd | Arabidopsis Genome Initiative | Target Description | Regulation | Name |
|---|---|---|---|---|---|
| 0.55 | 0.74 | AT1G04250 | Auxin-responsive protein/indole-3-acetic acid-induced protein 17 (IAA17) | BRM | AXR3IAA17 |
| 0.80 | 0.32 | AT1G15550 | Gibberellin 3-β-dioxygenase/gibberellin 3 β-hydroxylase (GA4) | SYD | GA4 |
| 0.19 | 0.36 | AT1G22690 | Gibberellin-responsive protein, putative | Both | GASA2-like |
| 0.44 | 1.04 | AT1G29440 | Auxin-responsive family protein | BRM | SAUR63 |
| 0.40 | 0.83 | AT1G29500 | Auxin-responsive protein, putative | BRM | SAUR66 |
| 0.63 | 0.62 | AT1G54820 | Protein kinase family protein | SYD* | |
| 0.33 | 0.17 | AT1G75750 | Gibberellin-regulated protein1 (GASA1)/gibberellin-responsive protein1 | Both | GASA1 |
| 0.55 | 0.46 | AT1G76190 | Auxin-responsive family protein | BRM* | SAUR56 |
| 1.69 | 0.46 | AT1G76520 | Auxin efflux carrier family protein | SYD | |
| 1.08 | 0.53 | AT1G80080 | Leu-rich repeat family protein | SYD | TMM |
| 0.57 | 0.21 | AT2G18890 | Protein kinase family protein | SYD* | |
| 0.43 | 0.67 | AT2G21650 | myb family transcription factor | BRM | |
| 1.19 | 1.24 | AT2G27920 | Ser carboxypeptidase S10 family protein | SYD | Did not repeat |
| 0.87 | 0.42 | AT2G40670 | Two-component responsive regulator/response regulator 16 (ARR16) | SYD | ARR16 |
| 0.47 | 0.38 | AT2G44740 | Cyclin family protein | Both | P-type cyclin |
| 0.71 | 0.36 | AT2G46070 | Mitogen-activated protein kinase, putative/MAPK, putative (MPK12) | SYD | At MPK12 |
| 0.58 | 1.48 | AT3G48360 | Speckle-type POZ protein-related | BRM | BT2 |
| 0.57 | 0.60 | AT3G58120 | bZIP transcription factor family protein | BRM* | |
| 0.43 | 0.64 | AT4G17245 | Zinc finger (C3HC4-type RING finger) family protein | BRM* | E3 ligase? |
| 0.54 | 1.05 | AT4G17810 | Zinc finger (C2H2 type) family protein | BRM | SUP-like |
| 1.82 | 0.41 | AT4G30270 | MERI-5 protein (MERI-5) (MERI5B)/endoxyloglucan transferase/xyloglucan endo-1,4-β-d-glucanase (SEN4) | SYD | MERI5B/SEN4 |
| 0.72 | 0.38 | AT4G30610 | Ser carboxypeptidase S10 family protein | SYD | BRS1 |
| 0.74 | 0.55 | AT4G34750 | Auxin-responsive protein, putative/small auxin up RNA (SAUR_E) | SYD | SAUR49 |
| 1.10 | 0.54 | AT4G35480 | Zinc finger (C3HC4-type RING finger) family protein | SYD | E3 ligase? |
| 0.58 | 1.94 | AT4G37540 | LOB domain protein 39/lateral organ boundaries domain protein 39 (LBD39) | BRM | LOB39 |
| 0.87 | 0.10 | AT5G02760 | Protein phosphatase 2C family protein/PP2C family protein | SYD | PP2C-like |
| 1.05 | 0.36 | AT5G08260 | Ser carboxypeptidase S10 family protein | SYD | |
| 0.59 | 0.63 | AT5G47540 | Auxin-responsive protein, putative/Mo25 family protein | BRM* | |
| 1.90 | 0.57 | AT5G49330 | myb family transcription factor | SYD | |
| 0.60 | 0.49 | AT5G50915 | Basic helix-loop-helix family protein | SYD* |
Regulatory genes identified as downregulated (FDR <10%) in syd and/or brm in our microarray analysis (SYD, BRM, or both in the Regulation column; also shown in bold in Supplemental Tables 1 to 4 online) that were tested by real-time PCR analysis. Two biological replicates with three technical replicates each were analyzed. The fold change (FC) was determined after normalization with signal values for the translation initiation factor EIF4A by dividing the signal of syd or brm with that observed in the wild type (Ler). Genes downregulated >1.5-fold compared with the wild type were considered misexpressed, and the observed fold change is shown in bold. Expression of these genes is also depicted in Figure 1B. For all but one gene (“Did not repeat” in Name column), we confirmed the misexpression by real-time PCR. Several genes identified as misregulated in one mutant on the array were found to be misregulated in both mutants by real-time PCR (indicated by an asterisk).
Table 3.
Regulatory Genes Upregulated in brm101 and/or syd-2 as Determined by Real-Time PCR
| FC brm | FC syd | Arabidopsis Genome Initiative | Target Description | Regulation | Name |
|---|---|---|---|---|---|
| 2.49 | 0.88 | AT1G21250 | Wall-associated kinase1 (WAK1) | BRM | WAK1 |
| 0.86 | 4.87 | AT1G24260 | MADS box protein (AGL9) | SYD | SEP3 |
| 1.43 | 1.64 | AT1G33060 | No apical meristem (NAM) family protein | SYD | ANAC014 |
| 2.63 | 1.44 | AT1G56120 | Leu-rich repeat family protein | BRM | |
| 1.72 | 1.54 | AT1G58025 | DNA binding bromodomain-containing protein | BRM* | BRD5 |
| 3.09 | 1.73 | AT1G76380 | DNA binding bromodomain-containing protein | BRM* | BRD2 |
| 5.33 | 1.77 | AT1G77450 | No apical meristem (NAM) family protein | BRM* | ANAC032 |
| 1.34 | 1.33 | AT2G41980 | Seven in absentia (SINA) family protein | SYD | Did not repeat |
| 1.66 | 2.34 | AT3G03450 | Gibberellin response modulator, putative/gibberellin-responsive modulator, putative | SYD* | RGL2 |
| 1.54 | 2.23 | AT4G01270 | Zinc finger (C3HC4-type RING finger) family protein | SYD* | E3 ligase? |
| 4.31 | 3.13 | AT4G01870 | tolB protein-related | BRM* | |
| 2.93 | 1.22 | AT4G14365 | Zinc finger (C3HC4-type RING finger) family protein/ankyrin repeat family protein | BRM | E3 ligase? |
| 2.48 | 2.84 | AT4G21100 | UV-damaged DNA binding protein, putative | Both | DDB1B |
| 2.58 | 1.17 | AT5G53450 | Protein kinase family protein | BRM | ORG1 |
| 2.35 | 1.27 | AT5G63790 | No apical meristem (NAM) family protein | BRM | ANAC102 |
Regulatory genes identified as upregulated (FDR <10%) in syd and/or brm in our microarray (SYD, BRM, or both in the Regulation column; also shown in bold in Supplemental Tables 1 to 4 online) that were tested by real-time PCR analysis. Two biological replicates with three technical replicates each were analyzed. The fold change (FC) was determined after normalization with signal values for the translation initiation factor EIF4A by dividing the signal of syd or brm with that observed in the wild type (Ler). Genes upregulated >1.5-fold compared with the wild type were considered misexpressed, and the observed fold change is shown in bold. Expression of these genes is also depicted in Figure 1C. For all but one gene (“Did not repeat” in the Name column), we confirmed the misexpression by real-time PCR. Several genes identified as misregulated in one mutant on the array were found to be misregulated in both mutants by real-time PCR (indicated by an asterisk).
Identification of Genes Dependent on One or on Both ATPases
We next asked whether SYD and BRM have common targets or control common processes. To address this question, we examined whether genes identified as misregulated in our analysis (Rank Product; FDR <10%) are dependent only on one or on both ATPases. On average >20% of all genes identified as misregulated in either syd-2 or brm-101 are misregulated in both mutants (Figure 1D). This is a significant enrichment (two-tailed Fisher's exact test; P < 10−9 and P < 10−29 for upregulated and downregulated genes, respectively). Furthermore, our real-time PCR analyses of 45 genes identified 13 additional genes as coregulated (Tables 2 and 3), suggesting that coordinate regulation of gene expression by SYD and BRM may be more common than suggested by our microarray analysis using a cutoff FDR value of <10%. Of the 43 genes validated, 12 were dependent on BRM and 14 on SYD, while 17 genes were dependent on both ATPases. No statistically significant regulation of expression was observed between genes upregulated in one mutant and downregulated in the other (Table 4). Thus, it appears that BRM and SYD are both required for coordinate regulation of a considerable number of the genes misexpressed in each mutant.
Table 4.
Overlap of Genes Misregulated in Chromatin Mutants Compared with the Wild Type
| Genotype | brm Up | brm Down | syd Up | syd Down | lhp1 Up | lhp1 Down | emf1 Up | emf1 Down | pkl Up | pkl Down |
|---|---|---|---|---|---|---|---|---|---|---|
| brm Up | 7.0/0.2 −9 | 5.0/0.6 −3 | ||||||||
| brm Down | 23.0/0.6 −29 | 4.0/0.2 −4 | ||||||||
| syd Up | ||||||||||
| syd Down | 4.0/0.2 −4 | |||||||||
| lhp1 Up | 3.0/0.1 −4 | 4.0/0.4 −3 | ||||||||
| lhp1 Down | 4.0/0.1 −5 | |||||||||
| emf1 Up | ||||||||||
| emf1 Down | ||||||||||
| pkl Up | ||||||||||
| pkl Down |
Genes misexpressed in chromatin regulator mutants were identified as described in Methods using an FDR cutoff of <10%. Expression values were derived from our data set (this work for brm and syd) or from the raw data of the experiments described by Nakahigashi et al. (2005) for lhp1/tfl2, by Moon et al. (2003) for emf1, and by Dean Rider et al. (2003) for pkl. The observed number of genes that are coordinately misregulated (left numeral) and the values that would be expected if there was no association between the two lists of genes (right numeral) are indicated. Statistical significance was determined using a Fisher's exact test. The negative log 10 for the two-tailed P value is shown below the observed/expected overlap.
Since both syd-2 and brm-101 have slightly slower growth rates than the wild type (Figure 1A; Wagner and Meyerowitz, 2002; Kwon et al., 2006), a trivial explanation for the observed overlap in gene expression defects could be a delay in the upregulation of genes expressed at later seedling stages in both mutants. To test this hypothesis, we identified genes preferentially expressed early and late in wild-type seedling development using a publicly available microarray data set (AtGenExpress; Schmid et al., 2005; see Methods for details). We compared genes expressed early and late during seedling development to the genes upregulated or downregulated in both syd and brm (Figure 1E). We did not observe a strong correlation between early genes and genes upregulated in both brm and syd or between late genes and genes downregulated in both brm and syd in the four pairwise comparisons. This suggests that the large overlap in genes dependent on both ATPases is not simply due to the delayed growth of the mutants. Even when we remove all genes that could potentially be due to growth bias (i.e., remove all early genes from the list of genes upregulated in both mutants and remove all late genes from the list of genes downregulated in both mutants), we still observe very strong coregulation by BRM and SYD (P < 10−5 and P < 10−18 for coordinate upregulation and downregulation, respectively). These data indicate that both ATPases are required for correct expression of a significant number of genes.
On the other hand, significant overlap was detected between early genes and those downregulated in both brm and syd (P < 10−7). Thus, despite their slow growth, brm and syd show reduced expression of genes typically expressed early in seedling development, suggesting that the two ATPases may display precocious developmental transitions.
To determine whether coordinate regulation of gene expression can also be observed between BRM or SYD and other chromatin remodeling proteins, we compared the changes in gene expression in mutants of the two SWI/SNF ATPases to those of (1) LIKE HETEROCHROMATIN PROTEIN1/TERMINAL FLOWER2 (LHP1/TFL2), the Arabidopsis Heterochromatin Protein 1 (Nakahigashi et al., 2005); (2) EMBRYONIC FLOWER1 (EMF1), a Polycomb group protein (Moon et al., 2003); and (3) PICKLE (PKL), a CHD-type chromatin remodeling ATPase (Dean Rider et al., 2003). We analyzed expression data from these published experiments using the same statistical tools we applied to the syd/brm data set (see Methods for details). None of the pairwise comparisons revealed the same degree of correlation as that observed for brm and syd. However, statistically significant correlations were observed in several cases (Table 4). Notably, there was significant overlap for both upregulated and downregulated genes in lhp1 and emf1, suggesting that the two corresponding proteins might share common targets. In addition, some genes downregulated in syd are also downregulated in emf, and some genes downregulated in brm are upregulated in lhp1 (Table 4). Finally, there is a weak correlation between genes upregulated in pkl and upregulated in brm or lhp1. The actual overlap in regulation of gene expression by BRM and SYD on one hand and LHP1, EMF1, and PKL on the other may be larger than we were able to detect here because of differences in the experimental design of the individual microarrays. However, we were able to detect coordinate regulation of gene expression for EMF1 and LHP1, suggesting that coregulation can be observed in these independent data sets.
brm syd Double Mutants
To further investigate the interdependence of SYD and BRM, we constructed brm syd double mutants in a Ler background using the brm-101 null allele (Kwon et al., 2006) and the syd-2 null allele (Wagner and Meyerowitz, 2002). We also generated double mutants in the Columbia ecotype using brm-1 (SALK_ 030046), a strong or null brm allele (Hurtado et al., 2006; Kwon et al., 2006), and syd-5 (SALK_023209), a SYD RNA null allele (see Supplemental Figure 1 online). Since brm-101 syd-2 and brm-1 syd-5 behaved similarly, we henceforth refer to the double mutants collectively as brm syd.
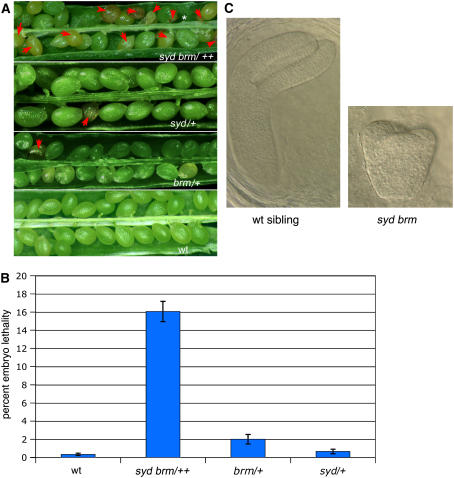
Based on physical map distance, SYD and BRM are ∼20 centimorgans apart from each other on chromosome 2. To obtain the double mutants, we first generated cis-heterozygotes by backcrossing trans-heterozygotes (syd +/+ brm) to the wild type and PCR genotyping the resulting progeny to identify recombinants. The F2 progeny of these (syd brm/++) recombinants were visually screened for novel phenotypes as seedlings and as adult plants. We were not able to identify novel phenotypes in >8000 F2 progeny plants examined, nor were we able to detect homozygous double mutants by PCR. However, inspection of the siliques of selfed cis-heterozygotes revealed many white and misshapen seeds (Figure 2A). These seeds turned dark brown in older siliques. We very rarely observed such defects in the selfed single mutant siliques (Figure 2A, Table 5). These data suggest that brm syd causes embryonic lethality. Based on the map distance (20 centimorgans), 16% of the progeny are expected to be syd brm/syd brm homozygotes. Quantitation of the seed phenotypes revealed a very close fit to the expected number (Figure 2B), suggesting that the double mutant is embryonic lethal. Embryo development was generally arrested by early heart stage (Figure 2C), but arrest was also observed as early as the eight cell stage (data not shown). These data indicate that presence or activity of at least one of the two ATPases is necessary for proper embryo development.
Figure 2.
Phenotypes of syd brm Double Mutants.
(A) Siliques of selfed syd brm/++, syd/+, brm/+, and the Ler wild-type plants. Parental genotypes are indicated in each panel. Several misshapen and shrunken seeds are indicated by a red arrow, and unfertilized ovules are marked with an asterisk.
(B) Average percentage (mean number plus se of the mean) of misshapen and shrunken seeds in each silique after selfing. Genotypes are indicated below the graph. The total number of seeds counted (n) is shown in Table 4.
(C) Representative cleared embryos from a misshapen seed (right) of selfed syd brm/++ siliques and a wild-type-looking sibling (left) from the same silique. Embryos are arrested at the heart stage (right) or earlier (data not shown).
Table 5.
Embryo and Gametophyte Defects in brm and syd Single and Double Mutants Compared with the Wild Type
| Unfertilizeda | Embryonic Lethal | Normal | Total | |
|---|---|---|---|---|
| brm/+ | 4.0 (0.6) | 2.0 (0.5) | 94.0 (0.9) | 1319 |
| syd/+ | 2.4 (1.2) | 0.6 (0.2) | 96.9 (1.1) | 1349 |
| syd brm/++ | 12.3 (1.1) | 16.1 (1.1) | 71.6 (1.0) | 1252 |
| Wild type | 3.7 (0.6) | 0.3 (0.1) | 95.9 (1.4) | 1772 |
Shown are the mean percent and se (in parentheses) of defects observed in the progeny of selfed plants. Parental genotypes are indicated in the left column. The total number of seeds scored (n) is indicated in the right column.
Unfertilized embryos are the result of male or female gametophytic defects.
In addition, a small number of unfertilized ovules were observed in syd brm/++ siliques (Table 5). The frequency of these unfertilized ovules was higher than that observed for the wild type or the single mutants, suggesting that the double mutant may result in a weakly penetrant gametophyte defect. We did not observe a strong gametophyte defect for brm-101/+ or brm-1/+ (Table 5; data not shown) in apparent disagreement with another recent study (Hurtado et al., 2006), nor did we observe distorted segregation ratios for either brm or syd mutants (data not shown). The reason for this difference is not understood. It is possible that the penetrance of the gametophyte defect in brm/+ is subject to environmental variability.
SYDN Interacts with a Subset of ATSWI3 Proteins
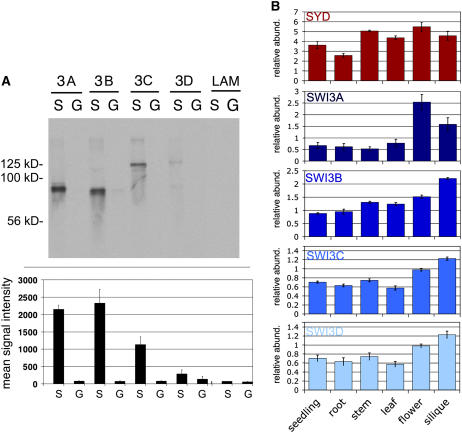
Studies in budding yeast and other organisms have demonstrated that the N-terminal domain of SWI/SNF ATPases interacts with a second SWI/SNF core complex component called Swi3 (yeast) or BAF155/170 (human) (Treich et al., 1995; Treich and Carlson, 1997; Phelan et al., 1999; Vignali et al., 2000). To test for interaction between SYDN and the four ATSWI3 proteins, we performed glutathione S-transferase (GST) pull-down experiments using recombinant GST-SYDN or GST alone together with in vitro transcribed and translated radioactively labeled ATSWI3 proteins (Figure 3A). Results of this assay indicate that three of the four Arabidopsis ATSWI3 proteins bind specifically to GST-SYDN in vitro. The strongest interactions were observed between ATSWI3A and SYDN and ATSWI3B and SYDN. A somewhat weaker but significant interaction was observed between ATSWI3C and SYDN. ATSWI3D did not interact strongly with SYDN (Figure 3A, bottom panel). We conclude that SYDN interacts with ATSWI3A, ATSWI3B, and ATSWI3C.
Figure 3.
SYDN Interaction with ATSWI3 Proteins.
(A) Top panel: GST pull-down assays using SYDN-GST (S) or GST alone (G) and in vitro transcribed and translated 35S Met–labeled candidate interacting proteins. Autoradiograph of a 12% polyacrylamide gel. Size marker migration is indicated at the left. Interacting proteins tested included ATSWI3A (3A), ATSWI3B (3B), ATSWI3C (3C), ATSWI3D (3D), and LAMIN (LAM). Equal amounts of these proteins were used in each reaction (data not shown). Bottom panel: Quantitation of three independent experiments of the type shown in the top panel, normalized by protein levels. Shown is the mean with se of the mean.
(B) Quantitative real-time PCR analysis of ATSWI3 genes compared with SYD in different tissues. The mean and se of the mean of one representative biological replicate with three technical replicates normalized by the value obtained for the ubiquitously expressed eukaryotic translation initiation factor EIF4A are shown. Tissues tested were from plants grown in long-day conditions in soil unless otherwise indicated. Stages harvested were 10-d-old seedlings, 5-d-old roots (vertical half-strength Murashige and Skoog agar plates), second internode from 28-d-old plants, expanding leaves (8th and 9th leaves) from 21-d-old plants, inflorescences (not including fully open flowers) from 35-d-old plants, and elongating siliques from 35-d-old plants.
To test whether SYD and the ATSWI3 proteins might be present in similar tissues, we investigated the expression of the corresponding genes by real-time PCR. Like SYD, all four ATSWI3 genes are expressed in most tissues in agreement with Zhou et al. (2003), although small differences in the expression levels were observed for the individual ATSWI3 genes within the six tissues tested (Figure 3B). Our results are in general agreement with available global expression data for all genes (AtGenExpress [Schmid et al., 2005] and Genevestigator [Zimmermann et al., 2004]). These results are consistent with in vivo interaction between SYD and ATSWI3 proteins.
DISCUSSION
SWI/SNF ATPases in Arabidopsis Are Specific Regulators of Transcription
Chromatin remodeling complexes have recently been shown to play a major role in patterning and differentiation of multicellular eukaryotes (Buszczak and Spradling, 2006; de la Serna et al., 2006; C.S. Kwon and D. Wagner, unpublished data). However, it is not understood whether the complexes act as general or specific regulators of transcription by regulating a large number of targets or very few genes, respectively. Previously, we showed that SYD plays a specific role (controls a limited number of targets) in one pathway in Arabidopsis, namely, maintenance of the stem cell pool in the shoot apical meristem (Kwon et al., 2005). This suggests that SYD can play a specific role. Our genome-wide transcriptome analysis of syd and brm single mutants compared with the wild type demonstrates that both SYD and BRM control very few genes (∼1% of all genes). Since the expression changes in the null mutants are comprised of direct and indirect (i.e., downstream) effects on gene expression, the data indicate that only a small number of genes require SYD or BRM for proper expression. A similar analysis of the effects of SWI/SNF ATPases on gene expression in the organism is not available for any other multicellular eukaryote. In yeast, 3 to 10% of all genes showed altered expression in the swi2/snf2 and sth1 yeast SWI/SNF ATPase mutants (Holstege et al., 1998; Krebs et al., 2000; Sudarsanam et al., 2000; Angus-Hill et al., 2001; Kasten et al., 2004; Soutourina et al., 2006). Thus, in Arabidopsis, SWI/SNF ATPases have increased regulatory specificity, perhaps due to the presence of two additional SWI/SNF ATPases (Flaus et al., 2006; http://www.chromdb.org/).
If BRM and SYD only control accessibility of a very small number of all promoters, this raises the issue of how chromatin-based constraints on cis-regulatory elements are controlled for the remaining genes. First, other types of chromatin remodeling ATPases (ISWI and CHD/Mi2) or complexes that covalently modify histones (for example, see Dean Rider et al., 2003; Noh and Amasino, 2003; Tian et al., 2005) may help overcome chromatin constraints for a different subset of promoters. Second, different genes may require SYD or BRM for proper expression at other developmental stages. For example, we did not identify the direct SYD target WUSCHEL (WUS) in this experiment. A reduction of WUS expression can only be observed in syd-2 mutants at a later developmental stage than that assayed here (day 19; Kwon et al., 2005). Finally, recent studies in yeast suggest that many promoters that are constitutively active have fewer nucleosomes and may not require chromatin remodeling for transcriptional activation (Ioshikhes et al., 2006).
The SWI/SNF ATPases BRM and SYD Have Overlapping Roles
It has been proposed that the increase in organismal complexity during evolution is due to more elaborate gene regulation (Levine and Tjian, 2003; Carroll, 2005) via functional diversification of regulatory proteins, such as those involved in chromatin remodeling (Levine and Tjian, 2003; Taatjes et al., 2004). Most metazoans have multiple chromatin remodeling SWI/SNF ATPases (Flaus et al., 2006; http://www.chromdb.org/). Here, we have investigated the functional relationship between two SWI/SNF chromatin remodeling ATPases in Arabidopsis. We have uncovered three types of roles for BRM and SYD: unique, in which only one of the two SWI/SNF ATPases is required for a certain target or process; shared, with several genes or processes dependent on both proteins; and redundant, where either is sufficient for regulation of a target gene or process. We will discuss our findings in light of possible diversification and specialization of SWI/SNF chromatin remodeling in Arabidopsis.
Our genomic expression studies of brm and syd mutant seedlings have identified several genes that are uniquely dependent on either SYD or BRM. This finding is consistent with the unique developmental defects observed in syd and brm null mutants: brm, but not syd, is male sterile and has root growth defects (Wagner and Meyerowitz, 2002; Hurtado et al., 2006; Kwon et al., 2006). In addition, BRM plays a unique role in control of expression of two regulators of cotyledon separation: the CUP-SHAPED COTYLEDON genes CUC1 and CUC3 (Kwon et al., 2006). Thus, the roles of the two closely related paralogs BRM and SYD have diverged.
On the other hand, a significant number of genes are coordinately misregulated in brm and syd mutants, suggesting that both ATPases are required for proper expression of these genes. This high degree of coregulation was specific to these two ATPases. We did not observe similar coordinate regulation of gene expression between SYD or BRM and other chromatin regulators: EMF1, a polycomb group protein; LHP1/TFL2, an Arabidopsis Heterochromatin Protein 1 homolog involved in epigenetic control of euchromatic transcription; and PKL, a CHD-type chromatin remodeling ATPase. A shared role of BRM and SYD is consistent with the finding that both brm and syd single mutants display strong pleiotropic phenotypes (suggesting the two ATPases do not simply act redundantly) and that many similar developmental defects are observed in brm and syd mutants. Both mutants are slow growing, have reduced apical dominance, are female sterile, and show precocious activation of FLOWERING LOCUS T in short day (Wagner and Meyerowitz, 2002; Farrona et al., 2004; Hurtado et al., 2006; Su et al., 2006). In addition, both BRM and SYD act upstream of the same embryonic patterning gene, CUC2 (Kwon et al., 2006), and both are required for proper floral homeotic gene expression (Wagner and Meyerowitz, 2002; Hurtado et al., 2006). It is possible that a similar functional overlap will be observed between BRM, SYD, and the other two Arabidopsis SWI/SNF ATPases, CHR12 and CHR23 (http://www.chromdb.org/), but mutants in these two have not yet been described.
Protein Interactions Predict the Presence of Multiple Core SWI/SNF Complexes in Arabidopsis
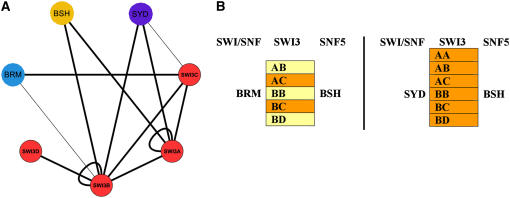
The interactions between individual subunits of the Arabidopsis core SWI/SNF chromatin remodeling complex were elucidated using protein interaction studies (Figure 4A; Sarnowski et al., 2005; Hurtado et al., 2006). If we assume that the stoichiometry of core Arabidopsis SWI/SNF complex components is similar to that found in metazoans (one catalytic subunit, two SWI3 proteins, and one SNF5 protein; Mohrmann and Verrijzer, 2005), we predict 11 possible core complexes in Arabidopsis based on known protein interactions (Figure 4B). This compares to only two core complexes described in yeast, Drosophila, and humans (Mohrmann et al., 2004). Even if only some of these core complexes exist in vivo, the number of SWI/SNF chromatin remodeling complexes in plants is likely to be greater than in metazoans. The slight differences observed in the spatial expression of individual ATSWI3 genes suggest that these subunits may contribute to presence and activity of distinct SWI/SNF complexes in different tissues. One exciting challenge for the future is identification and purification of tissue- and/or stage-specific SWI/SNF complexes from Arabidopsis followed by characterization of their individual biological roles.
Figure 4.
Hypothetical SWI/SNF Core Complexes in Arabidopsis.
(A) Schematic of all possible protein interactions based on individual protein–protein interactions identified in this study as well as by others (Sarnowski et al., 2002; Hurtado et al., 2006). The protein interaction network was visualized using Cytoscape 2.3.2. The edges (interactions) connecting the nodes (proteins tested) are represented by thick black lines for strong interactions and thin black lines for weak interactions. ATSWI3 proteins are referred to as SWI3A to SWI3D (red circles), BSH is the Arabidopsis SNF5 ortholog (yellow circle), and SYD and BRM are the two SWI/SNF ATPases analyzed (blue circles).
(B) The protein interactions depicted in (A) theoretically allow formation of 11 potential SWI/SNF core complexes if the subunit stochiometry is the same as in metazoans (Mohrmann and Verrijzer, 2005). Each core complex consists of the central chromatin remodeling ATPase (SYD or BRM), two SWI3 subunits (ATSWI3), and one SNF5 subunit (BSH). Strong interactions are shown in orange and weaker interactions in yellow. SYD is potentially able to form a unique core complex with ATSWI3A and BSH.
Redundant Roles for BRM and SYD during Embryo Development?
We show here that brm syd double mutants are embryonic lethal. By contrast, single brm and syd null mutants are viable, indicating that presence of either BRM or SYD is sufficient for proper embryo development (Wagner and Meyerowitz, 2002; Hurtado et al., 2006; Kwon et al., 2006). It is possible that the embryonic defect in double mutants is due to SYD and BRM regulating parallel pathways that cause synthetic lethality when simultaneously inactivated. Alternatively, BRM or SYD may redundantly regulate expression of an essential gene or process required for proper embryo development. We favor the second possibility for the following reasons. Mutations in ATSWI3A and ATSWI3B, two putative SWI/SNF core complex components, cause recessive embryonic lethality (Sarnowski et al., 2005). We show here that SYD interacts with both ATSWI3A and ATSWI3B. BRM can also interact with ATSWI3B (Hurtado et al., 2006). The simplest explanation for the observed embryonic lethality is therefore that a single chromatin remodeling complex containing both ATSWI3A and ATSWI3B, and either BRM or SYD (Figure 4B), is required to regulate an essential gene or process during embryo development.
BRM and SYD have overlapping roles and protein interactions, yet BRM is the only Arabidopsis SWI/SNF ATPase with a bromodomain. This characteristic SWI/SNF ATPase motif stabilizes binding to acetylated Lys residues on histone tails (Dhalluin et al., 1999). Our findings suggest that the bromodomain is not required for SWI/SNF function in Arabidopsis, at least with respect to embryo development, where SYD can apparently substitute for loss of BRM. One possible explanation for this phenomenon is that other SYD complex components may contain bromodomains that can compensate for the absence of this motif in SYD. Alternatively, this domain may not be absolutely required for SWI/SNF ATPase and complex function. Consistent with this hypothesis, deletion of the bromodomain had no adverse effect on SWI/SNF ATPase activity in several organisms (Laurent et al., 1993; Elfring et al., 1998; Inayoshi et al., 2006).
Functional Diversification of Paralogous SWI/SNF Chromatin Remodeling ATPases
Our combined transcription profiling, phenotypic, and protein interaction studies indicate that the two paralogous SWI/SNF ATPases BRM and SYD in Arabidopsis have diversified. In addition, we find an expansion of the number of BRM- and SYD-related proteins in poplar, a recently sequenced tree genome (Tuskan et al., 2006). Poplar has two SYD orthologs and two BRM orthologs (see Supplemental Figure 2A online; http://www.chromdb.org/), even though the large superfamily of SNF2 ATPases, to which the SWI/SNF subfamily belongs, is not similarly expanded (58 genes in poplar versus 42 genes in Arabidopsis). This suggests further diversification of these two ATPases in a more complex plant species, in support of the hypothesis that organismal complexity may result from more elaborate transcriptional regulation (Levine and Tjian, 2003; Taatjes et al., 2004).
SYD and BRM represent an ancient duplication event (Su et al., 2006) that occurred prior to the split between eudicots and monocots (∼200 million years ago). Diversification of gene function typically occurs after gene duplication (Ohno, 1970). Two paralogous genes like BRM and SYD might have diversified by neofunctionalization, where one paralog retains the ancestral function while the other acquires new functions, or by subfunctionalization, where the ancestral functions are divided between the two paralogs (Ohno, 1970; Hughes, 1994; Lynch and Conery, 2000; Lynch and Force, 2000; Lynch et al., 2001; He and Zhang, 2005). Presence of shared and possibly redundant roles for SYD and BRM suggest retention of at least a part of the ancestral roles for both proteins. While we cannot rule out subfunctionalization, the combined data (below) suggest that SYD and BRM diverged by partial neofunctionalization, where one paralog acquired some new functions and lost some ancestral functions as defined by He and Zhang (2005).
Duplicate genes often diverge through acquisition of differential expression patterns (Lynch and Force, 2000; Carroll, 2005). Our expression data (data not shown) and publicly available data (AtGenexpress; Schmid et al., 2005) suggest no or very little difference in tissue- or stage-specific expression between SYD and BRM (see Supplemental Figure 2B online). Duplicate genes can also diverge through changes in the protein coding sequence (Lynch and Force, 2000; Carroll, 2005). The domain architecture of BRM and SYD is quite divergent (Farrona et al., 2004; Su et al., 2006), especially the C-terminal domain downstream of the ATPase domain and portions of the N-terminal domain upstream of the ATPase domain. The unique C-terminal domain of SYD is not required for biochemical function but may modulate protein activity (Su et al., 2006). While the N-terminal domain of both proteins can still interact with the same proteins (ATSWI3B and ATSWI3C), only SYD can interact with ATSWI3A (this study; Hurtado et al., 2006). The novel domain architecture in SYD and conservation of the metazoan protein structure in BRM suggest that the latter might represent the ancestral paralog. Thus, the functional divergence of SYD and BRM is likely based on changes in their coding sequences that result in altered protein interactions.
These considerations provide a conceptual framework for further investigations. Several phenotypes, for example, defects in flower patterning, can be observed in both brm and syd single mutants, but they are subtle (Wagner and Meyerowitz, 2002; Hurtado et al., 2006; Kwon et al., 2006). We propose that the weak phenotypes may be due to partial compensation of one ATPase for loss of the other, which can be tested by tissue-specific inactivation of both SWI/SNF ATPases.
In summary, SYD and BRM, two SWI/SNF ATPases in Arabidopsis, likely form several tissue-specific chromatin remodeling complexes, and both have distinct and shared functions. They act in multiple developmental pathways, in which they function as specific regulators of transcription by controlling the expression of a small number of targets.
METHODS
Microarray Hybridization and Data Analysis
Wild-type (Ler) and mutant (brm-101 and syd-2) seedlings were grown for 10 d at 22°C in 16 h light at 120 μmol m−2 s−1 of cool white light. RNA was isolated from entire seedlings in two biological replicates as previously described (William et al., 2004) except that the tissue was ground to a fine powder using a mortar. RNA (5 μg) was used for cRNA synthesis. Labeling, hybridization, and detection were performed at the University of Pennsylvania Microarray Facility (http://www.med.upenn.edu/microarr/). All microarray data preparation and data analysis were performed in the statistical package R. Standard Affymetrix quality controls were performed using the Bioconductor package Simpleaffy (Wilson and Miller, 2005). The six samples passed all quality control tests (scaling factor, spike in controls, background, amplification, signal intensity, and 3′ to 5′ signal bias). A nonspecific filter was applied such that only those genes identified as “Present” using the MAS5.0 algorithm in at least one of the six arrays were used for further analysis, and 14,780 of 22,810 passed the filtering criteria.
Signal values were obtained using the gcRMA algorithm (Wu et al., 2004). Normalization was effective based on median signal intensities and overall signal distribution for each sample. Principle components analysis and hierarchical clustering (average linkage) of the Pearson's correlation coefficients for all genes revealed significant separation based on condition (genotype) that far exceeded experimental variation. A nonparametric approach, Rank Product (Breitling et al., 2004), was used to identify differentially expressed genes. This method performs well on experiments with a small number of replicates, is robust (Breitling and Herzyk, 2005; Jeffery et al., 2006), and has been used in a variety of recent analyses (Hufton et al., 2006; Ma et al., 2006; Nemhauser et al., 2006). FDR was calculated in R (Storey and Tibshirani, 2003). Genes with an FDR of <10% were considered significantly altered in expression in the mutants compared with the wild type.
Late versus Early Seedling Genes
We used a publicly available developmental microarray data set (AtGenExpress; Schmid et al., 2005) to identify genes upregulated during later stages in seedling development. The mean of the triplicate gcRMA values was determined for genes that were present at least once in the syd brm microarray for five samples: ATGE_5, ATGE_6, ATGE_8, ATGE_10, and ATGE_12 (leaves 1 plus 2 [day 7], shoot apex vegetative [day 7], shoot apex transition before bolting [day 14], rosette leaf 4 [day 10], and rosette leaf 2 [day 17]). Genes upregulated (late genes) or downregulated (early genes) during vegetative development were defined as genes increased or decreased twofold or more in expression in at least one of three pairwise comparisons: ATGE_8/ATGE_6, ATGE_10/ATGE_5, and ATGE_12/ATGE_5. A total of 1919 and 2548 unique genes fulfilled these criteria. After comparison with genes upregulated or downregulated in both brm and syd, statistical significance of the overlap was calculated using the Fisher's exact test as described below.
Genes Misregulated in Other Chromatin Regulatory Mutants
Microarray data sets based on the first generation Affymetrix array were kindly provided to us as *.cel files by Koji Goto (Nakahigashi et al., 2005) for LHP1(TFL2), by Renee Sung (Moon et al., 2003) and Tong Zhu (Syngenta) for EMF1, and by Joe Ogas (Dean Rider et al., 2003) for PKL. Plant age and growth differed for each array; however, all tissues were harvested during the vegetative stage. To compare the overlap between genes regulated, for example, by LHP1 and by BRM or SYD, the BRM and SYD gene lists (above) were filtered to include only those genes identified as “Present” in at least one of the lhcp1 microarrays using the MAS5.0 algorithm implemented in R. A total of 4171 genes (LHP1), 5382 genes (EMF1), and 41,250 genes (PKL) passed the nonspecific filter. gcRMA-normalized expression values were determined for each of the microarrays, and the data were analyzed using Rank Product (Breitling et al., 2004). Genes with an FDR of <10% were considered significantly altered in expression in the mutants compared with the wild type.
Significance of Overlap in Gene Expression
Significance of the overlap between genes differentially expressed in each of the mutants was determined using the Fisher's exact test. Two-tailed P values were calculated as defined by Agresti (1992) based on the following table, where X1 represents the number of genes regulated by factor 1, X2 represents the number of genes regulated by factor 2, and X12 is the number of genes regulated by both genes. Y represents the total number of genes that passed nonspecific filtering and that were included in the overlap analysis.
Table 6.
| Factor 2 | Not Factor 2 | |
|---|---|---|
| Factor 1 | X12 | X2 − X12 |
| Not factor 1 | X1 − X12 | Y − X1 − X2 + X12 |
Real-Time PCR
Real-time PCR was performed using two biological replicates. Reverse transcription of 5 μg RNA in a 20-μL reaction was as per the manufacturer's instructions using the Superscript III kit (Invitrogen). The RT reaction was diluted fourfold, and 1.3 μL of the RT reaction was used in triplicate 12 μL real-time PCR reactions using the QuantiTect SYBR Green PCR kit (Qiagen) on a DNA Engine Opticon Thermal cycler (MJ Research). Thermal cycling conditions were as follows: 15 min at 95°C and then 45 cycles of 15 s at 94°C, 30 s at 55°C, and 30 s at 72°C, followed by a melting curve analysis. The data obtained were analyzed with the Opticon Monitor Analysis Software (version 1.4). Relative amounts of all mRNA were calculated from threshold cycle values and standard curves and normalized with the signal values obtained for expression of the eukaryotic translation initiation factor 4A-1 (EIF4A). The mean and standard error were determined from the six samples (two biological replicates and three real-time PCR reactions). Specificity of real-time PCR products was confirmed by electrophoresis on a 2.5% agarose gel. Primers used are listed in Supplemental Table 5 online.
Mutant Lines and Reporter Studies
syd-2 and brm-101 were described previously (Wagner, 2003; Kwon et al., 2006).The syd-5 T-DNA insertion allele was obtained from the ABRC (SALK_023209) (Alonso et al., 2003). brm-1 was described by Hurtado et al. (2006) and Kwon et al. (2006). syd and brm alleles are summarized in Supplemental Table 6 online.
GST Pull-Down Experiments
For in vitro interaction tests, prey constructs were cloned into pGADT7 (Clontech) and in vitro transcribed and translated in the presence of 35S Met using the TnT rabbit reticulocyte system (Promega) as previously described (Zhu et al., 2000), followed by addition of protease inhibitors (20 μg/mL pepstatin, 20 μg/mL leupeptin, 8 trypsin inhibitor units of aprotinin, and 0.8 mM PMSF). Ten percent of the TnT reaction was separated by gel electrophoresis on a 12% polyacrylamide gel and quantitated after drying using a phosphor imager. The bait (SYDN) was cloned into pGEX (GE Healthcare/Amersham Biosciences). The resulting GST fusion protein and the pGEX vector alone were used to generate purified recombinant protein as per the manufacturer's instructions. Equal amounts (15 μg) of recombinant GST-SYDN and GST alone as determined by gel electrophoresis and Coomassie Brilliant Blue staining and after protein gel blot transfer detection with anti-GST antibody (1:100; Amersham) were incubated with 30 μL G Sepharose in BC500 (20 mM Tris, pH 8.3, 50 μM EDTA, pH 8, 500 mM KCl, 2% glycerol, 1% Nonidet P-40, 15 mM DTT, 1.2 mM PMSF, 12 trypsin inhibitor units of aprotinin, 20 μg/mL pepstatin, and 20 μg/mL leupeptin), and equal amounts of labeled prey (amount based on phosphor imager quantitation) were incubated in a 500-μL reaction overnight at 4°C with rotation. The Sepharose was washed three times with buffer BC150 (same as for BC500, except contains 150 mM KCl) followed by resuspension in 40 μL of protein loading buffer. Ten microliters of the reaction was run on a 12% gel and quantitated using a phosphor imager. Another 10 μL of the reaction was analyzed on a protein gel blot using anti-GST antiserum (1:1000; Amersham). The phosphor imager quantitations were normalized by the amount of bait protein precipitated as determined by densitometry of the chemiluminescence band from the protein gel blot.
Accession Numbers
Sequence data from this article have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through the GEO Series accession number GSE5806. Relevant accession numbers for the genes referred to in the text are as follows: At2g28290 (SYD), At2g46020 (BRM), At2g47620 (ATSWI3A), At2g33610 (ATSWI3B), At1g21700 (ATSWI3C), At4g34430 (ATSWI3D), At5g11530 (EMF1), At5g17690 (LHP1/TFL2), At2g25170 (PKL), At3g06010 (CHR12), At5g19310 (CHR23), LG_VIII 13515362:13533977 (CHR910), LG_X:4547683-4576184 (CHR925), LG_II:12295068-12309538 (CHR958), and LG_XIV:2589792-2604708 (CHR902).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypic and Molecular Analysis of syd-5.
Supplemental Figure 2. Phylogenetic and Expression Studies of BRAHMA and SPLAYED.
Supplemental Table 1. Genes Upregulated in brm-101.
Supplemental Table 2. Genes Downregulated in brm-101.
Supplemental Table 3. Genes Upregulated in syd-2.
Supplemental Table 4. Genes Downregulated in syd-2.
Supplemental Table 5. Primers Used in Real-Time PCR Analyses.
Supplemental Table 6. Alleles of brm and syd.
Supplementary Material
Acknowledgments
We thank John Tobias, Jennifer Nemhauser, and Fangxin Hong for advice and suggestions for the statistical analysis of the microarray data. We thank Scott Poethig, Kim Gallagher, Zongchi Liu, and Tony Cashmore for critical comments on the manuscript and the University of Pennsylvania Microarray Core Facility for performing the microarray probe synthesis and hybridization. This research was funded by National Institutes of Health Grant RO1 GM064650-01 to D.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Doris Wagner (wagnerdo@sas.upenn.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Agresti, A. (1992). A survey of exact inference for contingency tables. Stat. Sci. 7 131–153. [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Angus-Hill, M.L., Schlichter, A., Roberts, D., Erdjument-Bromage, H., Tempst, P., and Cairns, B.R. (2001). A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7 741–751. [DOI] [PubMed] [Google Scholar]
- Blazquez, M.A., and Weigel, D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404 889–892. [DOI] [PubMed] [Google Scholar]
- Breitling, R., Armengaud, P., Amtmann, A., and Herzyk, P. (2004). Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573 83–92. [DOI] [PubMed] [Google Scholar]
- Breitling, R., and Herzyk, P. (2005). Rank-based methods as a non-parametric alternative of the T-statistic for the analysis of biological microarray data. J. Bioinform. Comput. Biol. 3 1171–1189. [DOI] [PubMed] [Google Scholar]
- Brzeski, J., Podstolski, W., Olczak, K., and Jerzmanowski, A. (1999). Identification and analysis of the Arabidopsis thaliana BSH gene, a member of the SNF5 gene family. Nucleic Acids Res. 27 2393–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak, M., and Spradling, A.C. (2006). Searching chromatin for stem cell identity. Cell 125 233–236. [DOI] [PubMed] [Google Scholar]
- Carroll, S.B. (2005). Evolution at two levels: On genes and form. PLoS Biol. 3 e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo, C., Romera-Branchat, M., and Pelaz, S. (2005). A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J. 43 586–596. [DOI] [PubMed] [Google Scholar]
- Dean Rider, S., Jr., Henderson, J.T., Jerome, R.E., Edenberg, H.J., Romero-Severson, J., and Ogas, J. (2003). Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 35 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna, I.L., Ohkawa, Y., and Imbalzano, A.N. (2006). Chromatin remodelling in mammalian differentiation: Lessons from ATP-dependent remodellers. Nat. Rev. Genet. 7 461–473. [DOI] [PubMed] [Google Scholar]
- Dhalluin, C., Carlson, J.E., Zeng, L., He, C., Aggarwal, A.K., and Zhou, M.M. (1999). Structure and ligand of a histone acetyltransferase bromodomain. Nature 399 491–496. [DOI] [PubMed] [Google Scholar]
- Elfring, L.K., Daniel, C., Papoulas, O., Deuring, R., Sarte, M., Moseley, S., Beek, S.J., Waldrip, W.R., Daubresse, G., DePace, A., Kennison, J.A., and Tamkun, J.W. (1998). Genetic analysis of brahma: The Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 148 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona, S., Hurtado, L., Bowman, J.L., and Reyes, J.C. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131 4965–4975. [DOI] [PubMed] [Google Scholar]
- Flaus, A., Martin, D.M., Barton, G.J., and Owen-Hughes, T. (2006). Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34 2887–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., and Zhang, J. (2005). Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege, F.C., Jennings, E.G., Wyrick, J.J., Lee, T.I., Hengartner, C.J., Green, M.R., Golub, T.R., Lander, E.S., and Young, R.A. (1998). Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95 717–728. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529. [DOI] [PubMed] [Google Scholar]
- Hufton, A.L., Vinayagam, A., Suhai, S., and Baker, J.C. (2006). Genomic analysis of Xenopus organizer function. BMC Dev. Biol. 6 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A.L. (1994). The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 256 119–124. [DOI] [PubMed] [Google Scholar]
- Hurtado, L., Farrona, S., and Reyes, J.C. (2006). The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol Biol. 62 291–304. [DOI] [PubMed] [Google Scholar]
- Inayoshi, Y., Miyake, K., Machida, Y., Kaneoka, H., Terajima, M., Dohda, T., Takahashi, M., and Iijima, S. (2006). Mammalian chromatin remodeling complex SWI/SNF is essential for enhanced expression of the albumin gene during liver development. J. Biochem. (Tokyo) 139 177–188. [DOI] [PubMed] [Google Scholar]
- Ioshikhes, I.P., Albert, I., Zanton, S.J., and Pugh, B.F. (2006). Nucleosome positions predicted through comparative genomics. Nat. Genet. 38 1210–1215. [DOI] [PubMed] [Google Scholar]
- Jeffery, I.B., Higgins, D.G., and Culhane, A.C. (2006). Comparison and evaluation of methods for generating differentially expressed gene lists from microarray data. BMC Bioinformatics 7 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam, S., and Emerson, B.M. (2003). Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11 377–389. [DOI] [PubMed] [Google Scholar]
- Kasten, M., Szerlong, H., Erdjument-Bromage, H., Tempst, P., Werner, M., and Cairns, B.R. (2004). Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 23 1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston, R.E., and Narlikar, G.J. (1999). ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13 2339–2352. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962. [DOI] [PubMed] [Google Scholar]
- Krebs, J.E., Fry, C.J., Samuels, M.L., and Peterson, C.L. (2000). Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102 587–598. [DOI] [PubMed] [Google Scholar]
- Kwon, C.S., Chen, C., and Wagner, D. (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 19 992–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, C.S., Hibara, K.I., Pfluger, J., Bezhani, S., Metha, H., Aida, M., Tasaka, M., and Wagner, D. (2006). A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133 3223–3230. [DOI] [PubMed] [Google Scholar]
- Laurent, B.C., Treich, I., and Carlson, M. (1993). The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 7 583–591. [DOI] [PubMed] [Google Scholar]
- Levine, M., and Tjian, R. (2003). Transcription regulation and animal diversity. Nature 424 147–151. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and Conery, J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and Force, A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., O'Hely, M., Walsh, B., and Force, A. (2001). The probability of preservation of a newly arisen gene duplicate. Genetics 159 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., Morrow, D.J., Fernandes, J., and Walbot, V. (2006). Comparative profiling of the sense and antisense transcriptome of maize lines. Genome Biol. 7 R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann, L., Langenberg, K., Krijgsveld, J., Kal, A.J., Heck, A.J., and Verrijzer, C.P. (2004). Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 24 3077–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann, L., and Verrijzer, C.P. (2005). Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta 1681 59–73. [DOI] [PubMed] [Google Scholar]
- Moon, Y.H., Chen, L., Pan, R.L., Chang, H.S., Zhu, T., Maffeo, D.M., and Sung, Z.R. (2003). EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 15 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahigashi, K., Jasencakova, Z., Schubert, I., and Goto, K. (2005). The Arabidopsis heterochromatin protein1 homolog (TERMINAL FLOWER2) silences genes within the euchromatic region but not genes positioned in heterochromatin. Plant Cell Physiol. 46 1747–1756. [DOI] [PubMed] [Google Scholar]
- Nemhauser, J.L., Hong, F., and Chory, J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475. [DOI] [PubMed] [Google Scholar]
- Noh, Y.S., and Amasino, R.M. (2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. (1970). Evolution by Gene Duplication. (Berlin, New York: Springer-Verlag).
- Pelaz, S., Tapia-Lopez, R., Alvarez-Buylla, E.R., and Yanofsky, M.F. (2001. a). Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11 182–184. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Gustafson-Brown, C., Kohalmi, S.E., Crosby, W.L., and Yanofsky, M.F. (2001. b). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26 385–394. [DOI] [PubMed] [Google Scholar]
- Phelan, M.L., Sif, S., Narlikar, G.J., and Kingston, R.E. (1999). Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3 247–253. [DOI] [PubMed] [Google Scholar]
- Saha, A., Wittmeyer, J., and Cairns, B.R. (2006). Chromatin remodelling: The industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7 437–447. [DOI] [PubMed] [Google Scholar]
- Sarnowski, T.J., Rios, G., Jasik, J., Swiezewski, S., Kaczanowski, S., Li, Y., Kwiatkowska, A., Pawlikowska, K., Kozbial, M., Kozbial, P., Koncz, C., and Jerzmanowski, A. (2005). SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell 17 2454–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowski, T.J., Swiezewski, S., Pawlikowska, K., Kaczanowski, S., and Jerzmanowski, A. (2002). AtSWI3B, an Arabidopsis homolog of SWI3, a core subunit of yeast Swi/Snf chromatin remodeling complex, interacts with FCA, a regulator of flowering time. Nucleic Acids Res. 30 3412–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Simone, C. (2006). SWI/SNF: The crossroads where extracellular signaling pathways meet chromatin. J. Cell. Physiol. 207 309–314. [DOI] [PubMed] [Google Scholar]
- Smith, C.L., and Peterson, C.L. (2005). ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 65 115–148. [DOI] [PubMed] [Google Scholar]
- Soutourina, J., Bordas-Le Floch, V., Gendrel, G., Flores, A., Ducrot, C., Dumay-Odelot, H., Soularue, P., Navarro, F., Cairns, B.R., Lefebvre, O., and Werner, M. (2006). Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol. Cell. Biol. 26 4920–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar, V.V., Surendrarao, A., and Liu, Z. (2006). APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133 3159–3166. [DOI] [PubMed] [Google Scholar]
- Storey, J.D., and Tibshirani, R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y., Kwon, C.S., Bezhani, S., Huvermann, B., Chen, C., Peragine, A., Kennedy, J.F., and Wagner, D. (2006). The N-terminal ATPase AT-hook-containing region of the Arabidopsis chromatin-remodeling protein SPLAYED is sufficient for biological activity. Plant J. 46 685–699. [DOI] [PubMed] [Google Scholar]
- Sudarsanam, P., Iyer, V.R., Brown, P.O., and Winston, F. (2000). Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes, D.J., Marr, M.T., and Tjian, R. (2004). Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 5 403–410. [DOI] [PubMed] [Google Scholar]
- Teper-Bamnolker, P., and Samach, A. (2005). The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17 2661–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L., Fong, M.P., Wang, J.J., Wei, N.E., Jiang, H., Doerge, R.W., and Chen, Z.J. (2005). Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treich, I., Cairns, B.R., de los Santos, T., Brewster, E., and Carlson, M. (1995). SNF11, a new component of the yeast SNF-SWI complex that interacts with a conserved region of SNF2. Mol. Cell. Biol. 15 4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treich, I., and Carlson, M. (1997). Interaction of a Swi3 homolog with Sth1 provides evidence for a Swi/Snf-related complex with an essential function in Saccharomyces cerevisiae. Mol. Cell. Biol. 17 1768–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama, T. (2002). The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat. Rev. Mol. Cell Biol. 3 422–429. [DOI] [PubMed] [Google Scholar]
- Tuskan, G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604. [DOI] [PubMed] [Google Scholar]
- Vignali, M., Hassan, A.H., Neely, K.E., and Workman, J.L. (2000). ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20 1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D. (2003). Chromatin regulation of plant development. Curr. Opin. Plant Biol. 6 20–28. [DOI] [PubMed] [Google Scholar]
- Wagner, D., and Meyerowitz, E.M. (2002). SPLAYED, a novel SWI/SNF ATPase homolog controls reproductive development in Arabidopsis thaliana. Curr. Biol. 12 1–20. [DOI] [PubMed] [Google Scholar]
- William, D.A., Su, Y., Smith, M.R., Lu, M., Baldwin, D.A., and Wagner, D. (2004). Genomic identification of direct target genes of LEAFY. Proc. Natl. Acad. Sci. USA 101 1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, C.L., and Miller, C.J. (2005). Simpleaffy: A BioConductor package for Affymetrix quality control and data analysis. Bioinformatics 21 3683–3685. [DOI] [PubMed] [Google Scholar]
- Wu, Z., Irizarry, R., Gentleman, R., Martinez Murillo, F., and Spencer, F. (2004). A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. Johns Hopkins University, Department of Biostatistics Working Papers, Working Paper 1. http://www.bepress.com/jhubiostat/paper1.
- Zhou, C., Miki, B., and Wu, K. (2003). CHB2, a member of the SWI3 gene family, is a global regulator in Arabidopsis. Plant Mol. Biol. 52 1125–1134. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Tepperman, J.M., Fairchild, C.D., and Quail, P.H. (2000). Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc. Natl. Acad. Sci. USA 97 13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- Transcription profiling of Drosophila melanogaster pupae carrying dominant-negative mutations in SWI/SNF complex components revealed that 0.7 to 1.4% of all genes exhibited altered expression compared to wild-type pupae (Zraly et al., 2006). The extent of the alteration in gene expression is very similar to that which we observed in our study of expression changes in the SWI/SNF ATPase null mutant Arabidopsis thaliana seedlings compared to wild-type seedlings.
- Zraly, C.B., Middleton, F.A., and Dingwall, A.K. (2006). Hormone-response genes are direct in vivo regulatory targets of Brahma (SWI/SNF) complex function. J. Biol. Chem. 281 35305–35315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.