Abstract
Embryo patterning in Arabidopsis thaliana is highly affected when KANADI or Class III HD-Zip genes are compromised. Triple loss-of-function kan1 kan2 kan4 embryos exhibit striking defects in the peripheral–central axis, developing lateral leaf-like organs from the hypocotyls, whereas loss of Class III HD-Zip gene activity results in a loss of bilateral symmetry. Loss of KANADI activity in a Class III HD-Zip mutant background mitigates the defects in bilateral symmetry, implying that the two gene families act antagonistically during embryonic pattern formation. Dynamic patterns of auxin concentration and flux contribute to embryo patterning. Polar cellular distribution of PIN-FORMED1 (PIN1) mediates auxin flow throughout embryogenesis and is required for establishment of the apical–basal axis and bilateral symmetry. Defects in the pattern of PIN1 expression are evident when members of either the KANADI or Class III HD-Zip gene families are compromised. Abnormal expression patterns of PIN1 in KANADI or Class III HD-Zip multiple mutants and the phenotype of plants in which members of both gene families are mutated suggest that pattern formation along the central–peripheral axis results from interplay between auxin and the KANADI and Class III HD-Zip transcription factors, whose defined spatial and temporal expression patterns may also be influenced by auxin.
INTRODUCTION
The development of a multicellular organism from a single cell involves a combination of cell division and patterning events to initially specify axes and subsequently organ and cell types. In angiosperms, the apical–basal and peripheral–central axes are established early in embryogenesis, followed closely by a subdivision of the radially symmetric embryo into one that has a specific phyllotactic pattern of embryonic leaves. In Arabidopsis thaliana, the subdivision results in a bilaterally symmetric embryo, a pattern that dictates the subsequent phyllotactic patterning of postembryonic leaf formation. As the apical–basal and peripheral–central axes are established, the shoot apical meristem (SAM) and root apical meristem are initiated, completing the basic pattern formation of the sporophyte. Most of subsequent plant development consists of the elaboration of organ formation and cell and tissue type differentiation based on the asymmetric axes established during embryogenesis.
Three major patterning events in angiosperm embryogenesis—establishment of apical–basal and central–peripheral axes, and establishment of bilateral symmetry—are correlated with dynamic patterns of auxin concentration and flux. Thus, determining how these fluxes are established and how they are modulated is critical to understanding pattern formation in plants. Dynamic changes in auxin flux and maxima are mediated by both transcriptional control of the PIN-FORMED (PIN) family of proteins that mediate cellular auxin efflux and the regulation of polar localization of PIN proteins. At least three gene family members, PIN1, PIN4, and PIN7, are differentially expressed in the zygote to globular stage embryos, and their dynamic expression patterns lead to an initial accumulation of auxin in the embryo proper, followed by a subsequent basal efflux of auxin from the embryo, defining the future root. Subsequent reversals in the polar localization of epidermal PIN proteins in the late globular embryo result in auxin maxima, marking the site of cotyledon initiation (Benkova et al., 2003; Friml et al., 2003). Although single mutants of pin1, pin7, and pin4 exhibit subtle defects as a result of the compensatory action of related family members, severe embryonic phenotypes, including embryos lacking apical-basal polarity, are observed in pin1 pin3 pin4 pin7 quadruple mutants (Friml et al., 2002, 2003; Blilou et al., 2005; Vieten et al., 2005).
Mutations in genes regulating PIN protein localization result in abnormal auxin flow and embryonic patterning. GNOM (GN), a membrane-associated guanine-nucleotide exchange factor, regulates vesicle budding, an important process for recycling of PIN proteins between endosomal compartments and the plasma membrane. The gn mutant resembles the severe embryo phenotype of pin1 pin3 pin4 pin7, suggesting that dynamic recycling of PIN proteins is important for embryo development. However, the random changes in PIN1 distribution observed in gn mutants implies that GN does not determine the polarity of PIN localization (Steinmann et al., 1999; Geldner et al., 2003). By contrast, PINOID (PID), a Ser-Thr protein kinase, may more directly regulate PIN1 polarity, because loss of PID activity leads to basal PIN1 localization and gain of PID function leads to apical PIN1 localization in cells of cotyledon primordia (Friml et al., 2004).
One consequence of auxin maxima created by convergent auxin flow is the activation of AUXIN RESPONSE FACTORS (ARFs), transcription factors that activate and/or repress target genes. ARF proteins are negatively regulated by AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) proteins, which are targeted for degradation in response to auxin. Two members of the ARF gene family, MONOPTEROS (MP/ARF5) and NONPHOTOTROPIC HYPOCOTYL4 (NPH4/ARF7), have demonstrated roles in embryo axis patterning, with double mutants lacking both apical and basal embryonic development (Berleth and Jurgens, 1993; Hardtke and Berleth, 1998; Hardtke et al., 2004). Thus, patterning is the output of the interplay between auxin and transcription factors with defined spatial and temporal expression patterns, likely to be established in part by auxin flux. The robustness of the process may be attributable to extensive feedback mechanisms regulating the PIN proteins themselves as well as between auxin and transcription factors (Sieberer et al., 2000; Schrader et al., 2003; Paciorek et al., 2005; Vieten et al., 2005; Abas et al., 2006; Sauer et al., 2006).
Members of the Class III HD-Zip transcription factor family (PHABULOSA [PHB], PHAVOLUTA [PHV], REVOLUTA [REV], and CORONA [CNA]) are implicated in the establishment of bilateral symmetry and differentiation of the central–peripheral axis. The genes are expressed throughout the early globular embryo proper, with expression being restricted to the apical central region by the globular stage and to the SAM, the adaxial region of the cotyledons, and the vasculature during the heart stage (McConnell et al., 2001; Otsuga et al., 2001; Emery et al., 2003; Prigge et al., 2005). The apical regions of phb phv rev embryos consist of a single radial cotyledon and lack both bilateral symmetry and the SAM, indicating that Class III HD-Zip genes are required to properly pattern the apical region of the globular embryo (Emery et al., 2003; Prigge et al., 2005).
The KANADI genes (KAN1 to KAN4), members of the GARP transcription factor family, exhibit an embryonic expression pattern complementary to that of the Class III HD-Zip genes. KAN1 and KAN2 are expressed at the abaxial region of the hypocotyl and cotyledons, and KAN3 is expressed at the abaxial region of the cotyledons (Kerstetter et al., 2001; Eshed et al., 2004). As a result of genetic redundancy, conspicuous abnormal phenotypes are observed primarily in multiple loss-of-function combinations: kan1 kan2 and kan1 kan2 kan3 plants exhibit a loss of abaxial fates in lateral organs and radialized stem vascular bundles (Eshed et al., 2001, 2004; Emery et al., 2003). KAN4 (also known as ABERRANT TESTA SHAPE [ATS]) together with KAN1 and KAN2 direct the growth and polarity of integument development (Mcabee et al., 2006).
Complementary loss- and gain-of-function phenotypes of KANADI and Class III HD-Zip gene family members led to the hypothesis that KANADI and Class III HD-Zip genes function antagonistically to pattern tissues along the central–peripheral axis of the plant (McConnell and Barton, 1998; Eshed et al., 2001, 2004; Kerstetter et al., 2001; McConnell et al., 2001; Emery et al., 2003). For example, kan1 kan2 kan3 leaves are adaxialized and radialized, a phenotype reminiscent of that of the phb-1d gain-of-function allele (McConnell and Barton, 1998; Eshed et al., 2004). Likewise, the vasculature bundles in kan1 kan2 kan3 stems are radialized in a manner similar to that seen in the rev-10d gain-of-function allele (Emery et al., 2003).
Although the mechanistic nature of the mutually antagonistic activities of KANADI and Class III HD-Zip genes is not known, expression patterns and mutant phenotypes suggest an association with auxin. For instance, leaf phenotypes of kan1 kan2 plants are strikingly similar to those of ettin (arf3) arf4 plants (Pekker et al., 2005), and lateral root initiation, known to require auxin maxima, is altered by both KANADI loss-of-function and REV gain-of-function alleles (Hawker and Bowman, 2004). At least one Class III HD-Zip gene, ATHB8, is induced by auxin (Baima et al., 1995), and the expression of other members is associated with the differentiation of vascular tissues that are induced by auxin (Ohashi-Ito et al., 2005). Finally, Class III HD-Zip expression in both vascular and nonvascular plants mirrors the predicted flow of auxin in these plants (Floyd and Bowman, 2006; Floyd et al., 2006).
Here, we describe novel KANADI and Class III HD-Zip mutant phenotypes that exhibit defects in embryonic patterning and propose that members of the KANADI gene family pattern tissues within the plant by regulating the flow of auxin via the regulation of PIN1 polar expression.
RESULTS
The Roles of KAN1, KAN2, and KAN4 during Embryogenesis
To examine the role of KAN4 in plant development, we constructed double, triple, and quadruple mutant combinations of loss-of-function alleles of KAN4 together with the other three KANADI genes. kan1 kan2 kan4 plants exhibit a novel phenotype in which ectopic leaf-like organs develop from the hypocotyl and outgrowths develop on the abaxial side of the cotyledons (Figures 1B and 1D). The hypocotyl leaf-like organs are radialized (Figures 1I to 1K), with epidermal layers consisting of cells resembling leaf margin cells (Figure 1K). Approximately 10 d after germination, meristems develop in the axils of the hypocotyl leaf-like organs, similar to axillary meristems that develop in the axils of wild-type leaves (Figures 1J and 3G below). The leaves and flowers produced by both the SAM and the hypocotyl axillary meristems are indistinguishable from those of kan1 kan2 plants, suggesting that KAN4 does not play a major role in lateral organs other than the cotyledons (Figure 1K).
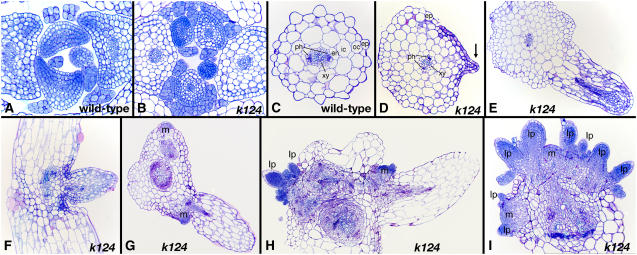
Figure 1.
The Phenotype of kan1 kan2 kan4.
(A) and (B) Fourteen-day-old wild-type (A) and kan1 kan2 kan4 (k124) (B) seedlings. Outgrowths develop on the abaxial side of kan1 kan2 kan4 cotyledons, and radialized leaf-like structures develop on the hypocotyl.
(C) and (D) Cotyledon (cot; asterisk) and hypocotyl (hyp) outgrowths (arrow) can be observed already in a 2-d-old kan1 kan2 kan4 seedling (D) but are lacking in a 2-d-old wild-type seedling (C).
(E) and (F) In a 6-d-old kan1 kan2 kan4 seedling (F), outgrowths form radialized structures (arrows) on the hypocotyl, and cotyledon outgrows (asterisk) continue to develop on the abaxial side. Compare with a 6-d-old wild-type seedling (E).
(G) and (H) The epidermal cell structure that in the wild type is characterized by an alternating pattern of large and small cell rows (G) is disrupted in the kan1 kan2 kan4 hypocotyl and consists of uniform small cells (H).
(I) to (K) Ten-day-old (I), 21-d-old (J), and 28-d-old (K) kan1 kan2 kan4 plants. The hypocotyl outgrowths elongate and form radialized leaf-like structures (white arrows) with an occasional trumpet-shaped leaf (black arrow). Ectopic meristems develop in the axils of the hypocotyl leaves, giving rise to leaves with polarity.
Figure 3.
Anatomical Features of kan1 kan2 kan4.
(A) and (B) Transverse sections through 12-d-old seedlings. Although in wild-type seedlings, young leaf primordia exhibit polarity in the abaxial–adaxial axis (A), kan1 kan2 kan4 (k124) leaf primordia lack polarity and are radialized (B).
(C) and (D) Transverse sections through wild-type (C) and kan1 kan2 kan4 (D) hypocotyls. The vascular anatomy of the kan1 kan2 kan4 hypocotyl (D) is indistinguishable from that of the wild type (C) with a single phloem (ph) strand on either side of the primary xylem (xy). The kan1 kan2 kan4 ground tissue has more cell layers than the wild-type tissue and lacks the distinct endodermis (en), inner cortex (ic), outer cortex (oc), and epidermis (ep) cell layers seen in the wild type. Early in development (D), the hypocotyl outgrowth (arrow) resembles leaf primordia.
(E) and (F) The leaf primordia develop into a radialized leaf lacking any signs of polarity, as seen in transverse (E) and longitudinal (F) sections. The vasculature differentiates initially at the distal end of the developing outgrowth (E).
(G) to (I) Meristems develop in the axils of hypocotyl leaves. A young hypocotyl leaf with a meristem (m) developing at its base is seen in a transverse section (G). A second meristem is associated with another hypocotyl leaf that is not included in the section. As the hypocotyl leaves mature, ectopic meristems continue to develop at their axils and are observed in longitudinal sections through the base of mature hypocotyls ([H] and [I]). The ectopic meristems give rise to leaf primordia (lp).
kan1 kan2 kan4 embryos are morphologically distinguishable from wild-type embryos by the heart stage. The cotyledons and the region that will give rise to the SAM are broader in kan1 kan2 kan4 embryos, and mutant cotyledon primordia are at a more obtuse angle than in wild-type embryos (Figures 2A to 2D). The region that will give rise to the abaxial side of the cotyledons and hypocotyl also exhibits an abnormal phenotype. In wild-type hypocotyls, cells primarily divide anticlinally, giving rise to organized cell files (Figures 2A and 2C). By contrast, periclinal planes of cell division are common in subepidermal layers of kan1 kan2 kan4 hypocotyls (Figures 2B and 2D). These periclinal cell divisions result in a broadening of the heart stage embryo. These, or subsequent, periclinal cell divisions may be the first manifestation of the outgrowths on the abaxial side of the cotyledons and the hypocotyl evident during the late heart and torpedo stages (Figure 2F).
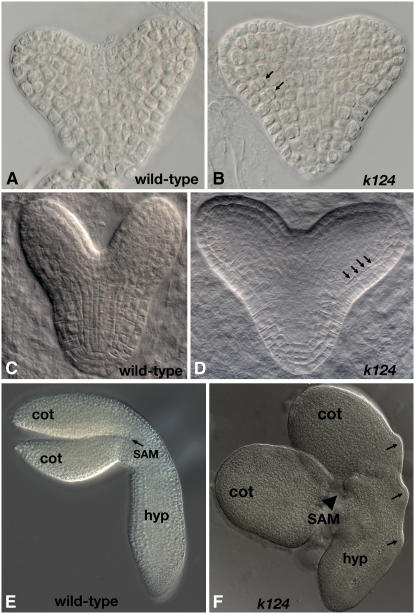
Figure 2.
The Embryo Phenotype of kan1 kan2 kan4.
(A) and (B) Wild-type (A) and kan1 kan2 kan4 (k124) (B) early heart stage embryos. Periclinal cell divisions are already visible in the kan1 kan2 kan4 subepidermal layer (arrows).
(C) and (D) Cotyledons are at a more obtuse angle and the region that will give rise to the SAM is broader in kan1 kan2 kan4 embryos (D) than in wild-type embryos (C). Arrows in (D) indicate abnormal periclinal cell divisions that will give rise to the cotyledon and hypocotyl outgrowths.
(E) and (F) Torpedo stage kan1 kan2 kan4 embryo (F) with cotyledon (cot) and hypocotyl (hyp) outgrowths (arrows), which are lacking in the wild-type torpedo stage embryo (E).
Anatomical Features of kan1 kan2 kan4 Seedlings
Wild-type leaves are characterized by anatomical differences between their adaxial (upper in Arabidopsis) and abaxial (lower in Arabidopsis) sides. The adaxial side consists of dense columnar palisade mesophyll, whereas the abaxial side consists of spongy mesophyll with intercellular air spaces that develop gradually. kan1 kan2 kan4 leaves produced from the SAM appear different from wild-type leaves and similar to the previously described kan1 kan2 leaves (Eshed et al., 2001, 2004), with nearly radialized leaves at emergence and delayed signs of polar anatomical features (Figures 3A and 3B). In contrast with leaves produced by the SAM, the ectopic leaf-like organs on the hypocotyl are radialized and lack evidence of polar anatomical cell structure (Figures 1J, 1K, 3E, and 3G). The hypocotyl outgrowths resemble leaf primordia early in development (Figure 3D), and as they mature, vasculature differentiates initially at the distal end of the developing outgrowth (Figure 3E). The developing vascular tissue joins the existing hypocotyl vasculature in a manner similar to the developing vasculature of a leaf primordium connecting to the stem vasculature. In contrast with wild-type leaves, which usually have a single axillary meristem, several meristems are usually present in the axil of each hypocotyl leaf-like organ (Figures 3G to 3I).
The vascular anatomy of kan1 kan2 kan4 hypocotyls (Figure 3D) is indistinguishable from that of the wild type (Figure 3C), with a single phloem strand on either side of the primary xylem (Busse and Evert, 1999). However, in kan1 kan2 kan4 hypocotyls, the ground tissue has more cell layers as a result of ectopic periclinal cell divisions throughout the developing hypocotyl, and the correspondence between the extra cell layers and those in the wild type is not clear from anatomical features. In addition, the epidermal patterning seen in wild-type hypocotyls is disrupted in kan1 kan2 kan4 hypocotyls (Figures 1G, 1H, 3C, and 3D). In Arabidopsis, hypocotyl and root epidermal cells in contact with two underlying cortical cells (cells over an anticlinal cortical cell wall [Figure 3C]) differentiate into stomata and root hairs in the hypocotyls and roots, respectively, whereas cells in contact with a single cortical cell (cells over a periclinal cortical cell wall [Figure 3C]) do not give rise to stomata or root hairs (Berger et al., 1998; Hung et al., 1998). kan1 kan2 kan4 hypocotyls may lack the regular epidermal cell pattern attributable to extensive cell divisions in the ground tissue, leading to altered patterns of stomata in the hypocotyl epidermis (Figures 1H and 3D) and root hairs (data not shown).
The KANADI Quadruple Mutant Is Additive
To further analyze the redundancy between the four KANADI genes, we generated the quadruple mutant kan1 kan2 kan3 kan4. The phenotype of the quadruple mutant appears additive, combining the two individual triple phenotypes, kan1 kan2 kan3 and kan1 kan2 kan4 (see Supplemental Figures 1A to 1D online). The stem vascular anatomy of the quadruple mutant is similar to that of kan1 kan2 kan3 (see Supplemental Figures 1E to 1I online) (Emery et al., 2003), implying that KAN4 does not play a significant role in vascular development.
YABBY Gene Activity in kan1 kan2 kan4 Embryos
YABBY gene family members, such as FIL, act in leaf primordia to promote abaxial identity and lamina expansion (Sawa et al., 1999; Siegfried et al., 1999; Eshed et al., 2001, 2004; Golz et al., 2004). FIL is expressed in the outgrowths developing on the abaxial side of kan1 kan2 leaves and is required for their development, because outgrowths do not develop from kan1 kan2 fil yabby3 (yab3) leaves (Eshed et al., 2004). To further examine the role of YABBY genes in the development of kan1 kan2 kan4 hypocotyl and cotyledon outgrowths, we examined FIL expression in kan1 kan2 kan4 embryos. During the heart stage, FIL expression in kan1 kan2 kan4 is confined to the abaxial side of developing cotyledons, similar to the wild-type expression pattern (Figures 4D and 4E). However, during the torpedo stage, FIL is ectopically expressed throughout leaf-like outgrowths on the hypocotyl (Figure 4G).
Figure 4.
YABBY and the kan1 kan2 kan4 Cotyledon and Hypocotyl Outgrowths.
(A) and (B) In kan1 kan2 kan4 fil yab3 yab5 (k124fily35) (B), outgrowths (arrows) forming on the hypocotyl are similar in shape to kan1 kan2 kan4 (k124) (A) outgrowths (arrows) but do not reach the full length of the kan1 kan2 kan4 outgrowths. Three narrow cotyledons (cot) develop in kan1 kan2 kan4 fil yab3 yab5 plants (B). The cotyledons lack the outgrowths (asterisks) observed on the abaxial side of kan1 kan2 kan4 cotyledons (A).
(C) to (G) mRNA expression pattern of FIL in kan1 kan2 kan4 embryos. At the early (D) and late (E) heart stages, FIL expression in kan1 kan2 kan4 is similar to that in the wild type (C) and confined to the abaxial side of developing cotyledons. During the torpedo stage, FIL expression is seen throughout hypocotyl outgrowth primordia (arrow) (G) and on the abaxial side of the cotyledons, as in the wild type (F).
Compromising the activity of three YABBY genes in the kan1 kan2 kan4 background does not eliminate radialized outgrowth formation on the hypocotyl (Figure 4B). However, these outgrowths develop more slowly and do not reach the full length of those of kan1 kan2 kan4 mutants. The hextuple mutants have radialized leaves without any abaxial outgrowths (Figure 4B), similar to kan1 kan2 fil yab3 leaves (Eshed et al., 2004). The cotyledons of the kan1 kan2 kan4 fil yab3 yab5 hextuple mutant exhibit unique features, with the development of three narrow cotyledons per plant, and the cotyledons lack the outgrowths observed on the abaxial side of kan1 kan2 kan4 cotyledons (Figure 4B). A similar defect in cotyledon number is also observed in the fil yab3 yab5 background (R. Sarojam and J.L. Bowman, unpublished data).
Do the Hypocotyl Leaves Arise from a Preexisting Meristem?
Because we observed similar features in ectopic hypocotyl outgrowths and leaves, we tested whether the hypocotyl outgrowths in kan1 kan2 kan4 are the outcome of a delocalized ectopic SAM by examining the expression of SHOOT MERISTEMLESS (STM) (Long and Barton, 1998) and CLAVATA3 (CLV3) (Fletcher et al., 1999) in kan1 kan2 kan4 embryos. Expression of STM in kan1 kan2 kan4 embryos is indistinguishable from that in the wild type, with the transcript restricted to the SAM at the heart stage, where the first signs of abnormal cell divisions could be seen, and later at the torpedo stage (see Supplemental Figures 2A to 2D online). To analyze the expression pattern of CLV3, we used a direct promoter fusion with the β-glucuronidase reporter gene, CLV3:GUS. In the kan1 kan2 kan4 background, CLV3 is expressed in heart stage embryos only in the SAM, similar to its expression pattern in wild-type embryos (see Supplemental Figures 2E and 2F online) (Fletcher et al., 1999). To summarize, cells in the kan1 kan2 kan4 hypocotyl did not ectopically express meristem markers.
Expression of PIN1 in kan1 kan2 kan4 and phb phv rev Embryos
Defects in auxin distribution during early stages of embryo development in kan1 kan2 kan4 mutants could lead to ectopic auxin maxima in the hypocotyl. To study the role of polar auxin transport in kan1 kan2 kan4 embryo development, we used a translational fusion of PIN1 to green fluorescent protein (GFP) (pPIN1:PIN1-GFP [Heisler et al., 2005]) and the synthetic reporter DR5 composed of auxin-responsive elements transcriptionally fused with GFP (DR5:GFP [Friml et al., 2003]).
PIN1 is expressed throughout the embryo at the early globular stage (Figure 5A), but by the transition and heart stages, expression is restricted to the upper half of the embryo, with regions of higher expression marking the sites of the incipient cotyledons and vasculature (Figures 5C and 5E). By the late heart stage, PIN1 expression continues in the vasculature of the future hypocotyl and root but is diminished in the cotyledons except at their tips and vasculature (Figure 5H). In addition, expression of PIN1 can be seen in the L1 layer of the incipient SAM, and this expression persists through the torpedo and later stages (Figures 5H and 5L). PIN1 expression in kan1 kan2 kan4 embryos is similar to that of wild-type embryos during the early stages of development (Figure 5B); however, by the transition stage, ectopic expression of PIN1 can be seen in cells that will give rise to the hypocotyl (Figure 5D), whereas PIN1 is not detected in these cells in wild-type embryos (Figure 5C). By the heart stage, PIN1 expression in wild-type embryos is excluded from the hypocotyl cells and restricted to the cotyledon primordia, whereas in kan1 kan2 kan4 embryos, ectopic expression in the hypocotyl cells is maintained (Figure 5F). Significantly, these changes in PIN1 expression appear before any alterations in cell division patterns can be seen (Figures 5D and 5G). Ectopic PIN1 expression in kan1 kan2 kan4 embryos remains in the hypocotyl through later stages of embryo development, in which the abnormal mutant morphology is apparent (Figures 5I to 5J and 5M). The cellular orientation of PIN1 in the hypocotyls of late heart stage kan1 kan2 kan4 embryos implies that a reversal in PIN1 polarity has occurred in those epidermal cells just below the cotyledons, from an initial apical localization to a basal localization. This reversal leads to an abnormal polar arrangement of PIN1 in hypocotyl cells such that ectopic auxin maxima are positioned within the hypocotyl (Figure 5K). DR5 expression was observed, indicating areas of auxin accumulation (Friml et al., 2003) and marking the mutant hypocotyl outgrowths (Figure 5T).
Figure 5.
PIN1 Expression and DR5 Auxin Response during Embryogenesis.
(A) and (B) PIN1 is expressed throughout both wild-type (A) and kan1 kan2 kan4 (k124) (B) globular embryos.
(C) and (D) In wild-type embryos (C), PIN1 is expressed during the transition stage at the sites of incipient cotyledons and developing vasculature, whereas in kan1 kan2 kan4 embryos (D), PIN1 expression can also be seen in the embryonic cells of the L1 layer that will give rise to the hypocotyl (arrows).
(E) to (G) At the heart stage, PIN1 is excluded from the hypocotyl cells in the wild type (E) but continues to be expressed in these cells in kan1 kan2 kan4 embryos ([F] and [G]). A transverse section through the hypocotyl region of the kan1 kan2 kan4 embryo at the heart stage (G) exhibits ectopic expression of PIN1 in the L1 cells, whereas no changes in cell division are evident at this stage.
(H) to (M) By the late heart (H) and later (L) stages, PIN1 expression in wild-type embryos is restricted to the SAM, vasculature, and tips of the cotyledons. In kan1 kan2 kan4, ectopic expression of PIN1 in the hypocotyl is maintained through the late heart ([I] and [J]) and later (M) stages, correlating with the aberrant phenotype. An abnormal polar arrangement of PIN1 in the hypocotyl cells with protein at the base of more apical cells and at the apex of more basal cells (K) leads to ectopic auxin maxima within the hypocotyl.
(N) and (P) In phb phv rev embryos, PIN1 is expressed throughout the apical part of the embryo and in the provasculature cells from the globular through the heart stages.
(Q) and (R) At later stages, PIN1 expression is lacking from the single radialized cotyledon (cot) and expressed only in the central and basal regions of the vasculature.
(S) and (T) GFP expression driven by the artificial DR5 promoter can be seen in kan1 kan2 kan4 embryos (T) during the torpedo stage in the hypocotyl and cotyledon outgrowths (arrows), indicating areas of auxin accumulation not present in wild-type embryos (S).
Given the phenotype of phb phv rev embryos, we predicted that PIN1 expression would also be altered in this genetic background. Triple mutants with a single radialized cotyledon and no SAM can be identified at the heart stage, when the apical portions of embryos lack the bilateral symmetry characteristic of wild-type embryos (Prigge et al., 2005). PIN1 is expressed throughout the apical half of phb phv rev embryos during the globular stage, similar to its expression in wild-type embryos (data not shown). The bilateral expression pattern of PIN1 observed at the heart stage in wild-type embryos (Figures 5C, 5E, and 5H) is lacking in phb phv rev embryos, in which PIN1 is expressed throughout the apical part of the embryo, the provasculature, and in the L1 layer, implying an auxin flow toward the apical tip of the single cotyledon (Figures 5N to 5P). PIN1 expression in phb phv rev embryos suggests auxin flow from all sides of the embryo through the L1 toward its central apical point, forming only a single auxin maximum and leading to the development of a single cotyledon primordium. By the torpedo stage, PIN1 expression is lacking from the single radialized cotyledon and expressed only in the central and basal part of the embryonic vasculature (Figures 5Q to 5R).
Activity of Class III HD-Zip in kan1 kan2 kan4 Plants
In wild-type embryos, PHB transcripts can be detected as early as the 16-cell stage of the developing embryo (McConnell et al., 2001). Initially, PHB is initially expressed throughout the embryo but is subsequently restricted to the central apical cells of the embryo, the cells that will give rise to the cotyledons and the SAM. By the heart stage, PHB expression is confined to the SAM and the adaxial regions of the cotyledons (McConnell et al., 2001). PHB expression in kan1 kan2 kan4 embryos is similar to that of wild-type embryos during the globular and heart stages (data not shown).
To examine the antagonistic roles of KANADI and Class III HD-Zip gene activity in embryogenesis, we generated kan1 kan2 kan4 phb phv rev plants. The hextuple kan1 kan2 kan4 phb phv rev has a novel phenotype, with loss of KANADI activity partially mitigating the phenotype conferred by phb phv rev. kan1 kan2 kan4 phb phv rev seedlings have two cotyledons with abaxial outgrowths and ectopic leaf-like outgrowths developing from the hypocotyl, similar to kan1 kan2 kan4 seedlings (Figures 4A and 6A). However, unlike kan1 kan2 kan4 and phb phv rev plants, the hextuple mutant produces a single radial leaf-like organ in the position normally occupied by the SAM in wild-type seedlings (Figure 6A). The initiation of the leaf-like organ is evident at the heart stage of embryogenesis, when subepidermal periclinal cell divisions in the position between the two cotyledons give rise to a leaf primordium (Figures 6B and 6C). The hypocotyl outgrowths of the hextuple mutant, also evident at the heart stage, are similar to those of kan1 kan2 kan4 plants in terms of their structure, size, length, and number per plant (Figures 4A and 6A).
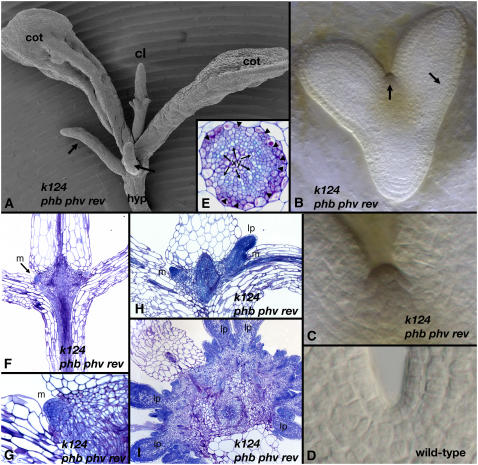
Figure 6.
The Phenotype of kan1 kan2 kan4 phb phv rev.
(A) A 14-d-old kan1 kan2 kan4 phb phv rev seedling (k124 phb phv rev) has two cotyledons (cot) and ectopic leaf-like outgrowths developing from the hypocotyl (arrows), similar to kan1 kan2 kan4 seedlings (cf. with Figure 4A, same age). In addition, in kan1 kan2 kan4 phb phv rev seedlings, a single radial central leaf (cl) develops in the position normally occupied by the SAM. The central leaf exhibits outgrowths on its distal end and is radialized toward its proximal end.
(B) to (D) The central leaf substituting for the SAM in kan1 kan2 kan4 phb phv rev is initiated during embryogenesis, with subepidermal periclinal cell divisions giving rise to a leaf primordium (C) instead of anticlinal cell divisions, as in wild-type SAMs (D). Arrows in (B) indicate areas of periclinal cell divisions.
(E) The vascular bundle at the proximal end of the central leaf is radialized with patches of phloem (arrows) surrounding a ring of xylem (xy) and parenchyma cells located at the center.
(F) to (I) Ectopic meristems (m) develop around the base of the central leaf, as seen in longitudinal sections through the central leaf ([F] to [H]; [G] is a closeup of [F]), and give rise to leaf primordia (lp) (I).
The central leaf-like organ has outgrowths toward its distal end and is radialized toward its proximal end (Figure 6A), with several vascular bundles arranged around the circumference distally that join into a single vascular bundle proximally. The proximal vascular bundle is radialized, with patches of phloem surrounding a ring of xylem and parenchyma cells located at the center (Figure 6E), similar to the vascular arrangement seen in radial phb phv rev cotyledons (Emery et al., 2003). The epidermal cell structure of the leaf-like organ consists primarily of long leaf margin–like cells. Ectopic meristems develop around its basal circumference at ∼10 d after germination (Figures 6F to 6H). In turn, these meristems give rise to leaves (Figure 6I).
DISCUSSION
Redundancies within the KANADI Clade of Genes
The four Arabidopsis KANADI genes display a complex pattern of genetic redundancy. KAN1, KAN2, and KAN3 promote polarity establishment in leaves, with KAN1 and KAN2 also contributing to polarity in floral organs, including the outer integument (Eshed et al., 2001, 2004). The same three genes also direct the proper polar differentiation of stem vascular bundles (Emery et al., 2003). By contrast, kan4 mutants display a novel ovule phenotype with a single integument consisting of fused outer and inner integuments (Mcabee et al., 2006). Although KAN4 plays little, if any, role in leaf and vascular development, during embryogenesis KAN4 acts redundantly with KAN1 and KAN2 in hypocotyl differentiation. The phenotype of kan1 kan2 kan3 kan4 plants supports the notion that KAN3 and KAN4 have distinct roles, because it is a combination of the phenotypes conferred by kan1 kan2 kan3 and kan1 kan2 kan4. Thus, although both KAN3 and KAN4 act redundantly with KAN1 and KAN2 in different aspects of plant development, KAN3 and KAN4 themselves have nonoverlapping roles. Although it is not clear at present when the gene duplications giving rise to the four Arabidopsis genes occurred relative to major taxonomic divergences, the presence of KANADI genes in the genomes of Selaginella and Physcomitrella suggests an ancestral function not associated specifically with leaf or vascular differentiation (Floyd and Bowman, 2007).
The Nature of the Leaves Developing from the Hypocotyl
The YABBY gene family in Arabidopsis consists of six members with at least four genes, FIL, YAB2, YAB3, and YAB5, expressed during embryogenesis and in developing leaves (Sawa et al., 1999; Siegfried et al., 1999; R. Sarojam and J.L. Bowman, unpublished data). Members of the YABBY gene family promote lamina expansion and abaxial cell fate in leaf primordia (Sawa et al., 1999; Siegfried et al., 1999; Eshed et al., 2001, 2004). The phenotype of kan1 kan2 kan4 fil yab3 yab5 plants (Figure 4B) suggests that the cotyledon outgrowths and the hypocotyl outgrowths are formed via two different developmental processes. The cotyledon outgrowths arise from a preexisting leaf lamina and are YABBY-dependent, similar to the previously described kan1 kan2 leaf laminar outgrowths (Eshed et al., 2001, 2004). By contrast, the hypocotyl leaves are YABBY-independent, supporting the idea that initiation of the hypocotyl outgrowths is analogous to leaf primordium formation.
The expression patterns of genes associated with the SAM, such as STM and CLV3, are similar in wild-type and kan1 kan2 kan4 plants (see Supplemental Figures 2B, 2D, and 2F online). Thus, no ectopic organized SAM formation is observed in kan1 kan2 kan4 embryos, with the hypocotyl leaves developing from cells that have never expressed STM or CLV3. The meristems that develop in the axils of the hypocotyl leaves are thus a consequence and not a cause of the ectopic leaf development. The leaf-like rather than cotyledon-like identity of the hypocotyl leaves may reflect the fact that most of their differentiation occurs after germination, whereas that of cotyledons occurs during embryogenesis.
Ectopic development of leaves from the hypocotyl was also observed in stm mutants (Barton and Poethig, 1993). However, the hypocotyl leaves of stm mutants develop postembryonically and can be first observed when the seedling is 14 d old. It was suggested that the formation of the hypocotyl leaves in stm mutants is a secondary effect caused by the absence of a SAM (Barton and Poethig, 1993). Hence, the developmental program leading to the formation of the hypocotyl leaf-like organs is different in kan1 kan2 kan4 and stm mutants.
Role of Class III HD-Zip Genes in the kan1 kan2 kan4 Embryonic Phenotype
Based on the antagonistic and complementary relationship between the KANADI and the Class III HD-Zip genes, it is possible that ectopic expression of the Class III HD-Zip genes is responsible for the kan1 kan2 kan4 embryo phenotype and, conversely, that ectopic expression of KANADI is responsible for the phb phv rev phenotype. Because bilateral symmetry is restored in phb phy rev kan1 kan2 kan4 embryos, Class III HD-Zip activity must repress KANADI activity in the central domain of the embryo, and ectopic expression of KANADI genes is responsible for the loss of bilateral symmetry in phb phv rev embryos. However, compromising the activity of three Class III HD-Zip genes (PHB, PHV, and REV) in the kan1 kan2 kan4 background does not lead to a loss of the ectopic hypocotyl leaves (Figure 6A), suggesting that either CNA (Prigge et al., 2005) is responsible for the phenotype or, alternatively, that ectopic Class III HD-Zip expression is not causal in the development of hypocotyl leaves in kan1 kan2 kan4 plants.
Patterns of Auxin Flux in KANADI and Class III HD-Zip Mutant Embryos
Regulation of auxin flux via PIN proteins plays an important patterning role during Arabidopsis embryogenesis, with reversals in PIN orientation correlating with major patterning events (Benkova et al., 2003; Friml et al., 2003; Blilou et al., 2005; Vieten et al., 2005). The loss of proper PIN polarity establishment, as in PIN multiple mutants and gn mutants (Steinmann et al., 1999; Friml et al., 2003; Geldner et al., 2003; Blilou et al., 2005; Vieten et al., 2005), or the loss of ARF action, as in mp mutants (Berleth and Jurgens, 1993; Hardtke and Berleth, 1998), results in defects in embryo patterning events, suggesting the reversals in auxin flow, as inferred by polar cellular PIN localization, may be instructive. Major changes in auxin flow occur at the transition stage, with PIN1 expressed such that bidirectional transport of auxin is established through the L1 layer: from the upper central region outward toward the periphery of the embryo (Figure 7A) (Benkova et al., 2003) and upward from the basal part of the embryo (Figure 7A) (Vieten et al., 2005). The result of the bidirectional auxin flow is to create auxin maxima at the periphery of the embryo, resulting in the induction of cotyledon primordia (likely through the activation of specific ARF genes such as MONOPTEROS) and bilateral symmetry, the establishment of which is enigmatic at present (Reinhardt et al., 2003; Hay et al., 2004; Jenik and Barton, 2005). It must be acknowledged that auxin biosynthesis also likely contributes to the establishment of auxin maxima (Cheng et al., 2006); however, the polar localization of PIN proteins correlates well with auxin maxima inferred by reporter genes and immunochemistry in both roots and shoots (Friml et al., 2002; Benkova et al., 2003; Reinhardt et al., 2003; Blilou et al., 2005). Thus, in the discussion that follows, we infer auxin maxima from the expression pattern and polar localization of PIN1, and we use the term “flow” to describe the presumed direction of auxin transport.
Figure 7.
A Model for Auxin Flow and the Establishment of Bilateral Symmetry during Embryo Patterning.
(A) and (B) In wild-type embryos at the transition stage (A), changes in auxin flow occur, with the reversal of auxin flow from the SAM central region outward toward the periphery of the embryo (red arrows). At the same time, the earlier flow toward the tip of the incipient cotyledons and down through the center of the globular embryo continues (black arrows). The reversal of auxin flow leads to auxin maxima at the incipient cotyledon tips and to cotyledon primordia development, as manifested in a heart-shaped embryo. During the heart stage (B), a second reversal of auxin flow from the cotyledon primordia toward the SAM region is proposed to occur (blue arrows). However, the effects of auxin maxima are repressed by genes expressed in the central domain, where the SAM will develop, such as Class III HD-Zip genes, with only the peripheral domain being responsive to auxin maxima.
(C) and (D) In phb phv rev embryos during the transition stage (C), auxin flows from the basal part of the embryo upward in the epidermal cells, toward the apical tip, and down through the center of the embryo (black arrows). The reversal of auxin flow from the SAM region to the incipient cotyledons does not occur, because of ectopic KANADI expression in that region; thus, only a single auxin maxima is formed at the apex of the embryo during the transition stage, leading to an embryo bearing a single cotyledon primordium (D).
(E) and (F) In kan1 kan2 kan4 embryos during the transition stage (E), auxin flows upward to the apical part of the embryo (black arrows). Apical reversal of the flow from the SAM region to the incipient cotyledons (red arrows) and back from the cotyledons to the SAM (blue arrows) takes place at the transition stage, similar to that in wild-type embryos. However, at the heart stage (F), an abnormal reversal of auxin flow occurs in the abaxial region of the cotyledon and hypocotyl cells (red arrows), resulting in bidirectional auxin flow. Auxin accumulation in cells located at the abaxial side of the cotyledons and hypocotyl promotes outgrowths and leaf primordia initiation, respectively.
(G) and (H) In kan1 kan2 kan4 phb phv rev embryos, apical reversal of auxin flow at the transition stage (G) occurs normally (red arrows), as a result of KANADI loss of function in this background, leading to the establishment of a bilateral symmetry. At the heart stage (H), the second reversal of auxin flow back toward the meristem once the incipient cotyledons are established also occurs (blue arrows), similar to that in the wild type. This reversal of auxin creates an auxin maximum in the position normally occupied by the SAM in the wild type. However, because PHB, PHV, and REV are needed for SAM establishment, the ectopic auxin maximum induces an ectopic central leaf instead of a SAM. Loss of KANADI activity in this background leads to the reversal of flow on the abaxial side of the cotyledons and the hypocotyl (red arrows), resulting in auxin maxima and the formation of outgrowths and leaf primordia in the cotyledons and hypocotyl, respectively.
The flow of auxin in mutant embryos as extrapolated from PIN1 expression patterns parallels the kan1 kan2 kan4 and phb phv rev embryonic phenotypes (Figures 7C to 7F). In kan1 kan2 kan4 embryos, cells normally destined to become the hypocotyl exhibit ectopic PIN1 expression, which precedes the early changes in cell divisions, suggesting that KANADI activity is required to exclude PIN1 expression from specific cells in the hypocotyl during the transition and heart stages of embryogenesis (Figures 5D, 5F, 5G, 7E, and 7F). Subsequently, a reversal in PIN1 polarity occurs below cotyledon primordia, resulting in bidirectional auxin flow and auxin maxima in the hypocotyl, promoting leaf primordia initiation (Figures 5J, 5K, and 7F). In phb phv rev embryos, auxin flow, as inferred from PIN1 expression, is a continuation of that observed in late globular embryos, with embryos failing to initiate the reversal in auxin flow outward from a region normally destined to become the SAM to the incipient cotyledon primordia (Figures 5N to 5P, 7C, and 7D). Thus, only a single auxin maximum is formed at the apex of the embryo, leading to a single cotyledon primordium. That the primary root of phb phv rev seedlings grows normally suggests that a constant supply of auxin from the apical portion of the plant is not required (Hawker and Bowman, 2004).
The phenotype of kan1 kan2 kan4 phv phb rev embryos indicates that the loss of bilateral symmetry in phb phv rev embryos is attributable to ectopic KANADI activity in the central region of the embryo and that PHB, PHV, and REV are not absolutely required for the establishment of bilateral symmetry. We infer the reversal in PIN1 at the apex of transition stage embryos to occur normally in the hextuple mutant, establishing the bilateral symmetry of heart stage embryos (Figure 7G). The development of the central leaf-like organ could be explained if a subsequent reversal of auxin flow back toward the meristem once the incipient cotyledons are established occurs in the embryo (Figure 7H), as was reported to take place in Arabidopsis inflorescence meristems, in which, once a flower primordium is established, the orientation of PIN1 reverses such that auxin is transported away from the newly initiated flower primordium back toward the meristem (Heisler et al., 2005). If this reversal is a general process not limited to the establishment of flower meristems, one might expect to see a reversal of PIN1 from leaf/cotyledon primordia toward the SAM (Figure 7B). Because PIN1 is positively regulated by auxin (Schrader et al., 2003; Vieten et al., 2005; Scarpella et al., 2006), such a reversal could account for the high expression levels of PIN1 in the L1 layer of the meristem in late heart and torpedo stage embryos (Figures 5H and 5L). This scenario also implies that the effects of auxin maxima differ between the central and peripheral regions, with only the peripheral domain being responsive to auxin maxima with respect to leaf primordium initiation, consistent with experiments in which leaf primordia are only initiated from the peripheral region when exogenous auxin is applied to shoot apices (Reinhardt et al., 2000; de Reuille et al., 2006; Smith et al., 2006). The flow of auxin back toward the center of the embryo in the hextuple kan1 kan2 kan4 phv phb rev mutant would create an auxin maximum in the position normally occupied by the SAM in the wild type, but because PHB, PHV, and REV are required for normal SAM development (Emery et al., 2003; Prigge et al., 2005), the ectopic auxin maximum induces an ectopic leaf-like organ in the hextuple mutant background (Figure 7H). Finally, because of the loss of KAN1, KAN2, and KAN4 activity in the hextuple background, ectopic expression of PIN1 in hypocotyl cells leads to ectopic auxin maxima and the formation of leaf primordia on the hypocotyl (Figure 7H).
Ectopic expression of PIN1 is observed in kan1 kan2 kan4 embryos before abnormal periclinal cell divisions, suggesting that KANADI genes could act to restrict auxin flow during embryogenesis by regulating PIN1 gene expression. Recent work demonstrated that the regulation of auxin flux and distribution consists of a complex feedback mechanism, with auxin controlling the activity of components directing their own transport at different regulatory levels. Auxin was shown to control PIN gene expression as well as PIN cellular polarity via the TIR1-AUX/IAA-ARF pathway (Schrader et al., 2003; Vieten et al., 2005; Sauer et al., 2006). Auxin was also shown to modulate PIN2 protein stability in roots and to control the rate of PIN recycling between endosomal compartments and the plasma membrane (Sieberer et al., 2000; Paciorek et al., 2005; Abas et al., 2006). Altering this complex feedback mechanism may indirectly lead to many of the phenotypic effects seen in KANADI mutants. In this scenario, the ectopic PIN1 expression and auxin maxima in the hypocotyls of kan1 kan2 kan4 embryos could be an indirect effect of the failure to properly regulate PIN1 expression during the globular to transition stage of embryogenesis. A reduction in PIN1 transcript level after the induction of KAN2, as observed in microarray experiments, supports this hypothesis (I. Pekker, Y. Eshed, and J.L. Bowman, unpublished data). By contrast, the loss of the establishment of bilateral symmetry as a result of ectopic KANADI expression in phb phv rev embryos indicates that KANADI activity can influence dynamic changes in auxin flux, either by altering the complex feedback of auxin-regulated processes described above or more directly. That ectopic PIN1 expression is observed around the entire circumference of kan1 kan2 kan4 embryos suggests that KANADI gene activity is required to repress PIN1 throughout the entire periphery of the embryo.
Other phenotypes of KANADI loss-of-function mutants are consistent with KANADI activity modulating auxin biology. For example, the phenotype of kan1 kan2 leaves resembles the phenotype of ett arf4 leaves (Pekker et al., 2005). ETT and ARF4 are members of the auxin response factor gene family and are proposed to act to repress the transcription of target genes in response to auxin (Tiwari et al., 2003). Mutations in ETT enhance the phenotypic effects of kan1 mutations and also suppress the effects of ectopic KANADI activity, leading Pekker et al. (2005) to speculate that KANADI and ARF proteins act together to regulate transcription. The ectopic abaxial leaf outgrowths of kan1 kan2 plants are similar in some respects to leaf primordia, and expression of PIN1 is higher in the outgrowths than in the surrounding leaf tissue, suggesting that the outgrowths may also be the result of ectopic auxin maxima forming in the lamina (Eshed et al., 2001, 2004; A. Izhaki and J.L. Bowman, unpublished data). Finally, loss- and gain-of-function KANADI mutants have altered vascular differentiation. Ectopic expression of KANADI results in a complete loss of vascular tissues, and loss of KANADI activity results in ectopic xylem differentiation (Eshed et al., 2001; Kerstetter et al., 2001; Emery et al., 2003). Both of these phenotypes are consistent with KANADI activity restricting auxin flow. Although the mechanistic details by which the KANADI genes control PIN proteins are unknown, because the KANADI genes likely encode transcription factors and may act in a pathway with two ARF transcription factors (ETT and ARF4), spatially regulated KANADI proteins together with ARF proteins activated by auxin could represent a feedback mechanism to regulate (reduce) the expression of PIN genes and hence the flow of auxin. A similar complex regulatory interaction between auxin and the Class I KNOX transcription factor BREVIPEDICELLUS (BP) was shown to occur during leaf development. Although PIN1-dependent auxin flow is required to downregulate BP during leaf primordial initiation, BP can also induce alterations in PIN expression during leaf margin development (Hay et al., 2006).
The KANADI and Class III HD-Zip genetic system regulates the central–peripheral differentiation of tissues in leaves and stems. By removing the activity of all KANADI genes and most Class III HD-Zip genes, we show that these genes are not absolutely required for the establishment of either bilateral symmetry or a central–peripheral axis. Rather, based on the patterns of PIN1 expression demonstrated and inferred in multiple mutant genotypes, we propose that KANADI genes pattern tissues within the plant by regulating auxin flow, thereby modulating where and when reversals in PIN polarity occur. Class III HD-Zip genes may modulate the response of cells to auxin maxima, promoting meristem fate and preventing leaf initiation in response to auxin in the central part of the embryo. Elucidation of cause and effect with respect to auxin and transcription factors controlling embryonic patterning will require the analysis of precise temporal and spatial patterns of gene expression relative to changes in auxin flux (Heisler et al., 2005) and the acknowledgment that both auxin and transcription factors may be both upstream and downstream in patterning events attributable to complex feedback mechanisms.
METHODS
Plant Growth and Genetics
All Arabidopsis thaliana mutants are in the Landsberg erecta background, except kan3-1 and kan4-3 (ats-3) (both in Wassilewskija), which were backcrossed into Landsberg erecta. Plants were grown under 18 h of cool-white fluorescent light at 20°C. Mutant lines have been described previously: kan1-2 (Eshed et al., 1999); kan2-1 (Eshed et al., 2001); kan3-1, phb-6, phv-5, and rev-9 (Emery et al., 2003); kan4-3 (Mcabee et al., 2006); and fil-8 and yab3-2 (Kumaran et al., 2002). yab-5 has a stop codon in the YABBY domain (Till et al., 2003).
To generate the kan1-2 kan2-1 kan3-1 kan4-3 quadruple mutant, F1 plants derived from the crosses kan1-2 kan2-1/+ × kan3-1 and kan1-2 kan2-1/+ × kan4-3 were self-fertilized. F2 families segregating (at 1:4) kan1-2 kan2-1 double mutants were genotyped for kan3-1 or kan4-3 alleles using the following primers: for kan3-1, forward (5′-GGATGTGCAAGATCTCACATTGGCTCATG-3′) and reverse (5′-CATTTTATAATAACGCTGCGGACATCTAC-3′); for kan4-3, forward (5′-CGTTCTCTTTCCACACATTCTTTGATCTCC-3′) and reverse (5′-CGCACCGATCGCCCTTCCCAACAGTTGCGC-3′). Positive F2 kan1-2 kan2-1/+ plants were self-fertilized, and F3 families segregating the triple mutants kan1-2 kan2-1 kan3-1 or kan1-2 kan2-1 kan4-3 (at 1:3) were identified as kan1-2 kan2-1/+ kan3-1 and kan1-2 kan2-1/+ kan4-3, respectively. F1 plants of the cross kan1-2 kan2-1/+ kan3-1 × kan1-2 kan2-1/+ kan4-3 were self-fertilized, and kan1-2 kan2-1/+ F2 plants from families segregating the kan1-2 kan2-1 phenotype at 1:3 were self-fertilized. F3 families segregating (at 1:4) the quadruple kan1-2 kan2-1 kan3-1 kan4-3 mutants were analyzed further.
Generation of the kan1-2 kan2-1 kan4-3 phb-6 phv-5 rev-9 hextuple mutant required putting the kan2-1 and phv-5 alleles in a cis configuration. Because KAN2 and PHV are 830 kb apart on chromosome 1 and using an estimate of a genome-wide average of 200 kb:1 centimorgan, we estimated that the two genes are ∼4 centimorgans apart. Thirty-six F2 kan1-2 kan2-1 plants from the cross kan1-2 kan2-1/+ × phb-6 phv-5 rev-9/+ were genotyped for phb-6, phv-5, and rev-9 using the following primers: for phb-6, forward (5′-CTCTTCGCTTCTCCTTCCTCTCC-3′) and reverse (5′-GGTTCCCGTCCGATTTCGACT-3′); for phv-5, forward (5′-GTTCCTTGTCCTTTCTCTCTCAG-3′) and reverse (5′-ACGGTCGGGAAACTAGCTCTAC-3′); for rev-9, forward (5′-CCTCTGTTCCAAAGTTCAGCAGAAGC-3′) and reverse (3′-GCCGCCGACTTCGGTTTGCGGTCGCG-3′). A single kan1-2 kan2-1 plant was identified that also harbored a phv-5 allele. This plant was also heterozygous for phb-6 and rev-9. Pollen from this plant was crossed with kan1-2 kan2-1/+ kan4-3. F1 kan1-2 kan2-1/+ plants positive for phv-5, phb-6, and rev-9 were identified and self-fertilized. F3 families segregating the hextuple kan1-2 kan2-1 kan4-3 phb-5 phv-6 rev-9 mutant at a ratio of 1:15 (kan1-2 kan2-1/+ kan4-3 phb-6 phv-5/+ rev-9/+; with the kan2-1 and phv-5 alleles in the coupling) were analyzed further.
To generate the kan1-2 kan2-1 kan4-3 fil-8 yab3-2 yab5-1 hextuple mutant, kan1-2 kan2-1/+ plants from F2 families from the cross kan1-2 kan2-1/+ kan4-3 × fil-8/+ yab3-2 yab5-1, in which the phenotypes conferred by kan1-2 kan2-1 kan4-3 and fil-8 yab3-2 yab5-1 were segregating at 1:3, were self-fertilized. F3 families segregating the hextuple kan1-2 kan2-1/+ kan4-3 fil-8/+ yab3-2 yab5-1 mutant at 1:15 were analyzed further.
Microtechniques and Microscopy
Scanning electron microscopy, GUS staining, and in situ hybridization were performed according to the methods described by Eshed et al. (1999) and Emery et al. (2003). FIL and STM probes were generated by linearizing cDNA plasmids and synthesizing digoxigenin-labeled antisense RNA using T7 and T3 RNA polymerase, respectively. Histological analyses were performed as described by Emery et al. (2003). For whole-mount analyses of embryos, developing seeds were excised from developing gynoecia with 27-gauge needles, cleared overnight in Hoyer's solution (Liu and Meinke 1998), and observed on a Zeiss Axioplan Imaging 2 microscope under differential interference contrast optics. Images were captured on an Axiocam HRCCCD camera (Zeiss) using the Axiovision program (version 3.1). PIN1:GFP and DR5:GFP have been described previously (Benkova et al., 2003; Friml et al., 2003; Heisler et al., 2005). For GFP analyses, embryos were removed from the developing seed coat with 27-gauge needles, mounted on slides with 50% glycerol, and observed on a confocal laser scanning microscope (Olympus FluoView 1000) with an argon laser at 448 nm for excitation and at 505 to 525 nm for GFP emission, or on a Nikon Eclipse E600 microscope using Chroma GFP and 4′,6-diamidino-2-phenylindole filter sets. Images were acquired using an Orca-100 CCD camera (Hamamatsu Photonics) driven by the ImagePro Plus 4.0 software package (Media Cybernetics).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: KANADI1, At5g16560; KANADI2, At1g32240; KANADI3, At4g17695; KANADI4/ATS, At5g42630; PHABULOSA, At2g34710; PHAVOLUTA, At1g30490; REVOLUTA, At5g60690; FILAMENTOUS FLOWER, At2g45190; YABBY3, At4g00180; and YABBY5, At2g26580.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. KANADI Mutants and Vasculature Anatomy.
Supplemental Figure 2. STM and CLV3 mRNA Expression in kan1 kan2 kan4 Embryos.
Supplementary Material
Acknowledgments
We thank Marcus Heisler, Yuval Eshed, Sandra Floyd, Pia Steiger, and members of the Bowman laboratory for insightful discussions and Bo Liu for help with microscopy. This work was supported by National Science Foundation Grant IOB 0332556 (J.L.B.), by Research Grant Award 3637-05 from the U.S.–Israel Binational Agricultural Research and Development Fund (J.L.B.), and by Postdoctoral Award FI-314-2001 from the U.S.–Israel Binational Agricultural Research and Development Fund (A.I.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: John L. Bowman (jlbowman@ucdavis.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abas, L., Benjamins, R., Malenica, N., Paciorek, T., Wirniewska, J., Moulinier-Anzola, J.C., Sieberer, T., Friml, J., and Luschnig, C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8 249–256. [DOI] [PubMed] [Google Scholar]
- Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121 4171–4182. [DOI] [PubMed] [Google Scholar]
- Barton, M.K., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119 823–831. [Google Scholar]
- Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Berger, F., Linstead, P., Dolan, L., and Haseloff, J. (1998). Stomata patterning on the hypocotyl of Arabidopsis thaliana is controlled by genes involved in the control of root epidermis patterning. Dev. Biol. 194 226–234. [DOI] [PubMed] [Google Scholar]
- Berleth, T., and Jurgens, G. (1993). The role of the Monopteros gene in organizing the basal body region of the Arabidopsis embryo. Development 118 575–587. [Google Scholar]
- Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., Heidstra, R., Aida, M., Palme, K., and Scheres, B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44. [DOI] [PubMed] [Google Scholar]
- Busse, J.S., and Evert, R.F. (1999). Vascular differentiation and transition in the seedling of Arabidopsis thaliana (Brassicaceae). Int. J. Plant Sci. 160 241–251. [Google Scholar]
- Cheng, Y., Dai, X., and Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Reuille, P.B., Bohn-Courseau, I., Ljung, K., Morin, H., Carraro, N., Godin, C., and Traas, J. (2006). Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class IIIHD-ZIP and KANADI genes. Curr. Biol. 13 1768–1774. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11 1251–1260. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Izhaki, A., Baum, S.F., Floyd, S.K., and Bowman, J.L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131 2997–3006. [DOI] [PubMed] [Google Scholar]
- Fletcher, L.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 1911–1914. [DOI] [PubMed] [Google Scholar]
- Floyd, S.K., and Bowman, J.L. (2006). Distinct developmental mechanisms reflect the independent origins of leaves in vascular plants. Curr. Biol. 16 1911–1917. [DOI] [PubMed] [Google Scholar]
- Floyd, S.K., and Bowman, J.L. (2007). The ancestral patterning toolkit of land plants. Int. J. Plant Sci. 168 1–35.
- Floyd, S.K., Zalewski, C.S., and Bowman, J.L. (2006). Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics 173 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J., Benkova, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., Woody, S., Sandberg, G., Scheres, B., Jurgens, G., and Palme, K. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108 661–673. [DOI] [PubMed] [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jurgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153. [DOI] [PubMed] [Google Scholar]
- Friml, J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jurgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230. [DOI] [PubMed] [Google Scholar]
- Golz, J.F., Roccaro, M., Kuzoff, R., and Hudson, A. (2004). GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development 131 3661–3670. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., Ckurshumova, W., Vidaurre, D.P., Singh, S.A., Stamatiou, G., Tiwari, S.B., Hagen, G., Guilfoyle, T.J., and Berleth, T. (2004). Overlapping and non-redundant functions of the Arabidopsis auxin response factor MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131 1089–1100. [DOI] [PubMed] [Google Scholar]
- Hawker, N.P., and Bowman, J.L. (2004). Roles for class IIIHD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 135 2261–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A., Barkoulas, M., and Tsiantis, M. (2004). PINning down the connections: Transcription factors and hormones in leaf morphogenesis. Curr. Opin. Plant Biol. 7 575–581. [DOI] [PubMed] [Google Scholar]
- Hay, A., Barkoulas, M., and Tsiantis, M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133 3955–3961. [DOI] [PubMed] [Google Scholar]
- Heisler, M.G., Ohno, C., Das, P., Sieber, P., Reddy, G.V., Long, J.A., and Meyerowitz, E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hung, C.Y., Lin, Y., Zhang, M., Pollock, S., Marks, M.D., and Schiefelbein, J. (1998). A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol. 117 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik, P.D., and Barton, M.K. (2005). Surge and destroy: the role of auxin in plant embryogenesis. Development 132 3577–3585. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, R.A., Bomblies, K., and Poethig, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411 706–709. [DOI] [PubMed] [Google Scholar]
- Kumaran, M.K., Bowman, J.L., and Sundaresan, V. (2002). YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14 2761–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.M., and Meinke, D.W. (1998). The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 16 21–31. [DOI] [PubMed] [Google Scholar]
- Long, J.A., and Barton, M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125 3027–3035. [DOI] [PubMed] [Google Scholar]
- Mcabee, J.M., Hill, T.A., Skinner, D.J., Izhaki, A., Hauser, B.A., Meister, R.J., Reddy, G.V., Meyerowitz, E.M., Bowman, J.L., and Gasser, C.S. (2006). ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 46 522–531. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 709–713. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito, K., Kubo, M., Demura, T., and Fukuda, H. (2005). Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol. 46 1646–1656. [DOI] [PubMed] [Google Scholar]
- Otsuga, D., DeGuzman, B., Prigge, M.J., Drews, G.N., and Clark, S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25 223–236. [DOI] [PubMed] [Google Scholar]
- Paciorek, T., Zazimalova, E., Ruthardt, N., Petrasek, J., Stierhof, Y.-D., Kleine-Vehn, J., Morris, D.A., Emans, N., Jurgens, G., Geldner, N., and Friml, J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435 1251–1256. [DOI] [PubMed] [Google Scholar]
- Pekker, I., Alvarez, J.P., and Eshed, Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17 2899–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge, M.J., Otsuga, D., Alonso, J.M., Ecker, J.R., Drews, G.N., and Clark, S.E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Mandel, T., and Kuhlemeier, C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Sauer, M., Balla, J., Luschnig, C., Wisniewska, J., Reinohl, V., Friml, J., and Benkova, E. (2006). Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 20 2902–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, E.H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella, E., Marcos, D., Friml, J., and Berleth, T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader, J., Baba, K., May, S.T., Palme, K., Bennett, M., Bhalerao, R.P., and Sandberg, G. (2003). Polar auxin transport in the wood-forming tissues of hybrid aspen is under simultaneous control of developmental and environmental signals. Proc. Natl. Acad. Sci. USA 100 10096–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer, T., Seifert, G.J., Hauser, M.-T., Grisafi, P., Fink, G.R., and Luschnig, C. (2000). Post-transcriptional control of the Arabidopsis auxin efflux carrier EIR1 requires AXR1. Curr. Biol. 10 1595–1598. [DOI] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128. [DOI] [PubMed] [Google Scholar]
- Smith, R.S., Guyomarc'h, S., Mandel, T., Reinhardt, D., Kuhlemeier, C., and Prusinkiewicz, P. (2006). A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. USA 103 1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Galweiler, L., Palme, K., and Jurgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318. [DOI] [PubMed] [Google Scholar]
- Till, B.J., et al. (2003). Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 13 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten, A., Vanneste, S., Wisniewska, J., Benkova, E., Benjamins, R., Beeckman, T., Luschnig, C., and Friml, J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132 4521–4531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.