Abstract
Cys synthesis in plants constitutes the entry of reduced sulfur from assimilatory sulfate reduction into metabolism. The catalyzing enzymes serine acetyltransferase (SAT) and O-acetylserine (OAS) thiol lyase (OAS-TL) reversibly form the heterooligomeric Cys synthase complex (CSC). Dominant-negative mutation of the CSC showed the crucial function for the regulation of Cys biosynthesis in vivo. An Arabidopsis thaliana SAT was overexpressed in the cytosol of transgenic tobacco (Nicotiana tabacum) plants in either enzymatically active or inactive forms that were both shown to interact efficiently with endogenous tobacco OAS-TL proteins. Active SAT expression resulted in a 40-fold increase in SAT activity and strong increases in the reaction intermediate OAS as well as Cys, glutathione, Met, and total sulfur contents. However, inactive SAT expression produced much greater enhancing effects, including 30-fold increased Cys levels, attributable, apparently, to the competition of inactive transgenic SAT with endogenous tobacco SAT for binding to OAS-TL. Expression levels of tobacco SAT and OAS-TL remained unaffected. Flux control coefficients suggested that the accumulation of OAS and Cys in both types of transgenic plants was accomplished by different mechanisms. These data provide evidence that the CSC and its subcellular compartmentation play a crucial role in the control of Cys biosynthesis, a unique function for a plant metabolic protein complex.
INTRODUCTION
Cys synthesis in plants represents the decisive process of integration of reduced sulfur from assimilatory sulfate reduction into the organic form of an amino acid. After uptake of sulfate by specific and transcriptionally tightly regulated sulfate/H+ cotransporters at the plasmalemma, reductive assimilation of sulfate takes place in most kinds of plant cells. It is located exclusively in plastids and predominates in chloroplasts from green tissues, where supplies of redox equivalents are plentiful (Hawkesford and Wray, 2000; Leustek et al., 2000). By contrast, Cys synthesis occurs in plastids, the cytosol, and mitochondria of most cells and in most plant species. It is performed by a protein system of compartment-specific isoforms with reversible quaternary structure (for reviews, see Droux, 2004; Wirtz et al., 2004; Johnson et al., 2005). Serine acetyltransferase (SAT) is assumed to consist of a dimer of homotrimers that only forms O-acetylserine (OAS) from Ser and acetyl-CoA with highest efficiency if it is associated with two homodimers of O-acetylserine (thiol) lyase (OAS-TL) in the Cys synthase complex (CSC). Monomer sizes of SAT in different species range between 29 and 34 kD, whereas OAS-TL monomer size is between 68 and 75 kD, giving rise to CSC quaternary sizes of ∼320 kD (Wirtz and Hell, 2006). Purified SAT has been shown to be catalytically active, but in vitro it requires a 400-fold excess of OAS-TL activity to achieve maximal Cys formation (Ruffet et al., 1994). Similar ratios have been observed in whole leaf and chloroplast protein extracts (for review, see Wirtz and Hell, 2006).
The CSC was first discovered by the groundbreaking work of Kredich et al. (1969) in Salmonella typhimurium. SAT and OAS-TL proteins are closely related between bacteria and plants, in terms of both primary sequence and three-dimensional structure, indicating a common evolutionary ancestry (Wirtz and Hell, 2006). Biochemical analyses of bacterial and plant CSC revealed several shared properties, but the function was generally assumed to consist of substrate channelling. Regulation of Cys synthesis in bacteria was attributed to the Cys regulon that is governed by a typical bacterial repressor/operator mechanism that controls effector-driven DNA binding of the CysB transcription factor (Kredich, 1996). Multienzyme complexes that are able to catalyze sequential reactions allow the transfer of metabolites within specific spatial reaction channels. Such microenvironments or subvolumes of the cellular space may allow the thermodynamic or kinetic character of a metabolic process to be different from the bulk cellular space. Reduced interference with other reactions as a result of diffusion equilibrium, enhanced specificity, and reaction velocity can be achieved (Srere, 1987; Ovadi, 1991; Winkel, 2004). Although Cook and Wedding (1976) demonstrated that at least part of the OAS diffused out of the S. typhimurium CSC, the channelling function was still discussed, because basically all known catalytically active multiprotein complexes in metabolism follow various channelling mechanisms (Srere, 1987; Ovadi, 1991; Winkel, 2004).
Kinetic analyses of the plant CSC revealed that the bound OAS-TL dimers are catalytically almost inactive in the complex, whereas SAT requires the presence of a large excess of OAS-TL to gain full activity (Droux et al., 1998; Wirtz et al., 2004). Therefore, it is conceivable that the intermediate OAS is released from the complex and serves, together with sulfide from assimilatory sulfate reduction in plastids, as a substrate for free and highly active OAS-TL dimers to synthesize Cys. SAT activity limits the rate of Cys formation and is feedback-sensitive to Cys to varying degrees, depending on subcellular isoforms and plant species (Droux, 2004; Saito, 2004). In vitro sulfide stabilizes the CSC, thus promoting SAT activity and Cys formation, whereas OAS destabilizes the CSC (Kredich et al., 1969). Under sulfide limitation in vivo, OAS accumulates first, which results in CSC dissociation, followed by the inactivation of SAT. This antagonism of metabolite effects and enzyme activity changes, together with other observations, gave rise to the model of a regulatory circuit based on reversible protein–protein interaction that functions in metabolic sensing of sulfide, flux control of Cys synthesis, and, potentially, the activation of expression of genes encoding proteins for sulfate uptake and reduction (reviewed in Hell and Hillebrand, 2001; Hell, 2003; Droux, 2004; Wirtz and Hell, 2006). In brief, situations of limited sulfate supply will result in decreased levels of sulfide availability in a cell. OAS concentrations, which are very low under regular sulfate supply, will now begin to increase, because sulfide for Cys formation is lacking. The combination of low sulfide and high OAS levels will dissociate the complex, resulting in the inactivation of SAT. Thus, SAT enzymatic activity and binding to OAS-TL contribute to the control of the rate of Cys synthesis. During sulfate limitation, the transcription of genes encoding sulfate transporters and enzymes of sulfate reduction is enhanced, providing the basis for the readjustment of sulfur homeostasis in the cell. This effect suggests that (1) suppression triggered by sulfide or other sulfur-containing compounds is released, (2) transcriptional activation is promoted via OAS, or (3) the equilibrium of free and bound CSC subunits is sensed and able to affect gene expression. By contrast, no such regulatory function has been attributed to the bacterial CSC, suggesting that the properties of the plant CSC may be unique.
The hypothesis of a regulatory circuit with the plant CSC in the center is based on evidence from numerous in vitro and physiological experiments. However, direct evidence for such a function in vivo was missing. To this end, we generated transgenic tobacco (Nicotiana tabacum) plants expressing either enzymatically active (i.e., fully active when complexed by OAS-TL in the CSC) or mutagenized inactive SAT from Arabidopsis thaliana in the cytosol. The chosen isoform, SAT3 (At3g13110), shows very little feedback sensitivity toward Cys and presumably is not inhibited by cellular Cys concentrations (Wirtz and Hell, 2003), allowing a direct comparison of both approaches without interference of allosteric regulation. The active and inactive versions of SAT3 were shown to be able to form a heterologous CSC with endogenous tobacco OAS-TL. The aim of our study was to prove the regulatory function of the CSC in planta by modification of the equilibrium of its subunits. Overexpression of active SAT in the cytosol of tobacco yielded the expected increase of Cys, as well as glutathione, which acts as a transient storage form of Cys. However, dominant-negative expression in tobacco of the mutagenized SAT without catalytic activity but still capable of binding to OAS-TL resulted in the accumulation of Cys, glutathione, and total sulfur, which greatly exceeded that observed for active SAT overexpression. Thus, the Cys synthesis system of CSC and free subunits was deregulated and caused dramatic overproduction of the end product. We conclude that the formation of CSC is an essential component of Cys synthesis and that protein–protein interaction of its subunits has a regulatory function in plants.
RESULTS
Production of Transgenic Tobacco Plants Expressing Active and Inactive SAT
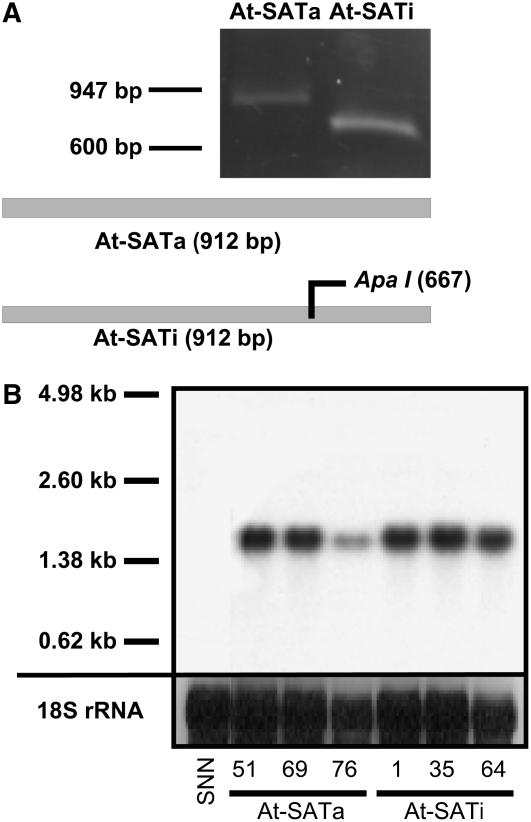
Tobacco plants of cv Samsun NN were transformed by Agrobacterium tumefaciens–mediated gene transfer with binary constructs expressing either enzymatically active SAT (At-SATa) or inactivated SAT (At-SATi) from Arabidopsis under the control of the cauliflower mosaic virus 35S promoter. The DNA fragment encoding the mitochondrial signal peptide of isoform SAT3 (U22964; gene number At3g13110) (Hell et al., 2002) was removed as described by Bogdanova et al. (1995) to achieve cytosolic localization upon expression in plant cells. The SAT3 enzyme was inactivated to yield At-SATi by site-directed mutagenesis of His-309 to Ala. The mutation prevents the transfer of the acetyl moiety of CoA to Ser in the catalytic center, but not the interaction with OAS-TL (Wirtz et al., 2001; Wirtz and Hell, 2006) (see Supplemental Figure 1A online). The enzymatic inactivation of SATi was verified by recombinant expression and purification and showed <1/1000th of SATa activity under standard assay conditions (see Supplemental Figure 1B online). At-SATa and At-SATi had been shown previously to interact with cytosolic Arabidopsis OAS-TL in vitro and in the yeast two-hybrid system (Wirtz et al., 2001). Fifteen of 80 primary tobacco transformants of each construct were further propagated after genomic PCR screening. Restriction of an analytical ApaI site that had been introduced along with the H309A mutagenesis allowed us to distinguish between At-SATa and At-SATi transgenics (Figure 1A). After genomic DNA gel blot analysis of the T3 generation to determine numbers of T-DNA insertion loci, the following three lines of each construct were selected for all further analyses: SATa51, SATa69, and SATa76 (all with one insertion) and SATi1 (one insertion), SATi35 (three insertions), and SATi64 (two insertions). These insertion numbers were supported by segregation ratios after selection in the presence of kanamycin (data not shown). Expression intensities of these six lines were determined by RNA gel blot hybridization using RNA from tobacco leaves (Figure 1B). The mRNA expression levels of At-SATa and At-SATi mRNAs were similar except for the lower levels of SATa76. Expression was also tested in roots and generally found to be weaker compared with leaves (see Supplemental Figure 2 online). Steady state mRNA contents matched the abundance of the encoded SAT proteins (Figure 2A).
Figure 1.
Molecular Characterization of Tobacco Lines Transformed with Constructs Harboring Active (At-SATa) and Inactive (At-SATi) At-SAT3 from Arabidopsis.
(A) PCR with At-SAT3–specific primers and genomic DNA from tobacco as template allowed the identification of transgenic lines. Products encoding active At-SATa and inactive At-SATi were distinguished by restriction of PCR products at the ApaI site that had been introduced together with active site mutagenesis.
(B) RNA gel blot hybridization of RNA from tobacco leaf with an At-SATa probe. Ethidium bromide–stained 18S rRNA served as a loading control. SNN, wild-type tobacco.
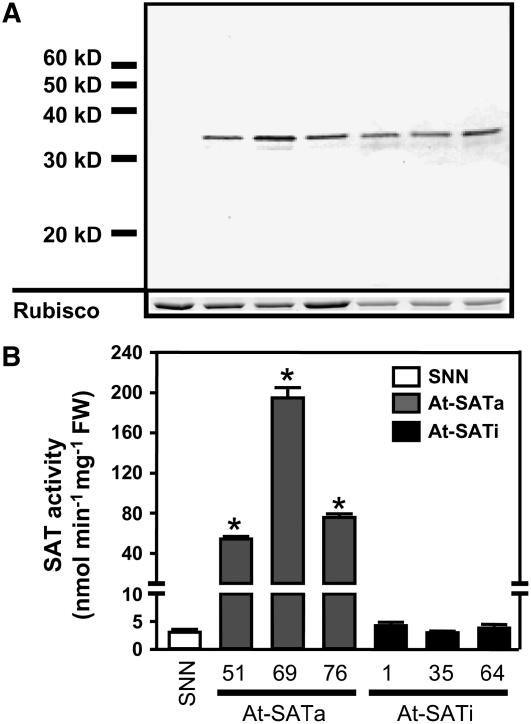
Figure 2.
SAT Activity and Abundance of At-SATa and At-SATi Protein in Transgenic Tobacco.
(A) Immunoblot of 7.5 μg of total leaf protein of each transgenic line decorated with polyclonal serum against Arabidopsis SAT3 protein. The Coomassie blue–stained band of Rubisco protein run in a parallel SDS-PAGE loaded with the same extracts served as a loading control.
(B) Total SAT activity was determined from protein extracts of leaves of wild-type tobacco (SNN; white bars) and three independent transgenic lines expressing At-SATa (gray bars) or At-SATi (black bars). For each transformed line, mean values and sd of enzyme assays from three individual plants with three measurements each are shown (n = 9). Error bars indicate sd, and asterisks mark significant differences determined using the unpaired t test.
Active and Inactive Transgenic SAT Were Expressed and Functional in Tobacco
Recombinant At-SATa and At-SATi were detected in leaf protein extracts using an anti-At-SAT3 polyclonal antiserum. No cross-reactivity with endogenous tobacco SATs was observed under these conditions (Figure 2A). The apparent molecular mass was determined as 32.8 kD for active and inactive At-SAT. A smaller band that occurred in most of the samples was identified as an N-terminal degradation product (data not shown). When staining intensities of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) protein in the same samples were considered, the abundance of At-SATa and At-SATi in the different lines was comparable. Alterations of <1.5-fold were observed between the three lines of each construct. Density gradient centrifugation to separate cytosol and organelles revealed no At-SAT signal in organelle fractions, indicating that At-SATa and At-SATi were located in the cytosol as expected (data not shown).
Determination of total SAT activity in protein extracts from wild-type tobacco leaves revealed a specific activity of 2.79 ± 0.45 nmol OAS·min−1·mg−1 protein. SAT activity in lines with overexpression of active At-SATa was enhanced by 40-fold on average (Figure 2B). In lines with overexpressed inactive At-SATi, no significant increase of total SAT activity was detected. Thus, both SAT proteins were overexpressed to substantial levels in transgenic tobacco and exhibited the anticipated catalytic properties. The phenotypes of both types of transgenic lines were similar compared with those of wild-type plants grown under the same light conditions (200 μE·m−2·s−1). Growth was slightly retarded in lines expressing At-SATi, and seed production in both types was reduced by ∼50% (see Supplemental Figure 3 online).
Expression of Active as Well as Inactive SAT Causes Strong Accumulation of Soluble Thiols
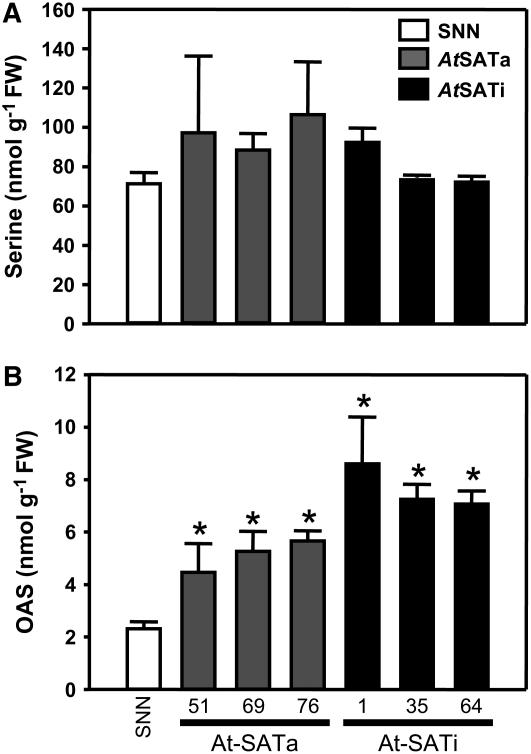
Ser provides the amino acid backbone for Cys synthesis that proceeds via the acetyl-activated intermediate OAS. Free Ser concentrations in wild-type tobacco leaves averaged 71 ± 6 nmol/g−1 fresh weight (FW) and were not affected by the enhanced requirement for Cys synthesis in the transgenic lines (Figure 3A). OAS concentrations were 2.3 ± 0.3 nmol/g−1 FW in wild-type leaves and increased significantly by 2.5-fold in At-SATa–expressing lines and even by 3.5-fold in At-SATi lines (Figure 3B). Such OAS levels could be expected to partially dissociate the CSC, but the abundant sulfate supply in the medium presumably kept the intracellular sulfide availability sufficiently high to override the OAS effect (Wirtz and Hell, 2006).
Figure 3.
Ser and OAS Contents in Leaves of Transgenic Tobacco Plants.
Three plants of wild-type tobacco (SNN; white bars) and three plants of each independent transgenic line expressing either At-SATa (gray bars) or At-SATi (black bars) were analyzed after metabolite extraction from two samples of each plant (n = 6) for Ser (A) and OAS (B). Error bars indicate sd, and asterisks mark significant differences determined using the unpaired t test.
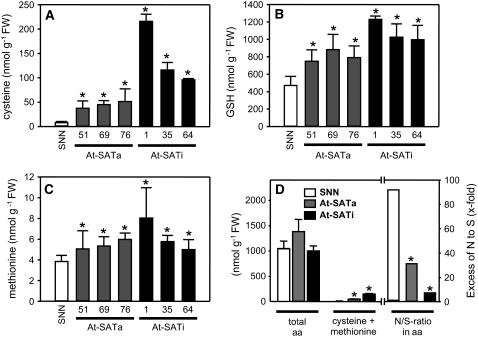
Cys concentrations in leaves of wild-type tobacco were 8.7 ± 1.7 nmol/g−1 FW and increased by fivefold in At-SATa lines, indicating sufficient availability of all substrates required (Figure 4A). Consequently, levels of γ-glutamylcysteine, although very low, increased by 2.4-fold (data not shown), and those of GSH and Met increased by 1.7- and 1.4-fold, respectively (Figures 4B and 4C). Thus, the strongly enhanced availability of Cys enhanced free levels of the major cellular sulfur metabolites. Surprisingly, however, the Cys contents of transgenic tobacco that expressed the inactive At-SATi increased by up to 30-fold, in agreement SATa1, and by 16-fold on average in all three lines. In addition, the levels of γ-glutamylcysteine (3.7-fold), GSH (2.3-fold), and Met (1.6-fold) increased more strongly in At-SATi lines.
Figure 4.
Cys, GSH, and Met Contents in Leaves of Transgenic Tobacco Plants.
(A) to (C) Three plants of wild-type tobacco (SNN; white bars) and three plants of each independent transgenic line expressing either At-SATa (gray bars) or At-SATi (black bars) were analyzed using two samples per plant (n = 6) for Cys (A), GSH (B), and Met (C).
(D) Total of free amino acids (aa) and sulfur-containing amino acids and the derived molar ratio of nitrogen to sulfur bound in the free amino acid fraction of wild-type (three plants) and At-SATa– or At-SATi–expressing plants (means ± sd from three plants of three transgenic lines for each construct; n = 9).
Error bars indicate sd, and asterisks mark significant differences determined using the unpaired t test.
The total amount of free amino acids without a sulfur moiety did not change significantly (Figure 4D), although the contents of several minor basic amino acids increased by 1.5- to 2-fold (see Supplemental Figure 4 online). However, the enhancement of sulfur amino acids was so strong that the ratio of nitrogen to sulfur bound in the total of all free amino acids changed dramatically. Although the nitrogen:sulfur ratio was 92:1 in wild-type tobacco, it decreased to 31:1 in At-SATa lines and to 7:1 in At-SATi lines. Therefore, the presence of the catalytically inactive At-SATi in the cytosol caused a consistent increase in Cys and thiol metabolites that exceeded by far the accumulation achieved by overexpression of similar amounts of active At-SATa in the same compartment.
SAT Overexpression Increases the Total Sulfur Content of Transgenic Plants
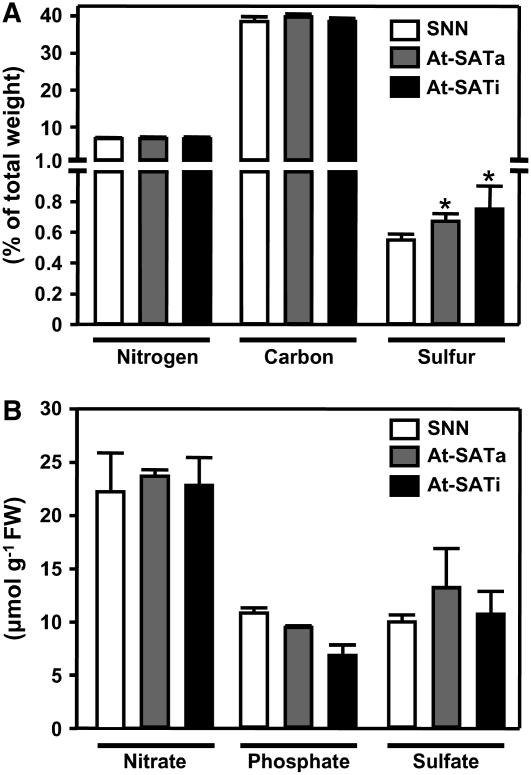
In general, most sulfur in a plant tissue is bound in proteins, followed by inorganic sulfate, sulfolipids, and the pool of free sulfur metabolites (De Kok et al., 1988). The strong increase in free Cys and downstream sulfur metabolites also could affect sulfur contents in other cellular pools. The total sulfur content of wild-type and transgenic plants was determined and compared with total carbon and nitrogen contents. Although the latter elements were apparently unaffected compared with wild-type leaves in percentage of total dry weight, the sulfur content increased by 23 and 36% in At-SATa and At-SATi lines, respectively (Figure 5A). Even stronger increases in the same direction were observed for roots (see Supplemental Figure 5 online). Determination of the major nutrient anions in leaves revealed no significant differences between the two transgenic genotypes and the wild type (Figure 5B). This was also the case for roots (see Supplemental Figure 6 online), indicating that none of the plants suffered from sulfate, phosphate, or nitrate deficiency as a consequence of the accumulation of organically bound sulfur under these conditions. Because the proportion of sulfur bound in low-molecular-mass compounds other than sulfate and sulfur amino acids including GSH is small, it is concluded that most of the increase in total sulfur in the transgenic lines can be attributed to protein-bound sulfur, most likely as Cys. Again, more sulfur accumulated in lines expressing inactive At-SATi than active At-SATa.
Figure 5.
Total Nitrogen, Carbon and Sulfur Contents, and Anion Concentrations in Leaves of Transgenic Tobacco Plants.
(A) Nitrogen, carbon, and sulfur contents.
(B) Anion concentrations.
Three plants of wild-type tobacco (SNN; white bars) and three plants of three independent transgenic lines expressing either At-SATa (gray bars) or At-SATi (black bars) were analyzed (n = 9). Error bars indicate sd, and asterisks mark significant differences determined using the unpaired t test.
At-SAT Expressed in Tobacco Interacts with Endogenous Tobacco OAS-TL
The only difference between At-SATa and At-SATi was the H309A mutation at the acetyl-CoA binding loop of the catalytic center. Binding to other SAT subunits and OAS-TL was not affected significantly, according to in vitro assays, interaction studies with the yeast two-hybrid system, and prediction from the structural At-SAT model (Wirtz et al., 2001; Pye et al., 2004). Furthermore, it is known that a general interaction capability between SAT and OAS-TL from different origins exists, because heterologous complexes are formed in vitro between plastidic SAT from Arabidopsis and plastidic OAS-TL from spinach (Spinacia oleracea) and even with OAS-TL from Escherichia coli (Droux et al., 1998). To show that wild-type and mutated SAT3 protein from Arabidopsis interacted with cytosolic OAS-TL in transgenic tobacco, the CSC consisting of these heterologous subunits was demonstrated by three independent methods.
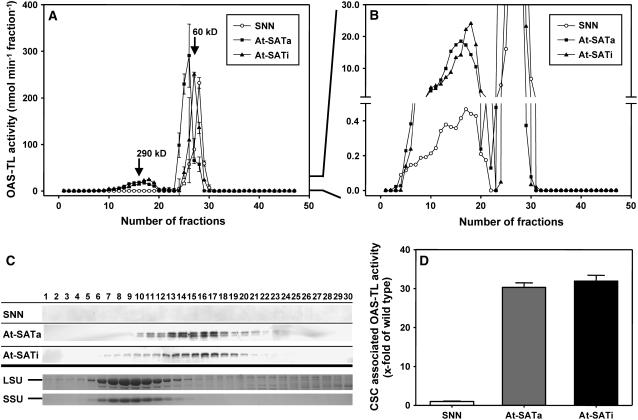
First, leaf protein extracts from wild-type, At-SATa, and At-SATi lines were separated by size-exclusion chromatography. Eluted fractions were tested for the presence of OAS-TL activity and At-SAT protein using polyclonal antiserum. SAT activity was too low to assay in the diluted fractions of wild-type and At-SATi extracts but was detectable in eluents of At-SATa plant extracts (data not shown). Nt-OAS-TL activity eluted in two peaks that corresponded to the apparent molecular masses of the CSC (290 ± 15 kD) and free dimers (60 ± 12 kD) (Figure 6A). These values corresponded well to the expected sizes of CSC and OAS-TL dimers from other plant species (Wirtz and Hell, 2006). The vast majority of Nt-OAS-TL eluted as dimers, even after preincubation of CSC fractions with OAS to dissociate the complex and regain OAS-TL activity. At-SATa and At-SATi each eluted in only a single peak that strictly correlated with fractions of ∼290 kD, where Nt-OAS-TL activity was detected (enlarged panel in Figure 6B and immunoblots in Figure 6C). No cross-reaction of anti-At-SAT antibody was observed in wild-type control extracts. The proportion of Nt-OAS-TL activity associated with the fractions at ∼290 kD increased by ∼30-fold compared with that in wild-type tobacco, reflecting the strongly increased abundance of At-SAT protein in the transgenic lines (Figure 6D). Thus, apparently all At-SAT protein in transgenic tobacco was associated with Nt-OAS-TL at a size corresponding to the CSC.
Figure 6.
Interaction of Transgenic At-SAT with Endogenous OAS-TL.
Whole leaf extracts from wild-type tobacco (SNN) and transgenic lines At-SATa-69 and At-SATi-1 were applied to a Superdex 200 HiLoad column (GE Healthcare).
(A) OAS-TL activity in eluted fractions was determined after a 10-min preincubation with 10 mM OAS to dissociate the CSC complex and determine all present OAS-TL activity.
(B) An enlarged portion of (A).
(C) The top three panels show immunoblots of SDS-PAGE gels of column fractions 1 to 30 using serum against Arabidopsis SAT3 protein. The bottom two panels show Coomassie blue stains of Rubisco large and small subunits (LSU and SSU) from the same gels as separation controls.
(D) Ratio of CSC-associated OAS-TL activity in transgenic and wild-type tobacco (calculated from [B]). Error bars represent sd, and data are based on three measurements of OAS-TL activity.
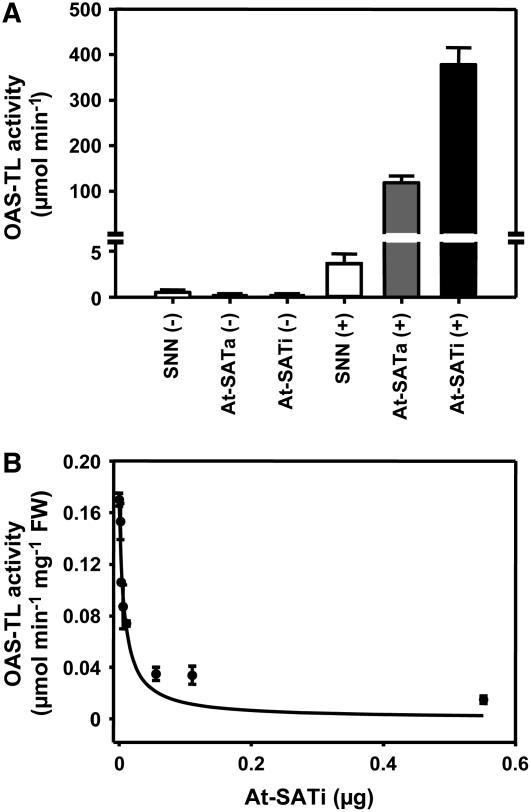
Second, pull-down experiments with anti-SAT3 polyclonal antibodies that targeted At-SATa and At-SATi demonstrated the interaction with Nt-OAS-TL (Figure 7A). When the depleted complexes of anti-At-SAT antibody, At-SAT protein, and Nt-OAS-TL were treated with OAS, the enzymatic activity of Nt-OAS-TL could be recovered. Because the antibody applied is specific for At-SAT, the associated OAS-TL activity must have been a result of heterologous SAT–OAS-TL interaction.
Figure 7.
Heterologous At-SAT and Nt-OAS-TL Interaction Maintains Features of Functional CSC.
(A) Transgenic At-SAT was pulled down from protein extracts of wild-type tobacco (SNN) and transgenic lines expressing either At-SATa or At-SATi using a polyclonal serum against Arabidopsis SAT3 protein (+). As a negative control, SAT3 antiserum was omitted (−). Endogenous OAS-TL bound to At-SAT was eluted by specific dissociation of CSC with 10 mM OAS for 5 min. The identity of copurified OAS-TL was verified by its activity.
(B) Endogenous Nt-OAS-TL from wild-type tobacco was inactivated by titration with At-SATi protein as a result of CSC formation.
Error bars represent sd and are based on three measurements of OAS-TL activity.
Third, the characteristic feature of the inactivation of OAS-TL during complex formation with SAT was used as a tool to indirectly show interaction in an activity titration experiment (Figure 7B). Purified recombinant At-SATi was added to leaf protein extracts from wild-type tobacco. Nt-OAS-TL activity in this extract decreased to ∼10% with increased addition of SAT, as a result of the formation of heterologous CSC and concomitant OAS-TL inactivation. Only the smallest additions of purified At-SAT (0.05 μg) to crude protein extract (100 μg) were required to achieve an 80% reduction of Nt-OAS-TL activity, reflecting the specificity of interaction and the low abundance of this protein complex.
In addition to this evidence, a His-tagged fusion of At-SATa was expressed in E. coli, immobilized on a metal affinity column, and used to affinity purify heterologous OAS-TL from wild-type tobacco whole leaf protein extracts (see Supplemental Figure 7 online). After cleavage and elution with OAS, two-dimensional gel electrophoresis revealed three protein spots that could be attributed to different tobacco OAS-TL isoforms, according to mass spectrometry analysis. These experiments demonstrate regulatory protein–protein interactions of all expressed Arabidopsis SAT proteins with endogenous Nt-OAS-TL in a functional CSC.
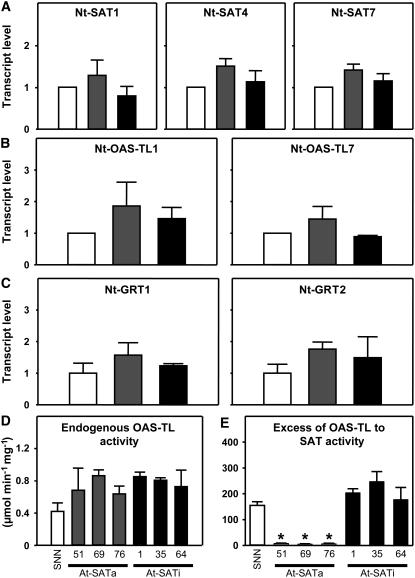
Cys Accumulation Is Not a Consequence of Endogenous Gene or Enzyme Activation
SAT activity was strongly enhanced upon active At-SATa overexpression, but no change in total SAT activity in lines overexpressing inactive At-SATi was observed (Figure 2B), although Cys accumulated to an even higher extent in those plants. Therefore, we investigated whether the upregulation of endogenous tobacco Cys synthesis genes could account for the enhanced formation of Cys in both transgenic genotypes. cDNAs encoding three SATs (Nt-SAT1, Nt-SAT4, and Nt-SAT7 [Wirtz and Hell, 2003]) and two cDNAs encoding OAS-TLs (Nt-OAS-TL1 and Nt-OAS-TL7) were isolated by functional complementation of cysE and cysK/cysM null mutants of E. coli. Sequence comparisons with known SAT and OAS-TL proteins assigned Nt-SAT7 and Nt-OAS-TL7 to the cytosol, whereas Nt-SAT1, Nt-SAT4, and Nt-OAS-TL1 were predicted to be localized either in plastids or mitochondria (data not shown). RNA gel blot hybridization using the three Nt-SAT cDNAs showed no significant increase of steady state mRNA contents, either in At-SATa or in At-SATi plants (Figure 8A). Expression of both Nt-OAS-TLs showed a minor increase without statistical significance in At-SATa lines but hardly any increase in At-SATi lines (Figure 8B). To extend these observations, specific activities of OAS-TL were determined from leaf extracts. They increased by ∼50 to 80% in At-SATa and At-SATi lines, although with rather large standard deviations (Figure 8D). Specific activities of OAS-TL are known to be much higher than those of SAT in crude protein extracts (Droux et al., 1998; Droux, 2004). The biological significance of the OAS-TL activity increases was evaluated using the ratios of OAS-TL to SAT specific activities (Figure 8E). Ratios were between 150- and 180-fold in wild-type and At-SATi lines and not altered significantly, suggesting that the measured OAS-TL increases were of minor significance. In At-SATa lines, the ratio decreased to <25-fold, as expected from the strong increase of transgene-linked SAT activity.
Figure 8.
Expression of Sulfur Metabolism–Related Genes in Leaves of Transgenic Tobacco Plants.
Transcript levels of endogenous SAT (A), OAS-TL (B), and glutathione reductase (GRT) (C) were analyzed by RNA gel blot hybridization of total mRNA from leaves of wild-type tobacco (SNN; white bars) and three independent transgenic lines expressing either At-SATa (gray bars) or At-SATi (black bars). In addition, total OAS-TL activity (D) was determined from protein extracts of leaves. Mean values and sd of enzyme assays from three individual plants with three measurements each are shown (n = 9). The excess of OAS-TL to SAT activity (E) was calculated using the SAT activity shown in Figure 4. Asterisks mark significant differences determined using the unpaired t test. Error bars represent sd.
Because Cys and GSH are redox-active metabolites that could give rise to stress reactions or regulatory redox signals, the expression of the known GSH reductase genes encoding plastidic (Nt-GR1) and cytosolic (Nt-GR2) isoforms (Creissen and Mullineaux, 1995) was determined. RNA contents of both genes were increased by 30 to 60% on average, but without statistical significance (Figure 8C). This could still reflect an enhanced requirement for the reduction of thiol moieties. However, HPLC-based determination of redox status revealed that Cys was reduced by ∼80% and GSH by 90% in leaves of wild-type plants and that the two transgenic genotypes showed approximately the same level of reduction (see Supplemental Figure 8 online). Therefore, compared with wild-type controls, the redox state of these cells was not altered with respect to the accumulation of thiol metabolites. We concluded that only the two transgenic proteins At-SATa and At-SATi contributed to the observed accumulation of Cys and other sulfur compounds, rather than the expression of tobacco genes responsible for Cys synthesis (Nt-SAT and Nt-OAS-TL) or redox regulation.
Cys and Thiol Accumulation Is Accomplished by Different Mechanisms in Active and Inactive At-SAT–Expressing Plants
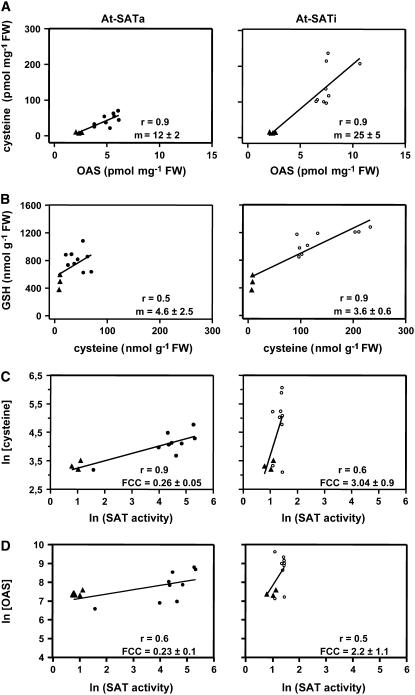
The correlation between steady state concentrations of OAS and Cys on the one hand and Cys and GSH on the other hand was compared. OAS and Cys concentrations found in wild-type and At-SATa and At-SATi plants, respectively, were plotted against each other (Figure 9A). The slope obtained for lines expressing At-SATi was doubled compared with that for At-SATa–expressing lines (i.e., twice as much Cys accumulated per mole of OAS present in tissue). When the slopes of Cys versus GSH were compared, the values were much lower but, more importantly, quite similar between At-SATa and At-SATi lines (Figure 9B). This finding suggests that the formation of GSH from Cys was less effective than that from OAS to Cys but proceeded by the same mechanism in the two transgenic genotypes. By contrast, the different slopes in the lines with active or inactive At-SAT expression suggest mechanisms operating in different ways.
Figure 9.
Flux Control Coefficients of SAT and Correlations of Metabolites Inside the Sulfur Network of Transgenic Tobacco Plants.
Steady state levels of OAS and Cys (A) as well as Cys and GSH (B) in leaves of wild-type tobacco (triangles) and three independent transgenic lines for At-SATa (closed circles) or At-SATi (open circles) were plotted (n = 12) and fitted by linear regression (y = m × x + b). The correlation coefficient (r) and the slope (m) for each fit are shown inside the panel. The flux control coefficient (FCC) of SAT for Cys (C) and OAS (D) was determined as described by Wu et al. (2004).
Therefore, the flux control coefficients of SAT activity for OAS and Cys accumulation in wild-type and transgenic lines were determined. The flux control coefficients for Cys were 0.26 ± 0.05 in At-SATa and 3.04 ± 0.90 in At-SATi (Figure 9C); those for OAS were 0.23 ± 0.10 in At-SATa and 2.21 ± 1.16 in At-SATi (Figure 9D) and thus similar compared with the Cys coefficients within the margin of regression. Importantly, however, they differed for both OAS and Cys by a factor of ∼10 when At-SATa and At-SATi lines were compared. This indicated different flux rates and corroborated the assumption that enhanced product formation in active or inactive At-SAT expression transgenic lines was achieved by different mechanisms.
DISCUSSION
Increased Contents of Sulfur-Related Compounds by Overexpression of Active SAT
The aim of this study was to investigate whether the CSC indeed plays a crucial role in the regulation of Cys biosynthesis, as hypothesized from collected biochemical evidence, by expressing either an active or an inactivated SAT protein from Arabidopsis in the cytosol of tobacco cells. At-SATa was not only shown to interact with free OAS-TL dimers in the cytosol but to be completely complexed, based on the documented high excess of OAS-TL (Kuske et al., 1994; Ruffet et al., 1994; Wirtz et al., 2004). Consequently, total SAT activity and free Cys concentrations increased strongly, as did levels of the primary reaction product OAS, the downstream product GSH, Met, and even total sulfur content in At-SATa expression lines. Similar experiments with constitutive overexpression of active SATs from E. coli or plants in the cytosol and plastids have been conducted with transgenic tobacco, potato (Solanum tuberosum), and Arabidopsis (Blaszczyk et al., 1999; Harms et al., 2000; Noji and Saito, 2002; Wirtz and Hell, 2003) and demonstrated that Cys synthesis could be enhanced in both compartments. In the case of At-SATa, the overexpression was so efficient that not only sulfur metabolites but also the sulfur:nitrogen ratio in free amino acids and total sulfur contents increased significantly. Considering that additional Cys was bound by increased GSH while sulfate content remained unchanged, the protein fraction must have received a considerable part of this increase as an important part of the total sulfur pool.
OAS concentrations in At-SATa lines 51, 69, and 76 increased significantly (∼2.3-fold on average), in agreement with increased SAT activity in whole protein extracts of these lines. Where assayed, OAS also increased to varying extents in other overexpression approaches (Noji and Saito, 2002; Wirtz and Hell, 2003; Hopkins et al., 2005). This may indicate that OAS is indeed not channelled within the Cys synthase complex, at least under these conditions. It also means that the local concentrations of OAS in the target compartments of SAT overexpression were either still below the dissociation constant of the CSC (KdOAS ≈ 60 to 80 μM) (Berkowitz et al., 2002) or that free sulfide that might have increased from enhanced activity of the reductive sulfate assimilation pathway stabilized the complex to promote SAT activity and OAS production.
The Biochemical Phenotype of Expression of Inactive SAT Is Based on OAS-TL Interaction
Surprisingly, constitutive overexpression of inactive SATi resulted in stronger increases of all parameters tested with respect to the metabolism of reduced sulfur compared with overexpression of unmodified active SATa. In fact, the increases in Cys and total downstream sulfur compounds were very large compared with other approaches used to enhance sulfur metabolism in plants (for review, see Sirko et al., 2004), although total SAT activity was not enhanced significantly in transgenic At-SATi. Even the concentrations of OAS, the direct reaction product of the SAT enzyme, increased by 3.3-fold on average. Consistent with earlier evidence (Wirtz et al., 2001), several independent experimental approaches demonstrated that both active At-SATa protein and the catalytically inactive At-SATi could bind to Nt-OAS-TL in the cytosol of transgenic tobacco lines. Both Arabidopsis SATs were shown to coelute with tobacco OAS-TL during size-exclusion chromatography (Figure 6). Pull-down assays provided further evidence for interaction in transgenic tobacco. An activity titration experiment with recombinant At-SATi protein and tobacco leaf extracts corroborated the interaction of the heterologous subunits and confirmed two functional features of the CSC: the OAS-dependent dissociation of the complex (Figure 7A) and the decrease in OAS-TL activity upon CSC formation (Figure 7B).
Heterologous CSC formation forms the basis for the conclusion that inactive At-SATi outcompeted endogenous active Nt-SAT for binding to Nt-OAS-TL in a dominant-negative manner. This was based on several observations. (1) Seventy to 80% of SAT activity in leaves is found in the mitochondria and only 5 to 20% is found in the plastids and cytosol, respectively, in pea (Pisum sativum) and spinach (Droux, 2003). (2) Strong expression of At-SATi was achieved by the 35S promoter and resulted in protein levels comparable to those of At-SATa, which was sufficient to cause a >40-fold increase in total SAT activity. (3) Wild-type SAT is assumed to be of relatively low abundance. The only successful purification of native SAT from a plant achieved a 300,000-fold purification (Ruffet et al., 1994). (4) Specific activities per milligram of purified OAS-TLs are ∼10-fold higher than those of purified SATs (Kuske et al., 1994; Ruffet et al., 1994; Wirtz et al., 2001, 2004), but activity ratios are on the order of 100-fold, suggesting at least a 10-fold molar excess of OAS-TL over SAT proteins in wild-type plants (Ruffet et al., 1994). Therefore, it seems reasonable to assume that At-SATi (and also At-SATa) effectively competed with cytosolic Nt-SAT, unless the latter had a significantly higher affinity for Nt-OAS-TL. This, however, seems unlikely, because all available biochemical and yeast two-hybrid data indicate fairly similar interaction intensities and a considerable promiscuity between SATs and OAS-TLs from different cellular compartments, plant species, and even E. coli (Bogdanova and Hell, 1997; Droux et al., 1998; Liszewska et al., 2005).
Thus, competition for binding to Nt-OAS-TL in the tobacco cytosol between Nt-SAT and the At-SATi polypeptide, which has no enzymatic activity but is capable of interaction with OAS-TL, is the trigger for the deregulation of the CSC.
Possible Mechanisms of Deregulation of the CSC
In the experiments described, the effects are caused by expression of the active and inactive SAT proteins. In both approaches, the availability of sulfide was apparently not limiting for this strong accumulation of reduced sulfur compounds. This implies sufficient availability and transport of sulfate as well as activity of the assimilatory reduction pathway in transgenic lines grown on soil. Furthermore, Nt-OAS-TL activities, being already 100 times greater than those of SAT in wild-type plants, provided no apparent limitation for the enhanced formation of Cys. Therefore, no substantial upregulation of OAS-TL gene expression or activity as a consequence of transgene expression in either transgenic genotype was required. Moreover, the Nt-SATs were also not upregulated, as shown by the unchanged total activities in the At-SATi–expressing lines and by RNA gel blot hybridization. Therefore, the strong accumulation of Cys and downstream sulfur compounds must have been based on the regulation of the CSC, either at the metabolic level or possibly by altered protein modification. If such an unknown regulatory mechanism was involved, one would have expected similar effects in both At-SATa and At-SATi plants, because these proteins differ in only one amino acid in the catalytic center.
However, it is important to note that At-SATa– and At-SATi–expressing lines achieved the observed effects by different mechanisms. The correlation between Cys and OAS accumulation differed markedly between the transgenic lines, with At-SATi lines achieving twice as much Cys per mole of OAS than At-SATa lines. By contrast, GSH biosynthesis proceeds by the same two-step mechanism in cytosol and plastids of tobacco, spinach, and pea (Hell and Bergmann, 1988, 1990), and the correlations between Cys and GSH concentrations were quite similar. These steady state–derived results were confirmed by the different flux control coefficients determined for SAT activities on the one hand and for OAS and Cys concentrations on the other. For both metabolites, the flux control coefficients were on the order of 10-fold higher in At-SATi compared with At-SATa lines. This means that, based on the wild-type situation, At-SATi expressors accumulated 10-fold more reaction products per unit of SAT activity than At-SATa lines. It is reasonable to assume that this effect was brought about by a mechanism other than the presence of more SAT protein, in particular when the complete absence of measurable changes in total SAT and OAS-TL activity and expression in the transgenic At-SATi tobacco was considered.
Although not observed in the transgenic lines, an increase in tobacco OAS-TL activity could provide more interaction partners for SAT to form the CSC but would not explain the Cys and downstream product accumulation, because this enzyme is already present in excess in the tobacco background. Accordingly, overexpression of OAS-TL in the cytosol and plastids of transgenic plants had only small consequences for Cys and GSH steady state levels under nonstress conditions (for review, see Sirko et al., 2004). Significant effects on thiol contents required feeding of OAS in such approaches (Saito et al., 1994). A posttranslational modification of tobacco SAT in cytosol, plastids, or mitochondria could lead to enhanced SAT activity. Without detailed analysis, this possibility cannot be completely excluded, but no significant activity changes were observed under Vmax assay conditions in the transgenic At-SATi lines. Alternatively, phosphorylation followed by reduced feedback sensitivity of SAT, as proposed by Saito (2000) for cytosolic SAT from soybean (Glycine max), could allow enhanced fluxes. This would require that Nt-SAT not be completely outcompeted from the CSC by At-SATi or be able to function more efficiently as a free hexamer for some reason. Such a scenario seems unlikely, because At-SATi and At-SATa differ by only one amino acid, H309A, and should give rise to the same mechanism that leads to Cys accumulation. Furthermore, the apparent molecular masses of At-SATa and At-SATi proteins in tobacco were not changed according to immunoblot analysis (Figure 2) and mass spectrometry of purified At-SAT (M. Wirtz, T. Mentzel, and R. Hell, unpublished data), making any kind of differential posttranslational modification unlikely.
Deregulation of the CSC in Chloroplasts and Mitochondria
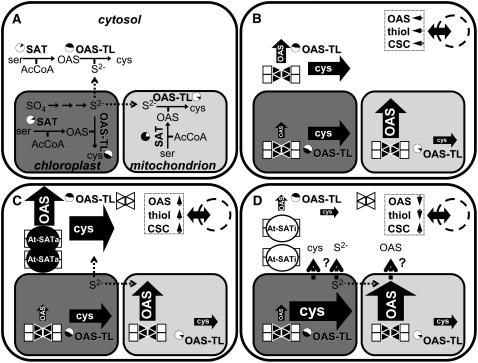
The presence of the Cys synthase system in the different compartments of the plant cell provides a possible explanation for the Cys increase in At-SATi lines without measurable changes in SAT and OAS-TL activities. Distribution and assumed fluxes as well as transport processes of Cys synthesis in wild-type and transgenic lines are schematically outlined in Figure 10. The efficient overexpression of SAT and its binding to Nt-OAS-TL had probably outcompeted endogenous Nt-SAT in the cytosol. This resulted in strongly decreased OAS and Cys production in this compartment, because SAT outside the complex shows no or only reduced activity (Droux et al., 1998; Wirtz et al., 2001). This deficiency in the cytosol could have been compensated for by Cys synthesis activity in the plastids or the mitochondria or both. The regulatory circuit of the CSC would be able to increase fluxes of Cys synthesis in vivo without measurable increase in Vmax of the contributing enzyme subunits (Hell and Hillebrand, 2001; Berkowitz et al., 2002; Wirtz and Hell, 2006). Thus, such a hypothesis would be based on the downregulation of the cytosolic CSC and limited communication with the plastidic and mitochondrial sites of Cys synthesis.
Figure 10.
Organization of Cys Biosynthesis Pathways in Plant Cells and Transgenic Tobacco Expressing At-SATa or At-SATi in the Cytosol.
(A) Schematic overview of enzymatic activities and metabolites involved in the synthesis of Cys in the cytosol (white panel), the mitochondrion (light gray panel), and the chloroplast (dark gray panel). Enzymes are indicated in boldface letters. Black fields of the pie charts show the ratio of enzymatic activities present inside the respective compartment as a percentage of total extractable activity calculated from subcellular fractionation experiments with pea and spinach leaves (Droux, 2004). Solid arrows represent enzymatic reactions, and broken arrows indicate transport of metabolites across membranes.
(B) Relative fluxes of OAS and Cys (indicated by the size of the solid arrows) in different subcellular compartments as deduced from the enzymatic activities of SAT and OAS-TL. Endogenous tobacco SAT (triangles) is associated with OAS-TL (squares) to form the CSC. Black triangles and squares indicate the active forms of enzymes, and white triangles indicate inactivated or less active enzymes. The solid double-headed arrow through the broken circle indicates that the nucleus as a control center of sulfur-related gene expression is in equilibrium with concentrations of thiols (sulfide, Cys, and GSH) and OAS in the cytosol. These effector concentrations and the degree of CSC association are believed to keep a certain balance to achieve coordinated transcriptional control of the genes of sulfur uptake and assimilation.
(C) and (D) Overexpression of At-SATa (black circles) (C) and At-SATi (white circles) (D) in the cytosol. The impact on subcellular fluxes and the transport of metabolites across membranes as a consequence of disturbed CSC formation in the cytosol and metabolite signaling in the nucleus are indicated by the size of the arrows.
Evidence for crosstalk among these three compartments with respect to Cys synthesis is rare. Direct evidence for Cys transport between the cytosol and the organelles is still lacking. Thus, the enzymes for Cys synthesis might be required in each of the three compartments in which proteins can be synthesized (Lunn et al., 1990; Rolland et al., 1992). Plasmalemma transport of Cys is evident in higher plants (Boorer et al., 1996; Miranda et al., 2001), but reports of Cys transport across plastidic or mitochondrial membranes are scarce. Virtually nothing is known about the uptake of Cys by mitochondria or the export of Cys from both organelles. It is interesting that the downstream products of Cys synthesis, Met and GSH, are formed in plastids as well as the cytosol (Hell and Bergmann, 1988, 1990; Ravanel et al., 2004). However, recent evidence showed that in Arabidopsis and Brassica juncea the first enzyme of GSH synthesis, GSH1, is restricted to the plastids, whereas GSH2 is present in plastids and cytosol (Wachter et al., 2005). This finding implies the export of the intermediate γ-glutamylcysteine from plastids to the cytosol, a finding also supported by physiological data (Meyer and Fricker, 2002). The exclusive synthesis of γ-glutamylcysteine in plastids would provide an explanation for similar Cys-to-GSH correlations in transgenic plants expressing At-SATi in the cytosol. Furthermore, this apparent correlation would imply that compensatory SAT activity is upregulated in mitochondria, because upregulation in the plastids would first enhance GSH production. In this context, it should be noted that 70 to 80% of total SAT activity is found in mitochondria of pea and spinach, whereas only 20 to 30% is associated with plastids and cytosol (Ruffet et al., 1995; Droux, 2004) (Figures 10A and 10B). Although nothing is known about the transport of OAS across any intracellular membrane, exchange of metabolites between cellular compartments appears to be important for intracellular sulfur homeostasis, because peroxisomes are known to be the major source of Gly during photorespiration, which serves as a substrate for GSH synthesis in the chloroplasts and the cytosol (Noctor et al., 1999). Finally, transport of reduced sulfur across all organelle membranes may occur as H2S, which is believed to penetrate biomembranes, followed by fixation into Cys and transfer to GSH (Jacques, 1936).
It may be speculated from these observations that the communication between cytosol and organelles with respect to Cys concentrations is impaired because of interference of these and other possibly related metabolic intermediates. In the case of At-SATi–mediated downregulation in the cytosol, Cys synthesis in mitochondria and/or plastids might have overcompensated for the local deficiency caused by limited transfer of Cys (or transport intermediates) to the cytosol (Figures 10C and 10D). Hence, bulk measurements of Cys, GSH, and total S in At-SATi plants showed strong overall accumulation of these compounds as a result of a completely different overexpression mechanism than the one observed in At-SATa plants with increased abundance of the active enzyme. These data also imply increased sulfate uptake and reduction of sulfate during the growth of both types of transgenic plants. The hypothesis of disrupted cytosolic CSC equilibrium and overcompensation by organellar CSC sites or other possible explanations can be tested in future experiments. Assuming similar mechanisms for the CSC in the three compartments, downregulation of the CSC in plastids or mitochondria should result in similar accumulations of sulfur compounds. This could be achieved by overexpression of SATi in these compartments or possibly by downregulation of endogenous SAT in these compartments. Simultaneous CSC disruption in two compartments should result in an even stronger phenotype.
Together, these data demonstrate in vivo that the CSC plays an integral role in the regulation of Cys synthesis. The dominant-negative overexpression of the enzymatically inactivated SAT subunit of the CSC in the cytosol resulted in a very strong accumulation of Cys and downstream sulfur compounds and demonstrates that the CSC is a metabolic enzyme complex with a regulatory function and not substrate channelling.
METHODS
General Cloning
Standard molecular biology technologies, such as plasmid isolation, PCR, and RNA gel blot hybridization, were performed as described by Sambrook et al. (1989) according to GLP standards. DNA fragments as probes for RNA gel blot analysis were obtained by PCR with random primers and subsequent labeling with [α-32P]dATP using the Megaprime DNA labeling system (Amersham). For plant transformation, the cDNA encoding SATH309A (At-SATi; Wirtz et al., 2001) was cloned into the BamHI-SalI restriction sites of pBinAR as described for At-SAT3 (At-SATa) by Wirtz and Hell (2003). Necessary restriction sites for cloning were introduced by amplification of the respective cDNA by PCR using the primers SAT 208 (5′-GGATCCCATGAACTACTTCCGTTATC-3′) and SAT 209 (5′-GTCGACTCAAATTACATAATCCGAC-3′).
Plant Growth and Transformation
Transformation of Agrobacterium tumefaciens C58 with binary vectors and subsequent transformation and selection of tobacco (Nicotiana tabacum cv Samsun NN) were performed as described by Henkes et al. (2001). Wild-type plants and plants expressing either At-SATa or At-SATi (F3 generation) were grown in climate chambers in growth medium containing half soil and half substrate 2 (Klasmann-Deilmann) under controlled conditions: 16 h of light; 200 μE light intensity; 24 and 18°C day and night, respectively; and 60% humidity. The second and third leaves from the top of 6-week-old plants were pooled and used to analyze metabolites as well as the expression of sulfur-related genes at the transcript and protein levels.
Determination of Metabolites
Hydrophilic metabolites were extracted from leaves of tobacco plants according to Wirtz and Hell (2003). Thiols and amino acids were quantified after derivatization with monobromobimane (Calbiochem) or AccQ-Tag reagent (Waters), respectively. The derivatization procedure and separation of thiol derivatives were performed as described by Wirtz et al. (2004) using the same HPLC system. Anions were separated after 10-fold dilution in water using an AS10, 4 × 250 mm column (Dionex) coupled to a Dionex HPLC system with an isocratic flow of 0.3 mL/min 0.1 M sodium hydroxide and detected by conductivity. Quantification of anions was achieved with Dionex Peaknet chromatography software using external standards as reference. Relative contents of total carbon, nitrogen, and sulfur in dried, powdered samples of leaf material were measured using an elemental analyzer (Vario EL; Elementaranalysensysteme).
Determination of Enzyme Activities
Total soluble proteins were isolated with 1.2 mL of 50 mM HEPES, pH 7.4, 10 mM KCl, 1 mM EDTA, 10% glycerine, 30 mM DTT, and 0.5% phenylmethylsulfonyl fluoride from 0.2 g of leaf material that was ground to a fine powder in liquid nitrogen. Cell debris was removed by centrifugation at 16,000g and 4°C for 10 min. The supernatant was desalted in 50 mM HEPES, pH 7.5, 1 mM EDTA, 1 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride with a PD-10 column (Amersham) according to the manufacturer's protocol. Proteins were quantified as described by Bradford (1976) using BSA as a standard. The enzymatic activities of SAT and OAS-TL were determined according to Nakamura et al. (1987): SAT activity was assayed by coupling to the OAS-TL reaction. All SAT activity determinations were supplemented with 2 units of purified recombinant OAS-TL (Wirtz et al., 2004) to ensure high excess of OAS-TL activity during the coupling of both reactions.
Immunological Detection of Proteins
Total proteins from leaves were separated according to Laemmli (1970) by discontinuous SDS-PAGE in Mini-Protean II cells (Bio-Rad). The monomer concentration (% T) and cross-linker concentration (% C) of focusing and resolving gels were 4% T/0.026% C and 12.5% T/0.026% C, respectively. Before separation, proteins were denatured by incubation for 10 min at 65°C in 20 mM Tris, pH 7.0, 0.1% SDS, 0.7% β-mercaptoethanol, and 4% glycerol. Blotting of separated proteins (7.5 μg) to nitrocellulose was achieved in a Bio-Rad transfer cell according to the manufacturer's protocol. After blocking of nitrocellulose with 5% BSA, an alkaline phosphatase–anti-rabbit IgG conjugate (Promega; 1: 10,000) in combination with the nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate substrate system (Roche) was used to detect primary antibody against At-SAT3 (1:10,000).
SAT–OAS-TL Interaction Studies by Size-Exclusion Chromatography
Association of the heterologous complex of endogenous OAS-TL (Nt-OAS-TL) with At-SAT was analyzed by size-exclusion chromatography of total soluble tobacco leaf protein isolated as described above. Approximately 40 mg of protein was loaded onto a Superdex 200 HiLoad 16/60 column (bed volume, 120 mL) purchased from GE Healthcare. The column was connected to an ÄKTA basic fast-protein liquid chromatography system at a flow rate of 1 mL/min buffer (50 mM Tris-HCl, pH 7.5, and 250 mM NaCl). In general, fractions of 2 mL were collected from 40 to 120 mL, whereas the fraction size was reduced to 1 mL between 52 and 68 mL to optimize the resolution of the CSC. Calibration of the column was performed using the low and high molecular weight standard kits from GE Healthcare.
The resulting fractions were analyzed for OAS-TL activity as described above except that 10 μL of each fraction was used instead of crude protein extract and the incubation time was increased to 15 min. The presence of At-SAT protein was tested by immunological detection of SAT using 10% SDS-PAGE gels loaded with 10 μL of each fraction.
Titration of endogenous OAS-TL (Nt-OAS-TL) activity with At-SATi was achieved by incubation of 0.1 mg of total protein from wild-type tobacco leaves with varying amounts of purified At-SATi protein (0 to 0.55 μg). The latter was purified by S-Tag affinity chromatography according to Wirtz et al. (2001) after overexpression of an S-Tag At-SATi fusion protein in Escherichia coli. Total soluble tobacco proteins were isolated as described above. After 10 min of coincubation at room temperature, the decrease of endogenous OAS-TL activity was measured according to Nakamura et al. (1987) as a marker of heterologous CSC formation, as shown previously (Berkowitz et al., 2002).
In addition, interaction of Nt-OAS-TL with At-SAT was verified by pull-down of heterologous CSC from total protein extracts of transgenic tobacco plants expressing either At-SATa or At-SATi using polyclonal antiserum against At-SAT3 (Wirtz and Hell, 2003). Fourteen milligrams of total protein from transgenic or wild-type tobacco plants was incubated with 0.3 mL of polyclonal serum against At-SAT3 for 3 h at 22°C. The polyclonal SAT3 antibodies were captured by incubation with 0.2 mL of protein A–agarose (Gibco BRL) and collected by centrifugation at 5000g and 22°C for 5 min. The protein A–agarose was washed for three times with 50 mM tris(hydroxymethyl)-aminomethane, pH 7.5, and 250 mM NaCl, and the heterologous CSC was dissociated with 10 mM OAS for 5 min. The protein A–agarose was removed by centrifugation, and the resulting supernatant was tested for OAS-TL activity.
Statistical Analysis and Calculation of Flux Correlation Coefficient
Regression and correlation analysis of data sets were performed with SigmaPlot 8.0, which uses the Marquardt-Levenberg algorithm for the determination of independent variables. The P value for the Pearson product moment correlation was <0.05 in all cases. Comparison of means from different data sets was analyzed for statistical significance with the unpaired t test. Constant variance and normal distribution of data were carefully checked with SigmaStat 3.0 before statistical analysis.
For calculation of the slope-based flux control coefficient, the natural logarithms of specific SAT activity and respective metabolite concentrations were plotted against each other according to Wu et al. (2004). Slopes resulting from data points of individual wild-type and transgenic plants in the same diagram reflect the flux control coefficient.
Accession Numbers
The following cDNAs were used in this study: At-SAT3 (X80938; Bogdanova et al., 1995), Nt-SAT1 (AJ414051), Nt-SAT4 (AJ414052), Nt-SAT7 (AJ414053) (all Wirtz and Hell, 2003), Nt-OAS-TL1 (AM087457), Nt-OAS-TL7 (AM087458), and the GRT cDNAs Nt-GRT1 (X76293) and Nt-GRT2 (X76455) (Creissen and Mullineaux, 1995).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. At-SATi Is >1000-Fold Less Active Than At-SATa.
Supplemental Figure 2. mRNA Levels of At-SAT in Roots of Wild-Type (SNN) and Transgenic Tobacco Expressing At-SATa or At-SATi.
Supplemental Figure 3. Production of Seeds in Wild-Type (SNN) and Transgenic Tobacco Expressing At-SATa or At-SATi.
Supplemental Figure 4. Contents of Basic Amino Acids in Leaves of Wild-Type and Transgenic Tobacco Plants Expressing At-SATa or At-SATi.
Supplemental Figure 5. Total Nitrogen, Carbon, and Sulfur Contents in Roots of Wild-Type and Transgenic Tobacco Plants Expressing At-SATa or At-SATi.
Supplemental Figure 6. Contents of Anions in Roots of Wild-Type and Transgenic Tobacco Plants Expressing At-SATa or At-SATi.
Supplemental Figure 7. At-SATa Can Interact with Endogenous Tobacco OAS-TLs.
Supplemental Figure 8. Ratio of GSH to GSSG in Leaves of Wild-Type and Transgenic Tobacco Plants Expressing At-SATa or At-SATi.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding by Grant SFB 363 from the German Science Foundation and the Leibniz-Institute for Plant Genetics and Crop Plant Research (IPK) at Gatersleben. We are indebted to Uwe Sonnewald (University of Erlangen) for plant transformation and M. Hajirezaei and H. Rolletschek (IPK Gatersleben) for analytical support. We thank P. Mullineaux (University of Essex) and G. Creissen (John Innes Centre) for tobacco DNA probes, Dagmar Böhmert (IPK Gatersleben) for excellent technical assistance, and A. Meyer and C. Heeg (University of Heidelberg) for critically reading the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Rüdiger Hell (rhell@hip.uni-heidelberg.de).
Online version contains Web-only data.
References
- Berkowitz, O., Wirtz, M., Wolf, A., Kuhlmann, J., and Hell, R. (2002). Use of biomolecular interaction analysis to elucidate the regulatory mechanism of the cysteine synthase complex from Arabidopsis thaliana. J. Biol. Chem. 277 30629–30634. [DOI] [PubMed] [Google Scholar]
- Blaszczyk, A., Brodzik, R., and Sirko, A. (1999). Increased resistance to oxidative stress in transgenic tobacco plants overexpressing bacterial serine acetyltransferase. Plant J. 20 237–243. [DOI] [PubMed] [Google Scholar]
- Bogdanova, N., Bork, C., and Hell, R. (1995). Cysteine biosynthesis in plants: Isolation and functional identification of a cDNA encoding a serine acetyltransferase from Arabidopsis thaliana. FEBS Lett. 358 43–47. [DOI] [PubMed] [Google Scholar]
- Bogdanova, N., and Hell, R. (1997). Cysteine synthesis in plants: Protein-protein interactions of serine acetyltransferase from Arabidopsis thaliana. Plant J. 11 251–262. [DOI] [PubMed] [Google Scholar]
- Boorer, K.J., Frommer, W.B., Bush, D.R., Kreman, M., Loo, D.D., and Wright, E.M. (1996). Kinetics and specificity of a H+/amino acid transporter from Arabidopsis thaliana. J. Biol. Chem. 271 2213–2220. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Cook, P.F., and Wedding, R.T. (1976). A reaction mechanism from steady state kinetic studies for O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2. J. Biol. Chem. 251 2023–2029. [PubMed] [Google Scholar]
- Creissen, G.P., and Mullineaux, P.M. (1995). Cloning and characterisation of glutathione reductase cDNAs and identification of two genes encoding the tobacco enzyme. Planta 197 422–425. [DOI] [PubMed] [Google Scholar]
- De Kok, L.-J., Buwalda, F., and Bosma, W. (1988). Determination of cysteine and its accumulation in spinach leaf tissue upon exposure to excess sulfur. J. Plant Physiol. 133 502–505. [Google Scholar]
- Droux, M. (2003). Plant serine acetyltransferase: New insights for regulation of sulfur metabolism in plant cells. Plant Physiol. Biochem. 41 619–627. [Google Scholar]
- Droux, M. (2004). Sulfur assimilation and the role of sulfur in plant metabolism: A survey. Photosynth. Res. 79 331–348. [DOI] [PubMed] [Google Scholar]
- Droux, M., Ruffet, M.L., Douce, R., and Job, D. (1998). Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants—Structural and kinetic properties of the free and bound enzymes. Eur. J. Biochem. 255 235–245. [DOI] [PubMed] [Google Scholar]
- Harms, K., von Ballmoos, P., Brunold, C., Hofgen, R., and Hesse, H. (2000). Expression of a bacterial serine acetyltransferase in transgenic potato plants leads to increased levels of cysteine and glutathione. Plant J. 22 335–343. [DOI] [PubMed] [Google Scholar]
- Hawkesford, M.J., and Wray, J.L. (2000). Molecular genetics of sulfate assimilation. Adv. Bot. Res. 33 159–223. [Google Scholar]
- Hell, R. (2003). Metabolic regulation of cysteine synthesis and sulfur assimilation. In Sulfate Transport and Assimilation in Plants, J.C. Davidian, D. Grill, L.J. De Kok, I. Stulen, M.J. Hawkesford, E. Schnug, and H. Rennenberg, eds (Leiden, The Netherlands: Backhuys Publishers), pp. 21–31.
- Hell, R., and Bergmann, L. (1988). Glutathione synthetase in tobacco suspension cultures: Catalytic properties and localization. Physiol. Plant. 72 70–76. [Google Scholar]
- Hell, R., and Bergmann, L. (1990). γ-Glutamylcysteine synthetase in higher plants: Catalytic properties and subcellular localization. Planta 180 603–612. [DOI] [PubMed] [Google Scholar]
- Hell, R., and Hillebrand, H. (2001). Plant concepts for mineral acquisition and allocation. Curr. Opin. Biotechnol. 12 161–168. [DOI] [PubMed] [Google Scholar]
- Hell, R., Jost, R., Berkowitz, O., and Wirtz, M. (2002). Molecular and biochemical analysis of the enzymes of cysteine biosynthesis in the plant Arabidopsis thaliana. Amino Acids 22 245–257. [DOI] [PubMed] [Google Scholar]
- Henkes, S., Sonnewald, U., Badur, R., Flachmann, R., and Stitt, M. (2001). A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 13 535–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, L., Parmar, S., Blaszczyk, A., Hesse, H., Hoefgen, R., and Hawkesford, M.J. (2005). O-acetylserine and the regulation of expression of genes encoding components for sulfate uptake and assimilation in potato. Plant Physiol. 138 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques, A.G. (1936). The kinetics of penetration. XII. Hydrogen sulfide. J. Gen. Physiol. 19 397–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C., Roderick, S., and Cook, P. (2005). The serine acetyltransferase reaction: Acetyl transfer from an acylpantothenyl donor to an alcohol. Arch. Biochem. Biophys. 433 85–95. [DOI] [PubMed] [Google Scholar]
- Kredich, N.M. (1996). Biosynthesis of cysteine. In Escherichia coli and Salmonella typhimurium. Cellular and Molecular Biology, F.C. Neidhardt, R. Curtiss, J.L. Ingraham, E.C.C. Lin, K.B. Low, B. Magasanik, W.S. Reznikoff, M. Riley, M. Schaechter, and E. Umberger, eds (Washington, DC: ASM Press), pp. 514–527.
- Kredich, N.M., Becker, M.A., and Tomkins, G.M. (1969). Purification and characterization of cysteine synthetase, a bifunctional protein complex, from Salmonella typhimurium. J. Biol. Chem. 244 2428–2439. [PubMed] [Google Scholar]
- Kuske, C.R., Ticknor, L.O., Guzman, E., Gurley, L.R., Valdez, J.G., Thompson, M.E., and Jackson, P.J. (1994). Purification and characterization of O-acetylserine sulfhydrylase isoenzymes from Datura innoxia. J. Biol. Chem. 269 6223–6232. [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Leustek, T., Martin, M.N., Bick, J.-A., and Davies, J.P. (2000). Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 141–165. [DOI] [PubMed] [Google Scholar]
- Liszewska, F., Gaganidze, D., and Sirko, A. (2005). Isolation of Nicotiana plumbaginifolia cDNAs encoding isoforms of serine acetyltransferase and O-acetylserine (thiol) lyase in a yeast two-hybrid system with Escherichia coli cysE and cysK genes as baits. Acta Biochim. Pol. 52 117–128. [PubMed] [Google Scholar]
- Lunn, J.E., Droux, M., Martin, J., and Douce, R. (1990). Localization of ATP-sulfurylase and O-acetylserine(thiol)lyase in spinach leaves. Plant Physiol. 94 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, A.J., and Fricker, M.D. (2002). Control of demand-driven biosynthesis of glutathione in green Arabidopsis suspension culture cells. Plant Physiol. 130 1927–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda, M., Borisjuk, L., Tewes, A., Heim, U., Sauer, N., Wobus, U., and Weber, H. (2001). Amino acid permeases in developing seeds of Vicia faba L.: Expression precedes storage protein synthesis and is regulated by amino acid supply. Plant J. 28 61–71. [DOI] [PubMed] [Google Scholar]
- Nakamura, K., Hayama, A., Masada, M., Fukushima, K., and Tamura, G. (1987). Measurement of serine acetyltransferase activity in crude plant extracts by a coupled assay system using cysteine synthase. Plant Cell Physiol. 28 885–891. [Google Scholar]
- Noctor, G., Arisi, A., Jouanin, L., and Foyer, C. (1999). Photorespiratory glycine enhances glutathione accumulation in both the chloroplastic and cytosolic compartments. J. Exp. Bot. 50 1157–1167. [Google Scholar]
- Noji, M., and Saito, K. (2002). Molecular and biochemical analysis of serine acetyltransferase and cysteine synthase towards sulfur metabolic engineering in plants. Amino Acids 22 231–243. [DOI] [PubMed] [Google Scholar]
- Ovadi, J. (1991). Physiological significance of metabolic channeling. J. Theor. Biol. 152 1–22. [PubMed] [Google Scholar]
- Pye, V.E., Tingey, A.P., Robson, R.L., and Moody, P.C. (2004). The structure and mechanism of serine acetyltransferase from Escherichia coli. J. Biol. Chem. 279 40729–40736. [DOI] [PubMed] [Google Scholar]
- Ravanel, S., Block, M.A., Rippert, P., Jabrin, S., Curien, G., Rebeille, F., and Douce, R. (2004). Methionine metabolism in plants: Chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J. Biol. Chem. 279 22548–22557. [DOI] [PubMed] [Google Scholar]
- Rolland, N., Droux, M., and Douce, R. (1992). Subcellular distribution of O-acetylserine(thiol)lyase in cauliflower (Brassica oleracea L.) inflorescence. Plant Physiol. 98 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffet, M.L., Droux, M., and Douce, R. (1994). Purification and kinetic properties of serine acetyltransferase free of O-acetylserine(thiol)lyase from spinach chloroplasts. Plant Physiol. 104 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffet, M.L., Lebrun, M., Droux, M., and Douce, R. (1995). Subcellular distribution of serine acetyltransferase from Pisum sativum and characterization of an Arabidopsis thaliana putative cytosolic isoform. Eur. J. Biochem. 227 500–509. [DOI] [PubMed] [Google Scholar]
- Saito, K. (2000). Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr. Opin. Plant Biol. 3 188–195. [PubMed] [Google Scholar]
- Saito, K. (2004). Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 136 2443–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K., Kurosawa, M., Tatsuguchi, K., Takagi, Y., and Murakoshi, I. (1994). Modulation of cysteine biosynthesis in chloroplasts of transgenic tobacco overexpressing cysteine synthase. O-Acetylserine(thiol)-lyase. Plant Physiol. 106 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sirko, A., Blaszczyk, A., and Liszewska, F. (2004). Overproduction of SAT and/or OASTL in transgenic plants: A survey of effects. J. Exp. Bot. 55 1881–1888. [DOI] [PubMed] [Google Scholar]
- Srere, P.A. (1987). Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 56 89–124. [DOI] [PubMed] [Google Scholar]
- Wachter, A., Wolf, S., Steininger, H., Bogs, J., and Rausch, T. (2005). Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: Implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J. 41 15–30. [DOI] [PubMed] [Google Scholar]
- Winkel, B. (2004). Metabolic channeling in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 55 85–107. [DOI] [PubMed] [Google Scholar]
- Wirtz, M., Berkowitz, O., Droux, M., and Hell, R. (2001). The cysteine synthase complex from plants. Mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein-protein interaction. Eur. J. Biochem. 268 686–693. [DOI] [PubMed] [Google Scholar]
- Wirtz, M., Droux, M., and Hell, R. (2004). O-Acetylserine (thiol) lyase: An enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J. Exp. Bot. 55 1785–1798. [DOI] [PubMed] [Google Scholar]
- Wirtz, M., and Hell, R. (2003). Production of cysteine for bacterial and plant biotechnology: Application of cysteine feedback-insensitive isoforms of serine acetyltransferase. Amino Acids 24 195–203. [DOI] [PubMed] [Google Scholar]
- Wirtz, M., and Hell, R. (2006). Functional analysis of the cysteine synthase protein complex from plants: Structural, biochemical and regulatory properties. J. Plant Physiol. 163 273–286. [DOI] [PubMed] [Google Scholar]
- Wu, L., Wang, W., van Winden, W.A., van Gulik, W.M., and Heijnen, J.J. (2004). A new framework for the estimation of control parameters in metabolic pathways using lin-log kinetics. Eur. J. Biochem. 271 3348–3359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.