Abstract
In Lotus japonicus, seven genetic loci have been identified thus far as components of a common symbiosis (Sym) pathway shared by rhizobia and arbuscular mycorrhizal fungi. We characterized the nup85 mutants (nup85-1, -2, and -3) required for both symbioses and cloned the corresponding gene. When inoculated with Glomus intraradices, the hyphae managed to enter between epidermal cells, but they were unable to penetrate the cortical cell layer. The nup85-2 mutation conferred a weak and temperature-sensitive symbiotic phenotype, which resulted in low arbuscule formation at 22°C but allowed significantly higher arbuscule formation in plant cortical cells at 18°C. On the other hand, the nup85 mutants either did not form nodules or formed few nodules. When treated with Nod factor of Mesorhizobium loti, nup85 roots showed a high degree of root hair branching but failed to induce calcium spiking. In seedlings grown under uninoculated conditions supplied with nitrate, nup85 did not arrest plant growth but significantly reduced seed production. NUP85 encodes a putative nucleoporin with extensive similarity to vertebrate NUP85. Together with symbiotic nucleoporin NUP133, L. japonicus NUP85 might be part of a specific nuclear pore subcomplex that is crucial for fungal and rhizobial colonization and seed production.
INTRODUCTION
More than 80% of land plant families have symbiotic relationships with arbuscular mycorrhizal (AM) fungi belonging to the fungal class Glomeromycota. The AM fungi absorb minerals, including phosphate, from the soil via the extraradical hyphae and provide them to the plants, possibly via arbuscules formed in the cortical cells (Smith and Gianinazzi-Pearson, 1988; Smith and Read, 1997; Harrison, 1999). The fossil record of the arbuscules indicates the origin of AM symbiosis in the early Devonian period ∼400 million yeas ago (Remy et al., 1994; Taylor et al., 1995). The cytology of AM colonization of Lotus japonicus is well documented (Bonfante et al., 2000; Genre and Bonfante, 2002; Novero et al., 2002; Demchenko et al., 2004). First, AM fungi form appressoria along the longitudinal border between epidermal cells and grow between the two adjacent epidermal cells. During the second step, hyphae penetrate one of the two adjacent epidermal cells, traverse the underlying cortical cells, and grow between or within cortical cells. The third step is formation of arbuscules within inner cortical cells, where hyphae branch profusely.
The nitrogen-fixing symbiosis between legumes and rhizobia, unlike the AM symbiosis, involves host-specific recognition and postembryonic development of a nitrogen-fixing organ, the nodule. The rhizobial symbiosis is initiated by reciprocal signal exchange. (Iso)flavonoids secreted from the plant roots are recognized by rhizobial NodD proteins that induce the expression of a set of bacterial genes essential for nodulation. A subset of these genes encodes enzymes involved in the biosynthesis of lipo-chitin oligosaccharides (Nod factors), which act as determinants of host specificity and as morphogens (Dénarié et al., 1996). Nod factors released by rhizobia elicit early responses in root hairs, such as a calcium influx, membrane depolarization, alkalization, calcium spiking, rearrangement of actin filaments, and root hair deformation. The Nod factors also act on the cortical cells, induce preinfection thread formation, and activate cortical cell division leading to differentiation of nodule tissues (Stougaard, 2000).
Despite marked differences between the fungal and bacterial symbioses, common genes required for both interactions were identified in pea (Pisum sativum) (Duc et al., 1989). This initial observation has now been extended to several legume species (Gianinazzi-Pearson, 1996), thus defining the so-called common symbiosis (Sym) pathway (Kistner and Parniske, 2002). Three genes, Does Not Make Infections1 (DMI1), DMI2, and DMI3, from Medicago truncatula (Endre et al., 2002; Ané et al., 2004; Lévy et al., 2004; Mitra et al., 2004) and at least seven loci, SymRK, CASTOR, POLLUX, NUP133, CCaMK, Lj Sym6/30/82, and Lj Sym24/73/85, from L. japonicus have been defined thus far as elements of the common Sym pathway (Kistner et al., 2005; Sandal et al., 2006).
NORK/DMI2/SymRK/Ps SYM19 has been identified as encoding a receptor-like kinase with three Leu-rich repeats in the extracellular domain (Endre et al., 2002; Stracke et al., 2002). CASTOR and POLLUX/DMI1 encode proteins with broad similarity to the NAD binding TrkA domain of bacterial K+ channels (Ané et al., 2004; Imaizumi-Anraku et al., 2005). NUP133 encodes a protein that has sequence similarity to human nucleoporin Nup133 (Kanamori et al., 2006). These genes are required for calcium spiking that is induced in response to Nod factors. By contrast, a Ca2+/calmodulin-dependent protein kinase (CCaMK) acts downstream of the calcium spiking (Lévy et al., 2004; Mitra et al., 2004; Gleason et al., 2006; Tirichine et al., 2006). The identification of additional common Sym genes promises to shed further light on the nature of ancient plant functions involved in symbiotic signaling. Here, we describe a putative nucleoporin gene identified through positional cloning and the phenotypic consequences of mutations in this gene for symbiosis.
RESULTS
Allelism Tests
The Ljsym24 and Ljsym85 loci were previously shown to be required for both rhizobial and AM symbioses (Szczyglowski et al., 1998; Kawaguchi et al., 2005; Kistner et al., 2005; Sandal et al., 2006). The Ljsym73 loci were reported as low nodulation mutants (Kawaguchi et al., 2002). Despite some phenotypic differences, these three mutants were mapped near the translocation site of the short arm of chromosome 1. We therefore performed allelism tests. When plants carrying Ljsym24, Ljysym73, and Ljsym85 were reciprocally crossed, none of the F1 progeny from crosses of Ljsym24 × Ljysym73 and Ljsym24 × Ljysym85 formed nodules (Table 1). In F1 plants of Ljsym73 × Ljysym85, 8 of 11 plants did not form nodules, and three plants had a phenotype like the Ljsym73 mutant, forming one or two large nodules per plant. The complementation tests showed that Ljsym24, Ljsym73, and Ljsym85 are allelic. Ljsym24, Ljsym73, and Ljsym85 are referred to hereafter as nup85-1, -2, and -3, respectively.
Table 1.
Nodulating Phenotype of F1 Plants Obtained from Reciprocal Crosses of Ljsym24 (nup85-1), Ljsym73 (nup85-2), and Ljsym85 (nup85-3)
| Parent Lines
|
Numbers of F1 Plants
|
|||
|---|---|---|---|---|
| Female | × | Male | Nod+ | Nod− |
| Ljsym24 | × | Ljsym73 | 0 | 2 |
| Ljsym73 | × | Ljsym85 | 1a | 1 |
| Ljsym73 | × | Ljsym85 | 0 | 2 |
| Ljsym85 | × | Ljsym24 | 0 | 2 |
| Ljsym85 | × | Ljsym24 | 0 | 2 |
| Ljsym85 | × | Ljsym73 | 0 | 2 |
| Ljsym85 | × | Ljsym73 | 2a | 3 |
The F1 plants formed only one or two nodules per plant.
nup85 Mutants Are Defective in Root Nodule Symbiosis and Nod Factor–Induced Calcium Spiking
The three alleles of nup85 caused different levels of defects in nodulation. The nup85-1 and nup85-2 mutants formed no nodules or a low number of effective nodules, as described previously (Szczyglowski et al., 1998; Kawaguchi et al., 2002; Kistner et al., 2005), whereas the nup85-3 mutant did not form nodules even 2 months after inoculation with Mesorhizobium loti. Hereafter, we used the nup85-2 and nup85-3 mutants as representatives of weak and strong phenotypic lines, respectively.
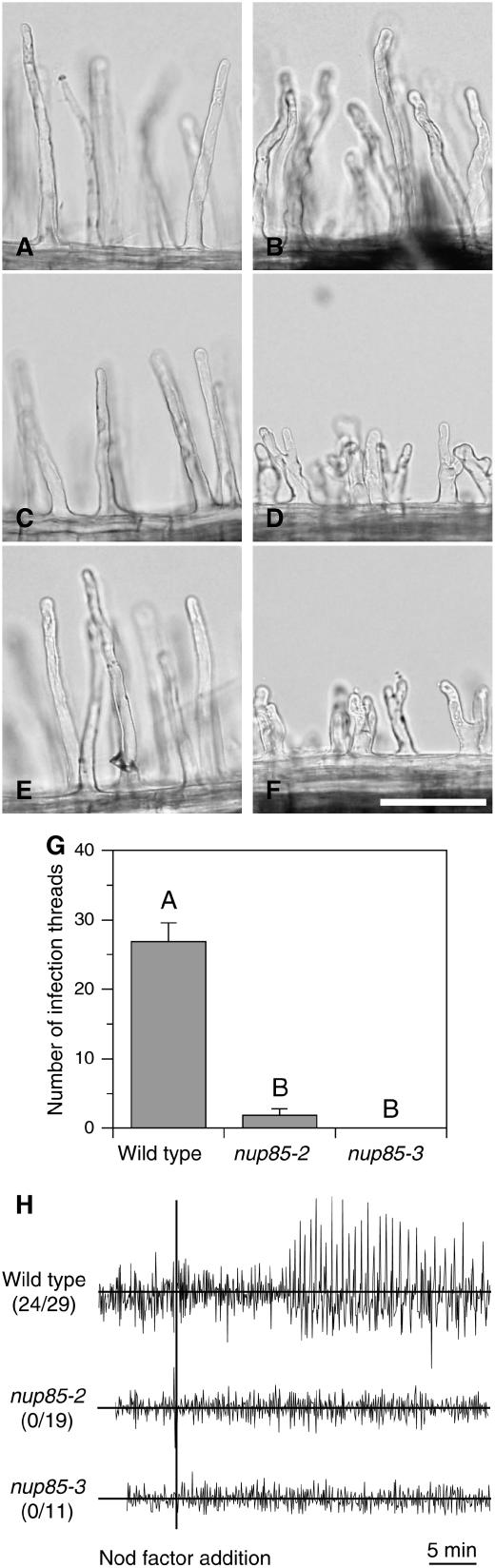
We examined root hair responses of the nup85 mutants to Nod factors. Root hairs of both nup85-2 and nup85-3 mutants responded to Nod factors in a similar manner (Figures 1D and 1F). Root hairs of both mutants exhibited a high degree of branching and appeared more stunted (Figures 1D and 1F) than those of the wild type (Figure 1B). Root hairs on seedlings without rhizobia or Nod factors were normal (Figures 1A, 1C, and 1E).
Figure 1.
Root Hair Deformation, Infection Thread Formation, and Calcium Spiking.
Root hair responses after treatment with 10−8 M Nod factor for 24 h.
(A) Wild-type control.
(B) Wild type with Nod factor treatment.
(C) nup85-2 control.
(D) nup85-2 Nod factor treatment.
(E) nup85-3 control.
(F) nup85-3 Nod factor treatment. The photos are representative of observations of at least 15 plants in each line. Bar = 100 μm.
(G) Number of infection thread of the wild type, nup85-2, and nup85-3. The data presented are means ± se (n = 6). Different letters above bars indicate significance among the means according to the Tukey multiple comparison test (P < 0.05). Analysis of calcium spiking in root hair.
(H) Each trace is from a single root hair using seedlings of the wild type, nup85-2, or nup85-3 mutant. Root hairs were injected with the calcium-sensitive dye Oregon green dextran, and after ∼20 min, Nod factor was added at 10−8 M. The data are graphed showing typical traces of the differences in fluorescence intensity between 5-s sequential time points. Only viable cells showing active cytoplasmic streaming were used in the analysis. These cells were observed for at least 60 min following Nod factor treatments. The fractions in parentheses show the numbers of root hairs in which calcium spiking was detected to the number of root hairs tested.
For β-galactosidase activity, roots were examined 5 d after infection with an M. loti strain carrying the lacZ reporter gene. In the wild type, an average of 27 infection threads per plant were observed. By contrast, the nup85-2 and nup85-3 mutants had a significant reduction in numbers of infection threads (Figure 1G).
Nod factors normally induce calcium spikes mostly around the nuclear region of legume root hairs (Ehrhardt et al., 1996; Cárdenas et al., 1999; Walker et al., 2000; Harris et al., 2003; Shaw and Long, 2003; Miwa et al., 2006). In root hairs of wild-type L. japonicus, calcium spiking initiated 10 to 30 min after Nod factor addition and persisted over the remainder of the experiment (n = 24/29 cells). However, no calcium spiking was observed in root hairs of the nup85-2 and nup85-3 mutants over the period of these experiments (Figure 1H). Similarly, the nup85-1 mutant was previously shown to have no calcium spiking in response to Nod factor application (Miwa et al., 2006).
AM Fungal Colonization Is Blocked at the Entry to the Outer Cortical Cells in nup85 Mutants
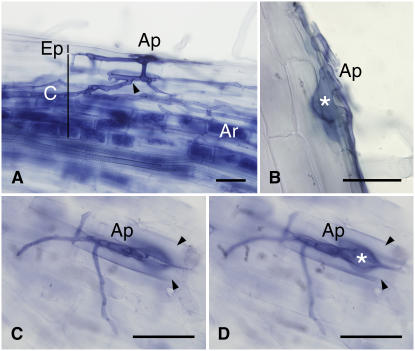
In the wild type, 9 weeks after inoculation with Glomus intraradices at 24°C, hyphal, arbuscular, and vesicular colonizations were 46.0% ± 1.2%, 46.0% ± 1.2%, and 14.8% ± 0.4%, respectively. Roots of nup85-3 were not colonized by the fungus, while in nup85-2, a very low level of mycorrhizal colonization (hyphal colonization, 0.4% ± 0.8%; arbuscular colonization, 0.2% ± 0.4%; and vesicular colonization, 0.0% ± 0.0%) was observed. The fungal colonization at low frequency in nup85-2 can be due to fungal entry from meristematically arrested roots, as observed in symRK and sym15 (ccamk) mutants (Demchenko et al., 2004). Nevertheless, the apparent block in the interaction was identical for all three mutant lines. The fungus formed appressoria along the longitudinal axis at the surface of nup85 roots. Although hyphal colonization was occasionally blocked at the epidermal surface, frequent successful entry between epidermal cells was observed (Figure 2C). Nevertheless, the fungus was unable to penetrate the outer cortical cells (Figure 2B). Instead, the hyphae swelled at the epidermis/cortex interface and did not progress further inside the root (Figures 2B and 2D). By contrast, wild-type plants were normally infected by G. intraradices. The fungus crossed the epidermis of wild-type roots and traversed the outer cortical cells, and this was followed by hyphal extension in the intercellular space of the cortex and arbuscule formation in the inner cortical root cells (Figure 2A).
Figure 2.
AM Colonization.
(A) Wild-type plant inoculated with G. intraradices. G. intraradices formed appressoria (Ap) and entered between epidermal (Ep) cells. The hyphae penetrated the cortical (C) cells. The arrowhead shows the entry point of the hyphae into plant cells. The hyphae spread into the intercellular space of the cortex and then penetrated into inner cortical cells, where arbuscules (Ar) formed.
(B) nup85-2 mutant inoculated with G. intraradices. The hyphae from appressoria could enter the epidermis but could not penetrate the outer cortical cells. The hypha (asterisk) has swollen at the epidermis/cortex interface.
(C) and (D) Different focal planes of the root surface of the nup85-3 mutant inoculated with G. intraradices: surface of epidermal cells (C) and cell layer of outer cortex (D). Fungus has formed an appressorium on the root surface and can enter between epidermal cells (arrowheads). However, the hypha has stopped its growth and swollen at the epidermis/cortex interface. The asterisk indicates the swelling structure. The photos are representative of observations of at least six plants from each plant line.
Bars = 50 μm.
nup85 Plants Are Temperature-Sensitive Mutants for Symbioses
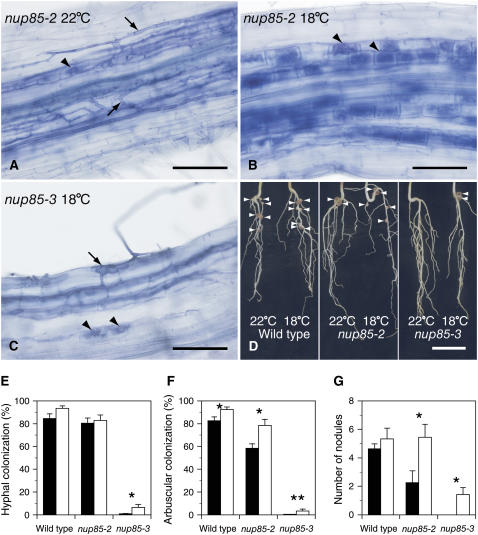
The symbiotic phenotypes in the roots of nup85 mutants seem to be affected by temperature. We observed the mycorrhization and nodulation of the nup85 mutants under precisely controlled temperatures of 22 ± 0.5°C or 18 ± 0.5°C. The nup85-2 mutant showing a weak symbiotic phenotype was highly colonized by the intraradical hyphae of G. intraradices at 22°C, whereas arbuscules in cortical cells were less frequently observed (Figure 3A). At 18°C, the nup85-2 mutant had significantly more arbuscules than at 22°C (Figures 3B and 3F). The nup85-3 mutant with a strong symbiotic phenotype also had significantly higher numbers of arbuscules at the lower growth temperature, even though the overall colonization level was very low (Figures 3E and 3F). At 18°C, the hyphae entered the nup85-3 epidermis and formed swollen structures at the interface of the epidermis and cortex; the hyphae emerging from these swollen structures spread into the cortex, and arbuscules occasionally formed in the inner cortical cells (Figure 3C). The number of nodules on the nup85-2 and nup85-3 mutants was also significantly increased at 18°C compared with at 22°C (Figures 3D and 3G), with the nup85-3 mutant showing no nodules at 22°C but some at 18°C.
Figure 3.
Temperature Effects on Mycorrhization and Nodulation.
(A) The nup85-2 mutant inoculated with G. intraradices and grown at 22°C. Hyphae (arrows) spread into an intercellular space of the cortex; however, few arbuscules (arrowhead) were formed in cortical cells. Bar = 100 μm.
(B) Mycorrhizal roots of the nup85-2 mutant grown at 18°C. A hypha spread into the intercellular space of the cortex and penetrated into inner cortical cells, where arbuscules (arrowheads) formed as infected in the wild type. Bar = 100 μm.
(C) nup85-3 mutant inoculated with G. intraradices and grown at 18°C. The hypha entered epidermis and formed swollen structures (arrow) at the epidermis/cortex interface. The hyphae emerging from the swollen structures penetrated to the outer cortical cells and spread into intercellular space of cortex. Arbuscules (arrowheads) formed in inner cortical cells. Bar = 100 μm.
(D) Nodule formation of the wild type, nup85-2, and nup85-3 mutants grown at 22°C and 18°C. The number of nodule varied at different temperatures. Arrowheads indicate visible nodules. Bar = 1 cm.
(E) to (G) Hyphal colonization (E), arbuscular colonization (F), and number of nodules (G) of the wild type, nup85-2, and nup85-3 mutants grown at 22°C (closed bars) and 18°C (open bars). Error bars indicate SE (n = 3). For assessing rates of mycorrhizal colonization, at least 100 points in the mycorrhizal roots were analyzed in each replicate. Asterisks show significant differences between 22°C and 18°C by t test: * P < 0.05 and ** P < 0.01.
NUP85 Is Predicted to Encode a Nucleoporin
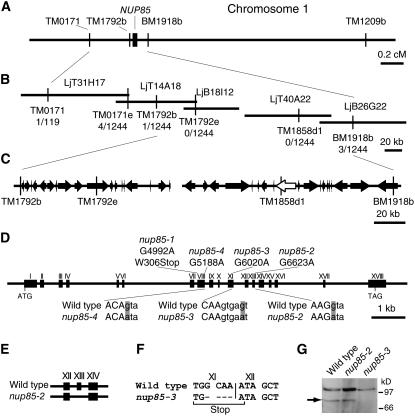
When the nup85-3 mutant was backcrossed to the parental line L. japonicus B-129 Gifu, 68 of 210 F2 plants tested did not form nodules, indicating that the strong non-nodulation phenotype segregated as a monogenic recessive trait (χ2 = 0.84, P > 0.05). To generate a mapping population, the nup85 mutants (B-129 Gifu background) were crossed with L. japonicus MG-20 Miyakojima. The resulting F2 progeny were scored using simple sequence length polymorphism (SSLP) markers. The NUP85 locus was located on the short arm of chromosome 1, where suppression of recombination occurs between Gifu and Miyakojima due to an exceptional translocation of the short arm of Gifu chromosome 1 to the bottom of chromosome 2 in Miyakojima (Hayashi et al., 2001). For fine mapping, Lotus burttii B-303 was used as an alternative crossing partner because this translocation is not present (Kawaguchi et al., 2005). Fine mapping revealed that the NUP85 locus was localized to the genetic interval flanked by the TM1792b and BM1918b markers (Figures 4A and 4B). This genetic region contains two physical contigs and was covered by four TAC/BAC clones (Figure 4B). Thirty-seven putative open reading frames were predicted in this region (Figure 4C), and sequence comparison of wild-type Gifu and the mutants revealed mutations in a gene encoding a nucleoporin-like protein (Figure 4D).
Figure 4.
Positional Cloning of the NUP85 Gene.
(A) Genetic map of the NUP85 gene on chromosome 1. Name and positions of the molecular markers are indicated.
(B) Physical map of the TAC/BAC clones. The names of the clones are indicated above the lines. The number of recombination events that occurred between the NUP85 locus and the molecular markers are indicated below the lines. Note that there is a gap between LjB18I12 and LjT40A22.
(C) Predicted open reading frames between molecular markers TM1792b and BM1918b. The region contains 37 open reading frames (arrows). The NUP85 gene is indicated as a white arrow.
(D) Representation of the exon and intron organization of the NUP85 gene. The gene consists of 18 exons (boxes) and 17 introns. The positions of the mutations in the nup85-1, -2, -3, and -4 alleles are indicated the line above. Roman numerals above the boxes indicate the exon order. nup85-1 has a nonsense mutation in exon VIII. The partial genomic sequence of the wild type, nup85-2, and nup85-3 are shown. Uppercase and lowercase characters indicate sequence in exons and introns, respectively. The positions of mutations are shaded in gray. nup85-2, -3, and -4 have a point mutation in the 13th, 11th, and eighth introns, respectively.
(E) Sequence analysis of NUP85 cDNA in the nup85-2 mutant. Boxes indicate transcribed sequences on the genomic sequence. The mutation in the nup85-2 mutant results in a deletion of exon XIII in the transcript.
(F) Sequence comparison between NUP85 cDNAs of the wild type and nup85-3 mutant. Roman numerals indicate the exon order. The transcript of the nup85-3 mutant has a four-base deletion, which results in a premature stop codon.
(G) The wild type, nup85-2, and nup85-3 were subjected to immunoblot analyses with anti-NUP85 antibodies. Molecular masses are given at the right in kilodaltons. Arrow indicates predicted molecular mass of NUP85.
To confirm that the nucleoporin-like gene is responsible for the observed mutant symbiotic phenotypes, the full-length cDNA was introduced into the nup85-3 mutant using Agrobacterium rhizogenes–mediated hairy root transformation according to Kumagai and Kouchi (2003). The nup85-3 roots transformed with the full-length cDNA developed nodules (Figure 5A) but roots transformed with an empty vector did not form nodules (Figure 5B). Nodulation by the nup85-3 mutant was also complemented with a 16.6-kb genomic DNA fragment containing the nucleoporin-like gene and a predicted promoter (data not shown). The complementation tests demonstrate that the nucleoporin-like gene is required for nodule formation.
Figure 5.
Complementation of the nup85-3 Mutant with the Putative Nucleoporin-Like Gene in TAC Clone LjT40A22.
(A) A full-length cDNA of the putative nucleoporin-like gene was introduced into nup85-3 using A. rhizogenes–mediated hairy root transformation. GFP fluorescence indicates transformed hairy roots. Transformed roots developed nodules after inoculation with M. loti.
(B) Roots of nup85-3 were transformed with the empty vector pGUN. The transformed plants do not form nodules.
Bars = 2 mm.
The nucleoporin-like gene encodes a protein of 712 residues with a predicted mass of 79.6 kD. The conceptual protein showed the highest level of similarity with a putative nucleoporin gene of Arabidopsis thaliana (At4g32910; 64.7% identity and 79.4% similarity). In addition, it also showed similarity along the entire sequence to human Nup75 (19.5% identity and 35.2% similarity) (Figure 6A), which is homologous to the Saccharomyces cerevisiae and vertebrate Nup85 protein (Cronshaw et al., 2002). Nucleoporins constitute the components of a nuclear pore complex, consisting of >30 nucleoporins and other elements (Rout et al., 2000; Cronshaw et al., 2002; Suntharalingam and Wente, 2003). Nearly two-thirds of the nucleoporins reveal some level of similarity between yeast and vertebrates, although for others, the amino acid sequences are not well conserved (Suntharalingam and Wente, 2003). For example, identity between yeast Nup133 and human Nup133 is only 18% in a limited ∼360–amino acid residue region (Vasu et al., 2001). The Nup85 homologs from Arabidopsis, Caenorhabditis elegans, Drosophila melanogaster, Oryza sativa, Schizosaccharomyces pombe, Xenopus, and human have sizes ranging from 68 to 85 kD (Harel et al., 2003). Because of the sequence similarity, we refer to the L. japonicus nucleoporin-like protein as NUP85.
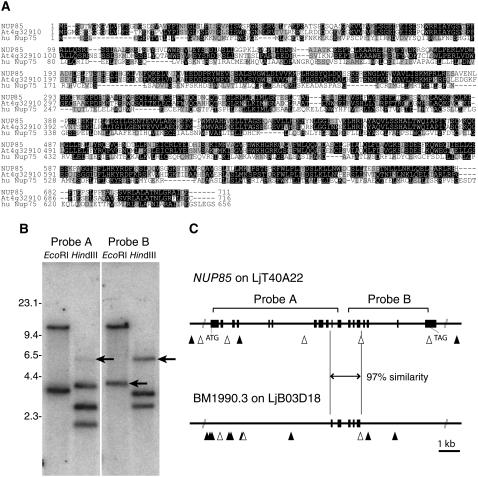
Figure 6.
Amino Acid Sequence Alignment and DNA Hybridization Analysis of NUP85.
(A) Amino acid sequence alignment of L. japonicus NUP85, Arabidopsis NUP85-like gene (At4g32910), and human nucleoporin 75 (huNup75). Identical and similar amino acids are shaded in black and gray, respectively. DNA hybridization analysis of L. japonicus Gifu genomic DNA hybridized with NUP85 probes.
(B) The DNA was digested with EcoRI or HindIII. NUP85 probes A and B recognize the 5′ and 3′ regions of the NUP85 gene, respectively. Arrows indicate extra fragments that were not predicted from the genomic sequence of the NUP85 gene.
(C) Genomic sequences of the NUP85 gene and a partial gene (BM1990.3) on clone LjB03D18. The region indicated by the arrow is highly conserved between the genes. Regions of genomic sequence hybridized with probes A and B were indicated. Filled boxes indicate predicted exons. Closed and open triangles show EcoRI and HindIII sites, respectively.
The L. japonicus NUP85 gene consists of 18 exons and 17 introns (Figure 4D). nup85-1 has a non-sense mutation (TGG [Trp] to TGA [Stop]) in the eighth exon. nup85-2 has a mutation at the 5′ intron acceptor site at the junction between the 13th exon and 13th intron, resulting in a deletion of the 13th exon in the mRNA as determined by sequencing the RT-PCR product (Figure 4E). nup85-3 carries a mutation at the putative 5′ splice site at the junction between the 11th exon and 11th intron, resulting in a deletion of four nucleotides at the 3′ end of the 11th exon in the cDNA (Figure 4F). This causes a premature stop codon at 1414 nucleotides from the translation initiation site. The nup85-4 allele (Murray et al., 2006) has a mutation at the 5′ intron acceptor site at the junction between the eighth exon and eighth intron (Figure 4D), which could affect splicing of the NUP85 gene. We raised anti-NUP85 antibody that binds to the C terminus of NUP85 protein. Immunoblots showed that NUP85 protein was detected in extracts of the wild type and nup85-2, but not nup85-3 (Figure 4G). The predicted size difference of NUP85 from the wild type and the nup85-2 mutant was probably not distinguishable by SDS-PAGE because the difference is only 2.3 kD.
A Highly Conserved Partial Sequence of NUP85 Exists in the L. japonicus Genome
Two cDNA probes of L. japonicus NUP85 were hybridized with genomic DNA of L. japonicus Gifu digested with EcoRI or HindIII (Figure 6B). The DNA bands detected by probe A (5′ region of NUP85 gene) and probe B (3′ region of NUP85 gene) corresponded with the fragments predicted from the genomic sequence of NUP85. Additional bands clearly distinguishable from the NUP85 gene were also detected, indicating other conserved NUP85 sequences in the genome of L. japonicus. Analysis of the available genomic sequence identified a highly conserved sequence (97% sequence similarity) corresponding to the genome sequence of NUP85 from +5517 to +6918 (taking the translation initiation site as +1) on the clone LjB03D18 (Figure 6C). This highly conserved sequence was predicted as a partial gene, in which candidates for start and stop codons were not predicted. The genome sequences in the vicinity of NUP85 exhibited no similarity to LjB03D18. This partial gene might have been generated by gene duplication in the partial sequence of NUP85.
NUP85 Is Expressed Constitutively
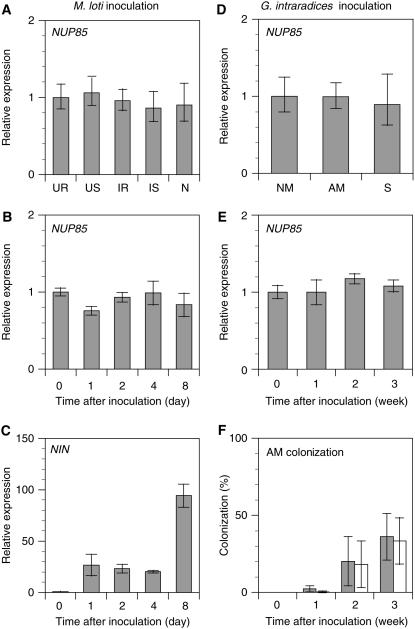
The expression level of NUP85 was examined by RT-PCR (Figure 7). NUP85 was expressed at similar levels in all organs tested (Figures 7A and 7D). M. loti inoculation did not affect the steady state level of NUP85 mRNA in infected roots, whereas the NIN gene was strongly upregulated (Figures 7B and 7C). G. intraradices inoculation also did not affect NUP85 expression in mycorrhizal roots (Figures 7E and 7F). Expression levels of symbiotic genes, including NUP85, NFR1 (Radutoiu et al., 2003), NFR5 (Madsen et al., 2003), SymRK (Stracke et al., 2002), CASTOR (Imaizumi-Anraku et al., 2005), POLLUX (Imaizumi-Anraku et al., 2005), CCaMK (Tirichine et al., 2006), and NIN (Schauser et al., 1999), were analyzed in nup85-3 mutants (Table 2). The abundance of NUP85 transcript was reduced in nup85-3 compared with the wild type, whereas the steady state level of other symbiotic transcripts was not different from that seen in the wild type.
Figure 7.
Gene Expression.
(A) to (C) M. loti inoculation.
(A) Expression of NUP85 in uninfected roots (UR), shoots of uninoculated plants (US), infected roots (IR), shoots of inoculated plants (IS), and nodules (N) at 3 weeks after inoculation treatment. Expression levels are normalized on the basis of the amount of EF-1 and expressed relative to uninfected roots.
(B) and (C) Expression of NUP85 and NIN from 0 to 8 d following inoculation. Expression levels are expressed relative to day 0.
(D) to (F) G. intraradices inoculation.
(D) Expression of NUP85 in nonmycorrhizal roots (NM), mycorrhizal roots (AM), and shoots of mycorrhizal plants (S) at 3 weeks after inoculation treatment. Expression levels are normalized on the basis of the amount of EF-1 and expressed relative to nonmycorrhizal roots.
(E) Expression of NUP85 from 0 to 3 weeks following inoculation. Expression levels are expressed relative to day 0.
(F) AM colonization. Gray and white bars show hyphal colonization (HC%) and arbuscular colonization (AC%), respectively. Values are the means of three experiments of different samples. sd is indicated.
Table 2.
Symbiotic Gene Expression in Uninoculated Roots of the nup85-3 Mutant
| Gene | Relative Expression (nup85-3/Wild Type)a |
|---|---|
| NUP85b | 0.20 ± 0.05 |
| NFR1 | 1.00 ± 0.10 |
| NFR5 | 0.93 ± 0.09 |
| SymRK | 0.84 ± 0.12 |
| CASTOR | 1.11 ± 0.21 |
| POLLUX | 1.39 ± 0.36 |
| CCaMK | 1.07 ± 0.14 |
| NIN | 1.06 ± 0.39 |
Expression levels are normalized on the basis of the amount of EF-1 and shown relative to the wild type. Mean ± sd (n = 3).
Ratio of the expression of NUP85 to EF-1 was 0.06 in the wild-type plants.
nup85 Does Not Arrest Plant Growth but Does Reduce Seed Yield
To ascertain whether the mutations of NUP85 affect physiological functions of L. japonicus as well as symbiotic associations, we examined the vegetative and reproductive growth of the nup85 mutants. The growth of the nup85-2 and nup85-3 mutants was analyzed under uninoculated conditions by growing the plants aseptically in a plastic container with 5 mM KNO3 added to the BandD solution (Figure 8A). Shoot and root growth of the nup85-2 mutant was slightly slower than that of both the wild type and the nup85-3 mutant, but analysis of variance results confirmed that there was no significant effect of plant line (F = 2.55; P = 0.095) on shoot dry weight or an interaction between plant line and sampling time (F = 0.51; P = 0.729), indicating no difference in growth between plant lines during 6 weeks. With M. loti–inoculated plants grown on nitrogen-free BandD solution, the shoot growth of nup85-2 and nup85-3 mutants was less than that of the wild type, whereas there was no significant effect on root growth (Figure 8B). When grown in a complete nutrient-rich medium, the seedpod length of the nup85-3 mutant was significantly shorter than that of the wild type and the nup85-2 mutant (Figure 8C), whereas average plant lengths and leaf sizes were not significantly different among plant lines (data not shown). The number of seeds per pod in both nup85-2 and nup85-3 mutants was significantly decreased compared with the wild type (Figure 8D). Notably, the number of seeds in the nup85-3 mutant was significantly lower than in the nup85-2 mutant (Figure 8D). Seed weight of the nup85-2 mutant was significantly greater than that of the wild type, whereas that of nup85-3 was not different from the wild type (Figure 8E). In vitro growth rates of pollen tubes in nup85 mutants were markedly reduced compared with the wild type (Figure 8F).
Figure 8.
Plant Growth and Seed Production.
(A) Dry weight of shoots and roots in the wild type (closed circles), nup85-2 (open squares), and nup85-3 (open triangles) mutants supplied with BandD solution containing 5 mM KNO3, without M. loti inoculation.
(B) Dry weight of the plants inoculated with M. loti and supplied with nitrogen-free BandD solution. Error bars show se of the means (n = 3).
(C) Pod length of the wild type, nup85-2, and nup85-3 grown in a complete nutrient-rich medium.
(D) Number of mature seeds per pod.
(E) Seed weight. The data presented are means ± se (n = 37 to 40). Different letters above bars indicate significance among the means according to the Tukey multiple comparison test (P < 0.05).
(F) Length of pollen tubes in the wild type (closed circles), nup85-2 (open squares), and nup85-3 (open triangles) at 1 and 2 h after pollen germination. At least 13 pollen tubes were observed. Error bars show se.
DISCUSSION
The L. japonicus nucleoporin gene NUP85 was identified as a common Sym gene. Nucleoporins are components of the nuclear pore complex, consisting of >30 nucleoporins and other elements (Rout et al., 2000; Cronshaw et al., 2002; Suntharalingam and Wente, 2003). The nuclear pore complex mediates macromolecular transport, such as mRNA export and protein import across the nuclear envelope. In vertebrates, Nup85 is part of the Nup107-160 subcomplex, which constitutes a central part of the nuclear pore complex, including Nup107, Nup160, Nup133, Nup85, Nup96, Sec13, Nup43, Nup37, and Seh1 (Walther et al., 2003). The vertebrate Nup107-160 complex resides on both sides of the central framework of the nuclear pore complex and represents the core element of the central framework (Lim and Fahrenkrog, 2006). Recently, L. japonicus NUP133, a likely component of the Nup107-160 subcomplex, was also shown to be a common Sym gene required for AM and nodulation (Kanamori et al., 2006). In Arabidopsis, Nup96 has been postulated to be involved in basic defense and constitutive resistance responses to pathogens mediated by a resistance gene (R gene) (Zhang and Li, 2005). In addition, mouse Nup96 has been shown to be involved in antiviral functions (Faria et al., 2006). These data indicate that the Nup107-160 subcomplex containing NUP85, NUP133, and NUP96 is an important component in not only plant–microbe interactions but also in animal–microbe interactions.
Nup85p of yeast has several cellular functions, such as mRNA export (Goldstein et al., 1996), nuclear pore complex assembly (Siniossoglou et al., 2000), and localization of the small GTPase Gsp1p (Gao et al., 2003). Such functions of yeast Nup85p seem to be related to general phenomena. In addition, L. japonicus NUP85 constitutively expressed at similar levels in all organs and was not up- or downregulated by Nod factor application. These results led us to speculate that mutations in NUP85 could affect overall plant growth. However, NUP85 appears to be mainly involved in symbiotic functions and seed yield. In Drosophila, a mutation in Nup88 did not affect mRNA export or global protein import but selectively impaired import of a Rel family protein, which is a transcription factor required for innate immune responses via the Toll-like receptors (Uv et al., 2000). The authors pointed out that selective protein import depends on individual nucleoporins. Recently, the yeast Nup84 subcomplex, functionally equivalent to the vertebrate Nup107-160 subcomplex, has been reported to be functionally and biochemically linked to the Rap1/Gcr1/Gcr2 transcriptional activation assemblage, during which gene regulation could occur at the nuclear periphery (Menon et al., 2005). Transcription factor–like proteins involved in nodulation, such as NSP1 (Smit et al., 2005), NSP2 of M. truncatula (Kaló et al., 2005), and NIN of L. japonicus (Schauser et al., 1999), have been identified. Nod factor treatment results in release of NSP2 from the nuclear periphery to the nucleoplasm (Kaló et al., 2005). It will be of interest to determine whether L. japonicus NUP85 and NUP133 are involved in this process.
NUP85 as well as SymRK, CASTOR, POLLUX, NUP133, NFR1, and NFR5 are required for the induction of calcium spiking in L. japonicus root hairs (Imaizumi-Anraku et al., 2005; Kanamori et al., 2006; Miwa et al., 2006); however, the underlying molecular mechanism is presently unknown. A possible role for a nuclear pore in calcium spiking might be to act as a gate for calcium ions or as a factor interacting with calcium channels. However, calcium ions are thought to be freely translocated via nuclear pores (Gerasimenko and Gerasimenko, 2004). A structural study of Xenopus nuclear pores by atomic force microscopy has demonstrated calcium-mediated opening and closing of nuclear baskets, suggesting that the distal ring of the basket acts as a calcium-sensitive iris-like diaphragm (Stoffler et al., 1999). However, there is no evidence that the structural changes actually affect the calcium permeability of nuclear pores. Observations of the structural changes of nuclear pores after treatment with Nod factor and measurement of nuclear pore size in nup85 and nup133 mutants are required to clarify the role of symbiotic NUP proteins in calcium spiking.
Despite the fact that Nod factor failed to induce calcium spiking in the nup85 mutants, there was clear rhizobial infection through infection threads, albeit at a lower level, as well as subsequent nodule formation. This could be explained by the decrease in responsible cells or a gradual increase in nuclear calcium. As has been proposed by Miwa et al. (2006), infection thread formation requires two distinct signaling steps: (1) morphological change in root hair cells that functions in entrapment of rhizobia at the root hair tip, and (2) calcium signaling and subsequent induction of ENOD genes associated with infection threads (Scheres et al., 1990; Journet et al., 2001). One possibility is that a transient but not repetitive calcium increase is enough to activate the activity of CCaMK, which is responsible for the subsequent ENOD gene expression and nodule formation (Gleason et al., 2006; Tirichine et al., 2006). Alternatively, there is a possibility that calcium spiking might be induced much later than the wild type beyond the time-course measurement of the calcium spiking assay, as nodule formation was delayed in nup85 mutants.
The nup85 mutants showed temperature-dependent symbiotic phenotypes, and the same was observed in the nup133 nucleoporin mutant of L. japonicus (Kanamori et al., 2006). In yeast, mutants defective in some nucleoporin genes are known to have temperature-sensitive growth (Wente and Blobel, 1993; Li et al., 1995; Goldstein et al., 1996). The underlying molecular mechanisms of the temperature-sensitive phenotype is unknown even in yeast, but there is evidence based on time-lapse atomic force microscopy that temperature-dependent plugging and unplugging of nuclear pores can occur in Xenopus (Stoffler et al., 2003). The temperature-dependent plasticity of nuclear pore plugging may be related to temperature-sensitive symbiotic phenotypes in nup85. In addition to the effects of mutation in NUP85 on the temperature-dependent phenotypes, temperature sensitivity of AM fungi might also be involved because arbuscule colonization of the wild-type plant was affected by growth temperature. Several studies have shown that there was a positive correlation between AM colonization and temperature (Heinemeyer and Fitter, 2004; Gavito et al., 2005), although our results show the opposite relationship.
In a mutant carrying the weak allele nup85-2, checkpoints for mycorrhizal development were dependent on growth temperature. The first checkpoint was entry in the outer cortical cells because fungal colonization was blocked at the epidermal-cortical interface of the nup85-2 mutant grown at 24°C. At 22°C, some hyphae could penetrate the outer cortical cells, which was followed by growth of hyphae between cortical cells. However, arbuscules within inner cortical cells were only formed at low frequency. This indicates that the second checkpoint is at the entry within inner cortical cells. At 18°C, arbuscules were formed normally within inner cortical cells. These results suggest that NUP85 helps with an accommodation of the hyphae within cortical cells. Novero et al. (2002) demonstrated that CASTOR is required not only for fungal entry into epidermal and outer cortical cells but also for arbuscule formation in inner cortical cells through analysis of later colonization stages of a weak castor-1 allele (previously Ljsym4-1). CASTOR and NUP85 are thought to function as mediators of fungal entry in plant cells.
We reported that in addition to defects in symbiotic interactions, nup85 mutants decreased seed yield. A similar seed yield decrease was seen with the nup133 mutants (Kanamori et al., 2006). Interestingly in the case of the nup85 mutants, pollen tube growth was markedly reduced (Figure 8F). Defects in male gametophyte development during fertilization as well as seriously reduced seed production are similar to that seen in the L. japonicus crinkle mutant (Tansengco et al., 2004). Careful characterization of male gametophyte development in the nup and crinkle mutants in the future may provide new insights into the molecular basis of plant reproductive biology.
METHODS
Biological Materials
Lotus japonicus B-129 Gifu was used as a wild type. nup85-1 (Ljsym24), nup85-2 (Ljsym73), and nup85-3 (Ljsym85) are alleles affecting AM and nodule symbioses in mutants isolated from ethyl methanesulfonate–mutagenized L. japonicus B-129 (Szczyglowski et al., 1998; Kawaguchi et al., 2002, 2005; Sandal et al., 2006). For allelism test, plants were crossed reciprocally according to Jiang and Gresshoff (1997). A possible allelic mutant, B46-D (Murray et al., 2006), is assigned as nup85-4. L. japonicus MG-20 Miyakojima and Lotus burttii B-303 were used for the positional cloning as a crossing partner. L. japonicus was inoculated with Mesorhizobium loti MAFF 303099, M. loti NZP2235 expressing lacZ reporter gene, or AM fungus Glomus intraradices DAOM 197198 (Mycorise ASP; Premier Tech). For hairy root–based transformation of L. japonicus, Agrobacterium rhizogenes LBA1334 (Visser et al., 1989) carrying a binary vector was used.
Root Hair Deformation Assay
Seeds of the wild type, nup85-2, and nup85-3 were germinated on 1% agar with half-strength Broughton and Dilworth (B&D) solution (Broughton and Dilworth, 1971) containing 0.1 μM l-α-(2-aminoethoxy vinyl)glycine (Sigma-Aldrich). Plates were wrapped with aluminum foil and placed in an incubator for 2 d in the dark. After 24 h, seedlings with abnormal growth were removed and plates were placed in an incubator for another 24 h. The seedlings, which then had an average 7-mm taproot length, were used for analysis. Nod factor was purified from M. loti harboring pMP2112 according to the method described by Niwa et al. (2001) and dissolved in distilled water (10−6 M). The nod factor was diluted to 10−8 M with half strength strength of B&D solution and was gently applied to 2-d-old seedlings to avoid disturbing root hairs. One half strength of B&D solution was applied as control. Seedlings were collected 24 h after treatment. Samples were fixed for 15 min with 2% paraformaldehyde dissolved in half-strength B&D solution. Following gentle mount on a glass slide (76 × 26 mm; Matsunami Glass Ind.) and covering with a cover glass (18 × 18 mm; Matsunami Glass Ind.), samples were examined by a bright-field microscope (BX-50; Olympus). Images were captured with a CCD camera (DP70; Olympus).
Infection Thread Formation
To visualize infection threads, roots were inoculated with M. loti NZP2235 expressing the lacZ reporter gene. Seeds of the wild type, nup85-2, and nup85-3 were sown in autoclaved vermiculite (Tekunon) supplied with B&D solution containing 0.5 mM KNO3. Seedlings (7 d old) were inoculated with the M. loti strains at a density of 108 cells mL−1. Five days later, infection threads were visualized by staining for β-galactosidase activity, as described by Tansengco et al. (2003). Infection threads were observed using an Olympus BX51 microscope under bright-field illumination. Significant differences of number of infection threads between the plant lines were analyzed with the Turkey's multiple comparison test (P < 0.05) using JMP (SAS).
Calcium Spiking
Seedlings were mounted and microinjected with Oregon green dextran dye and Texas red dextran as described (Harris et al., 2003; Miwa et al., 2006). Fluorescence was imaged using an inverted microscope coupled to a digital CCD camera. After microinjection, root hairs were left at least 20 min before Nod factor addition, and only cells showing active cytoplasmic streaming were used for the analysis. Nod factor was added directly to an incubation chamber to give an estimated final concentration of 10−8 M.
AM Colonization
L. japonicus plants were inoculated with 500 spores of G. intraradices per plant in a mixture (5:4:1 by volume) of autoclaved humus-rich Ando soil, sand, and a commercial horticulture medium (Kureha). The plants were grown in a growth cabinet at 24 ± 2°C with 16 h of light and 8 h dark. Harvested roots were cleared with 10% KOH and stained with 0.05% trypan blue using a phenol-free modification of the method of Phillips and Hayman (1970). Hyphal (HC%), arbuscular (AC%), and vesicular colonization (VC%) was determined as the percentage of root length colonization using a magnified intersection method at ×200 (McGonigle et al., 1990). Trypan blue–stained structures were observed under a bright-field microscope (BX51; Olympus), and images were acquired using a CCD camera (DP70; Olympus).
Experiments of Temperature Effect on Mycorrhization and Nodulation
The wild type, nup85-2, and nup85-3 were inoculated with spores of G. intraradices in the soil mixture described or M. loti in autoclaved vermiculite (Tekunon) supplied with nitrogen-free BandD solution. The plants were grown in growth cabinets at 22 ± 0.5°C or 18 ± 0.5°C for 1 month. The arcsine-transformed mycorrhizal colonization (HC% and AC%) and the number of nodules were compared with growth temperatures using t test of JMP (SAS) for each plant line.
Positional Cloning
NUP85 was mapped with three independent F2 mapping populations, resulting from a cross of nup85-1 and MG-20 Miyakojima and crosses between nup85-2 or nup85-3 and L. burttii. The genomic DNA was extracted from leaves and analyzed by SSLP markers and derived cleaved amplified polymorphic sequence (dCAPS) markers. Primer sequences used for molecular markers were as follows: SSLP markers, TM0171 (5′-ATCTCCCTCAGTCACGTTTC-3′ and 5′-GGTTTATTTTCCTGATTTGG-3′), TM0171e (5′-GCATTTCGAGCCCATAAAAA-3′ and 5′-TTTTTAGACGTTGGCAACAATTT-3′), TM1792b (5′-CACAATTTTCGGGGGTTATG-3′ and 5′-CCTTGGTTAAGGCTCGATGT-3′), TM1792e (5′-AGCCAGGTCAGTAGGAAGACC-3′ and 5′-ATTTTGCAGCGGAGGAAGTA-3′), BM1918b (5′-GTCGCTAGGTTGAGGCTTTG-3′ and 5′-TCCAATTCAAAAACCACATCA-3′), TM1209b (5′-CCACAAGCTGGTATCACACG-3′ and 5′-TCTCACAATAAGACACTCCTCTCA-3′); dCAPS marker, TM1858d1 (5′-GCACAATGACTTGCGAAGAA-3′ and 5′-TTCCGTTGGAACTACGTCAA-3′) (restriction enzyme: HincII). Sequences of BAC/TAC clones were analyzed by GENSCAN version 1.0 (http://genes.mit.edu/GENSCAN.html) and BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). ClustalW was used for multiple alignments.
Complementation Tests
For complementation of the nup85-3 mutation, full-length cDNA was amplified from cDNA clone MFB015g09 (Asamizu et al., 2004) and cloned into pGUN. The pGUN is a derivative of pCAMBIA1300 (CAMBIA). The hygromycin phosphotransferase gene of pCAMBIA under the control of the 35S promoter was replaced with sGFP (S65T; Niwa et al., 1999), and then the promoter region of Lotus polyubiquitin gene was inserted in the multiple cloning site of pCAMBIA, with a new multiple cloning site and a NOS terminator from pBI121 to confer strong and stable expression of a gene (M. Hayashi, unpublished results). Hairy roots were transformed using A. rhizogenes according to Kumagai and Kouchi (2003). The transformed plants were inoculated with M. loti and grown in growth cabinets at 22°C. Transformed hairy roots were selected by observing GFP expression using epifluorescent stereomicroscopy and confirmed nodule formation after M. loti inoculation.
DNA Hybridization
Two probes, NUP85 probes A and B, were prepared for DNA hybridization. Probe A and probe B recognized the 5′ (84 to 1314) and 3′ (1445 to 2073) regions of the coding sequence, respectively (Figure 6C). A fragment of NUP85 was amplified with the primers 5′-CCATGGCCTCTCCCTTCCAATTTCT-3′ and 5′-CTCTGCTTGCTCACTCCCTGCAGTC-3′ for probe A and with 5′-GCATGAAGCAAGGAATGGGCTTGTT-3′ and 5′-TTCAGGGGGTAAGCTTGGCTCAGTG-3′ for probe B from the cDNA clone MFB015g09. The fragments were labeled with [α-32P]dCTP using Megaprime (Amersham) following the manufacturer's protocols. Genomic DNA was extracted from leaves of L. japonicus Gifu digested with EcoRI or HindIII, transferred to nylon membrane, and hybridized with the probe. The membrane was exposed to an imaging plate (Fujifilm), and signals were detected with FLA-2000 (Fujifilm).
Immunoblot Analysis
To prepare anti-NUP85 antibody, a peptide derived from the C terminus of the NUP85 protein, CTNLGRAILDE, was chosen as an immunogen by Sigma-Aldrich Japan's in-house software. Sigma-Aldrich Japan synthesized the peptide, immunized two rabbits by injecting the immunogen every 2 weeks over 2 months, and prepared antisera by standard protocols. Antibody titer of a serum against the peptide was evaluated by an enzyme-linked immunosorbent assay. An anti-NUP85 antibody was purified from a serum showing higher titer by peptide affinity chromatography. Wild-type, nup85-2, and nup85-3 roots were homogenized in liquid nitrogen with a pestle. Cell lysates were extracted in 50 mM Tris-HCl, pH 6.8, 6% 2-mercaptoethanol, 10% glycerol, 2% sodium dodecyl sulfate, and 0.005% bromphenol blue. Proteins in extracts were resolved by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Millipore) and then reacted with anti-NUP85 antiserum followed by horseradish peroxidase–conjugated anti-rabbit IgG antibody (Jackson ImmunoResearch Laboratories). Immunodetection was performed with Immobilon Western (Millipore). Chemiluminescence was detected with an imaging system (LAS-1000 plus; Fujifilm).
Gene Expression Analyses
For the analyses of gene expression in M. loti–inoculated plants, 3-d-old seedlings of L. japonicus Gifu were grown on 1% agar medium (BandD containing 10 μM KNO3) for 2 d. The seedlings were inoculated with M. loti (108 cells per plate). For the analysis of AM plants, L. japonicus Gifu were inoculated with 500 spores of G. intraradices per plant in autoclaved vermiculite (Asahikogyo) supplied with modified BandD solution containing 5 mM KNO3 and 50 μM KH2PO4. For expression analysis of genes in the wild type and the nup85-3 mutant, seedlings were grown on 1% agar plates supplied with BandD solution and 10 μM KNO3 for 2 d, and total RNA was extracted from roots. Total RNA was extracted with the RNeasy Plant Mini kit (Qiagen) and treated with DNase I (Takara Biotechnology). Levels of expression were analyzed by semiquantitative RT-PCR with the primers NUP85 (5′-CAATTGCCACGAAATGTGAG-3′ and 5′-TCAGGTAGCCATGCTTGAGA-3′), elongation factor-1 (EF-1) (5′-GCAGGTCTTTGTGTCAAGTCTT-3′ and 5′-CGATCCAGAACCCAGTTCT-3′), NIN (5′-TGGATCAGCTAGCATGGAAT-3′ and 5′-TCTGCTTCTGCTGTTGTCAC-3′), NFR1 (5′-TGTTGCTCTTATGACCCTTTCA-3′ and 5′-GCATTTGCATGGAGAACCTT-3′), NFR5 (5′-CACGTTAAGCAAGGGAAGGT-3′ and 5′-TTCAATTCCAGTGTCTGACAAA-3′), SymRK (5′-CACAAGGGAACGGTTGAACT-3′ and 5′-AACTTGAAGCCATGCCAACT-3′), CASTOR (5′-ATGGTGGCCTTGACATAAG-3′ and 5′-AGTGACGACGTATAACAGCA-3′), POLLUX (5′-TTAGCGAAATTTTGGATTCT-3′ and 5′-CTAGTGCCATGCTTACCAGT-3′), and CCaMK (5′-GGAGACAATGCAACTCTGTCTGA-3′ and 5′-CGGTGCTAGAGGGATCAATGAG-3′). Semiquantitative RT-PCR was performed using ABI GeneAmp 5700 or ABI Prism 7000 (Applied Biosystems) with a Quantitect SYBR Green RT-PCR kit (Qiagen). Expression levels were normalized on the basis of EF-1 quantity.
Growth Analysis under Uninoculated and M. loti–Inoculated Conditions
Seeds of the wild type, nup85-2, and nup85-3 were treated with sulfuric acid and sown in autoclaved vermiculite (Tekunon). For the growth analysis under an M. loti–inoculated condition, the plants were supplied with nitrogen-free BandD solution and inoculated with M. loti. The plants under an uninoculated condition were supplied with BandD solution containing 5 mM KNO3. The plants were grown in a growth chamber at 22°C, harvested at 2, 4, and 6 weeks after sowing, and cut into shoot and root. These parts were dried separately at 65°C for 48 h and then weighed. To test the differences of the growth among the plant lines, the dry weight transformed as log (Y + 1) was analyzed with two-way analysis of variance of the effects of plant lines, sampling days and their interaction by JMP (SAS). Number of seeds per pod, seed weight, and pod length of plants growing in a nutrient-rich horticulture soil (Kureha) in a greenhouse were measured. Significant differences between means of the plant line were analyzed with Turkey's multiple comparison test (P < 0.05) using JMP (SAS). Pollen grains were collected from fully opened flowers and put on a thin layer of pollen germination medium prepared on a microscope slide. We used the following germination medium described by Tansengco et al. (2004) with some modifications: 10% sucrose, 0.01% H3BO3, 0.05% Ca(NO3)2·4H2O, and 0.7% Bacto agar. Samples were incubated in Petri dishes with moist filter papers and incubated for 1 or 2 h at 22°C. Pollen tube growth was examined at different time intervals after germination. At least 13 pollen tubes were chosen randomly for pollen tube length measurements.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL/DDBJ data libraries under accession numbers AB284835, AP009251, AP009252, AP009253, AP009254, and AP009255.
Acknowledgments
We thank Giles Oldroyd and Jongho Sun (Sainsbury Laboratory) for help with establishing and using microinjection and epifluorescence microscopy. We also thank Jodie Pike and Kate Vickers (Sainsbury Laboratory), Tomomi Nakagawa (National Institute of Agrobiological Sciences), Asuka Kuwabara (University of Tokyo), and Jeremy Murray (Agriculture and Agri-Food Canada) for experimental supports. The work was supported by the Biotechnology and Biological Science Research Council for J.A.D., the Core Research for Evolutional Science and Technology of Japan Science and Technology Agency, Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan for M.K., and the Kazusa DNA Research Foundation. H.M. was supported by a John Innes Foundation Studentship and by an award from the Universities UK, Overseas Research Students Awards Scheme. Research at the Sainsbury Laboratory is funded by the Gatsby Charitable Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Masayoshi Kawaguchi (masayosi@biol.s.u-tokyo.ac.jp).
References
- Ané, J.-M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367. [DOI] [PubMed] [Google Scholar]
- Asamizu, E., Nakamura, Y., Sato, S., and Tabata, S. (2004). Characteristics of the Lotus japonicus gene repertoire deduced from large-scale expressed sequence tag (EST) analysis. Plant Mol. Biol. 54 405–414. [DOI] [PubMed] [Google Scholar]
- Bonfante, P., Genre, A., Faccio, A., Martini, I., Schauser, L., Stougaard, J., Webb, J., and Parniske, M. (2000). The Lotus japonicus LjSym4 gene is required for the successful symbiotic infection of root epidermal cells. Mol. Plant Microbe Interact. 13 1109–1120. [DOI] [PubMed] [Google Scholar]
- Broughton, W.J., and Dilworth, M.J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas, L., Feijó, J.A., Kunkel, J.G., Sánchez, F., Holdaway-Clarke, T., Hepler, P.K., and Quinto, C. (1999). Rhizobium Nod factors induce increases in intracellular free calcium and extracellular calcium influxes in bean root hairs. Plant J. 19 347–352. [DOI] [PubMed] [Google Scholar]
- Cronshaw, J.M., Krutchinsky, A.N., Zhang, W., Chait, B.T., and Matunis, M.J. (2002). Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko, K., Winzer, T., Stougaard, J., Parniske, M., and Pawlowski, K. (2004). Distinct roles of Lotus japonicus SYMRK and SYM15 in root colonization and arbuscule formation. New Phytol. 163 381–392. [DOI] [PubMed] [Google Scholar]
- Dénarié, J., Debellé, F., and Promé, J.-C. (1996). Rhizobium lipo-chitooligosaccharide nodulation factors: Signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65 503–535. [DOI] [PubMed] [Google Scholar]
- Duc, G., Trouvelot, A., Gianinazzi-Pearson, V., and Gianinazzi, S. (1989). First report of non-mycorrhizal plant mutants (myc−) obtained in pea (Pisum sativum L.) and fababean (Vicia faba L.). Plant Sci. 60 215–222. [Google Scholar]
- Ehrhardt, D.W., Wais, R., and Long, S.R. (1996). Calcium spiking in plant root hairs responding to rhizobium nodulation signals. Cell 85 673–681. [DOI] [PubMed] [Google Scholar]
- Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kaló, P., and Kiss, G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966. [DOI] [PubMed] [Google Scholar]
- Faria, A.M.C., Levay, A., Wang, Y., Kamphorst, A.O., Rosa, M.L.P., Nussenzveig, D.R., Balkan, W., Chook, Y.M., Levy, D.E., and Fontoura, B.M.A. (2006). The nucleoporin Nup96 is required for proper expression of interferon-regulated proteins and functions. Immunity 24 295–304. [DOI] [PubMed] [Google Scholar]
- Gao, H., Sumanaweera, N., Bailer, S.M., and Stochaj, U. (2003). Nuclear accumulation of the small GTPase Gsp1p depends on nucleoporins Nup133p, Rat2p/Nup120p, Nup85p, Nic96p, and the acetyl-CoA carboxylase Acc1p. J. Biol. Chem. 278 25331–25340. [DOI] [PubMed] [Google Scholar]
- Gavito, M.E., Olsson, P.A., Rouhier, H., Medina-Peñafiel, A., Jakobsen, I., Bago, A., and Azcón-Aguilar, C. (2005). Temperature constraints on the growth and functioning of root organ cultures with arbuscular mycorrhizal fungi. New Phytol. 168 179–188. [DOI] [PubMed] [Google Scholar]
- Genre, A., and Bonfante, P. (2002). Epidermal cells of a symbiosis-defective mutant of Lotus japonicus show altered cytoskeleton organisation in the presence of a mycorrhizal fungus. Protoplasma 219 43–50. [DOI] [PubMed] [Google Scholar]
- Gerasimenko, O., and Gerasimenko, J. (2004). New aspects of nuclear calcium signalling. J. Cell Sci. 117 3087–3094. [DOI] [PubMed] [Google Scholar]
- Gianinazzi-Pearson, V. (1996). Plant cell responses to arbuscular mycorrhizal fungi: Getting to the roots of the symbiosis. Plant Cell 8 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason, C., Chaudhuri, S., Yang, T., Muñoz, A., Poovaiah, B.W., and Oldroyd, G.E.D. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441 1149–1152. [DOI] [PubMed] [Google Scholar]
- Goldstein, A.L., Snay, C.A., Heath, C.V., and Cole, C.N. (1996). Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol. Biol. Cell 7 917–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel, A., Orjalo, A.V., Vincent, T., Lachish-Zalait, A., Vasu, S., Shah, S., Zimmerman, E., Elbaum, M., and Forbes, D.J. (2003). Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11 853–864. [DOI] [PubMed] [Google Scholar]
- Harris, J.M., Wais, R., and Long, S.R. (2003). Rhizobium-induced calcium spiking in Lotus japonicus. Mol. Plant Microbe Interact. 16 335–341. [DOI] [PubMed] [Google Scholar]
- Harrison, M.J. (1999). Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 361–389. [DOI] [PubMed] [Google Scholar]
- Hayashi, M., et al. (2001). Construction of a genetic linkage map of the model legume Lotus japonicus using an intraspecific F2 population. DNA Res. 8 301–310. [DOI] [PubMed] [Google Scholar]
- Heinemeyer, A., and Fitter, A.H. (2004). Impact of temperature on the arbuscular mycorrhizal (AM) symbiosis: Growth responses of the host plant and its AM fungal partner. J. Exp. Bot. 55 525–534. [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku, H., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433 527–531. [DOI] [PubMed] [Google Scholar]
- Jiang, Q., and Gresshoff, P.M. (1997). Classical and molecular genetics of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 10 59–68. [DOI] [PubMed] [Google Scholar]
- Journet, E.-P., El-Gachtouli, N., Vernoud, V., de Billy, F., Pichon, M., Dedieu, A., Arnould, C., Morandi, D., Barker, D.G., and Gianinazzi-Pearson, V. (2001). Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14 737–748. [DOI] [PubMed] [Google Scholar]
- Kaló, P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789. [DOI] [PubMed] [Google Scholar]
- Kanamori, N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi, M., Imaizumi-Anraku, H., Koiwa, H., Niwa, S., Ikuta, A., Syono, K., and Akao, S. (2002). Root, root hair, and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 15 17–26. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, M., et al. (2005). Lotus burttii takes a position of the third corner in the Lotus molecular genetics triangle. DNA Res. 12 69–77. [DOI] [PubMed] [Google Scholar]
- Kistner, C., and Parniske, M. (2002). Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci. 7 511–518. [DOI] [PubMed] [Google Scholar]
- Kistner, C., Winzer, T., Pitzschke, A., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Webb, J., Szczyglowski, K., and Parniske, M. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17 2217–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai, H., and Kouchi, H. (2003). Gene silencing by expression of hairpin RNA in Lotus japonicus roots and root nodules. Mol. Plant Microbe Interact. 16 663–668. [DOI] [PubMed] [Google Scholar]
- Lévy, J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364. [DOI] [PubMed] [Google Scholar]
- Li, O., Heath, C.V., Amberg, D.C., Dockendorff, T.C., Copeland, C.S., Snyder, M., and Cole, C.N. (1995). Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol. Biol. Cell 6 401–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, R.Y.H., and Fahrenkrog, B. (2006). The nuclear pore complex up close. Curr. Opin. Cell Biol. 18 342–347. [DOI] [PubMed] [Google Scholar]
- Madsen, E.B., Madsen, L.H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., Sato, S., Kaneko, T., Tabata, S., Sandal, N., and Stougaard, J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640. [DOI] [PubMed] [Google Scholar]
- McGonigle, T.P., Miller, M.H., Evans, D.G., Fairchild, G.L., and Swan, J.A. (1990). A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 115 495–501. [DOI] [PubMed] [Google Scholar]
- Menon, B.B., Sarma, N.J., Pasula, S., Deminoff, S.J., Willis, K.A., Barbara, K.E., Andrews, B., and Santangelo, G.M. (2005). Reverse recruitment: The Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. USA 102 5749–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, R.M., Gleason, C.A., Edwards, A., Hadfield, J., Downie, J.A., Oldroyd, G.E.D., and Long, S.R. (2004). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa, H., Sun, J., Oldroyd, G.E.D., and Downie, J.A. (2006). Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol. Plant Microbe Interact. 19 914–923. [DOI] [PubMed] [Google Scholar]
- Murray, J., et al. (2006). Genetic suppressors of the Lotus japonicus har1-1 hypernodulation phenotype. Mol. Plant Microbe Interact. 19 1082–1091. [DOI] [PubMed] [Google Scholar]
- Niwa, S., Kawaguchi, M., Imaizumi-Anraku, H., Chechetka, S.A., Ishizaka, M., Ikuta, A., and Kouchi, H. (2001). Responses of a model legume Lotus japonicus to lipochitin oligosaccharide nodulation factors purified from Mesorhizobium loti JRL501. Mol. Plant Microbe Interact. 14 848–856. [DOI] [PubMed] [Google Scholar]
- Niwa, Y., Hirano, T., Yoshimoto, K., Shimizu, M., and Kobayashi, H. (1999). Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 18 455–463. [DOI] [PubMed] [Google Scholar]
- Novero, M., Faccio, A., Genre, A., Stougaard, J., Webb, K.J., Mulder, L., Parniske, M., and Bonfante, P. (2002). Dual requirement of the LjSym4 gene for mycorrhizal development in epidermal and cortical cells of Lotus japonicus roots. New Phytol. 154 741–749. [DOI] [PubMed] [Google Scholar]
- Phillips, J.M., and Hayman, D.S. (1970). Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 55 158–161. [Google Scholar]
- Radutoiu, S., Madsen, L.H., Madsen, E.B., Felle, H.H., Umehara, Y., Grønlund, M., Sato, S., Nakamura, Y., Tabata, S., Sandal, N., and Stougaard, J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592. [DOI] [PubMed] [Google Scholar]
- Remy, W., Taylor, T.N., Hass, H., and Kerp, H. (1994). Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. USA 91 11841–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., Aitchison, J.D., Suprapto, A., Hjertaas, K., Zhao, Y., and Chait, B.T. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandal, N., et al. (2006). Genetics of symbiosis in Lotus japonicus: Recombinant inbred lines, comparative genetic maps, and map position of 35 symbiotic loci. Mol. Plant Microbe Interact. 19 80–91. [DOI] [PubMed] [Google Scholar]
- Schauser, L., Roussis, A., Stiller, J., and Stougaard, J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195. [DOI] [PubMed] [Google Scholar]
- Scheres, B., van Engelen, F., van der Knaap, E., van de Wiel, C., van Kammen, A., and Bisseling, T. (1990). Sequential induction of nodulin gene expression in the developing pea nodule. Plant Cell 2 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, S.L., and Long, S.R. (2003). Nod factor elicits two separable calcium responses in Medicago truncatula root hair cells. Plant Physiol. 131 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., Lutzmann, M., Santos-Rosa, H., Leonard, K., Mueller, S., Aebi, U., and Hurt, E. (2000). Structure and assembly of the Nup84p complex. J. Cell Biol. 149 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, P., Raedts, J., Portyanko, V., Debellé, F., Gough, C., Bisseling, T., and Geurts, R. (2005). NSP1 of the GRAS protein family is essential for Rhizobial Nod factor–induced transcription. Science 308 1789–1791. [DOI] [PubMed] [Google Scholar]
- Smith, S.E., and Gianinazzi-Pearson, V. (1988). Physiological interactions between symbionts in vesicular- arbuscular mycorrhizal plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39 221–244. [Google Scholar]
- Smith, S.E., and Read, D.J. (1997). Mycorrhizal Symbiosis. (San Diego, CA: Academic Press).
- Stoffler, D., Feja, B., Fahrenkrog, B., Walz, J., Typke, D., and Aebi, U. (2003). Cryo-electron tomography provides novel insights into nuclear pore architecture: Implications for nucleocytoplasmic transport. J. Mol. Biol. 328 119–130. [DOI] [PubMed] [Google Scholar]
- Stoffler, D., Goldie, K.N., Feja, B., and Aebi, U. (1999). Calcium-mediated structural changes of native nuclear pore complexes monitored by time-lapse atomic force microscopy. J. Mol. Biol. 287 741–752. [DOI] [PubMed] [Google Scholar]
- Stougaard, J. (2000). Regulators and regulation of legume root nodule development. Plant Physiol. 124 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., and Parniske, M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962. [DOI] [PubMed] [Google Scholar]
- Suntharalingam, M., and Wente, S.R. (2003). Peering through the pore: Nuclear pore complex structure, assembly, and function. Dev. Cell 4 775–789. [DOI] [PubMed] [Google Scholar]
- Szczyglowski, K., Shaw, R.S., Wopereis, J., Copeland, S., Hamburger, D., Kasiborski, B., Dazzo, F.B., and de Bruijn, F.J. (1998). Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 11 684–697. [Google Scholar]
- Tansengco, M.L., Hayashi, M., Kawaguchi, M., Imaizumi-Anraku, H., and Murooka, Y. (2003). crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome, and seed development in Lotus japonicus. Plant Physiol. 131 1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansengco, M.L., Imaizumi-Anraku, H., Yoshikawa, M., Takagi, S., Kawaguchi, M., Hayashi, M., and Murooka, Y. (2004). Pollen development and tube growth are affected in the symbiotic mutant of Lotus japonicus, crinkle. Plant Cell Physiol. 45 511–520. [DOI] [PubMed] [Google Scholar]
- Taylor, T.N., Remy, W., Hass, H., and Kerp, H. (1995). Fossil arbuscular mycorrhizae from the early Devonian. Mycologia 87 560–573. [Google Scholar]
- Tirichine, L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441 1153–1156. [DOI] [PubMed] [Google Scholar]
- Uv, A.E., Roth, P., Xylourgidis, N., Wickberg, A., Cantera, R., and Samakovlis, C. (2000). members only encodes a Drosophila nucleoporin required for Rel protein import and immune response activation. Genes Dev. 14 1945–1957. [PMC free article] [PubMed] [Google Scholar]
- Vasu, S., Shah, S., Orjalo, A., Park, M., Fischer, W.H., and Forbes, D.J. (2001). Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155 339–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser, R.G.F., Jacobsen, E., Witholt, B., and Feenstra, W.J. (1989). Efficient transformation of potato (Solanum tuberosum L.) using a binary vector in Agrobacterium rhizogenes. Theor. Appl. Genet. 78 594–600. [DOI] [PubMed] [Google Scholar]
- Walker, S.A., Viprey, V., and Downie, J.A. (2000). Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by Nod factors and chitin oligomers. Proc. Natl. Acad. Sci. USA 97 13413–13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.C., Alves, A., Pickersgill, H., Loïodice, I., Hetzer, M., Galy, V., Hülsmann, B.B., Köcher, T., Wilm, M., Allen, T., Mattaj, I.W., and Doye, V. (2003). The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113 195–206. [DOI] [PubMed] [Google Scholar]
- Wente, S.R., and Blobel, G. (1993). A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol. 123 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and Li, X. (2005). A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell 17 1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]