Abstract
Plants form shoot meristems in the so-called boundary region, and these meristems are necessary for normal morphogenesis of aerial parts of plants. However, the molecular mechanisms that regulate the formation of shoot meristems are not fully understood. We report here that expression of a chimeric repressor from TCP3 (TCP3SRDX), a member of TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (TCP) transcription factors in Arabidopsis thaliana, resulted in the formation of ectopic shoots on cotyledons and various defects in organ development. Expression of TCP3SRDX induced ectopic expression of boundary-specific genes, namely the CUP-SHAPED COTYLEDON (CUC) genes, and suppressed the expression of miR164, whose product cleaves the transcripts of CUC genes. This abnormal phenotype was substantially reversed on the cuc1 mutant background. By contrast, gain of function of TCP3 suppressed the expression of CUC genes and resulted in the fusion of cotyledons and defects in formation of shoots. The pattern of expression of TCP3 did not overlap with that of the CUC genes. In addition, we found that eight TCPs had functions similar to that of TCP3. Our results demonstrate that the TCP transcription factors play a pivotal role in the control of morphogenesis of shoot organs by negatively regulating the expression of boundary-specific genes.
INTRODUCTION
Meristems are composed of small populations of undifferentiated cells. Plants can regenerate entire organs from shoot meristems via the production of organ primordia. Once organ primordia have been generated, they differentiate into various organs according to their specified fates and cannot return to the meristematic phase during normal development. Lateral organ primordia are established in the peripheral region of a shoot meristem, which is associated with the formation of boundaries that separate organ primordia from the shoot meristem (Aida and Tasaka, 2006a, 2006b). In dicotyledonous plants, a shoot apical meristem (SAM) is formed in the boundary region of two cotyledonary primordia during embryogenesis, and secondary shoot meristems are formed at the boundary of stems and leaves, namely, at leaf axils.
Several factors have been identified as regulators of the formation of shoot meristems in boundary regions. The expression of boundary-specific genes for NAC domain transcription factors, namely, NO APICAL MERISTEM, CUP-SHAPED COTYLEDON (CUC), and CUPULIFORMIS in petunia (Petunia hybrida), Arabidposis thaliana, and Antirrhinum majus, respectively, is necessary for the initiation of formation of shoot meristems (Souer et al., 1996; Aida et al., 1997; Vroemen et al., 2003; Weir et al., 2004). Moreover, loss of expression of two of the three CUC genes in Arabidopsis results in the fusion of cotyledons and defects in the formation of the SAM (Aida et al., 1997; Vroemen et al., 2003). By contrast, ectopic expression of CUC1 enhances the expression of class I KNOTTED1-like homeobox (KNOX) genes and induces the formation of ectopic shoots on the adaxial surface of Arabidopsis cotyledons (Takada et al., 2001; Hibara et al., 2003). This observation indicates that the expression of the CUC1 gene is sufficient for the induction of formation of ectopic shoots on cotyledons. The expression of CUC genes is detected only at the boundaries of embryonic cotyledonary primordia and postembryonic organs, such as leaf axils and floral organs (Aida et al., 1999; Takada et al., 2001; Vroemen et al., 2003; Hibara et al., 2006). Thus, the spatially restricted expression of CUC genes is important for the formation of a shoot meristem at a defined position. Although it has been suggested that the auxin response pathway and microRNA (miRNA) might be involved in the control of the expression of CUC genes (Vernoux et al., 2000; Aida et al., 2002; Furutani et al., 2004; Laufs et al., 2004; Mallory et al., 2004; Baker et al., 2005), the molecular mechanisms that regulate the spatial expression of CUC genes, which are involved in the formation of shoot meristems, remain unknown.
The TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (TCP) family of transcription factors has been reported to play roles in various aspects of plant development (Luo et al., 1996; Doebley et al., 1997; Kosugi and Ohashi, 1997; Cubas et al., 1999). Loss of function of a TCP gene, the CINCINNATA (CIN) gene, in Antirrhinum results in the abnormal curvature of leaves and petals (Nath et al., 2003; Crawford et al., 2004). In addition, suppression of the expression of TCP genes by the ectopic expression of miR319/JAW, which targets TCP2, TCP3, TCP4, TCP10, and TCP24 genes, induced a cin-like phenotype in Arabidopsis (Palatnik et al., 2003). The Arabidopsis genome contains 24 TCP genes, which have been classified into the CYC/TB and PCF subfamilies (see Supplemental Figure 1 online; Cubas, 2000). However, the functional roles of members of the TCP family, including CIN, remain to be clarified.
We developed a gene silencing system, designated chimeric repressor gene-silencing technology (CRES-T), in which a transcription factor fused to the EAR-motif repression domain (SRDX) dominantly represses the transcription of its target genes, even in the presence of endogenous and functionally redundant transcription factors (Hiratsu et al., 2003). Using the CRES-T system, which involves generation of a dominant repressor, we have obtained some insights into the function of TCPs in the regulation of morphogenesis of shoot lateral organs, including the formation of the shoot meristems, via negative control of the expression of boundary-specific genes.
RESULTS
The Chimeric TCP3 Repressor Induces the Formation of Ectopic Shoots
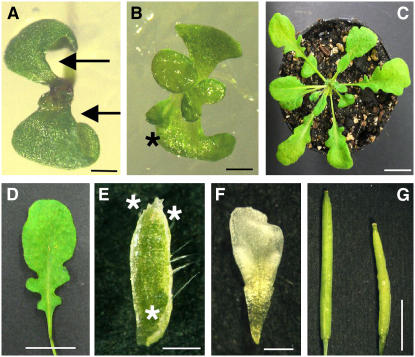
As part of our efforts to identify the biological functions of TCP transcription factors, we applied our CRES-T system to TCP transcription factors because we observed no visible abnormal phenotypic features in either Arabidopsis T-DNA–tagged lines for the TCP genes (data not shown). We converted TCP3, which is phylogenetically close to CIN (see Supplemental Figure 1 online), into a chimeric repressor by fusing it with the SRDX repression domain (Hiratsu et al., 2003), and we expressed the fused gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter (35S:TCP3SRDX; Figure 1A) in Arabidopsis. Expression of TCP3SRDX induced various morphological abnormalities, which were specifically evident on the surface and at the margins of various organs. Cotyledons of 35S:TCP3SRDX plants were wavy, serrated, and much smaller than those of the wild type, and a number of ectopic shoots were generated on the adaxial side of the cotyledons (Figures 1B to 1E). The abnormal phenotypes of 35S:TCP3SRDX seedlings could be grouped into three classes according to the severity of abnormalities. Seedlings with a mildly abnormal phenotype had epinastic cotyledons with indistinctly differentiated petioles and blades but no ectopic shoots (Figure 1C). Seedlings with a moderate phenotype had cotyledons with serrations at their margins and many ectopic shoots (Figure 1D). The seedlings with a severe phenotype had multiple ectopic shoots on their cotyledons, which resulted in defects in the expansion of the cotyledon (Figures 1E and 2B). In addition, we often observed ectopic trichomes on the surface of the cotyledons (Figure 2B). Severe defects in the elongation of the main root were also frequent (see Supplemental Figure 2 online), and the pattern of vein formation was severely disrupted in the cotyledons of 35S:TCP3SRDX plants (Figures 1F and 1G). Because the defects in the cotyledons were also evident in mature embryos (Figures 1H and 1I), we postulated that the abnormal development of the cotyledons in 35S:TCP3SRDX plants occurred during embryogenesis.
Figure 1.
Abnormal Phenotype of Various Organs Induced by TCP3SRDX.
(A) Schematic representation of the 35S:TCP3SRDX gene. CaMV 35S, Ω, SRDX, and nos represent the CaMV 35S promoter, the translational enhancer of Tobacco mosaic virus, the repression domain of 12 amino acids, and the terminator sequence of the NOS gene, respectively.
(B) to (E) Seedlings of wild-type (B) and 35S:TCP3SRDX plants with the mild (C), moderate (D), and severe (E) phenotypes. The asterisks and arrows in (D) indicate lobes and ectopic shoots of cotyledons, respectively.
(F) and (G) The patterns of vasculature in seedlings of wild-type (F) and 35S:TCP3SRDX (G) plants.
(H) and (I) Mature embryos of the wild-type (H) and the T2 generation of 35S:TCP3SRDX (I) plants.
(J) to (M) A rosette and leaf of a wild-type plant (J) and of 35S:TCP3SRDX plants with the mild (K), moderate (L), and severe (M) phenotypes. The plants shown were 3 weeks old.
(N) and (O) Flowers of wild-type (N) and 35S:TCP3SRDX (O) plants.
(P) and (Q) The adaxial surface of sepals of wild-type (P) and 35S:TCP3SRDX (Q) plants. Asterisks indicate serration of the margin.
(R) and (S) Petals of wild-type (R) and 35S:TCP3SRDX (S) plants.
(T) and (U) Stamens of wild-type (T) and 35S:TCP3SRDX (U) plants. Arrows in (U) indicate outgrowths on the anther.
(V) Siliques of wild-type (bottom panel) and 35S:TCP3SRDX (top panel) plants. st, style.
Bars = 0.5 mm in (B) to (G) and (M) to (U), 0.1 mm in (H) and (I), and 10 mm in (J) to (L) and (V).
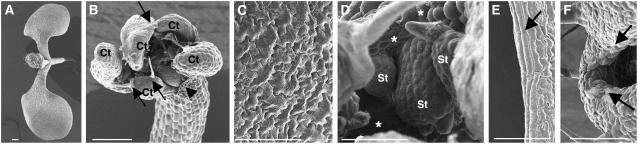
Figure 2.
Scanning Electron Microscopy Analysis of Wild-Type and 35S:TCP3SRDX Shoot Lateral Organs.
(A) and (B) Seedlings of a wild-type plant (A) and a 35S:TCP3SRDX plant with the severe phenotype (B). Ct, cotyledonary blades that had emerged from the same layer of the hypocotyl. Arrows indicate ectopic trichomes. A cotyledonary blade at the position of the arrowhead was detached to allow visualization of the interior. Bars = 200 μm.
(C) and (D) The adaxial surface of cotyledons of wild-type (C) and 35S:TCP3SRDX (D) plants. Asterisks in (D) indicate clusters of rounded cells. St, ectopic shoots.
(E) and (F) Marginal regions of wild-type (E) and 35S:TCP3SRDX (F) leaves. Arrows in (E) indicate rod-shaped cells that were typically observed in marginal regions and those in (F) indicate curling of the marginal region. Bars = 50 μm in (C) to (F).
The seedlings of 35S:TCP3SRDX plants with a mild phenotype grew relatively normally, but their rosette leaves were wavy and serrated (Figures 1J and 1K), resembling those of jaw-D plants (Palatnik et al., 2003). The 35S:TCP3SRDX plants with a moderate or severe phenotype often failed to grow when transferred to soil, and those that developed to rosette plants usually had a bushy phenotype, which was probably due to the presence of ectopic shoots and suggested that the ectopic shoots induced by TCP3SRDX had functional meristems (Figures 1L and 1M). Ectopic shoots were occasionally generated, but at low frequency, on the leaves of 35S:TCP3SRDX plants (data not shown). The morphology of the floral organs of 35S:TCP3SRDX plants was severely abnormal (Figures 1N to 1U). The sepals and petals were wavy and serrated (Figures 1Q and 1S), and the stamens often had clumped outgrowths of cells on their anthers (Figure 1U). Carpels of 35S:TCP3SRDX plants appeared normal, but the surface of siliques was crinkled and the siliques were significantly shorter than those of the wild type (Figure 1V). These results indicated that TCP3SRDX was able to induce abnormal development in various organs, regardless of their identity.
Observations by scanning electron microscopy revealed that wild-type cells in the epidermis and the marginal region were organized in a specific pavement-like pattern and were rod-shaped, respectively, whereas the cells of 35S:TCP3SRDX plants were rounded both in the epidermis and in the marginal regions, with features of undifferentiated cell clusters (Figures 2C to 2F; Donnelly et al., 1999; Ori et al., 2000). Because the differentiation of cells is regulated by their relative position within an organ (Donnelly et al., 1999; Ori et al., 2000), these observations suggested that the epidermal cells of 35S:TCP3SRDX plants do not undergo position-dependent differentiation.
A transient expression assay revealed that TCP3 had transactivation activity and that TCP3SRDX acted as a repressor in Arabidopsis leaves (see Supplemental Figure 3 online). In addition, TCP3mSRDX, in which the encoded amino acid sequence of the SRDX repression domain was mutated (mSRDX; Hiratsu et al., 2004), had no repressive activity (see Supplemental Figure 3 online). We confirmed that the expression of 35S:TCP3mSRDX in transgenic Arabidopsis was unable to induce morphological defects in cotyledons and in leaves (data not shown). These results indicated that the phenotype of 35S:TCP3SRDX plants was induced by the repressive activity of TCP3SRDX and not by some nonspecific negative effect(s), such as squelching (Cahill et al., 1994).
We expressed TCP3SRDX under the control of the 5′-upstream region of the TCP3 gene (ProTCP3:TCPS3RDX), instead of the CaMV 35S promoter, to examine the activity of TCP3SRDX in a condition similar to that of native TCP3. We found that ProTCP3:TCPS3RDX plants had the same phenotype as that of 35S:TCP3SRDX plants, although the frequency of the moderate and severer phenotypes was lower than in the case of 35S:TCP3SRDX plants (Figure 3). ProTCP3:TCPS3RDX plants with a mild or a moderate phenotype had leaves sepals, petals, and siliques with wavey surfaces and serrated margins (Figures 3C to 3G). These results indicate that TCP3SRDX, which is present at a concentration more similar to that of the corresponding native transcription factor, could induce the defective phenotype.
Figure 3.
Phenotype of ProTCP3:TCP3SRDX Plants.
(A) and (B) Seedlings with irregular differentiation of the petiole, as indicated by arrows (A), and with ectopic shoots on the cotyledon, as indicated by an asterisk (B).
(C) The rosette of a ProTCP3:TCP3SRDX plant.
(D) A leaf of a ProTCP3:TCP3SRDX plant.
(E) A sepal of a ProTCP3:TCP3SRDX plant. Asterisks indicate serrations.
(F) A petal of a ProTCP3:TCP3SRDX plant.
(G) Siliques of a wild-type (left) and a ProTCP3:TCP3SRDX (right) plant. The silique on the right is shorter and has a crinkled surface.
Bars = 0.5 mm in (A), (B), (E), and (F) and 10 mm in (C), (D), and (G).
TCP3 Regulates the Expression of Boundary-Specific Genes
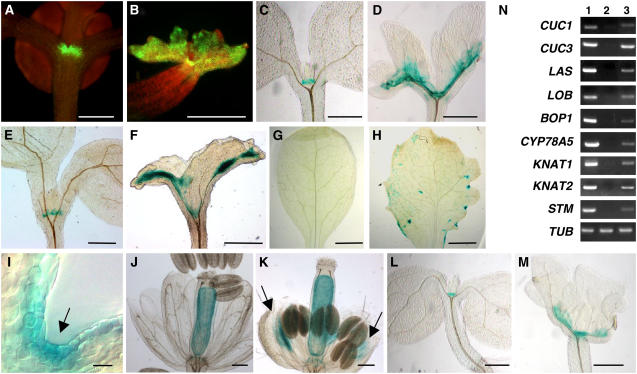
The 35S:TCP3SRDX plants were morphologically similar to transgenic plants that expressed CUC1 (35S:CUC1) ectopically with respect to the formation of ectopic shoots on the adaxial surface of cotyledons, wavy margins, irregular formation of vasculature, and the undifferentiated rounded shape of epidermal cells (Takada et al., 2001; Hibara et al., 2003). To analyze the effects of TCP3 on the regulation of the spatial expression of CUC1, we introduced TCP3SRDX into an enhancer trap line of CUC1, namely, M0223 (Cary et al., 2002). In M0223 plants, we only detected the promoter activity, as displayed by the fluorescence of green fluorescent protein, in the boundary region between two cotyledons (Figure 4A). By contrast, the region in which CUC1 was expressed had expanded broadly in cotyledons of M0223 plants that had been transformed with the TCP3SRDX construct (Figure 4B). Resembling results for the CUC1 gene, the areas of expression of two other boundary-specific genes, CUC3 and LATERAL ORGAN BOUNDARIES (LOB) (Shuai et al., 2002; Vroemen et al., 2003), as represented by signals due to β-glucuronidase (GUS) in the enhancer trap WET368 and ET22 lines, respectively, were broadly expanded in the cotyledons of the respective enhancer trap lines, when TCP3SRDX was expressed after it had been introduced by transformation (Figure 4C to 4F). In addition, inappropriate expression of CUC3 was also apparent in mature leaves and sepals of the WET368 line in association with the expression of TCP3SRDX, while no expression of CUC3 was evident in leaves and sepals in the absence of the TCP3SRDX transgene (Figures 4G to 4K). Cells in leaves in which the CUC3 gene was ectopically expressed were often rounded (Figure 4I), suggesting that the expression of the boundary-specific genes induced an undifferentiated state in these cells. Analysis of the expression of transcripts revealed that the boundary-specific genes CUC1, CUC3, CYP78A5, LOB, LATERAL SUPPRESSOR, and BLADE ON PETIOLE1 (BOP1) (Aida et al., 1997; Zondlo and Irish, 1999; Shuai et al., 2002; Greb et al., 2003; Vroemen et al., 2003; Ha et al., 2004) were expressed in leaves of 35S:TCP3SRDX plants, whereas these genes were not expressed in leaves of wild-type plants (Figure 4N). Similarly, the CUC1, CUC3, and LOB genes were also expressed ectopically in the cotyledons of ProTCP3:TCP3SRDX plants (see Supplemental Figure 4 online). These results indicate that TCP3SRDX induced the ectopic expression of a variety of boundary-specific genes in a variety of organs.
Figure 4.
The Effects of TCP3SRDX on the Pattern of Expression of Boundary-Specific Genes and Class I KNOX Genes.
(A) and (B) Expression of CUC1 in a seedling of the M0223 line, an enhancer trap line of CUC1 (C24 background) (A), and that in a similar seedling that expressed TCP3SRDX (B).
(C) and (D) Expression of CUC3 in a seedling of the WET368 line, an enhancer trap line of CUC3 (Landsberg erecta [Ler] background) (C), and that of the same line that expressed TCP3SRDX (D).
(E) and (F) Expression of LOB in a seedling of the ET22 line, an enhancer trap line of LOB (Ler background) (E), and that of the same line that expressed TCP3SRDX (F).
(G) and (H) Expression of CUC3 in rosette leaves of the WET368 line (G) and of the WET368 line that expressed TCP3SRDX (H).
(I) A magnified view of curling of the leaf margin in (H).
(J) and (K) Expression of CUC3 in sepals of the WET368 line (J) and in sepals of the same line that expressed TCP3SRDX (K). A strong GUS signal was detected between ovules, as reported previously (Vroemen et al., 2003).
(L) and (M) Expression of KNAT1 in a ProKNAT1:GUS seedling (L) and in a similar seedling that expressed TCP3SRDX (M).
(N) Analysis of the expression of boundary-specific genes and class I KNOX genes by RT-PCR. Lane 1, RNA isolated from wild-type seedlings as a positive control for boundary-specific genes; lane 2, RNA from wild-type leaves; and lane 3, RNA from 35S:TCP3SRDX leaves. Expression of the gene for Tubulin (TUB) was monitored as an internal control.
Bars = 0.5 mm in (A) to (F) and (J) to (M) and 5 mm in (H) and (I).
In addition to its effect on boundary-specific genes, we examined the effect of TCPSRDX on the expression of class I KNOX genes, namely, KNAT1, KNAT2, and SHOOT MERISTEMLESS (STM), which is required for the formation of a functional meristem (Chuck et al., 1996; Ori et al., 2000; Hake et al., 2004). The promoter activity of KNAT1 was detected only in the SAM of ProKNAT1:GUS plants, while the region in which KNAT1 was expressed was much more extensive in the presence of TCP3SRDX (Figures 4L and 4M). In addition, analysis by RT-PCR showed that KNAT1, KNAT2, and STM were expressed ectopically in leaves of 35S:TCP3SRDX plants (Figure 4N). Since the product of the CUC1 gene is a positive regulator of the expression of class I KNOX genes (Takada et al., 2001; Hibara et al., 2003), it seems likely that enhanced expression of boundary-specific genes, including the CUC1 gene, in response to TCP3SRDX induced the inappropriate expression of the class I KNOX genes in 35S:TCP3SRDX plants.
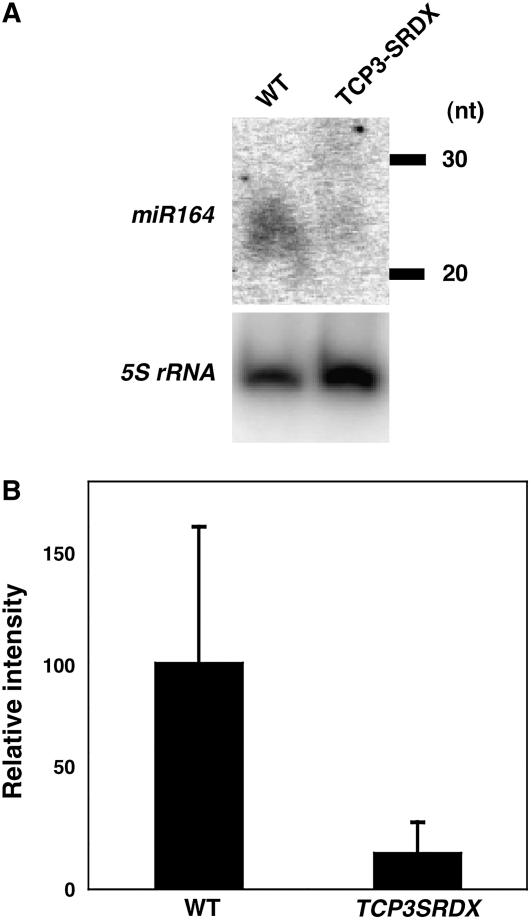
The expression of CUC genes is negatively regulated by miR164 (Laufs et al., 2004; Mallory et al., 2004; Baker et al., 2005). To investigate whether TCP3 is involved in the accumulation of miR164, we examined levels of miR164 in 35S:TCP3SRDX plants. RNA gel blot analysis revealed a significant reduction in the accumulation of miR164 in 35S:TCP3SRDX plants (Figure 5), suggesting the involvement of TCP3 in the regulation of the accumulation of this miRNA.
Figure 5.
TCP3SRDX Suppressed the Accumulation of miR164.
(A) RNA gel blot analysis for the detection of miR164 in wild-type and 35S:TCP3SRDX plants. 5S rRNA was used as an internal control. nt, nucleotides.
(B) Quantitative analysis of the accumulation of miR164 in wild-type and 35S:TCP3SRDX plants. The intensity of the signal due to miR164 is shown relative to that due to 5S rRNA. The relative value for the wild type was set at 100. The error bar indicates the sd of results from three independent experiments.
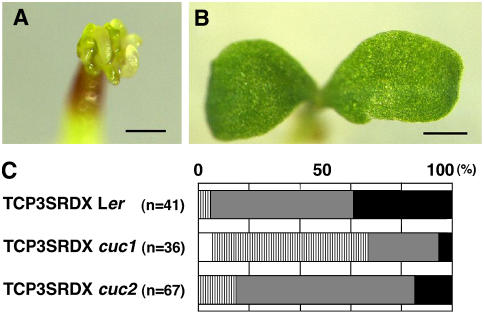
Loss of CUC Activity Suppresses the Function of the Chimeric TCP3 Repressor
To confirm that the abnormal phenotype of 35S:TCP3SRDX plants was due to the inappropriate expression of boundary-specific genes, we expressed TCP3SRDX in plants with a loss-of-function mutation in a boundary-specific gene. When TCP3SRDX was expressed on the cuc1 mutant background, the defective phenotype of cotyledons of 35S:TCP3SRDX plants was suppressed to a considerable extent and most of the seedlings had normal cotyledons with a flat surface and smooth margins, with no ectopic shoots (Figures 6A and 6B). Such recovery was observed similarly on the cuc2 mutant background, but the frequency of recovery was lower than on the cuc1 mutant background (Figure 6C). By contrast, no recovery was observed on the bop1-4 mutant background (data not shown). These results indicated that the abnormal phenotype of 35S:TCP3SRDX plants was most likely due to the inappropriate expression of the CUC genes.
Figure 6.
Mutations in CUC Genes Suppressed the Activity of TCP3SRDX.
(A) A 35S:TCP3SRDX Ler seedling with abnormal cotyledons and ectopic shoots.
(B) A 35S:TCP3SRDX cuc1 seedling, showing cotyledons with normal morphology. Bars = 0.5 mm in (A) and (B).
(C) Schematic representation of the frequency of reversal of the abnormal phenotype of 35S:TCP3SRDX seedlings by mutations in CUC genes. Phenotypic severity was classified as indicated in Figures 1C to 1E. Open box, similar to the wild type; striped box, mild phenotype; gray box, moderate phenotype; and closed box, severe phenotype. The number of seedlings examined is given in parenthesis in each case. The data are given as percentages. The background of the cuc mutants was the Ler ecotype.
Gain of Function of TCP3 Suppresses the Formation of Shoot Meristems
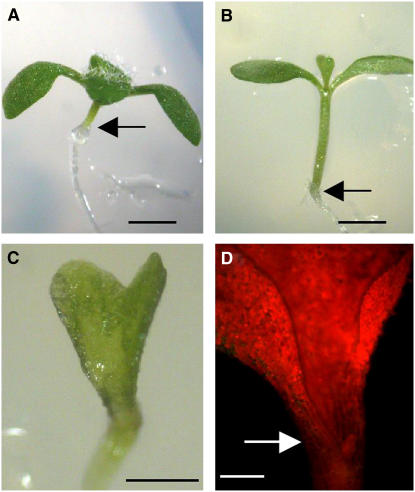
Transgenic plants that expressed TCP3 ectopically (35S:TCP3) had no visible abnormalities, probably as a result of the activity of miR319/JAW (data not shown). Therefore, we expressed a mutant form of TCP3 (mTCP3) in which the target site of miR319/JAW had been replaced by a nontarget sequence, without any change in the encoded amino acid sequence, as described previously in the case of TCP2 and TCP4 (Palatnik et al., 2003). We found that 35S:mTCP3 induced the fusion of cotyledons and defects in the formation of shoots, in addition to enhanced elongation of hypocotyls (Figures 7A to 7C). This phenotype was somewhat similar to that of the cuc1 cuc2 double mutant. Similar fusion of cotyledons was also observed in 35S:mTCP2 and 35S:mTCP4 plants to varying degrees (Palatnik et al., 2003; see Supplemental Figure 5 online). In seedlings of these plants, the expression of CUC1 and CUC3 was significantly suppressed or undetectable (Figure 7D; see Supplemental Figure 5 online). Analysis by RT-PCR confirmed that the level of the expression of these CUC genes was clearly reduced in 35S:mTCP3 plants (data not shown). These results demonstrated that TCP3, in addition to TCP2 and TCP4, can suppress the expression of CUC genes.
Figure 7.
Gain of Function of TCP3 Activity Inhibited Formation of Shoots.
(A) and (B) Side views of a wild-type seedling (A) and of a 35S:mTCP3 seedling that had a longer hypocotyl (B). Arrows indicate the junction of the hypocotyl and the main root.
(C) Fused cotyledons lacking shoots in a 35S:mTCP3 seedling.
(D) Expression of the CUC1 gene in the M0223 line that expressed mTCP3. Expression of CUC1 was suppressed in the presumptive boundary region of the M0223 seedling, as indicated by an arrow.
Bars = 0.5 mm in (A) to (C) and 0.1 mm in (D).
Redundant Functions of the Members of the TCP Family
As compared with transgenic plants that expressed TCP3SRDX, we found that tcp3-1 plants, namely, the TCP3 T-DNA–tagged homozygous line (CS855978), and transgenic plants that expressed the double-stranded RNA for RNA interference (RNAi) of the TCP3 gene had basically normal cotyledons (see Supplemental Figure 6 online). By contrast, ectopic expression of a genomic DNA fragment that encoded miR319/JAW, which should cleave transcripts of the TCP2, TCP3, TCP4, TCP10, and TCP24 genes, resulted in cotyledons that resembled those of 35S:TCP3SRDX plants with the mild phenotype (see Supplemental Figure 6 online), in addition to an effect on leaf phenotype, as reported previously (Palatnik et al., 2003). These analyses suggest that suppression of the expression of five TCP genes might be required for induction of cotyledons with the abnormal phenotype because of the functional redundancy of TCP genes.
We produced seven lines of transgenic Arabidopsis plants that expressed individual chimeric repressors derived from each of seven other TCPs in the CYC/TB subfamily, namely, TCP2, TCP4, TCP5, TCP10, TCP13, TCP17, and TCP24. We found that all seven chimeric repressors induced phenotypes similar to that induced by TCP3SRDX, although the severity of the phenotypes differed even when gene expression was driven by the CaMV 35S promoter (see Supplemental Figures 6A and 6B online). In these transgenic plants, we confirmed that the boundary-specific genes were expressed ectopically in cotyledons and leaves, as they had been also in 35S:TCP3SRDX plants (see Supplemental Figures 7A and 7B online). Moreover, expression of TCP10SRDX driven by its homologous promoter (ProTCP10:TCP10SRDX) induced defects in cotyledons and the ectopic expression of boundary-specific genes similar to those observed when transcription was driven by the CaMV 35S promoter (see Supplemental Figure 8 online). These observations suggested that the TCP transcription factors examined in this study have similar molecular functions and are possible regulators of the expression of boundary-specific genes.
The Patterns of Expression of the TCP Genes
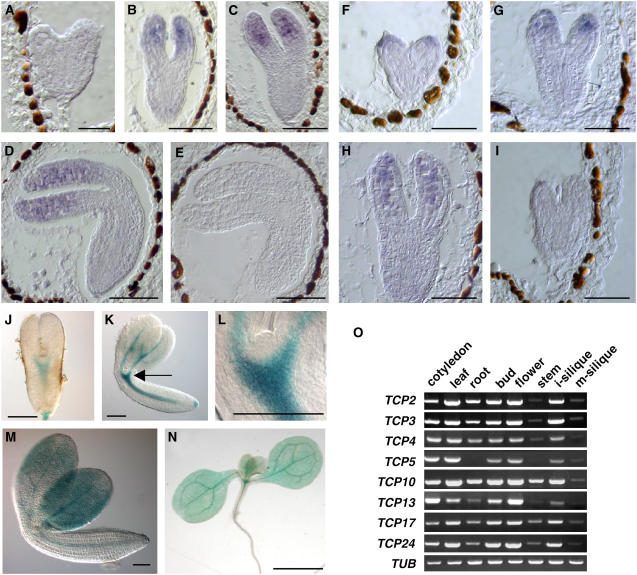
In situ hybridization revealed that the TCP3 transcript was localized in the distal and middle regions of cotyledons of torpedo, bending cotyledon, and mature embryos, but no signals were detected in the presumptive SAM and the boundary region during embryogenesis (Figures 8A to 8E). Resembling the TCP3 transcript, the TCP10 transcript was also detected in regions of the developing embryo except the region of the presumptive SAM and the boundary region (Figures 8F to 8I). In contrast with the TCP3 transcript, however, the TCP10 transcript was detectable in the cotyledons of heart-stage embryos (Figure 8F). Since expression of the TCP4 gene is detectable in embryonic cotyledons (Palatnik et al., 2003), these three TCP genes have similar patterns of expression in cotyledons.
Figure 8.
Expression of TCP Genes.
(A) to (E) Detection by in situ hybridization of transcripts of the TCP3 gene. The signal due to TCP3 transcripts was absent from heart-shaped embryos (A) but was present in cotyledons of torpedo (B), bending cotyledon (C), and mature (D) embryos. No signal was detected when the sense probe of the TCP3 gene was used (E).
(F) to (I) Detection by in situ hybridization of transcripts of the TCP10 gene. The signal due to TCP10 transcripts was detected in cotyledons of heart-shaped (F), torpedo (G), and bending cotyledon (H) embryos. No signal was observed when the sense probe for the TCP10 gene was used (I).
(J) to (N) Expression of the ProTCP3:GUS reporter gene in embryos at the torpedo stage (J) and the bending cotyledon stage (K). In (L), a magnified view of the SAM region is shown that is indicated by an arrow in (K). A mature embryo (M) and a seedling (N) are also shown.
(O) Analysis of the expression of eight TCP genes in various organs by RT-PCR. Expression of the gene for Tubulin (TUB) was monitored as an internal control. i-silique, immature silique; m-silique, mature silique.
Bars = 50 μm in (A) to (M) and 1 mm in (N).
We also examined the patterns of expression of TCP genes using a GUS reporter gene that was fused with the 5′-upstream region of each respective TCP gene (ProTCP:GUS). The promoter activity of the TCP3 gene, as represented by the GUS activity due to the ProTCP3:GUS gene, was strong in the vascular regions of the cotyledons and hypocotyl in torpedo and bending cotyledon embryos, as well as in developing seedlings (Figures 8J to 8N). The difference between the patterns of expression of TCP3 during early embryogenesis obtained with ProTCP3:GUS and by in situ analysis might have been due to posttranscriptional suppression by the miR319/JAW-mediated cleavage of the TCP3 transcript or to transcriptional regulation via sequences beyond the promoter region used in this study. The promoters of the TCP2, TCP4, TCP5, TCP10, TCP13, TCP17, and TCP24 genes were active in cotyledons and, in particular, in the vascular region, as was the case for the promoter of the TCP3 gene, even though each gene had a somewhat different pattern of expression (see Supplemental Figure 9 online). Analysis by RT-PCR confirmed that these eight TCP genes are expressed differentially in various organs in an overlapping manner, although no signal for TCP5 was detected in roots (Figure 8O).
DISCUSSION
In this study, we found that the expression of a chimeric TCP3 repressor induced the ectopic expression of boundary-specific genes, with the resultant formation of ectopic shoots, while overexpression of mTCP3 suppressed the expression of such genes, with resultant inhibition of the formation of shoots and of the separation of cotyledons. Our results demonstrate that TCP3 plays a pivotal role in the control of morphogenesis of shoot lateral organs via the negative control of the expression of boundary-specific genes, namely, the CUC genes, which control the initiation of the shoot meristem and the morphogenesis of organ boundaries (Aida et al., 1997). Given that TCP3 has transactivational activity, we can postulate that TCP3 might activate some unidentified factors that suppress the transcription of the CUC genes. The proposed function of TCP3 is consistent with the finding that the pattern of expression of the TCP3 gene does not overlap with that of CUC genes.
The expression of the CUC genes is regulated at the transcriptional level but is also regulated negatively by miR164 (Laufs et al., 2004; Mallory et al., 2004; Baker et al., 2005). miR164 is encoded by three genes, miR164A, miR164B, and miR164C, and it accumulates in various tissues, which include seedlings, leaves, and floral organs (Laufs et al., 2004; Mallory et al., 2004; Baker et al., 2005). The suppression of expression of miR164 in a T-DNA–tagged line for the miR164A gene deepened the serration of leaf margin (Nikovics et al., 2006), and this phenotype was somewhat similar to that of 35S:TCP3SRDX plants. Thus, control of the accumulation of miR164 might be important for appropriate development of leaves. We showed in this report that the level of miR164 was significantly reduced in 35S:TCP3SRDX plants (Figure 5). It is possible that suppression of the expression of the miR164 gene is also involved in the ectopic expression of CUC genes in 35S:TCP3SRDX plants. However, considering that mutation of the miR164A gene did not induce ectopic shoots, we postulate that TCP3 might control the expression of the CUC genes at both the transcriptional and posttranscriptional levels.
Our results suggest that elimination from cotyledons of the activities of proteins encoded by the CUC and other boundary-specific genes might be required for the normal development of shoot lateral organs. Since the CUC genes and several boundary-specific genes appear to control morphogenesis at boundaries and the formation of shoot meristems (Aida and Tasaka, 2006a, 2006b), inappropriate expression of these genes might prevent the specific differentiation of cells that is necessary for the formation of smooth surfaces and margins of organs. Indeed, ectopic expression of the CUC genes and other boundary-specific genes has been reported to disturb the normal development of the respective transgenic plants (Zondlo and Irish, 1999; Takada et al., 2001; Shuai et al., 2002; Hibara et al., 2003; Laufs et al., 2004). The abnormal distribution of or response to auxin induces the inappropriate expression of CUC genes in cotyledons and lateral organ primordia, preventing organ development (Vernoux et al., 2000; Furutani et al., 2004; Treml et al., 2005). In addition, the abnormal radial information generated by inappropriate expression of PINHEAD in the peripheral region of an embryo induces ectopic expression of CUC2 and the formation of shoot meristems on cotyledons (Newman et al., 2002). Thus, the spatial regulation of the expression of the CUC genes and other boundary-specific genes might be a critical determinant in the control of morphogenesis of shoot lateral organs. TCPs appear to be novel factors that are involved in this regulation.
By contrast, it appears that initiation of the formation of shoot meristems requires suppression of the activities of TCPs in the boundary region and the SAM. The accumulation of transcripts of the TCP3, TCP4, and TCP10 genes and the activities of the respective promoters were not evident either in the SAM or at cotyledonary boundaries (Figure 8; Palatnik et al., 2003). In addition, we found that ectopic expression of mTCP3, in which the target site of miR319/JAW had been mutated, inhibited formation of shoots, probably as a result of the expression of TCP3 in the boundary region. Consistent with these observations, the precursor to miR319/JAW is abundant in the shoot apex, which includes the SAM (Palatnik et al., 2003). Thus, the miR319/JAW-dependent cleavage of TCP transcripts might be required for the formation of the shoot meristem. Our results suggest that suppression of the activities of TCPs in the boundary region might be controlled at both the transcriptional and the posttranscriptional level.
We showed that eight TCPs have similar molecular functions by examining the effects of the chimeric repressors derived from each one, and we found that the corresponding TCP genes were expressed in an overlapping manner in various tissues. The TCP3, TCP4, and TCP10 genes have similar patterns of expression in cotyledons during embryogenesis, and seven of the eight TCP genes are expressed during the early development of flowers (Figure 8; Palatnik et al., 2003; Wellmer et al., 2006). In addition, the functional redundancy of these TCPs was apparent from the observation that neither T-DNA–tagged lines nor RNAi for TCP3 induced an informative phenotype in Arabidopsis. Although 35S:miR319/JAW plants have mildly abnormal cotyledons, the functions of the TCP5, TCP13, and TCP17 genes, whose transcripts lack a target sequence for miR319/JAW, might prevent formation of more severely abnormal cotyledons, with, for example, ectopic shoots. These observations suggest that the TCP transcription factors of Arabidopsis act redundantly to regulate the spatial expression of boundary-specific genes and to control the morphogenesis of shoot lateral organs. Arabidopsis appears to have a greater number of redundant TCP genes than does Antirrhinum because disruption of the CIN gene alone is sufficient to induce abnormal leaves in Antirrhinum (Nath et al., 2003).
Each TCP gene had a somewhat different pattern of expression, and each chimeric TCP repressor induced an abnormal phenotype with a different degree of severity, even when transcription of its gene was driven by the CaMV 35S promoter. The chimeric repressors derived from TCP3, TCP4, TCP5, and TCP10 induced the severe phenotype at higher frequency, perhaps because of higher activities of DNA binding and transactivation. In addition, the activities of the promoter regions of the TCP3, TCP4, TCP5, and TCP10 genes used in this study were more prominent in cotyledons. These genes might make a larger contribution to the development of cotyledons than do other TCP genes. Thus, the TCP transcription factors examined in this study appear to function similarly as regulators of the expression of boundary-specific genes, but the spatial activity of each TCP seems to be regulated differently.
Because of the close relationships among TCPs, in terms of phylogeny, and their functional similarities, we have grouped these eight TCPs as a subfamily of CIN-like TCPs. TCP genes are conserved in monocots, as well as in dicots, and there are 23 TCP genes in the rice (Oryza sativa) genome, just as there are in the Arabidopsis genome. Ten of these rice genes, which include genes for class II PCFs (Kosugi and Ohashi, 2002), can be considered to be CIN-like TCP genes. We found that five of the CIN-like TCP genes in rice have a target sequence for miR319/JAW in addition to those mentioned in a report by Palatnik et al. (2003). Thus, it is likely that not only the family of TCP genes but also the mechanism for regulation of the expression of genes for TCPs by the miR319/JAW gene evolved prior to the divergence of monocots and dicots. It has been suggested that CUC genes might be involved in the formation of the SAM in monocots (Zimmermann and Werr, 2005). Thus, the acquisition of a mechanism for regulation of the spatial activities of CUCs by TCPs might have been a key event in the evolution of plants.
In this study, we showed that TCP transcription factors are critical for the morphogenesis of shoot lateral organs. Extending the previous reports that TCPs are involved in the control of the development of leaves (Nath et al., 2003; Palatnik et al., 2003), we showed that TCPs are involved in the formation of the shoot meristem and the development of shoot lateral organs. When TCPs fail to suppress the expression of the CUC genes, ectopic expression of CUC genes results in the generation of multiple shoot meristems and inhibition of organ growth. Plant cells have extraordinary totipotency and have the ability to generate new organs continuously from shoot meristems and to form shoot meristems at organ boundaries throughout their life cycle (Weigel and Jurgens, 2002; Willemsen and Scheres, 2004; Schmitz and Theres, 2005; Aida and Tasaka, 2006a, 2006b). This process requires the coordination of the functions of the shoot meristem with those of the differentiating cells of a given organ. Thus, the TCP-dependent regulation of the expression of CUC genes, which controls morphogenesis of the shoot lateral organs and regulates formation of the shoot meristem, is of critical importance to plant development.
METHODS
Construction of Plasmids
The protein-coding regions of TCP genes were amplified from genomic DNA or from cDNAs provided by the RIKEN Bio Resource Center (BRC; Seki et al., 1998, 2002) with appropriate primers, as shown in Supplemental Table 1 online. The chimeric 35S:TCPSRDX gene was constructed as described previously (Mitsuda et al., 2006). The mSRDX, mTCP2, mTCP3, and mTCP4 genes were generated by site-directed mutagenesis with appropriate primers. The GUS reporter genes under the control of the promoters of individual TCP genes were constructed using 5′-upstream regions relative to sites of initiation of translation of the respective TCP genes (TCP2, 2978 bp; TCP3, 2593 bp; TCP4, 3027 bp; TCP5, 2879 bp; TCP10, 1054 bp; TCP13, 2863 bp; TCP17, 2276 bp; and TCP24, 2776 bp). The GUS genes of ProTCP3:GUS and ProTCP10:GUS were replaced by TCP3SRDX and TCP10SRDX for the construction of ProTCP3:TCP3SRDX and ProTCP10:TCP10SRDX, respectively. The construct for RNAi of TCP3 was generated from the 458-bp region at the 3′ end of the gene (nucleotides 715 to 1173) and pHELLSGATE8 (Wesley et al., 2001).
Plant Materials and Transformation
Arabidopsis thaliana ecotype Col-0 was used throughout this study unless otherwise indicated. Growth conditions and the strategy for transformation of Arabidopsis were described previously (Mitsuda et al., 2006). The exogenous expression of each transgene in transgenic plants was confirmed by RT-PCR with appropriate primers (see Supplemental Figure 10 and Supplemental Table 1 online). The tcp3-1 line had a T-DNA tag in the protein-coding region of the TCP3 gene at a site 261 bp from the site of initiation of translation.
Light and Scanning Electron Microscopy
Analysis of promoter activities using the GUS reporter gene was performed with T1 or T2 transgenic lines as described previously (Mitsuda et al., 2005). Light microscopy and fluorescence microscopy for detection of green fluorescent protein were performed with the Axioskop2 plus system (Carl Zeiss). Plant tissues were rendered transparent for the observations of vasculature as described previously (Aida et al., 1997). For scanning electron microscopy, samples were prepared and analyzed as described previously (Mitsuda et al., 2005).
Isolation of RNA and RT-PCR
Total RNA was isolated from tissues with Trizol as described previously (Fujimoto et al., 2000). For analysis by RT-PCR, aliquots of 50 ng of total RNA were subjected to first-strand cDNA synthesis (Hiratsu et al., 2003). PCR was performed with gene-specific primers (see Supplemental Table 1 online) for 30 to 39 cycles.
Small RNA was prepared with a mirVana miRNA isolation kit (Ambion) from 3-week-old plants. For detection of miR164, aliquots of 40 μg of small RNA were fractionated on a 15% polyacrylamide gel that contained 7 M urea, blotted onto a nylon membrane, and allowed to hybridize to the 32P-labeled nucleotide probe in ULTRAhyb-Oligo solution (Ambion). The synthetic RNA corresponding to the sense strand of miR164 was used as a positive control for hybridization. The intensity of signals was quantified with ImageQuant (Molecular Dynamics).
In Situ Hybridization
Preparation of samples and in situ hybridization were performed as described previously (Furutani et al., 2004). DNA fragments corresponding to positions 48 to 1176 from the site of initiation of translation of TCP3 and to the full-length coding sequence of TCP10 were used as templates for probes, respectively.
Assays of Transient Gene Expression
The coding sequences of TCP3, TCP3SRDX, and TCP3mSRDX were fused separately to that for GAL4DB (Ohta et al., 2000), and assays of transient gene expression were performed as described previously (Fujimoto et al., 2000). We used 0.8 μg of reporter plasmid and 0.6 μg of effector plasmid for each bombardment. For normalization of the activity of the reporter gene, we used 0.8 μg of a reference plasmid, pPTRL (Fujimoto et al., 2000). After bombardment, samples were incubated for 16 h in darkness, and then luciferase activity was quantified.
Accession Numbers
The Arabidopsis Genome Initiative (http://www.arabidopsis.org) identifiers for the genes and gene products were designated as follows: TCP2, At4g18390; TCP3, At1g53230; TCP4, At3g15030; TCP5, At5g60970; TCP10, At2g31070; TCP13, At3g02150; TCP17, At5g08070; and TCP24, At1g30210.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Primers Used in This Study.
Supplemental Figure 1. Phylogeny of TCPs in the CYC/TB Subfamily.
Supplemental Figure 2. Inhibition of Elongation of the Main Root by the Chimeric TCP Repressor.
Supplemental Figure 3. Transient Expression Assay for TCP3.
Supplemental Figure 4. Expression of Boundary-Specific Genes in ProTCP3:TCP3SRDX Plants.
Supplemental Figure 5. Phenotype Induced by the Ectopic Expression of the mTCP Gene.
Supplemental Figure 6. Phenotype Induced by Suppression of the Expression of the TCP Gene.
Supplemental Figure 7. Phenotypes Induced by Seven Different Chimeric TCP Repressors.
Supplemental Figure 8. Phenotype of ProTCP10:TCP10SRDX Plants.
Supplemental Figure 9. Analysis of the Promoter Activities of TCP Genes in Young Seedlings.
Supplemental Figure 10. Expression of the TCP3SRDX Gene in 35S:TCP3SRDX Plants.
Supplementary Material
Acknowledgments
We thank the ABRC for seeds of tcp3-1 (CS855978), ET-22, and ProKNAT1:GUS plants; C. de Vries for seeds of WET368 plants; C.M. Ha for seeds of bop1-4 plants; RIKEN BRC for cDNA clones for TCP genes; and P.M. Waterhouse for pHELLSGATE8. We thank Akita Prefectural University for sequencing plasmids and S. Miyamura and A. Iwase for scanning electron microscopy analysis. We also thank K. Hiratsu, N. Mitsuda, and M. Aida for helpful discussions and K. Yamaguchi, A. Kushida, N. Kawanami, and Y. Takiguchi for their skilled technical assistance.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Masaru Ohme-Takagi (m-takagi@aist.go.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126 1563–1570. [DOI] [PubMed] [Google Scholar]
- Aida, M., Vernoux, T., Furutani, M., Traas, J., and Tasaka, M. (2002). Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129 3965–3974. [DOI] [PubMed] [Google Scholar]
- Aida, M., and Tasaka, M. (2006. a). Genetic control of shoot organ boundaries. Curr. Opin. Plant Biol. 9 72–77. [DOI] [PubMed] [Google Scholar]
- Aida, M., and Tasaka, M. (2006. b). Morphogenesis and patterning at the organ boundaries in the higher plant shoot apex. Plant Mol. Biol. 60 915–928. [DOI] [PubMed] [Google Scholar]
- Baker, C.C., Sieber, P., Wellmer, F., and Meyerowitz, E.M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15 303–315. [DOI] [PubMed] [Google Scholar]
- Cahill, M.A., Ernst, W.H., Janknecht, R., and Nordheim, A. (1994). Regulatory squelching. FEBS Lett. 344 105–108. [DOI] [PubMed] [Google Scholar]
- Cary, A.J., Che, P., and Howell, S.H. (2002). Developmental events and shoot apical meristem gene expression patterns during shoot development in Arabidopsis thaliana. Plant J. 32 867–877. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, B.C., Nath, U., Carpenter, R., and Coen, E.S. (2004). CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 135 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas, P. (2000). Role of genes in the evolution of morphological characters in angiosperm. In Developmental Genetics and Plant Evolution, Q.C.B. Cronk, R.M. Bateman, and J.A. Hawkins, eds (London: Taylar and Francis), pp. 247–266.
- Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 18 215–222. [DOI] [PubMed] [Google Scholar]
- Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386 485–488. [DOI] [PubMed] [Google Scholar]
- Donnelly, P.M., Bonetta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215 407–419. [DOI] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani, M., Vernoux, T., Traas, J., Kato, T., Tasaka, M., and Aida, M. (2004). PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131 5021–5030. [DOI] [PubMed] [Google Scholar]
- Greb, T., Clarenz, O., Schafer, E., Muller, D., Herrero, R., Schmitz, G., and Theres, K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, C.M., Jun, J.H., Nam, H.G., and Fletcher, J.C. (2004). BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 45 1361–1370. [DOI] [PubMed] [Google Scholar]
- Hake, S., Smith, H.M., Holtan, H., Magnani, E., Mele, G., and Ramirez, J. (2004). The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 20 125–151. [DOI] [PubMed] [Google Scholar]
- Hibara, K., Karim, M.R., Takada, S., Taoka, K., Furutani, M., Aida, M., and Tasaka, M. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara, K., Takada, S., and Tasaka, M. (2003). CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 36 687–696. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Mitsuda, N., Matsui, K., and Ohme-Takagi, M. (2004). Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem. Biophys. Res. Commun. 321 172–178. [DOI] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30 337–348. [DOI] [PubMed] [Google Scholar]
- Laufs, P., Peaucelle, A., Morin, H., and Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131 4311–4322. [DOI] [PubMed] [Google Scholar]
- Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in antirrhinum. Nature 383 794–799. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Dugas, D.V., Bartel, D.P., and Bartel, B. (2004). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14 1035–1046. [DOI] [PubMed] [Google Scholar]
- Mitsuda, N., Hiratsu, K., Todaka, D., Nakashima, K., Yamaguchi-Shinozaki, K., and Ohme-Takagi, M. (2006). Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol. J. 4 325–332. [DOI] [PubMed] [Google Scholar]
- Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath, U., Crawford, B.C., Carpenter, R., and Coen, E. (2003). Genetic control of surface curvature. Science 299 1404–1407. [DOI] [PubMed] [Google Scholar]
- Newman, K.L., Fernandez, A.G., and Barton, M.K. (2002). Regulation of axis determinacy by the Arabidopsis PINHEAD gene. Plant Cell 14 3029–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., and Laufs, P. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18 2929–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, M., Ohme-Takagi, M., and Shinshi, H. (2000). Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J. 22 29–38. [DOI] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 257–263. [DOI] [PubMed] [Google Scholar]
- Schmitz, G., and Theres, K. (2005). Shoot and inflorescence branching. Curr. Opin. Plant Biol. 8 506–511. [DOI] [PubMed] [Google Scholar]
- Seki, M., Carninci, P., Nishiyama, Y., Hayashizaki, Y., and Shinozaki, K. (1998). High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 15 707–720. [DOI] [PubMed] [Google Scholar]
- Seki, M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296 141–145. [DOI] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Pena, C.G., and Springer, P.S. (2002). The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer, E., van Houwelingen, A., Kloos, D., Mol, J., and Koes, R. (1996). The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85 159–170. [DOI] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128 1127–1135. [DOI] [PubMed] [Google Scholar]
- Treml, B.S., Winderl, S., Radykewicz, R., Herz, M., Schweizer, G., Hutzler, P., Glawischnig, E., and Ruiz, R.A. (2005). The gene ENHANCER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development 132 4063–4074. [DOI] [PubMed] [Google Scholar]
- Vernoux, T., Kronenberger, J., Grandjean, O., Laufs, P., and Traas, J. (2000). PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127 5157–5165. [DOI] [PubMed] [Google Scholar]
- Vroemen, C.W., Mordhorst, A.P., Albrecht, C., Kwaaitaal, M.A., and de Vries, S.C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Jurgens, G. (2002). Stem cells that make stems. Nature 415 751–754. [DOI] [PubMed] [Google Scholar]
- Weir, I., Lu, J., Cook, H., Causier, B., Schwarz-Sommer, Z., and Davies, B. (2004). CUPULIFORMIS establishes lateral organ boundaries in antirrhinum. Development 131 915–922. [DOI] [PubMed] [Google Scholar]
- Wellmer, F., Alves-Ferreira, M., Dubois, A., Riechmann, J.L., and Meyerowitz, E.M. (2006). Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet. 2 1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Willemsen, V., and Scheres, B. (2004). Mechanisms of pattern formation in plant embryogenesis. Annu. Rev. Genet. 38 587–614. [DOI] [PubMed] [Google Scholar]
- Zimmermann, R., and Werr, W. (2005). Pattern formation in the monocot embryo as revealed by NAM and CUC3 orthologues from Zea mays L. Plant Mol. Biol. 58 669–685. [DOI] [PubMed] [Google Scholar]
- Zondlo, S.C., and Irish, V.F. (1999). CYP78A5 encodes a cytochrome P450 that marks the shoot apical meristem boundary in Arabidopsis. Plant J. 19 259–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.