Abstract
The exceptional toxicity of arsenate [As(V)] is derived from its close chemical similarity to phosphate (Pi), which allows the metalloid to be easily incorporated into plant cells through the high-affinity Pi transport system. In this study, we identified an As(V)-tolerant mutant of Arabidopsis thaliana named pht1;1-3, which harbors a semidominant allele coding for the high-affinity Pi transporter PHT1;1. pht1;1-3 displays a slow rate of As(V) uptake that ultimately enables the mutant to accumulate double the arsenic found in wild-type plants. Overexpression of the mutant protein in wild-type plants provokes phenotypic effects similar to pht1;1-3 with regard to As(V) uptake and accumulation. In addition, gene expression analysis of wild-type and mutant plants revealed that, in Arabidopsis, As(V) represses the activation of genes specifically involved in Pi uptake, while inducing others transcriptionally regulated by As(V), suggesting that converse signaling pathways are involved in plant responses to As(V) and low Pi availability. Furthermore, the repression effect of As(V) on Pi starvation responses may reflect a regulatory mechanism to protect plants from the extreme toxicity of arsenic.
INTRODUCTION
Arsenic, one of the most toxic metals found in soils, is derived from both natural and anthropogenic sources (Tamaki and Frankenberger, 1992; Fitz and Wenzel, 2002; Nordstrom, 2002; Oremland and Stolz, 2003). Arsenic can be solubilized in ground water, exposing animals and humans to potentially toxic effects (Nickson et al., 1998; Meharg and Hartley-Whitaker, 2002; Gómez et al., 2004; Katz and Salem, 2005).
In soils, the most abundant arsenic species is arsenate [As(V)] (Tamaki and Frankenberger, 1992; Brown et al., 1999). As(V) toxicity is derived from its close chemical similarity to phosphate (Pi); this mimicry enables As(V) to alter Pi metabolism (Clarkson and Hanson, 1980; Raghothama, 1999; Fitz and Wenzel, 2002). Indeed, the similarity between these two anions makes plants highly sensitive to As(V) because it is easily incorporated into cells through the high-affinity Pi transport system (Meharg and Macnair, 1990, 1991b, 1992b). Since this transport system is induced by Pi starvation, As(V) uptake is highly dependent upon the amount of Pi available in the soil (Bieleski, 1973; Raghothama, 1999). Arabidopsis thaliana mutants exhibiting As(V) tolerance harbor null alleles coding for the high affinity Pi transporters PHT1;1 or PHT1;4 (Shin et al., 2004), indicating that these transporters play a major role in As(V) uptake. In addition, a mutation in PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1), which is required for efficient trafficking of Pi transporters to the plasma membrane, also results in a strong tolerance to As(V) (González et al., 2005). Moreover, tolerance to As(V) in a variety of species, such as Holcus lanatus, is achieved through a reduction in As(V) uptake due to a suppression of the high-affinity Pi uptake system (Meharg and Macnair, 1990, 1991b, 1992b; Meharg and Hartley-Whitaker, 2002; Bleeker et al., 2003). These plants also exhibit enhanced arsenic accumulation (Meharg and Macnair, 1991a), and genetic analysis revealed that a single dominant locus could be responsible for both phenotypes (Macnair et al., 1992). Due to the apparent contradiction between reduced As(V) uptake and enhanced arsenic accumulation, it has been speculated that a complex rather than a simple locus was responsible for both traits (Meharg and Macnair, 1992a).
Once As(V) enters the cell, it is promptly reduced to arsenite [As(III)] (Pickering et al., 2000; Meharg and Hartley-Whitaker, 2002), which is highly toxic but is rapidly complexed with soluble thiols, in most cases phytochelatins (Clemens et al., 1999; Pickering et al., 2000; Meharg and Hartley-Whitaker, 2002), and then sequestered into vacuoles (Salt and Rauser, 1995; Lombi et al., 2002). This strategy has been widely used by plants to cope with arsenic and heavy metals. However, studies performed with the arsenic hyperaccumulator plant Pteris vittata indicate that there must be additional arsenic accumulation mechanisms that have yet to be identified (Zhao et al., 2003; Raab et al., 2004). Recently, the first screening for As(V)-tolerant mutants in Arabidopsis yielded the identification of the mutant arsenic resistant1 (ars1) (Lee et al., 2003), but the identity of the ars1 gene is still unknown.
Here, we report the identification and characterization of a new semidominant mutant allele of the high-affinity Pi transporter PHT1;1. This allele, named pht1;1-3, exhibits an enhanced ability to accumulate arsenic while Pi and As(V) uptake rate is reduced, suggesting that this may be the single mechanism operating in naturally selected arsenic-tolerant plants. Additionally, we show that As(V) suppresses the Pi starvation response while activating other genes potentially involved in As(V) detoxification/tolerance, suggesting that an As(V) Pi interacting pathway operates in plants to reduce arsenic uptake.
RESULTS
Screening for As(V)-Tolerant Mutants
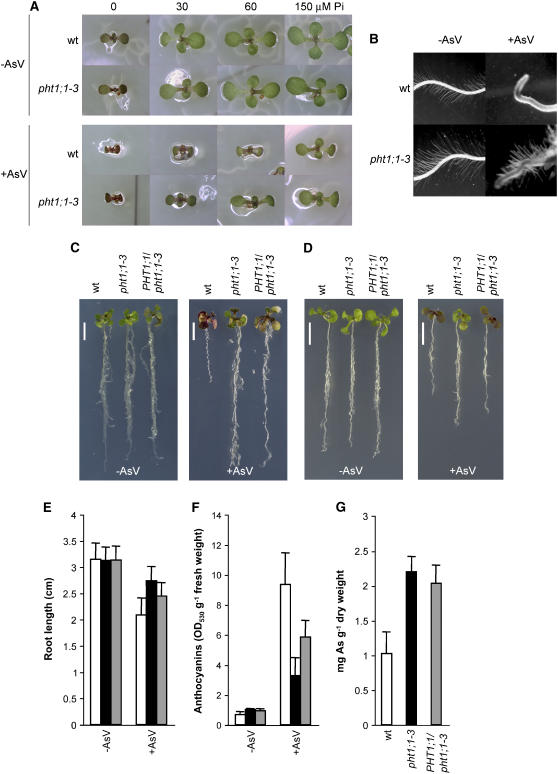
To identify As(V)-tolerant mutants, we first studied the phenotypic changes of wild-type Arabidopsis seedlings in response to the metalloid. Since As(V) competes with Pi for the Pi uptake system, we performed a morphological analysis of seedlings directly sown on medium containing 30 μM As(V) supplemented with different Pi concentrations. As expected, As(V) toxicity symptoms increased as Pi concentrations decreased (Figure 1A). In the above-ground (aerial) tissues, the most emblematic symptoms observed were growth arrest and anthocyanin accumulation. In roots, growth arrest was also characteristic of As(V) toxicity, and root hair elongation appeared to be completely inhibited (Figure 1B). Arabidopsis seedlings grown in the presence of 30 μM As(V) and 30 μM Pi demonstrated intermediate toxicity symptoms, indicating that this concentration range may be suitable for screening As(V)-tolerant mutants. Under these conditions, we screened 100,000 M2 seedlings from a population of ethyl methanesulfonate–mutagenized Columbia lines and ultimately identified nine mutants. One of the selected mutants developed a larger aerial part, with less growth arrest than that observed in wild-type plants when grown in the presence of 30 μM As(V) (Figure 1A). In addition, this mutant was able to elongate root hairs when grown on Pi-lacking medium supplemented with 30 μM As(V) (Figure 1B). Moreover, after an extended exposure to As(V), the mutant clearly accumulated less anthocyanins in the aerial portion and exhibited longer roots than wild-type plants (Figure 1C). We named this mutant pht1;1-3 in accordance with its molecular characterization (described below).
Figure 1.
As(V) Tolerance Phenotype Displayed by pht1;1-3.
(A) Above-ground phenotype of plants grown for 8 d on media with 30 μM As(V) (+AsV) or without (−AsV) at different Pi concentrations.
(B) Root hair elongation after 8 d of growth on media containing 30 μM Pi (−AsV) or 30 μM Pi plus 30 μM As(V) (+AsV).
(C) to (G) As(V) tolerance phenotype ([C] and [D]), root length (n ≥ 12; P < 0.01) (E), anthocyanin accumulation (n > 3; P < 0.01) (F), and total arsenic accumulation (n ≥ 3; P < 0.01) (G) of plants grown for 7 d on 30 μM Pi. Plants in (D) to (F) were grown for an additional 4 d on the same medium supplemented with 50 μM As(V) (+AsV) or without As(V) (−AsV). Plants in (C) and (G) were grown for an additional 12 d on the same medium supplemented with 50 μM As(V). Wild-type (white bars), pht1;1-3 (black bars), and heterozygous PHT1;1/pht1;1-3 (gray bars). Error bars represent sd. Bars in (C) and (D) = 0.5 cm.
pht1;1-3 Shows Enhanced Arsenic Accumulation
Genetic analysis revealed that the tolerant phenotype displayed by pht1;1-3 is caused by a single mutation and that heterozygous plants showed an intermediate As(V) tolerance phenotype (Figure 1C). When plants were exposed to As(V) for a shorter time (Figure 1D), quantification of root length (Figure 1E) and anthocyanin accumulation (Figure 1F) confirmed the intermediate tolerant phenotype of the heterozygotes. Therefore, in the conditions used here, the mutation behaved as semidominant. To further characterize the pht1;1-3 tolerance phenotype and to establish its potential for arsenic phytoremediation, we determined the arsenic concentration in mutant and wild-type plants. As shown in Figure 1G, pht1;1-3 plants accumulate at least twice the arsenic than that accumulated by wild-type plants after 12 d of growth on As(V)-containing medium. Based on these phenotypes, pht1;1-3 was chosen for further characterization.
pht1;1-3 Harbors a Missense Mutation in the Pi Transporter PHT1;1
To identify the pht1;1-3 mutant locus, we first selected plants that were able to elongate root hairs after 8 d of growth on 30 μM As(V) and Pi-lacking medium from an F2 mapping population obtained from crosses with the ecotype Landsberg erecta (Ler). Due to the semidominant phenotype displayed by pht1;1-3, we selected homozygous F2 plants at the PHT1;1 locus based on the segregation of the As(V) tolerance phenotype in their respective F3 offspring. Using 62 of those selected F2 plants, we mapped the PHT1;1 locus to chromosome V, close to the marker DFR. There is a cluster of four genes encoding Pi transporters linked to this marker, which represent potential candidate genes. Direct sequencing of the PHT1;1 locus revealed that this gene is mutated in pht1;1-3 . This mutation results from a single nucleotide exchange, which encodes a nonconservative amino acid substitution (Gly-378 to Glu) at the predicted ninth transmembrane domain of the transporter. Therefore, pht1;1-3 is a new semidominant allele for the Pi transporter PHT1;1 (Shin et al., 2004).
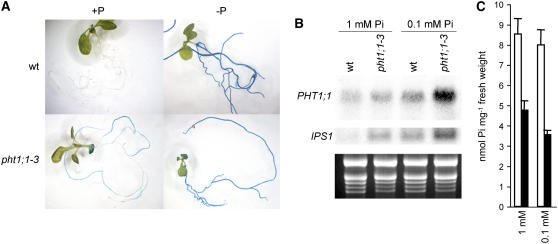
To evaluate the effect of the pht1;1-3 allele on the Pi starvation response, we first took advantage of the fact that the mutagenized collection from where pht1;1-3 was isolated harbors the Pi starvation responsive reporter gene IPS1:β-glucuronidase (GUS) (Martín et al., 2000; Rubio et al., 2001). Hence, the Pi starvation response can be easily monitored in these plants through histochemical GUS staining. As shown in Figure 2A, either wild-type or pht1;1-3 plants exhibit GUS staining when grown on Pi lacking (−P) medium. However, in contrast with what was observed for wild-type plants, GUS staining was also present in pht1;1-3 plants grown on medium containing 1 mM Pi (+P). This result was confirmed by RNA gel blot analysis of the IPS1 expression in plants grown under high Pi (Figure 2B). In this experiment, we also evaluated the expression of the Pi transporter gene PHT1;1, which is responsive to Pi starvation. As shown in Figure 2B, transcript accumulation for any of these genes was higher in the ph1;1-3 mutant than in wild-type plants when grown both in complete (1 mM) and intermediate (0.1 mM) concentrations of Pi. In line with this, quantification of soluble Pi on either Pi condition revealed that pht1;1-3 accumulates less than half the Pi accumulated by wild-type plants (Figure 2C). Therefore, pht1;1-3 exhibited a reduction in Pi content while arsenic accumulation was enhanced.
Figure 2.
pht1;1-3 Exhibits a Constitutive Pi Starvation Response.
(A) Histochemical GUS analysis of IPS1:GUS expression in wild-type and pht1;1-3 seedlings grown on Pi-lacking medium (−P) and on medium supplemented with 1 mM Pi (+P).
(B) and (C) RNA gel blot analysis of PHT1;1 and IPS1 (B) and soluble Pi content (n ≥ 3; P < 0.01) (C) in wild-type (white bars) and pht1;1-3 (black bars) seedlings grown in the presence of 1 mM or 0.1 mM Pi. Error bars represent sd.
Overexpression of pht1;1-3 Results in Decreased Pi Content and Enhanced Arsenic Accumulation
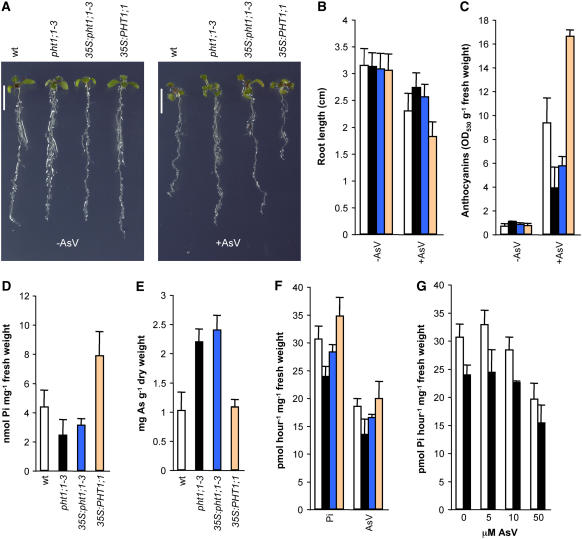
To confirm whether the pht1;1-3 allele is responsible for the observed semidominant mutant phenotypes, we obtained transgenic Arabidopsis lines in which either pht1;1-3 or PHT1;1 alleles were expressed in wild-type plants under the control of the constitutive 35S promoter (Figure 3A). No obvious phenotypic differences were observed between wild-type and any of the expressor lines in medium without As(V). However, in the presence of As(V), wild-type plants expressing pht1;1-3 displayed an As(V)-tolerant phenotype, while expression of the wild-type allele conferred hypersensitivity to the metalloid (Figure 3A). Quantification of root length and anthocyanin accumulation in these plants confirmed that the As(V) tolerance phenotypes were enhanced in the pht1;1-3 expressor line, while plants expressing the wild-type allele exhibited hypersensitivity to As(V) (Figures 3B and 3C). Analysis of soluble Pi and arsenic content in these lines showed that the expression of the mutant protein results in Pi content reduction and increased arsenic accumulation (Figures 3D and 3E, respectively). Pi and As(V) uptake experiments revealed that both Pi and As(V) influx were reduced in the mutant and in the pht1;1-3 expressor line, indicating that differential Pi versus As(V) uptake rates were not the cause of the opposite behavior in Pi and As(V) accumulation displayed by the mutant (Figure 3F). Moreover, competition Pi uptake experiments performed in wild-type and mutant plants showed that Pi uptake rate decreases in a similar proportion both in wild-type and mutant plants when exposed to increasing As(V) concentrations (Figure 3G). Therefore, differential affinity in Pi and As(V) transport was not the cause for the tolerance phenotypes observed in pht1;1-3.
Figure 3.
Phenotypic Characterization of Wild-Type Plants Overexpressing pht1;1-3.
As(V) tolerance phenotype (A), root length (n ≥ 15; P < 0.01) (B), anthocyanin accumulation (n ≥ 4; P < 0.01) (C), soluble Pi content (n ≥ 3; P < 0.1) (D), arsenic accumulation (n ≥ 3; P < 0.01) (E), Pi and As(V) uptake rates (n ≥ 3; P < 0.1) (F), and Pi uptake rates in competition with increasing As(V) concentrations (n ≥ 3; P < 0.01) (G). All plants were grown for 7 d on 30 μM Pi; plants in (D), (F), and (G) were analyzed at that point; plants in (A) to (C) were grown for an additional 4 d on the same medium supplemented with 50 μM As(V) (+AsV) or without As(V) (−AsV); plants in (E) were grown for an additional 12 d on the same medium supplemented with 50 μM As(V). Wild-type (white bars), pht1;1-3 (black bars), pht1;1-3 overexpressor (35S:pht1;1-3; blue bars), and PHT1;1 overexpressor (35S:PHT1;1; orange bars). Error bars represent sd. Bars in (A) = 0.5 cm.
These results also indicated that the expression of the mutated protein in wild-type plants accurately mimics the mutant phenotype with regard to As(V) tolerance and both Pi and As(V) accumulation.
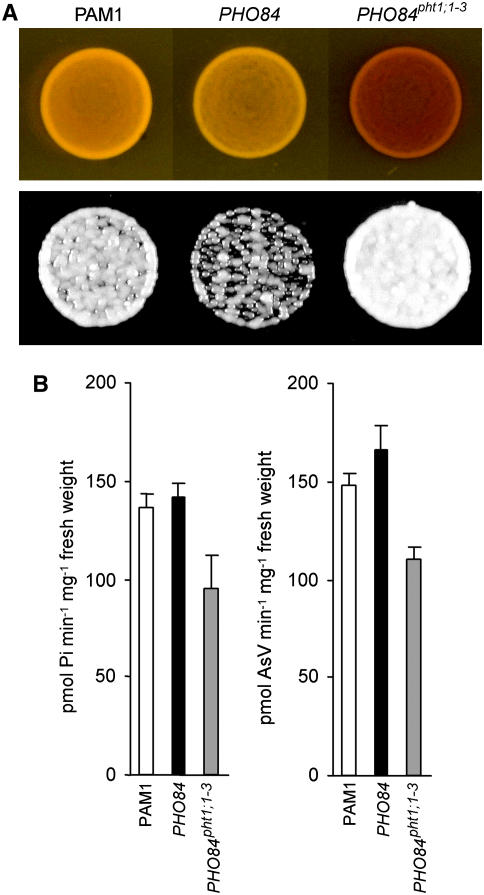
Mimicry of the pht1;1-3 Mutation in the Yeast Pho84p Transporter
The Gly residue mutated in the semidominant mutation pht1;1-3 is highly conserved not only in all the high-affinity Pi transporters from Arabidopsis, but also in other plants and from yeast. Based on this information, we decided to evaluate whether the expression of the native yeast Pi transporter carrying an equivalent mutation to that of pht1;1-3 might result in similar phenotypes to the ones observed in plants. In yeast, the Pi starvation response is easily monitored through staining for acid phosphatase activity, which drives the production of a dark-red precipitate and is highly induced when grown on low Pi medium. In this experiment, we used as the wild type the Saccharomyces cerevisae strain PAM1 (Martinez and Persson, 1998), which harbors a native copy of the high-affinity Pi transporter gene PHO84, a PHT1;1 yeast homolog. This strain was transformed with PHO84pht1;1-3, a mutagenized version of the PHO84 cDNA, encoding a Gly-to-Glu mutation identical to the one present in the pht1;1-3 allele. In the presence of 550 μM Pi, cells expressing PHO84pht1;1-3 exhibited more phosphatase activity than cells transformed with either PHO84 cDNA or the empty vector PAM1 (Figure 4A, top panel). Furthermore, Pi and As(V) uptake experiments showed that the rate of Pi and As(V) transport in cells expressing PHO84pht1;1-3 was significantly lower than in the original PAM1 (Figure 4B). No significant differences in Pi and As(V) uptake rates were observed between PAM1 and the cells expressing the wild-type PHO84. Expression of PHO84pht1;1-3 also conferred a slight increase in the tolerance to As(V) (Figure 4A, bottom panel). This increase is not as noteworthy as the one seen in pht1;1-3 overexpressors, probably due to the fact that wild-type S. cerevisiae itself exhibits a high degree of intrinsic tolerance to As(V). Therefore, we can conclude that the introduction of an equivalent pht1;1-3 Gly-to-Glu substitution in Pho84p provokes, in yeast, effects similar to those seen in Arabidopsis overexpressing pht1;1-3.
Figure 4.
Expression in Yeast of the Pi Transporter Pho84p Carrying an Equivalent Mutation to pht1;1-3.
(A) Phosphatase activity (top panel) and growth assay on As(V)-containing medium (bottom panel) of yeast PAM1 mutant cells transformed with the expression vector harboring either no insert (PAM1), PHO84 cDNA (PHO84), or the PHO84 cDNA carrying an equivalent mutation to pht1;1-3 (PHO84pht1;1-3).
(B) Pi uptake (left) and As(V) uptake (right) determined in the S. cerevisiae strains described in (A) (n ≥ 4; P < 0.01 for PHO84pht1;1-3). See Methods. Bars represent sd.
As(V) Represses the Pi Starvation Response while Activating Arsenic-Responsive Genes
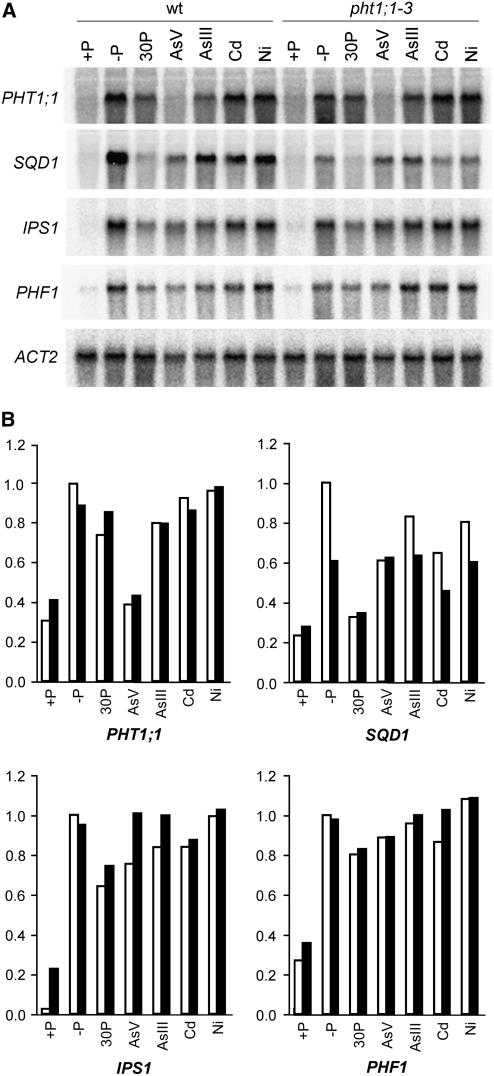
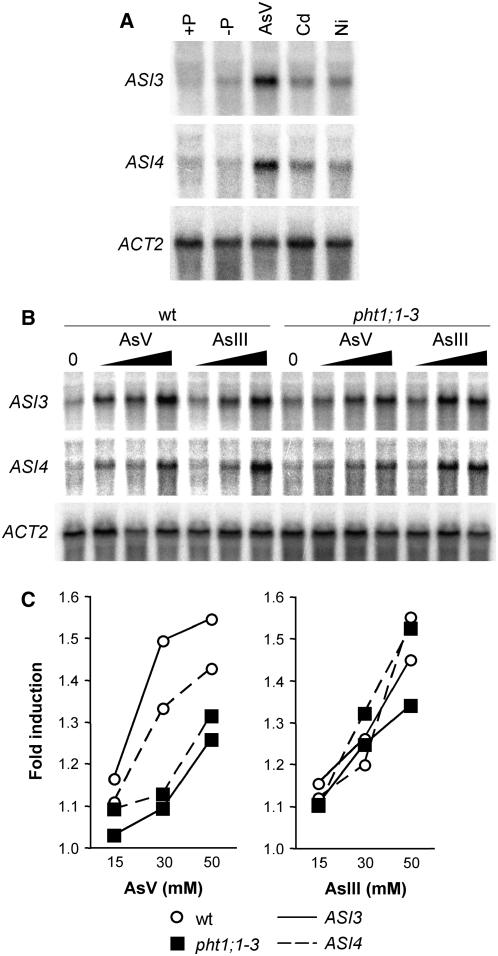
To further characterize the pht1;1-3 mutant phenotype, and because of the similarity between Pi and As(V), we next investigated the effect of As(V) on the Pi starvation response in pht1;1-3 and in wild-type plants. We performed RNA gel blot analysis of the Pi-responsive genes PHT1;1, SQD1, IPS1, and PHF1 in wild-type and pht1;1-3 plants grown in the presence of As(V). Additionally, we included in this experiment plants treated with Pi, As(III), cadmium (Cd), and nickel (Ni). As expected, all Pi-responsive genes analyzed were induced by Pi starvation (Figure 5A). When wild-type plants were then transferred for 8 h to the same medium supplemented with either 30 μM As(V) or Pi, the amount of transcript corresponding to each of the Pi-responsive genes was reduced (Figure 5B). As(V) was less efficient than Pi in the repression of SQD1, IPS1, and PHF1. By contrast, As(V) was more efficient than Pi in the repression of the Pi transporter PHT1;1. In pht1;1-3 plants, gene responsiveness to Pi starvation was reduced with respect to that in wild-type plants (Figure 5B). The reason for this behavior remains to be elucidated, but reduced gene responsiveness to Pi starvation was also observed in other mutants displaying partially constitutive Pi starvation response, such as phf1 and siz1 (González et al., 2005; Miura et al., 2005). Despite reduced responsiveness of SQD1 and IPS1 in Pi-starved mutant plants, a reduced repression of these genes was observed by Pi and particularly by As(V), for which repression was almost negligible. By contrast, PHT1;1 was completely downregulated by As(V) but not by Pi (Figure 5B). Therefore, each gene exhibits a different sensitivity to the repression by As(V) or Pi. We also examined the significance of Pi/As(V) uptake in the As(V) response. In the laboratory we have identified two As(V)-inducible genes named ASI3 and ASI4 that encode a short-chain dehydrogenase/reductase (At4g13180) and glyoxalase II (At4g33540), respectively. Regardless of its putative role in arsenic detoxification, both genes were induced in response to As(V) but not by Cd or Ni (Figure 6A), suggesting that the induction is not part of a general stress response. Actually, both genes respond to As(V) and As(III) in a dose-dependent manner (Figure 6B), although the amount of transcript detected in response to As(V) was higher in wild-type plants than in pht1;1-3 (Figure 6C). By contrast, no differences were observed in the response to As(III) of both genes between the wild-type and the mutant backgrounds (Figure 6C). These observations indicate that As(V) requires the Pi transport system to induce ASI3 and ASI4, whereas As(III) may use an independent pathway.
Figure 5.
Expression Analysis of Pi Starvation–Responsive Genes in Wild-Type and pht1;1-3 Plants.
RNA gel blot analysis (A) and densitometry analysis (B) of the expression of Pi-responsive marker genes PHT1;1, SQD1, IPS1, and PHF1. Plants were grown for 7 d on medium containing 1 mM Pi (+P), transferred to Pi-deficient medium (−P) for 3 d, and finally transferred to −P medium supplemented with either 30 μM Pi (30P), 30 μM As(V) (AsV), 30 μM As(III) (AsIII), 50 μM Cd (Cd), or 50 μM Ni (Ni) for 8 h. All intensity levels in (B) are represented as relative to −P levels in wild-type plants. White bars, wild-type plants; black bars, pht1;1-3 plants.
Figure 6.
Expression Analysis of the As(V)-Responsive Genes ASI3 and ASI4.
(A) RNA gel blot analysis of plants grown for 7 d on medium containing 1 mM Pi (+P), transferred to Pi-deficient medium (−P) for 3 d, and finally transferred to −P medium supplemented with either 30 μM As(V) (AsV), 50 μM Cd (Cd), or 50 μM Ni (Ni) for 8 h.
(B) and (C) Gel blot (B) and densitometry analysis (C) of dose-dependent expression of ASI3 (continuous line) and ASI4 (dashed line) in wild-type (white circles) and pht1;1-3 (black squares) plants. Seedlings were grown for 5 d on medium with 30 μM Pi (0) and then transferred to the same medium supplemented with 15, 30, or 50 μM (increasing slope) As(V) (AsV) or As(III) (AsIII) for 8 h. All intensity levels in (C) are represented as relative to 0 levels for each gene and genotype.
Therefore, we conclude that As(V) downregulates genes transcriptionally regulated by Pi starvation, being particularly efficient in the repression of the Pi/As(V) uptake system. The repression occurs conversely to the activation of As(V)-responsive genes.
DISCUSSION
In plants, restriction of As(V) uptake is the major strategy used by naturally selected As(V)-hypertolerant ecotypes. Actually, As(V) toxicity is based on the similarity between Pi and As(V), which allows the metalloid to be easily incorporated into plants cells through the high-affinity Pi transport system (Meharg and Macnair, 1990, 1991b, 1992b; Meharg and Hartley-Whitaker, 2002; Bleeker et al., 2003). Therefore, it is reasonable that plant adaptation to As(V)-contaminated soils has been achieved through alteration of the Pi/As(V) uptake system.
In this study, we identified an arsenic-tolerant mutant in Arabidopsis, named pht1;1-3, which harbors a new allele coding for the high-affinity Pi transporter PHT1;1. Characterization of pht1;1-3 revealed that decreased As(V) uptake contributes to enhanced arsenic content. Our investigations also uncovered the existence of an integrated Pi/As(V) signaling pathway, which modulates As(V) uptake.
The constitutive Pi starvation response displayed by pht1;1-3 is consistent with the observed reduction in Pi content, indicating that pht1;1-3 may be functionally impaired. However, despite As(V) uptake reduction, pht1;1-3 accumulates at least twice the amount of arsenic found in wild-type plants. The semidominant nature of the pht1;1-3 mutation allowed the confirmation of this association between reduced arsenic uptake and increased arsenic accumulation using transgenic plants overexpressing pht1;1-3. Indeed, pht1;1-3 overexpression affected both traits.
These two phenotypes conferred by the pht1;1-3 semidominant mutation help to explain the genetic data on arsenic tolerance of naturally selected variants of H. lanatus involving a single chromosomal region. In fact, it is unnecessary to invoke that complex loci are responsible for both apparently contradictory phenotypes.
One possible explanation for the apparent paradox represented by the association between decreased As(V) uptake and enhanced arsenic accumulation is that lowering As(V) content in the cytoplasm may allow the arsenic detoxification machinery to cope more efficiently with the metalloid, allowing for greater accumulation of arsenic into the vacuole. As(V) reduction to As(III) is a prerequisite for compartmentalization into the vacuole, and it has been recently shown that enhanced As(V) reductase activity is also a major determinant in As(V) hypertolerance (Bleeker et al., 2006). Overexpression of the As(V) reductase gene in Arabidopsis confers tolerance to low As(V). Therefore, decreasing As(V) uptake may allow As(V) reductase to process most of the As(V) present in the cytoplasm of pht1;1-3, thus enhancing As(III) sequestration into the vacuole. The fact that the response of ASI3 and ASI4 to As(V) is reduced in pht1;1-3, while As(III) responsiveness appears to be unaffected, provides further evidence that the increased tolerance of pht1;1-3 to the metalloid is due to reduced As(V) uptake in the mutant rather than enhanced arsenic responses.
The semidominant character of the pht1;1-3 mutation also has mechanistic implications. Dominant negative mutations have been reported in a wide variety of transporters (Zhou and Christie, 1997; Brockmann et al., 2001; Elumalai et al., 2002; Hahn et al., 2003; Rungroj et al., 2004), suggesting that transporters may work as dimers or higher-order oligomers (Monahan et al., 2002; Reinders et al., 2002; Ludewig et al., 2003). In fact, there is evidence that Pi transporters in Arabidopsis and Medicago truncatula may form functional dimers (Chiou et al., 2001; Shin et al., 2004). Our finding that a pht1;1-3 equivalent allele of the PHO84 yeast Pi transporter interferes with Pi/As(V) uptake in yeast suggests that oligomer formation in Pi transporters is universal.
The results presented here indicate that As(V) rapidly repressed genes involved in the Pi starvation response and induced the expression of other As(V)-responsive genes. While the repressor function on Pi starvation–responsive genes is specific for As(V), induction of arsenic-responsive genes is also mediated by As(III). This leads us to propose a model whereby arsenic acts through two different signaling pathways. Based on the analogy between Pi and As(V), we propose that As(V) could mislead the Pi sensor, thus triggering the repression of the Pi starvation–responsive genes. In that case, As(V) should act as a nonmetabolizable Pi analog as it was seen for phosphite (Ticconi et al., 2001; Varadarajan et al., 2002; Giots et al., 2003; Pratt et al., 2004). It is noticeable that repression by As(V) is not equally efficient for all Pi starvation–responsive genes, being most efficient in the repression of the Pi transporter even more than Pi itself, which is incorporated faster than As(V). The outstanding sensitivity of the Pi transporter to As(V) prompts us to speculate that the repression of Pi starvation responses by arsenic is not only an obvious consequence of the chemical similarity between As(V) and Pi but also reflects a natural selection process that capitalizes on this situation to ensure efficient repression of Pi starvation responses, particularly of the Pi transporter. On the other hand, given the fact that once As(V) enters the cell it is rapidly reduced to As(III) (Pickering et al., 2000; Meharg and Hartley-Whitaker, 2002), which has no chemical similarity to Pi and therefore no chance to confound the Pi sensor, we propose that activation of ASI3 and ASI4 occurs via an As(III) signaling pathway.
In conclusion, we propose that, due to the chemical similarity between As(V) and Pi, plants have evolved an integrated sensing mechanism in which As(V) and Pi signaling pathways act in opposition to preserve plant integrity from arsenic toxicity. Our data open the possibility of further evaluating the As(V) response to identify exclusive, or overlapping, elements in As(V), As(III), and Pi starvation responses that may be relevant to arsenic perception and accumulation.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotypes used in this study were Columbia (Col-0) and Ler. Seeds were surface-sterilized and plated onto Bates and Lynch medium (Bates and Lynch, 1996) and solidified with 0.6% bacto-agar (Difco), which was supplemented with appropriate amounts of Pi (KH2PO4) or As(V) (NaH2AsO4·7H2O), unless otherwise specified. Growth conditions were the same as described previously (Franco-Zorrilla et al., 2002). The mutant pht1;1-3 was selected from an ethyl methanesulfonate–mutagenized population derived from a Col transgenic line harboring the Pi starvation reporter construct IPS1:GUS (Rubio et al., 2001). asi (arsenic-induced) lines were identified in the laboratory from an F3 progeny of a transposant line collection generated in the Exon trapping insert consortium (EXOTIC QLG2-CT-1999-000351; http://www.jic.bbsrc.ac.uk/science/cdb/exotic/). Flanking DNA from asi lines was cloned by thermal asymmetric interlaced PCR (Liu et al., 1995).
Plant Measurements and Histochemical Staining
The method by Ames (1966) was used to quantify soluble Pi, and histochemical GUS stainings were performed as described by Martín et al. (2000). Roots were analyzed after 7 d (soluble Pi) or 8 d (GUS and root hairs) of culture in the desired Pi concentration. All P values were obtained through a Student's t test.
For measuring root length, anthocyanin accumulation, and As(V) accumulation, plants were cultured for 7 d on 30 μM Pi medium and then transferred to fresh medium supplemented with 50 μM As(V). Samples were collected 4 d (root length and anthocyanin accumulation) or 12 d (arsenic content) later. To measure As content, plants were dried at 60°C for 5 d, mineralized with HNO4-H2O2 in a pressure digester, and analyzed for total arsenic content through inductively coupled plasma–mass spectrometry (ICP-MS) at the Centro de Espectrometría Atómica of the Universidad Complutense de Madrid. Anthocyanins were measured according to Swain and Hillis (1959).
Plant Uptake Experiments
Plates used in preculture for both Pi and As(V) uptake experiments contained half-strength Bates and Lynch medium solidified with 0.4% bacto-agar supplemented with 30 μM Pi and were covered with 0.4-mm-pore nylon mesh. This medium permits solid culture, long root production, and easy root cleaning. Plants were sown onto the mesh and cultured for 7 d before being carefully extracted and washed with sterile water. Plants were then treated with PI and PII buffers as by Narang et al. (2000). Pi uptake experiments were performed in 3-mL wells at 5 μM KH232PO4 (ICN) and competing As(V) when suitable. As(V) uptake experiments were performed in 400-mL pots at 5 μM As(V). The duration of each uptake experiment was 1 h. Plants were introduced in either scintillation vials for 32Pi measurements or in 50-mL tubes for As(V) measurements and dried at 60°C for 5 d. 32Pi was measured in a scintillation counter with 10 mL of scintillation liquid (ICN), and arsenic content was analyzed trough ICP-MS as described above.
Genetic Analysis and Positional Cloning of pht1;1-3
pht1;1-3 plants were backcrossed three times to wild-type plants. Backcrossed seeds were crossed to the Ler ecotype. Positional cloning was performed by selecting F2 seedlings displaying the mutant phenotype and homozygous for the PHT1 locus according to the segregation of the mutant phenotype in their respective F3 offspring. DNA from these F2 selected plants was prepared (Dellaporta et al., 1983) and used to analyze the linkage of the pht1;1-3 mutation to previously described genetic markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). To identify the mutant gene, we used the Cereon collection of Col-Ler genetic polymorphisms (http://www.Arabidopsis.org/cereon/index.html).
cDNA Isolation and Overexpression
pht1;1-3 and PHT1;1 cDNA were amplified by RT-PCR using total RNA extracted from Pi-starved plants using primers 5′-CCTCAACTCTCCAGAGAAGTTC-3′ and 5′-ACATCATAACTTAAGGTCAACGAG-3′. These cDNAs were cloned into plasmid pBIBA7, a 35S promoter–containing vector derived from pBIB (Becker, 1990). The second intron of PHT1;1, amplified from genomic DNA using primers 5′-CAAGTTGTTCTATGGTCCAATGTTCG-3′ and 5′-CTGTTTTCAATCTTCTACGTACGA-3′, was inserted in its place in both cDNAs. This construct was necessary to avoid toxicity to Agrobacterium tumefaciens. Plants were transformed as by Bechtold et al. (1993).
RNA Extraction and Gel Blot Analysis
Total RNA was extracted with RNAwiz (Ambion), and 20 μg were loaded per sample and blotted onto Hybond N+ membranes (Amersham) as indicated by the manufacturer. Hybridizations were performed as by Church and Gilbert (1984), and membranes were washed with 0.5× SSC before exposing. Probes corresponding to IPS1 (Martín et al., 2000), SQD1 (Essigmann et al., 1998), PHF1 (González et al., 2005), and PHT1;1 (Smith et al., 1997) were obtained as by Franco-Zorrilla et al. (2005); probes corresponding to ASI3 and ASI4 were obtained by PCR amplification of the coding region on plant DNA.
Yeast Transformation and Constructs
The PHO84pht1;1-3 point mutation was introduced in PHO84 cDNA (kindly provided by Georg Leggewie) through directed mutagenesis using primers 5′-CAAGATGATGAATTCGGCTAATTGAATTG-3′ and 5′-TCAATTAGCCGAATTCATCATCTTGAC-3′ containing the equivalent point mutation found in pht1;1-3 (highlighted in boldface). Both PHO84pht1;1-3 and PHO84 cDNAs were cloned into the yeast expression vector p181A1NE (Leggewie et al., 1997). Both constructs were transformed into Saccharomyces cerevisiae PAM1 (kindly provided by Bengt Persson) (Martinez and Persson, 1998), according to Gietz and Woods (2002). Yeast was grown on YNB medium (Difco) supplemented with 2% sucrose (Calbiochem), unless stated otherwise. For acid phosphatase activity, we designed a Pi-deficient medium (SW medium) following the Wicherham formula (Sigma-Aldrich), in which appropriate amounts of KCl were substituted for KH2PO4, as done by Lau et al. (1998).
Yeast Acid Phosphatase Activity and Tolerance Experiments
Acid phosphatase activity was assayed in growing yeast cells in liquid YNB medium overnight, which was then diluted to OD660 = 0.1. Droplets (3 μL) of the diluted culture were spotted onto SW plates with 550 μM KH2PO4, in which sucrose was replaced by 3% glycerol (Bun-ya et al., 1991). Acid phosphatase activity was assayed as by Toh-e et al. (1973). Tolerance to As(V) was assayed in growing yeast cells in liquid YNB medium overnight. Droplets (3 μL) of media diluted to OD660 = 0.1 were spotted onto YPD media supplemented with 3.5 mM As(V) as described by Bun-ya et al. (1996).
Yeast Uptake Experiments
Pi uptake experiments were performed as by Daram et al. (1999) using carrier-free H332PO4 (ICN). Uptake took 4 minutes at 30°C in a 40 μL mix; Pi final concentration was 500 μM, and pH was adjusted to 6.5. Samples were then diluted to 10 mL with chilled water and filtered trough Me25 membranes held by 3-cm polysulphone holders (both from Schleicher and Schuell). Membranes were washed twice with chilled water before being dried on 3M paper and introduced into scintillation vials for counting. As(V) uptake experiments were scaled to a 40-mL uptake mixture in 50-mL screw cap tubes (Falcon), and uptake was stopped by immersing tubes in chilled water, followed quickly by centrifugation. Cells were then washed twice with chilled water, and pellets were dried at 60°C for 5 d. Total As was measured through ICP-MS as described above.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are At5g43350 (PHT1;1), At3g52190 (PHF1), At3g09922 (IPS1), At4g33030 (SQD1), At4g13180 (ASI3), and At33540 (ASI4).
Acknowledgments
We thank Georg Leggewie for kindly providing the PHO84 cDNA in the yeast expression vector p181A1NE and Bengt Persson for providing the S. cerevisiae strain PAM1. We also thank C.L. Torán for critical reading of the manuscript. The excellent technical assistance of María Jesús Benito and Yolanda Leo del Puerto is also acknowledged. P.C. and M.L. were supported by Spanish Ministry of Education Grants BIO99-0229 and BIO2001-1204, respectively; B.G.-P. was supported by a postdoctoral contract of the Consejo Nacional de Ciencia y Tecnología (010116). This research was supported by the EXOTIC grant (Contract QLG2-CT-1999-000351) funded by the 5th European Framework Program, by the Comunidad de Madrid (Contract 07B/0037/2002), and by the Spanish Comisión Interministerial de Ciencia y Tecnología (Contract BIO2001-1204).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Antonio Leyva (aleyva@cnb.uam.es).
References
- Ames, B.N. (1966). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8 115–118. [Google Scholar]
- Bates, T.R., and Lynch, J.P. (1996). Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 19 529–538. [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. III, Sci Vie 316 1194–1199. [Google Scholar]
- Becker, D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Bieleski, R.L. (1973). Phosphate pools, phosphate transport, and phosphate availability. Annu. Rev. Plant Physiol. 27 225–252. [Google Scholar]
- Bleeker, P.M., Hakvoort, H.W., Bliek, M., Souer, E., and Schat, H. (2006). Enhanced arsenate reduction by a CDC25-like tyrosine phosphatase explains increased phytochelatin accumulation in arsenate-tolerant Holcus lanatus. Plant J. 45 917–929. [DOI] [PubMed] [Google Scholar]
- Bleeker, P.M., Schat, H., Vooijs, R., Verkleij, J.A.C., and Ernst, W.H.O. (2003). Mechanisms of arsenate tolerance in Cytisus striatus. New Phytol. 157 33–38. [DOI] [PubMed] [Google Scholar]
- Brockmann, K., et al. (2001). Autosomal dominant glut-1 deficiency syndrome and familial epilepsy. Ann. Neurol. 50 476–485. [DOI] [PubMed] [Google Scholar]
- Brown, G.E., Jr., Foster, A.L., and Ostergren, J.D. (1999). Mineral surfaces and bioavailability of heavy metals: A molecular-scale perspective. Proc. Natl. Acad. Sci. USA 96 3388–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bun-ya, M., Nishimura, M., Harashima, S., and Oshima, Y. (1991). The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell. Biol. 11 3229–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bun-ya, M., Shikata, K., Nakade, S., Yompakdee, C., Harashima, S., and Oshima, Y. (1996). Two new genes, PHO86 and PHO87, involved in inorganic phosphate uptake in Saccharomyces cerevisiae. Curr. Genet. 29 344–351. [PubMed] [Google Scholar]
- Chiou, T.J., Liu, H., and Harrison, M.J. (2001). The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J. 25 281–293. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson, D.T., and Hanson, J.B. (1980). The mineral nutrition of higher plants. Annu. Rev. Plant Physiol. 31 239–298. [Google Scholar]
- Clemens, S., Kim, E.J., Neumann, D., and Schroeder, J.I. (1999). Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J. 18 3325–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daram, P., Brunner, S., Rausch, C., Steiner, C., Amrhein, N., and Bucher, M. (1999). Pht2;1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell 11 2153–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. R1 19–21. [Google Scholar]
- Elumalai, R.P., Nagpal, P., and Reed, J.W. (2002). A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 14 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann, B., Guler, S., Narang, R.A., Linke, D., and Benning, C. (1998). Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95 1950–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz, W.J., and Wenzel, W.W. (2002). Arsenic transformations in the soil-rhizosphere-plant system: Fundamentals and potential application to phytoremediation. J. Biotechnol. 99 259–278. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla, J.M., Martín, A.C., Leyva, A., and Paz-Ares, J. (2005). Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 138 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla, J.M., Martín, A.C., Solano, R., Rubio, V., Leyva, A., and Paz-Ares, J. (2002). Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation responses in Arabidopsis. Plant J. 32 353–360. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and Woods, R.A. (2002). Transformation of yeast by the LiAc/SS carrier DNA/PEG method. Methods Enzymol. 350 87–96. [DOI] [PubMed] [Google Scholar]
- Giots, F., Donaton, M.C., and Thevelein, J.M. (2003). Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 47 1163–1181. [DOI] [PubMed] [Google Scholar]
- Gómez, G., Baos, R., Gómara, B., Jiménez, B., Benito, V., Montoro, R., Hiraldo, F., and González, M.J. (2004). Influence of a mine tailing accident near Doñana National Park (Spain) on heavy metals and arsenic accumulation in 14 species of waterfowl (1998 to 2000). Arch. Environ. Contam. Toxicol. 47 521–529. [DOI] [PubMed] [Google Scholar]
- González, E., Solano, R., Rubio, V., Leyva, A., and Paz-Ares, J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17 3500–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M.K., Robertson, D., and Blakely, R.D. (2003). A mutation in the human norepinephrine transporter gene (SLC6A2) associated with orthostatic intolerance disrupts surface expression of mutant and wild-type transporters. J. Neurosci. 23 4470–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz, S.A., and Salem, H. (2005). Chemistry and toxicology of building timbers pressure-treated with chromated copper arsenate: A review. J. Appl. Toxicol. 25 1–7. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Lau, W.W., Schneider, K.R., and O'Shea, E.K. (1998). A genetic study of signaling processes for repression of PHO5 transcription in Saccharomyces cerevisiae. Genetics 150 1349–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.A., Chen, A., and Schroeder, J.I. (2003). ars1, an Arabidopsis mutant exhibiting increased tolerance to arsenate and increased phosphate uptake. Plant J. 35 637–646. [DOI] [PubMed] [Google Scholar]
- Leggewie, G., Willmitzer, L., and Riesmeier, J.W. (1997). Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: Identification of phosphate transporters from higher plants. Plant Cell 9 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8 457–463. [DOI] [PubMed] [Google Scholar]
- Lombi, E., Zhao, F.J., Fuhrmann, M., Ma, L.Q., and McGrath, S.P. (2002). Arsenic distribution and speciation in the fronds of the hyperaccumulator Pteris vittata. New Phytol. 156 195–203. [DOI] [PubMed] [Google Scholar]
- Ludewig, U., Wilken, S., Wu, B., Jost, W., Obrdlik, P., El Bakkoury, M., Marini, A.M., André, B., Hamacher, T., Boles, E., von Wirén, N., and Frommer, W.B. (2003). Homo- and hetero-oligomerization of ammonium transporter-1 NH4+ uniporters. J. Biol. Chem. 278 45603–45610. [DOI] [PubMed] [Google Scholar]
- Macnair, M.R., Cumbes, Q.J., and Meharg, A.A. (1992). The genetics of arsenate tolerance in Yorkshire fog Holcus lanatus L. Heredity 69 325–335. [DOI] [PubMed] [Google Scholar]
- Martín, A.C., del Pozo, J.C., Iglesias, J., Rubio, V., Solano, R., de La Peña, A., Leyva, A., and Paz-Ares, J. (2000). Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 24 559–567. [DOI] [PubMed] [Google Scholar]
- Martinez, P., and Persson, B.L. (1998). Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol. Gen. Genet. 258 628–638. [DOI] [PubMed] [Google Scholar]
- Meharg, A.A., and Hartley-Whitaker, J. (2002). Arsenic uptake and metabolism in arsenic resistant and nonresistant plant species. New Phytol. 154 29–43. [Google Scholar]
- Meharg, A.A., and Macnair, M.R. (1990). An altered phosphate uptake system in arsenate-tolerant Holcus lanatus L. New Phytol. 116 29–35. [Google Scholar]
- Meharg, A.A., and Macnair, M.R. (1991. a). Uptake, accumulation and translocation of arsenate in arsenate-tolerant and non-tolerant Holcus lanatus L. New Phytol. 117 225–231. [Google Scholar]
- Meharg, A.A., and Macnair, M.R. (1991. b). The mechanism of arsenate tolerance in Deschampsia cespitosa (L.) Beauv. and Agrostis capillaris L. New Phytol. 119 291–297. [DOI] [PubMed] [Google Scholar]
- Meharg, A.A., and Macnair, M.R. (1992. a). Genetic correlation between arsenate tolerance and the rate of influx of arsenate and phosphate in Holcus lanatus L. Heredity 69 336–341. [Google Scholar]
- Meharg, A.A., and Macnair, M.R. (1992. b). Suppression of the high affinity phosphate uptake system: A mechanism of arsenate tolerance in Holcus lanatus L. J. Exp. Bot. 43 519–524. [Google Scholar]
- Miura, K., Rus, A., Sharkhuu, A., Yokoi, S., Karthikeyan, A.S., Raghothama, K.G., Baek, D., Koo, Y.D., Jin, J.B., Bressan, R.A., Yun, D.J., and Hasegawa, P.M. (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA 102 7760–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan, B.J., Unkles, S.E., Tsing, I.T., Kinghorn, J.R., Hynes, M.J., and Davis, M.A. (2002). Mutation and functional analysis of the Aspergillus nidulans ammonium permease MeaA and evidence for interaction with itself and MepA. Fungal Genet. Biol. 36 35–46. [DOI] [PubMed] [Google Scholar]
- Narang, R.A., Bruene, A., and Altmann, T. (2000). Analysis of phosphate acquisition efficiency in different Arabidopsis accessions. Plant Physiol. 124 1786–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickson, R., McArthur, J., Burgess, W., Ahmed, K.M., Ravenscroft, P., and Rahman, M. (1998). Arsenic poisoning of Bangladesh groundwater. Nature 395 338. [DOI] [PubMed] [Google Scholar]
- Nordstrom, D.K. (2002). Public health. Worldwide occurrences of arsenic in ground water. Science 296 2143–2145. [DOI] [PubMed] [Google Scholar]
- Oremland, R.S., and Stolz, J.F. (2003). The ecology of arsenic. Science 300 939–944. [DOI] [PubMed] [Google Scholar]
- Pickering, I.J., Prince, R.C., George, M.J., Smith, R.D., George, G.N., and Salt, D.E. (2000). Reduction and coordination of arsenic in Indian mustard. Plant Physiol. 122 1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, J.R., Mouillon, J.M., Lagerstedt, J.O., Pattison-Granberg, J., Lundh, K.I., and Persson, B.L. (2004). Effects of methylphosphonate, a phosphate analogue, on the expression and degradation of the high-affinity phosphate transporter Pho84, in Saccharomyces cerevisiae. Biochemistry 43 14444–14453. [DOI] [PubMed] [Google Scholar]
- Raab, A., Feldmann, J., and Meharg, A.A. (2004). The nature of arsenic-phytochelatin complexes in Holcus lanatus and Pteris cretica. Plant Physiol. 134 1113–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama, K.G. (1999). Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 665–693. [DOI] [PubMed] [Google Scholar]
- Reinders, A., Schulze, W., Kühn, C., Barker, L., Schulz, A., Ward, J.M., and Frommer, W.B. (2002). Protein-protein interactions between sucrose transporters of different affinities colocalized in the same enucleate sieve element. Plant Cell 14 1567–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio, V., Linhares, F., Solano, R., Martín, A.C., Iglesias, J., Leyva, A., and Paz-Ares, J. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungroj, N., Devonald, M.A., Cuthbert, A.W., Reimann, F., Akkarapatumwong, V., Yenchitsomanus, P.T., Bennett, W.M., and Karet, F.E. (2004). A novel missense mutation in AE1 causing autosomal dominant distal renal tubular acidosis retains normal transport function but is mistargeted in polarized epithelial cells. J. Biol. Chem. 279 13833–13838. [DOI] [PubMed] [Google Scholar]
- Salt, D.E., and Rauser, W.E. (1995). MgATP-dependent transport of phytochelatins across the tonoplast of oat roots. Plant Physiol. 107 1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, H., Shin, H.S., Dewbre, G.R., and Harrison, M.J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39 629–642. [DOI] [PubMed] [Google Scholar]
- Smith, F.W., Ealing, P.M., Dong, B., and Delhaize, E. (1997). The cloning of two Arabidopsis genes belonging to a phosphate transporter family. Plant J. 11 83–92. [DOI] [PubMed] [Google Scholar]
- Swain, T., and Hillis, H.E. (1959). Phenolic constituents of Prunus domestica. I. Quantitative analysis of phenolic constituents. J. Sci. Food Agric. 10 63–68. [Google Scholar]
- Tamaki, S., and Frankenberger, W.T., Jr. (1992). Environmental biochemistry of arsenic. Rev. Environ. Contam. Toxicol. 124 79–110. [DOI] [PubMed] [Google Scholar]
- Ticconi, C.A., Delatorre, C.A., and Abel, S. (2001). Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol. 127 963–972. [PMC free article] [PubMed] [Google Scholar]
- Toh-e, A., Ueda, Y., Kakimoto, S.I., and Oshima, Y. (1973). Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J. Bacteriol. 113 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan, D.K., Karthikeyan, A.S., Matilda, P.D., and Raghothama, K.G. (2002). Phosphite, an analog of phosphate, suppresses the coordinated expression of genes under phosphate starvation. Plant Physiol. 129 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, F.J., Wang, J.R., Barker, J.H.A., Schat, H., Bleeker, P.M., and McGrath, S.P. (2003). The role of phytochelatins in arsenic tolerance in the hyperaccumulator Pteris vittata. New Phytol. 159 403–410. [DOI] [PubMed] [Google Scholar]
- Zhou, X.R., and Christie, P.J. (1997). Suppression of mutant phenotypes of the Agrobacterium tumefaciens VirB11 ATPase by overproduction of VirB proteins. J. Bacteriol. 179 5835–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]