Abstract
Deg1 is a Ser protease peripherally attached to the lumenal side of the thylakoid membrane. Its physiological function is unknown, but its localization makes it a suitable candidate for participation in photoinhibition repair by degradation of the photosystem II reaction center protein D1. We transformed Arabidopsis thaliana with an RNA interference construct and obtained plants with reduced levels of Deg1. These plants were smaller than wild-type plants, flowered earlier, were more sensitive to photoinhibition, and accumulated more of the D1 protein, probably in an inactive form. Two C-terminal degradation products of the D1 protein, of 16 and 5.2 kD, accumulated at lower levels compared with the wild type. Moreover, addition of recombinant Deg1 to inside-out thylakoid membranes isolated from the mutant could induce the formation of the 5.2-kD D1 C-terminal fragment, whereas the unrelated proteases trypsin and thermolysin could not. Immunoblot analysis revealed that mutants containing less Deg1 also contain less FtsH protease, and FtsH mutants contain less Deg1. These results suggest that Deg1 cooperates with the stroma-exposed proteases FtsH and Deg2 in degrading D1 protein during repair from photoinhibition by cleaving lumen-exposed regions of the protein. In addition, they suggest that accumulation of Deg1 and FtsH proteases may be coordinated.
INTRODUCTION
Bacterial DegP (or HtrA) is a Ser protease complex peripherally attached to the periplasmic side of the plasma membrane. It is best characterized in Escherichia coli, in which it is essential for survival at increased temperatures (for review, see Clausen et al., 2002). Determination of its three-dimensional structure has revealed that it forms a hexamer made of two staggered trimers. The 48-kD monomer is composed of two domains: an N-terminal domain, in which the typical catalytic triad of Ser proteases, Ser-Asp-His, is found; and a C-terminal domain with two PDZ domains, implicated in protein–protein interactions. E. coli has two DegP homologs, DegQ (HhoA) and DegS (HhoB) (Clausen et al., 2002). In eukaryotes, a homolog of the DegP protease, designated HtrA2, is found in mitochondria, where it is involved in apoptosis. This enzyme forms a trimer whose three-dimensional structure has also been determined (Li et al., 2002). Although the proteolytic activity of bacterial DegP is independent of ATP, it exhibits chaperone activity as well. Interestingly, at normal growth temperature, DegP is active as a chaperone, whereas the proteolytic activity dominates at higher temperatures (Spiess et al., 1999). This temperature-dependent switch between the two different activities can now be explained by the structure of the protein. At normal growth temperature, the active site is blocked by segments of the protein itself, and it is only upon a thermal-induced conformational change that it becomes accessible to substrates (Clausen et al., 2002; Krojer et al., 2002).
The first plant homolog of DegP, designated Deg1 (formerly DegP or DegP1), was found peripherally attached to the lumenal side of the thylakoid membrane (Itzhaki et al., 1998). Unlike the bacterial DegP, which has two PDZ domains in tandem, Deg1 contains only one such domain. Similar to the bacterial enzyme, it forms hexamers and its activity is stimulated by high temperature (Chassin et al., 2002). Another two homologs, designated Deg5 and Deg8, were also predicted to reside in the lumen, and proteomic analyses confirmed this prediction (Peltier et al., 2002; Schubert et al., 2002). Deg5 is very similar to Deg1 but does not contain a PDZ domain, whereas Deg8 is somewhat less similar but does have a PDZ domain; hence, its mature size is almost identical to that of Deg1, ∼35 kD. Chloroplasts contain at least one more distantly related homolog, Deg2, peripherally attached to the stromal side of the thylakoid membrane (Haussuhl et al., 2001). Ten other plant homologs are predicted to reside in chloroplasts, mitochondria, and maybe other cellular sites (Huesgen et al., 2005). It should be noted that all Arabidopsis thaliana Deg proteins were initially designated DegPs. However, because Deg1, Deg5, and Deg8 are more closely related to each other than to their E. coli homologs, it was recently suggested that they be renamed Degs (Huesgen et al., 2005).
Expression studies of Deg1, Deg2, and Deg8 have revealed that their transcript levels increase threefold to fivefold in response to exposure of Arabidopsis seedlings to high light intensity, but they do not change in response to exposure to either high or low temperature (Sinvany-Villalobo et al., 2004). Similar trends have been observed in data from Affymetrix experiments, the results of which are deposited in publicly available databases (Zimmermann et al., 2004). Nevertheless, very little is known about the function of Deg proteases in chloroplasts. Recombinant Deg1 was capable of degrading the model substrate β-casein in an in vitro assay in a temperature- and pH-dependent manner, as well as potential lumenal substrates such as in vitro–translated plastocyanin and OE33 (Chassin et al., 2002). In an in vitro study, recombinant Deg2 was demonstrated to cleave the D1 protein of photosystem II (PSII) after exposure to high light intensity (Haussuhl et al., 2001). However, a recent in vivo study with mutants lacking Deg2 demonstrated that their rate of D1 degradation under light stress conditions was comparable to that of wild-type plants, suggesting that Deg2 is not essential for D1 degradation (Huesgen et al., 2006). No other in vivo studies of the function of plant Deg proteases have been reported to date.
PSII captures the energy of sunlight and catalyzes the oxidation of water and the reduction of plastoquinone in the photosynthetic electron-transport chain. These reactions involve a dangerous stepwise accumulation of highly oxidizing species within PSII, as well as oxygen radicals, which cause photodamage to the photosynthetic machinery, especially PSII (Andersson and Aro, 2001; Yamamoto, 2001). Damage to PSII occurs at all light intensities, but net loss of PSII activity occurs only when the rate of damage exceeds the rate of repair. This situation is referred to as photoinhibition (Aro et al., 1993b). The repair cycle of PSII involves transcription and translation of the chloroplast psbA gene, encoding the D1 protein, but a prerequisite for successful repair is the degradation of oxidatively damaged copies of the protein. Although D1 degradation has been characterized for the past two decades, the first insights into the identity of the proteases involved have only been obtained in recent years. In vitro and in vivo studies have suggested that the FtsH protease participates in this process (Lindahl et al., 2000; Bailey et al., 2002; Sakamoto et al., 2002).
In the absence of any information on the function of thylakoid lumen proteases, Deg1 protease in particular, we sought to determine its physiological function. We hypothesized that its localization to the inner side of the thylakoid membrane makes it a suitable candidate for participation in the degradation of the D1 protein of PSII, in the context of repair from photoinhibition. We demonstrate here that deg1 mutant lines that accumulate reduced levels of Deg1 are more sensitive to photoinhibition than the wild type, that they accumulate higher levels of the D1 protein and less of its C-terminal degradation products, and that one of these fragments can be generated in vitro upon the addition of recombinant Deg1 to inverted thylakoid membranes. These results suggest that Deg1 is indeed involved in degradation of the D1 protein during the process of PSII repair from photoinhibition.
RESULTS
RNA Interference Mutants Contain Reduced Levels of Deg1
Our attempts to obtain homozygous T-DNA insertion lines have failed. Thus, to obtain transgenic plants with reduced levels of Deg1, we generated a transformation construct in which an intron sequence was flanked by a 186-bp specific fragment of the Deg1 gene in the sense and antisense orientations. This construct, under the control of the constitutive promoter 35S and containing a Basta resistance gene, was introduced into Arabidopsis plants by Agrobacterium tumefaciens–mediated transformation. Transformants were selected by virtue of their resistance to Basta and further analyzed by PCR to confirm the presence of the transgene. To assess the level of Deg1 in the transgenic plants, immunoblot analysis was performed, normalizing the level of Deg1 to the level of OE33, a subunit of the oxygen-evolving complex, which like Deg1 is also peripherally attached to the lumenal side of the thylakoid membrane. This analysis revealed that different mutant lines contained 10 to 80% Deg1 protein relative to the wild type (Figure 1A). It should be noted that the level of Deg1 differed not only between lines but also between different plants of the same line grown under different conditions. Thus, all further experiments were performed on plants containing ∼50% Deg1 relative to the wild type.
Figure 1.
Phenotype of deg1 Mutants.
(A) Immunoblot analysis. Protein samples from wild-type and deg1 mutant plants were subjected to immunoblot analysis with antibodies against Deg1 and OE33. Chl, chlorophyll.
(B) Visual phenotypes of 4-week-old representative wild-type and mutant plants.
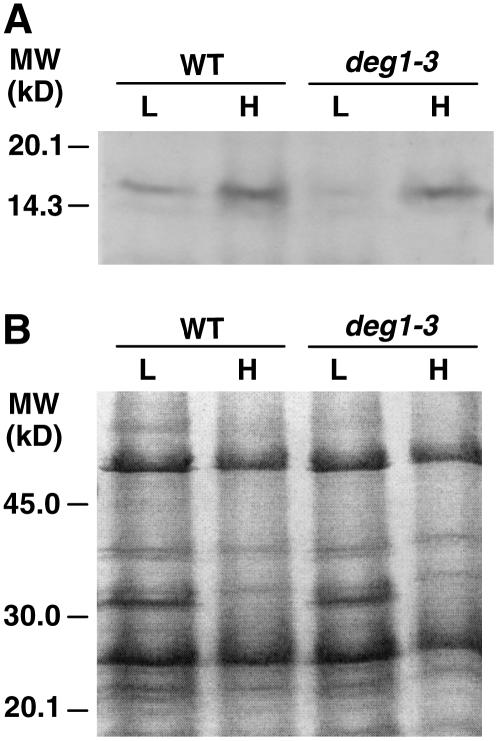
A reduced level of Deg1 protein in the mutants correlated with reduced vegetative biomass. These plants were smaller than wild-type plants, their leaves were thin, and their color was pale green (Figure 1B). Another significant visual difference between wild-type and mutant plants was the time required for flowering; as often occurs in stressed plants, deg1 mutant plants flowered earlier than wild-type plants, with fewer leaves. Consistent with the pale phenotype, mutant plants contained less chlorophyll b (see Supplemental Table 1 online). However, protein profiles of mutant plants containing less Deg1, as judged by SDS-PAGE, were similar to those of wild-type plants (see Supplemental Figure 1 online).
deg1 Mutants Are More Sensitive to Photoinhibition
The chlorophyll a fluorescence parameter Fv/Fm measures the maximum efficiency of PSII photochemistry. It correlates with both functional PSII reaction centers and the quantum yield of light-induced oxygen evolution; thus, a decrease in this parameter is indicative of photoinhibition. To test whether Deg1 is involved in the repair cycle of PSII during photoinhibition, deg1 and wild-type plants, grown at 75 to 80 μmol photons·m−2·s−1, were exposed to 17-fold higher light intensity for 3 h. As expected, the potential of PSII in wild-type plants decreased gradually, but the loss of activity in the deg1-2 and deg1-3 plants was more dramatic (Figure 2A). (All other lines containing reduced levels of Deg1 demonstrated similar sensitivity to photoinhibition.) Similar experiments were performed in the presence of lincomycin, an inhibitor of protein translation in chloroplasts. As shown in Figure 2B, wild-type leaves preloaded with the inhibitor were more sensitive to exposure to high light than leaves treated with water. This effect is attributed to a decreased rate of synthesis of the D1 protein of the PSII reaction center and, thus, an inability to replace the damaged and degraded D1 copies. A similar, albeit much more pronounced, effect was observed in the deg1 mutants. Here, the decrease in PSII potential in the absence of lincomycin was even greater than the decrease observed in wild-type leaves in the presence of the inhibitor (Figure 2B). These results suggest that the decrease in the level of Deg1 in the deg1 plants renders them more sensitive to photoinhibition, possibly as a result of a decreased ability to degrade the photodamaged D1 protein.
Figure 2.
PSII Potential in Mutant and Wild-Type Plants.
(A) Four-week-old plants grown at 75 to 80 μmol photons·m−2·s−1 were exposed to 1400 μmol photons·m−2·s−1 for up to 3 h. PSII potential was calculated from chlorophyll fluorescence values. Values are means ± sd of five replicates.
(B) Wild-type (white bars) and mutant (black bars) leaves were preloaded with either water or 1 mM lincomycin before exposure to high light (HL) for 2 h. PSII potential was calculated from chlorophyll fluorescence values before and after the exposure. Values are means ± sd of five replicates.
(C) Mutant (black bars) and wild-type (white bars) plants were grown for 3 weeks at 80 μmol photons·m−2·s−1. They were then transferred for 1 week to different light intensities, and their PSII potential was measured before and after exposure to 1400 μmol photons·m−2·s−1 for 2.5 h. The decrease in PSII potential was calculated using the equation [(Fv/Fm before HL) – (Fv/Fm after HL)]/(Fv/Fm before HL) × 100. Values are means ± se of five replicates.
To test how light intensity affects sensitivity to photoinhibition, we grew plants for 3 weeks at 80 to 100 mmol photons·m−2·s−1. They were transferred for 1 week to five different light regimes, ranging from 15 to 470 mmol photons·m−2·s−1, and their sensitivity to photoinhibition was assayed. As expected, wild-type plants grown at higher light intensities acclimated better and demonstrated lower sensitivity to photoinhibition than those grown at lower intensities. When exposed to high light intensity (1400 mmol photons·m−2·s−1), wild-type plants grown at 470 mmol photons·m−2·s−1 lost only 5% of their PSII activity, compared with a 25% loss observed in plants grown at 15 mmol photons·m−2·s−1 (Figure 2C). This trend was retained in the mutant plants as well; however, their sensitivity was greater than that of wild-type plants under all tested growth conditions (16 and 38% loss of PSII activity at the highest and lowest light intensities, respectively). Thus, it appears that Deg1 contributes to PSII repair during photoinhibition in plants acclimated to different light intensities.
The D1 Protein Is Stabilized in deg1 Mutants
If Deg1 is indeed involved in degradation of the D1 protein of PSII during photoinhibition, one would expect this protein to be stabilized in the mutants containing reduced levels of Deg1. Immunoblot analysis, with an antibody generated against the lumen-exposed C terminus of the D1 protein, revealed a higher steady state level of D1 in the mutant than in the wild type (Figure 3A; see Supplemental Figure 1 online). Monomeric D1 protein could be found in either the phosphorylated (upper band) or the nonphosphorylated (lower band) form (Aro et al., 1993a). Both forms were found in wild-type thylakoids, but the nonphosphorylated form was more abundant than the phosphorylated form in the mutant and was more abundant than either of the two forms in the wild type as well (Figure 3A). The D1/D2 heterodimer was also more abundant in the mutant than in the wild type, as were D1 aggregates that do not enter the resolving gel. These results suggest that the D1 protein is indeed stabilized when the level of the D1 protein is reduced.
Figure 3.
D1 Protein Level in Mutant Plants.
(A) Wild-type and mutant plants were grown under low-light conditions (75 to 80 μmol photons·m−2·s−1) for 4 weeks. Thylakoids were then isolated, and samples containing 4 μg of chlorophyll were subjected to SDS-PAGE on a Tricine gel, followed by immunoblot analysis with an antibody against the C terminus of the D1 protein. The different forms of the D1 protein are indicated at right. MW, molecular mass.
(B) Wild-type and mutant thylakoids were exposed to 1200 μmol photons·m−2·s−1 for 45 min. Protein samples, equivalent to 4 μg of chlorophyll, were then subjected to immunoblot analysis with antibodies against the C terminus of the D1 protein or the OE33 protein of PSII.
(C) Bands in (B) were quantified, and the level of the D1 fragment was normalized to that of OE33. Mean values ± sd of three replicates are presented.
In contrast with the higher levels of the full-length forms of D1 found in mutant lines, a higher level of a 5.2-kD degradation product was found in the wild type (Figure 3A). To better quantify the differences in the abundance of the 5.2-kD degradation product, thylakoids were prepared from wild-type and mutant plants, exposed to high light intensity, and then subjected to immunoblot analysis. As shown in Figures 3B and 3C, this degradation products is approximately fourfold more abundant in wild-type plants than in mutant plants.
Because the analysis in Figure 3A revealed only a single degradation product, we attempted to identify less abundant D1 fragments. To this end, thylakoids were isolated from plants growing under low light and exposed to either low or high light intensities for another 45 min, and then gels were loaded with five times more protein samples. This overloading allowed the detection of another C-terminal degradation product of ∼16 kD (Figure 4A). Similar to the 5.2-kD fragment, this fragment was more abundant in thylakoids from wild-type plants than mutant plants. Although the differences were lower (44 and 32% after exposure to low or high light, respectively), similar trends were observed in three replicate experiments as well. By contrast, differences in the protein profiles were observed only in response to the light treatment and were not attributable to the reduction in the level of Deg1 (Figure 4B). Together, these results suggest that the 16-kD fragment is generated by Deg1, but its low abundance suggests that it is rapidly degraded further. Moreover, the smaller differences between the wild type and deg1 mutants in the abundance of the longer and shorter degradation products suggest that the 16-kD fragment may be generated by other proteases in addition to Deg1.
Figure 4.
Levels of a 16-kD D1 Degradation Product in Wild-Type and Mutant Plants.
(A) Wild-type and mutant plants were grown under low-light conditions (75 to 80 μmol photons·m−2·s−1) for 4 weeks. Thylakoids were isolated and either exposed to 1200 μmol photons·m−2·s−1 for 45 min (H) or not (L). Protein samples, equivalent to 20 μg of chlorophyll, were then subjected to immunoblot analysis with antibodies against the C terminus of the D1 protein or the OE33 protein of PSII. MW, molecular mass.
(B) A Coomassie blue–stained gel of the samples shown in (A).
The higher level of D1 protein found in the mutant could be partially explained by the reduced level of chlorophyll b (see Supplemental Table 1 online). If a lower level of chlorophyll b is translated into lower levels of LHCII, it could result in the loading of higher amounts of chlorophyll a binding proteins, including the D1 protein. However, two observations argue against this possibility: unlike the full-length forms of D1, the levels of its fragments are lower in the mutant compared with the wild type (Figures 3 and 4); and the protein profiles, including the level of LHCII, are similar in the wild type and deg mutants (see Supplemental Figure 1 online).
A C-Terminal Degradation Product of D1 Is Generated by Deg1
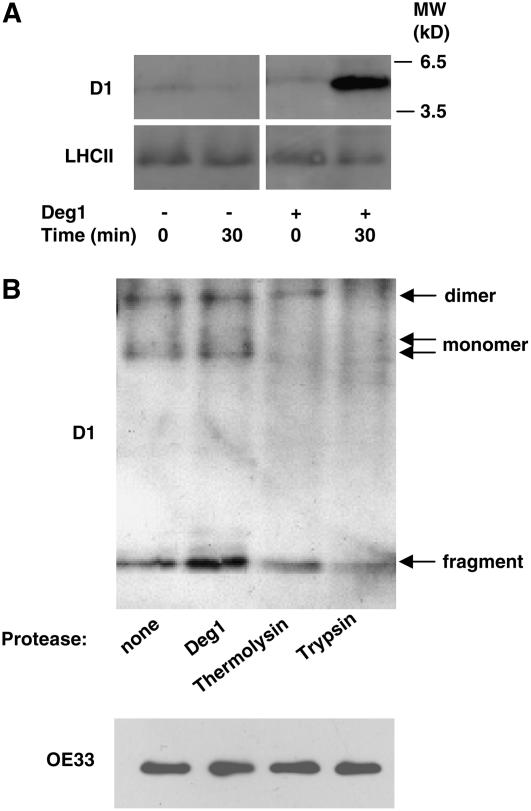
The apparent correlation between reduced levels of Deg1 in mutant lines and lower levels of C-terminal degradation products prompted us to test whether recombinant Deg1 is indeed capable of generating a D1 degradation product. Thus, thylakoid membranes were isolated from mutant plants, treated with a French press to obtain inside-out vesicles, and then subjected to photoinhibitory illumination. At the end of this procedure, the membranes were incubated with or without recombinant Deg1 for 30 min at 37°C. As shown in Figure 5A, addition of the recombinant protease led to an increase in the amount of the D1 C-terminal fragment, suggesting that this fragment is indeed a product of Deg1 activity.
Figure 5.
Generation of a D1 Degradation Product by Recombinant Deg1.
(A) Mutant thylakoids were subjected to treatment with a French press to obtain inside-out thylakoid membrane vesicles. These membranes were exposed to 1200 μmol photons·m−2·s−1 for 45 min and then further incubated with (+) or without (−) recombinant Deg1 for 30 min at 37°C in the dark. These reaction mixtures were then subjected to immunoblot analysis with antibodies against the D1 protein or LHCII. MW, molecular mass.
(B) A similar experiment was performed with recombinant Deg1, thermolysin, or trypsin to compare their ability to generate the D1 degradation product in wild-type thylakoids. An immunoblot with an antibody against the D1 protein is shown in the top panel. Migration of the different forms of the protein is indicated at right. The bottom panel shows an immunoblot of samples from the reaction mixtures with antibody against OE33 before the addition of proteases.
To further test whether the production of this fragment is specific to Deg1 protease, we repeated this experiment with wild-type thylakoids and Deg1, thermolysin, and trypsin in equimolar concentrations. Figure 5B shows that recombinant Deg1 could cleave the fragment also from wild-type thylakoids. However, the amount of fragment was lower in the presence of thermolysin and trypsin. Moreover, the full-length forms of the D1 protein were highly sensitive to the presence of either trypsin or thermolysin, further supporting the notion that Deg1 specifically cleaves the C terminus of the D1 protein, thereby probably facilitating the complete degradation of this protein.
Coordinated Reduction in the Levels of Deg1, Deg2, and FtsH Proteases in deg1 Mutants
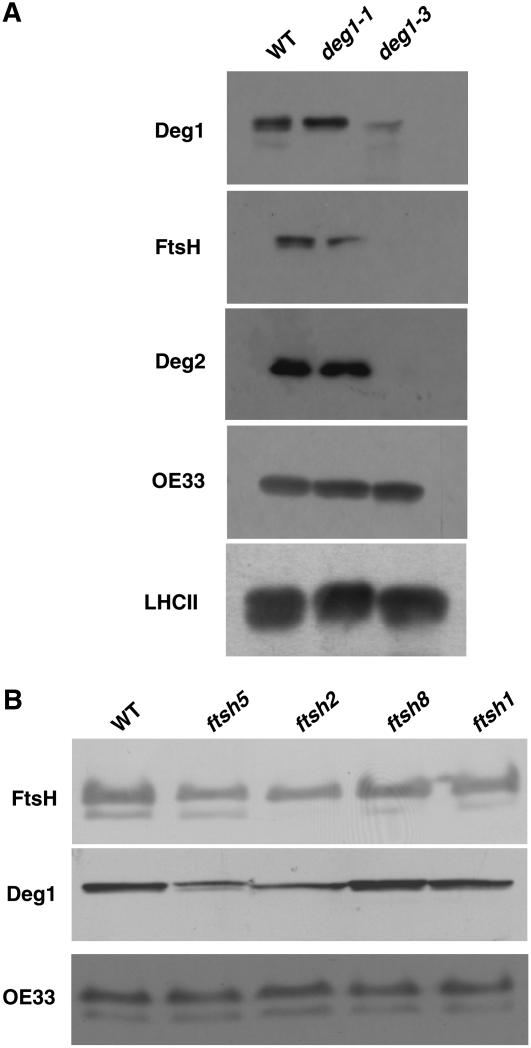
We recently observed that a knockout line of the FtsH2 protease also contains very low levels of Deg1 (Zaltsman et al., 2005a). Thus, we were interested in testing whether reduced levels of Deg1 would result in a concomitant reduction in the level of FtsH. To this aim, we compared the levels of different proteins in deg1 plants with reduced levels of Deg1 (e.g., deg1-3) with those of transgenic plants with Deg1 levels comparable to the wild type (e.g., deg1-1) by immunoblot analysis (Figure 6A). The three lines exhibited the same levels of the PSII-associated proteins LHCII and OE33 (Figure 6A). However, deg1-3, containing less Deg1, also had much reduced levels of FtsH protease and the stroma-exposed Deg2 protease. We also tested the level of Deg1 in different FtsH mutants. As shown in Figure 6B, FtsH1 and FtsH8 mutants, which have a wild-type phenotype (Sakamoto et al., 2003; Zaltsman et al., 2005b), had wild-type levels of both FtsH and Deg1. However, the FtsH2 and FtsH5 mutants, which have a variegated phenotype and are more sensitive to photoinhibition, also showed reduced levels of Deg1. These results suggest a coordinated accumulation of thylakoid proteases, although how this coordination is regulated has yet to be determined.
Figure 6.
Levels of Thylakoid Proteases in Deg1 and FtsH Mutants.
Proteins were extracted from 4-week-old wild-type and mutant plants, and equal amounts of chlorophyll were loaded and separated by PAGE. Immunoblot analysis was performed with antibodies against Deg1, Deg2, GST-FtsH1 (which recognizes the different chloroplast FtsHs equally well), OE33, and LHCII.
(A) Deg1 mutants.
(B) FtsH mutants.
DISCUSSION
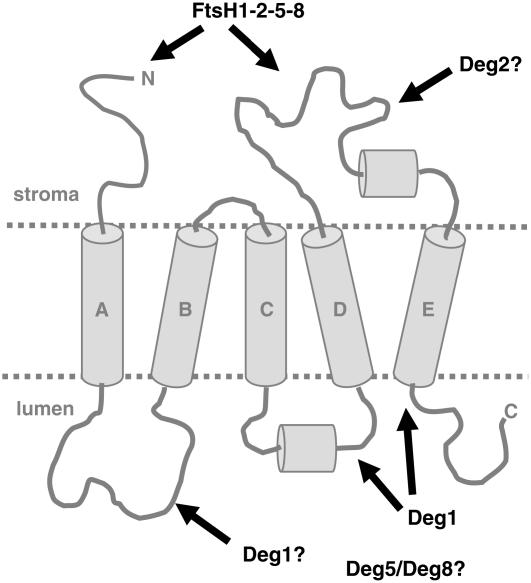
It has long been established that degradation of the D1 protein is inherent to the repair cycle of PSII from photoinhibition. Removal of photooxidatively damaged copies of this protein is a prerequisite for reassembly of the PSII complex with newly synthesized copies of D1. However, the mechanistic details of this rate-limiting step in the repair of PSII are far from being understood. Previous in vitro studies suggested that the FtsH (Lindahl et al., 2000) and Deg2 (Haussuhl et al., 2001) proteases are involved in degrading D1 protein. In vivo studies later confirmed this suggestion, at least for the FtsH protease, in higher plants (Bailey et al., 2002; Sakamoto et al., 2002) and cyanobacteria (Silva et al., 2003). The role of these two proteases is consistent with their localization to the stromal side of the thylakoid membrane (Lindahl et al., 1996; Haussuhl et al., 2001). Deg2, or other stromal peptidases, cleave at the large stromal DE-loop (Figure 7), creating new termini on this side of the membrane that can be recognized by FtsH, which in E. coli can degrade its substrates processively from either their N or C terminus (Chiba et al., 2002). The creation of two new termini on the stromal side of the membrane, in addition to the endogenous N terminus, is likely to enhance the rate of D1 degradation because these can serve as additional sites for the initiation of proteolysis.
Figure 7.
A Model for Degradation of the D1 Protein during Repair from Photoinhibition.
The transmembrane helices of the D1 proteins are marked A to E. The locations of the N and C termini are indicated. The previously described cleavage by Deg2, or other stromally located peptidases, exposes two additional termini for the initiation of proteolysis by the FtsH complex on the stromal side. Cleavage at the C terminus and the CD-loop by Deg1, and possibly at the AB-loop as well, facilitates extraction and proteolysis of the hydrophobic regions from the membrane.
The degradation of E. coli integral membrane proteins that span the membrane several times by FtsH protease involves dislocation of regions of the substrate from one side of the membrane to the other (Ito and Akiyama, 2005). However, how such dislocation is done in any biological system is not yet fully understood. Yeast mitochondria contain two oppositely oriented protease complexes in their inner membrane, m-AAA and i-AAA (Arnold and Langer, 2002), which are homologs of the FtsH protease. These two complexes cooperate in degrading integral membrane proteins. Work on a model substrate, containing a single transmembrane helix, revealed that proteolysis can proceed from either side of the membrane and that the trans region of the substrate is translocated across the membrane, provided that unfolding activity exists on the trans side of the membrane to unfold bulky hydrophilic regions of the substrate (Leonhard et al., 2000). However, the situation in bacteria and chloroplasts should be somewhat different. As ATP and ATP-dependent proteases are found only on one side of the membrane, translocation and proteolysis of membrane substrates are expected to be unidirectional. In this case, proteases and peptidases on the trans side of the membrane could facilitate the complete digestion of membrane substrates by ATP-dependent proteases found only on the other side of the membrane.
We hypothesized that proteases located in the lumen could facilitate D1 degradation simply by cleaving regions of the protein that are exposed to the lumen, and in doing so, shorten the segments that need to be pulled across the lipid bilayer by FtsH (Figure 7). Several results of this study are consistent with this hypothesis and Deg1 protease's possible role in it: (1) transgenic plants containing reduced levels of Deg1 are more sensitive to photoinhibition than are wild-type plants (Figure 2); (2) these plants contain higher levels of full-length D1 protein but lower levels of its degradation products (Figures 3 and 4); and (3) one of the C-terminal degradation products, which are observed in vivo, can be generated in vitro by recombinant Deg1 (Figure 5). However, it should be noted that photosynthetic parameters were affected in other mutants with reduced levels of chlorophyll b, such as the ch1 mutant (Li et al., 2000), suggesting that pleiotropic effects also could occur in the deg1 mutants. Nevertheless, further evidence for the importance of Deg1 at large was our inability to obtain homozygous knockout lines, suggesting that Deg1 is essential for plant viability.
Two C-terminal degradation products were detected here: the well-characterized 16-kD fragment (Andersson and Aro, 2001; Kanervo et al., 2003) and a much less characterized, but more stable, 5.2-kD fragment (Figures 3 and 4). The sizes of these fragments suggest that they are products of cleavage events in the lumen at the CD-loop and immediately downstream of transmembrane helix E, respectively (Figure 7). The much lower abundance of the 16-kD fragment, compared with that of the 5.2-kD fragment, suggests that it is less stable and undergoes further cleavage and degradation. This could be achieved by additional cleavage at the stromal DE-loop and the processive activity of the FtsH protease. Such translocation of protein segments across the membrane by FtsH has been shown directly in E. coli (Kihara et al., 1999). However, cooperation of periplasmic proteases in the degradation of membrane substrates has not been studied in E. coli, and in this sense the results presented here not only contribute to our understanding of D1 degradation but may have implications for the degradation of integral membrane proteins in other biological systems as well.
The results accumulated to date suggest that degradation of the D1 protein should be viewed as a two-step process: single cleavage events at hydrophilic regions of the protein on both sides of the thylakoid membrane, by Deg1, Deg2, and possibly other peptidases, to yield a limited number of distinct fragments, followed by their complete proteolysis by the processive ATP-dependent FtsH protease (Figure 7). Nevertheless, these steps do not necessarily occur in a sequential manner; in fact, it is highly likely that they occur simultaneously. Such a model implies that FtsH needs to extract no more than a single transmembrane helix at a time from the membrane, which is thermodynamically more favorable than threading the full-length protein in and out of the membrane to achieve complete degradation.
Why the D1 protein becomes susceptible to proteolysis after photoinhibition is not fully clear, but it is assumed that oxidative stress leads to the oxidation of amino acid residues, which in turn results in conformational changes that render the protein sensitive to preexisting proteases. In this context, it is interesting that three clusters of oxidized residues are found in the D1 protein after exposure to light: at the top of helix B (the pheophytin binding domain), at the bottom of helix D (the P680 domain), and at the lumen-exposed C terminus (Sharma et al., 1997). It is possible that oxidation of residues in the last two domains results in conformational changes nearby, rendering them susceptible to cleavage by Deg1 located on the lumenal surface of the thylakoid membrane and resulting in the 5.2- and 16-kD fragments observed here. Because the D2 protein is less oxidized than D1 (Sharma et al., 1997), it is not surprising that its turnover rate is lower.
Deg1 is not the only protease located in the thylakoid lumen. Two homologs of this protein, Deg5 and Deg8, have been identified there in proteomic analyses (Peltier et al., 2002; Schubert et al., 2002). As DegP in E. coli (Krojer et al., 2002) and Deg1 in chloroplasts (Chassin et al., 2002) form oligomers, lumenal Deg5 and Deg8 may form them as well. These can be homooligomers or heterooligomers composed of Deg1, Deg5, and Deg8. It should be noted that two other chloroplast proteases, Clp and FtsH, are also heterooligomers (for recent reviews, see Adam et al., 2006; Sakamoto, 2006). The core of the Clp protease is composed of four different ClpP subunits and four of their proteolytically inactive ClpR homologs, with at least some of them being indispensable. The FtsH protease is composed of two essential types of subunits, each encoded by redundant duplicated genes (Zaltsman et al., 2005b). Why these complexes require the presence of different highly related gene products is not clear, but in light of this fact, it will not be surprising if the Deg complex turns out to be heteromeric. Nevertheless, to date, there are no experimental data to support this possibility. Another protease found in the lumen, CtpA, is probably not involved in D1 degradation, as its activity is most likely limited to C-terminal processing of the pre-D1 protein as a prerequisite for its assembly into the functional PSII complex (Oelmuller et al., 1996).
Another interesting and as yet unexplained observation is the coordinated reduction in the levels of Deg2 and FtsH in plants that accumulate lower levels of Deg1 and the reduction in the level of Deg1 in two FtsH mutants (Figure 6). As already mentioned, both FtsH and Deg1 form oligomeric complexes. However, there is no evidence whatsoever for the existence of a complex between the two. As these proteases reside on opposite sides of the thylakoid membrane, it is difficult to envision a physical interaction between them, unless the lumenal loop connecting the two transmembrane helices that anchor FtsH to the membrane interacts with Deg1. However, if such an interaction does occur, this might suggest the existence of a transmembrane multiproteolytic complex operating simultaneously and in a coordinated manner from both sides of the membrane. As remote as it may sound, this possibility will have to await direct testing.
METHODS
Plant Material
All wild-type and mutant Arabidopsis thaliana plants were in the ecotype Columbia background. Plants were grown in Kekkila peat at 22°C with 16 h of illumination at 70 to 80 μmol photons·m−2·s−1. To generate Deg1 RNA interference mutants, a construct was created by cloning a cDNA fragment, specific to Deg1, corresponding to nucleotides 783 to 967 in the sense and antisense orientations flanking the intron sequence in the pRNA69 plasmid (Waterhouse et al., 1998). This plasmid contains, in addition to the 610-bp intron sequence, a 35S promoter and an Ocs terminator. The construct was then subcloned into the pCB302 binary vector (Xiang et al., 1999) containing the Basta resistance gene Bar. Transformation was performed by vacuum infiltration (Bechtold et al., 1993). The progeny from the transformed plants were germinated on peat, and transformants were selected by spraying the seedlings with 0.1% Basta. Resistant plants were selfed, and the progeny were subjected to PCR analysis to confirm the presence of the transgene.
All analyses were performed on rosette leaves from 4-week-old plants. To block the synthesis of chloroplast-encoded proteins, detached leaves were floated on 1 mM lincomycin for 3 h at a light intensity of 80 μmol photons·m−2·s−1 at 22°C. Photoinhibition treatment was performed by exposing detached leaves to 1400 μmol photons·m−2·s−1 at 22°C, and chlorophyll fluorescence measurements were performed as recently described (Zaltsman et al., 2005a).
Immunoblot Analysis
Total protein extracts were obtained by grinding 100 mg of leaf tissue in 300 μL of protein extraction buffer (0.5 M Tris-HCl, pH 6.8, 5 M urea, 8% [w/v] SDS, and 20% β-mercaptoethanol). Samples were centrifuged at 13,000g for 10 min, and the supernatant was subjected to SDS-PAGE. For detection of the D1 protein, the samples were separated on a 15% polyacrylamide gel in Tris-Gly buffer and electroblotted onto a nitrocellulose membrane. Blots were reacted with a commercially available antibody generated against the lumen-exposed C terminus of the D1 protein (Agrisera), diluted 1:5000, and an anti-chicken horseradish peroxidase–conjugated secondary antibody, diluted 1:5000. For detection of D1 degradation fragments, we used a 16% polyacrylamide gel in Tricine buffer (Schagger and Von Jagow, 1987), which was blotted onto polyvinylidene difluoride membranes and reacted with the primary and secondary antibodies described above at 1:3000 dilution. Other antibodies used were against OE33 (Zaltsman et al., 2005a), LHCII (Agrisera), Deg1 (Chassin et al., 2002), Deg2 (Haussuhl et al., 2001), and GST-FtsH1 (Zaltsman et al., 2005a). All blots were developed using the EZ-ECL chemiluminescence detection kit for horseradish peroxidase (Biological Industries).
Thylakoid Preparation and in Vitro Degradation of the D1 Protein
Leaf tissue (10 g) was ground in 100 mL of 10 mM HEPES-KOH, pH 8.0, filtered through six layers of cheesecloth, and centrifuged at 16,500g for 15 min at 4°C. The pellet was resuspended in the buffer described above and loaded on 40% (v/v) Percoll, then centrifuged at 4000g for 7 min at 4°C. Thylakoids were collected from the interphase of the buffer and the Percoll cushion and washed with 10 mM HEPES-KOH, pH 8.0. The thylakoid pellet was resuspended in 1 mL of the same buffer containing 300 mM sorbitol (SH), and the chlorophyll concentration was determined.
To prepare inside-out thylakoid membrane vesicles for the in vitro degradation assay, thylakoids (20 μg chlorophyll/mL) were passed through a French press at 1500 p.s.i. Intact thylakoids were precipitated by centrifugation at 16,500g for 30 min at 4°C. The resulting supernatant, containing the inside-out membrane vesicles, was illuminated at 1200 μmol photons·m−2·s−1 for 45 min at 22°C. The membranes were then centrifuged at 100,000g for 20 min and resuspended in SH to 500 μg chlorophyll/mL.
For the in vitro degradation assay, thylakoid membranes (13 mg of chlorophyll) were diluted to 1 mL with 10 mM HEPES, pH 6.8, and either supplemented or not with 10 pmol of recombinant Deg1, prepared as described previously (Chassin et al., 2002). The samples were incubated at 37°C for 30 min and then centrifuged at 100,000g for 20 min. The pellet was washed with 80% acetone to remove chlorophyll, resuspended in sample buffer equivalent to 0.33 mg chlorophyll/mL, and incubated at 40°C for 15 min. The solubilized proteins were centrifuged at 13,000g for 10 min, and the supernatant was subjected to PAGE and immunoblot analysis.
Accession Number
The Arabidopsis Genome Initiative locus identifier for Deg1 is At3g27925.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. D1 Protein Level in Mutant Plants.
Supplemental Table 1. Characteristics of deg1 Mutants.
Supplementary Material
Acknowledgments
We thank Iwona Adamska for the generous gift of Deg2 antibody. This work was supported by grants from the Israel Science Foundation and the U.S.–Israel Binational Science Foundation to Z.A. E.K.-P. was partially supported by a fellowship from the Otto Warburg–Minerva Center for Agricultural Biotechnology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zach Adam (zach@agri.huji.ac.il).
Online version contains Web-only data.
References
- Adam, Z., Rudella, A., and van Wijk, K.J. (2006). Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9 234–240. [DOI] [PubMed] [Google Scholar]
- Andersson, B., and Aro, E.-M. (2001). Photodamage and D1 protein turnover in photosystem II. In Regulation of Photosynthesis, B. Andersson and E.-M. Aro, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 377–393.
- Arnold, I., and Langer, T. (2002). Membrane protein degradation by AAA proteases in mitochondria. Biochim. Biophys. Acta 1592 89–96. [DOI] [PubMed] [Google Scholar]
- Aro, E.M., McCaffery, S., and Anderson, J.M. (1993. a). Photoinhibition and D1 protein degradation in peas acclimated to different growth irradiances. Plant Physiol. 103 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro, E.M., Virgin, I., and Andersson, B. (1993. b). Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143 113–134. [DOI] [PubMed] [Google Scholar]
- Bailey, S., Thompson, E., Nixon, P.J., Horton, P., Mullineaux, C.W., Robinson, C., and Mann, N.H. (2002). A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277 2006–2011. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316 1194–1199. [Google Scholar]
- Chassin, Y., Kapri-Pardes, E., Sinvany, G., Arad, T., and Adam, Z. (2002). Expression and characterization of the thylakoid lumen protease DegP1 from Arabidopsis thaliana. Plant Physiol. 130 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, S., Akiyama, Y., and Ito, K. (2002). Membrane protein degradation by FtsH can be initiated from either end. J. Bacteriol. 184 4775–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, T., Southan, C., and Ehrmann, M. (2002). The HtrA family of proteases: Implications for protein composition and cell fate. Mol. Cell 10 443–455. [DOI] [PubMed] [Google Scholar]
- Haussuhl, K., Andersson, B., and Adamska, I. (2001). A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 20 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesgen, P., Schumann, H., and Adamska, I. (2006). Photodamaged D1 protein is degraded in Arabidopsis mutants lacking the Deg2 protease. FEBS Lett. 580 6929–6932. [DOI] [PubMed] [Google Scholar]
- Huesgen, P.H., Schuhmann, H., and Adamska, I. (2005). The family of Deg proteases in cyanobacteria and chloroplasts of higher plants. Physiol. Plant. 123 413–420. [Google Scholar]
- Ito, K., and Akiyama, Y. (2005). Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59 211–231. [DOI] [PubMed] [Google Scholar]
- Itzhaki, H., Naveh, L., Lindahl, M., Cook, M., and Adam, Z. (1998). Identification and characterization of DegP, a serine protease associated with the luminal side of the thylakoid membrane. J. Biol. Chem. 273 7094–7098. [DOI] [PubMed] [Google Scholar]
- Kanervo, E., Spetea, C., Nishiyama, Y., Murata, N., Andersson, B., and Aro, E.M. (2003). Dissecting a cyanobacterial proteolytic system: Efficiency in inducing degradation of the D1 protein of photosystem II in cyanobacteria and plants. Biochim. Biophys. Acta 1607 131–140. [DOI] [PubMed] [Google Scholar]
- Kihara, A., Akiyama, Y., and Ito, K. (1999). Dislocation of membrane proteins in FtsH-mediated proteolysis. EMBO J. 18 2970–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krojer, T., Garrido-Franco, M., Huber, R., Ehrmann, M., and Clausen, T. (2002). Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416 455–459. [DOI] [PubMed] [Google Scholar]
- Leonhard, K., Guiard, B., Pellecchia, G., Tzagoloff, A., Neupert, W., and Langer, T. (2000). Membrane protein degradation by AAA proteases in mitochondria: Extraction of substrates from either membrane surface. Mol. Cell 5 629–638. [DOI] [PubMed] [Google Scholar]
- Li, W., Srinivasula, S.M., Chai, J., Li, P., Wu, J.W., Zhang, Z., Alnemri, E.S., and Shi, Y. (2002). Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol. 9 436–441. [DOI] [PubMed] [Google Scholar]
- Li, X.-P., Bjorkman, O., Shih, C., Grossman, A.R., Rosenquist, M., Jansson, S., and Niyogi, K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403 391–395. [DOI] [PubMed] [Google Scholar]
- Lindahl, M., Spetea, C., Hundal, T., Oppenheim, A.B., Adam, Z., and Andersson, B. (2000). The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, M., Tabak, S., Cseke, L., Pichersky, E., Andersson, B., and Adam, Z. (1996). Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J. Biol. Chem. 271 29329–29334. [DOI] [PubMed] [Google Scholar]
- Oelmuller, R., Herrmann, R.G., and Pakrasi, H.B. (1996). Molecular studies of CtpA, the carboxyl-terminal processing protease for the D1 protein of the photosystem II reaction center in higher plants. J. Biol. Chem. 271 21848–21852. [DOI] [PubMed] [Google Scholar]
- Peltier, J.-B., Emanuelsson, O., Kalume, D.E., Ytterberg, J., Friso, G., Rudella, A., Liberles, D.A., Soderberg, L., Roepstorff, P., von Heijne, G., and van Wijk, K.J. (2002). Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14 211–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, W. (2006). Protein degradation machineries in plastids. Annu. Rev. Plant Biol. 57 599–621. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Tamura, T., Hanba-Tomita, Y., Murata, M, and Sodmergen. (2002). The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 7 769–780. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Zaltsman, A., Adam, Z., and Takahashi, Y. (2003). Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger, H., and Von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166 368–379. [DOI] [PubMed] [Google Scholar]
- Schubert, M., Petersson, U.A., Haas, B.J., Funk, C., Schroder, W.P., and Kieselbach, T. (2002). Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 277 8354–8365. [DOI] [PubMed] [Google Scholar]
- Sharma, J., Panico, M., Shipton, C.A., Nilsson, F., Morris, H.R., and Barber, J. (1997). Primary structure characterization of the photosystem II D1 and D2 subunits. J. Biol. Chem. 272 33158–33166. [DOI] [PubMed] [Google Scholar]
- Silva, P., Thompson, E., Bailey, S., Kruse, O., Mullineaux, C.W., Robinson, C., Mann, N.H., and Nixon, P.J. (2003). FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp. PCC 6803. Plant Cell 15 2152–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinvany-Villalobo, G., Davydov, O., Ben-Ari, G., Zaltsman, A., Raskind, A., and Adam, Z. (2004). Expression in multigene families. Analysis of chloroplast and mitochondrial proteases. Plant Physiol. 135 1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess, C., Beil, A., and Ehrmann, M. (1999). A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97 339–347. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Graham, M.W., and Wang, M.B. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C., Han, P., Lutziger, I., Wang, K., and Oliver, D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40 711–717. [DOI] [PubMed] [Google Scholar]
- Yamamoto, Y. (2001). Quality control of photosystem II. Plant Cell Physiol. 42 121–128. [DOI] [PubMed] [Google Scholar]
- Zaltsman, A., Feder, A., and Adam, Z. (2005. a). Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts—Implications for thylakoid formation and photosystem II maintenance. Plant J. 42 609–617. [DOI] [PubMed] [Google Scholar]
- Zaltsman, A., Ori, N., and Adam, Z. (2005. b). Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopsis. Plant Cell 17 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.