Abstract
Cell survival depends on the cell's ability to acclimate to phosphorus (P) limitation. We studied the chloroplast ribonuclease polynucleotide phosphorylase (PNPase), which consumes and generates phosphate, by comparing wild-type Chlamydomonas reinhardtii cells with strains with reduced PNPase expression. In the wild type, chloroplast RNA (cpRNA) accumulates under P limitation, correlating with reduced PNPase expression. PNPase-deficient strains do not exhibit cpRNA variation under these conditions, suggesting that in the wild type PNPase limits cpRNA accumulation under P stress. PNPase levels appear to be mediated by the P response regulator PHOSPHORUS STARVATION RESPONSE1 (PSR1), because in psr1 mutant cells, cpRNA declines under P limitation and PNPase expression is not reduced. PNPase-deficient cells begin to lose viability after 24 h of P depletion, suggesting that PNPase is important for cellular acclimation. PNPase-deficient strains do not have enhanced sensitivity to other physiological or nutrient stresses, and their RNA and cell growth phenotypes are not observed under P stress with phosphite, a phosphate analog that blocks the stress signal. In contrast with RNA metabolism, chloroplast DNA (cpDNA) levels declined under P deprivation, suggesting that P mobilization occurs from DNA rather than RNA. This unusual phenomenon, which is phosphite- and PSR1-insensitive, may have evolved as a result of the polyploid nature of cpDNA and the requirement of P for cpRNA degradation by PNPase.
INTRODUCTION
Phosphorus (P) is an essential macronutrient whose uptake, storage, and transport are dynamically regulated during organismal development. P limitation is frequently encountered, because P is required in relatively high amounts but often sequestered in chemical forms that are not readily useable. This condition induces tightly regulated scavenging and reallocation mechanisms and a series of metabolic adjustments.
Photosynthetic eukaryotes have developed specialized responses to P limitation, because carbon fixation requires a continual supply of P within the chloroplast. Chloroplast P is imported from the cytosol and eventually reexported as triose or hexose phosphates (reviewed in Weber, 2004). When limiting, P can be replenished by mechanisms including translocation from chloroplasts of older to younger leaves (Versaw and Harrison, 2002), mobilization via starch synthesis (Hausler et al., 1998; Ciereszko et al., 2001), activation of transporters (Shimogawara et al., 1999), and secretion of phosphatases and ribonucleases (Grossman, 2000; Abel et al., 2002; Poirier and Bucher, 2002). Furthermore, numerous metabolic pathways intersecting carbohydrate and nitrogen metabolism are adjusted in P-limited tissues (Abel et al., 2002; Wu et al., 2003).
Central regulators play a key role in coordinating the P limitation response. Best described is the PHOSPHATASE (PHO) regulon of yeast (Persson et al., 2003), which integrates intracellular and extracellular adjustments. In photosynthetic eukaryotes, two PHO-like regulators have been identified, PHOSPHORUS STARVATION RESPONSE1 (PSR1) in the green alga Chlamydomonas reinhardtii (Shimogawara et al., 1999) and its ortholog PHOSPHATE STARVATION RESPONSE1 (PHR1) in Arabidopsis thaliana (Rubio et al., 2001). The psr1 mutant was isolated by virtue of its resistance to high levels of radioactive Pi and subsequently was found to be defective in multiple aspects of the P limitation response. The PSR1 gene was shown to encode a putative transcription factor, as PSR1 possesses a MYB domain and is localized to the nucleus (Wykoff et al., 1999). A screen for Arabidopsis mutants that failed to induce the expression of a reporter gene in response to P stress led to the cloning of PHR1, which also possesses a MYB domain. Recent evidence has shown that PHR1 acts in part by inducing the microRNA mi399 under P stress (Bari et al., 2006). One target of mi399 is the ubiquitination pathway, in particular the ubiquitin-conjugating enzyme UBC24, which is defined genetically as the PHO2 locus (Fujii et al., 2005; Miura et al., 2005; Aung et al., 2006; Chiou et al., 2006). The pho2 mutant is a phosphate overaccumulator (Delhaize and Randall, 1995); thus, PHO2 likely acts by repressing P uptake through the proteolysis of transporters or other proteins, an activity that is relieved when PHO2 mRNA is targeted by mi399.
Although the importance of chloroplast P metabolism is clear, how it is regulated under nutrient stress is largely unknown. This study focuses on polynucleotide phosphorylase (PNPase), a nucleus-encoded and chloroplast-localized ribonuclease that has a role in polyadenylation-stimulated RNA degradation (Kudla et al., 1996; Lisitsky et al., 1997; Nishimura et al., 2004). A readily reversible enzyme (Yehudai-Resheff et al., 2001), PNPase polymerizes poly(A)-rich tails on mRNA fragments generated by endonucleolytic cleavage, a reaction that primarily consumes nucleotide diphosphates (NDPs) and generates Pi. Because of its high affinity for poly(A), PNPase binds to these same tails and rapidly degrades the fragment in the 3′ to 5′ direction through a phosphorylytic, or P-requiring, reaction. Instability conferred by polyadenylation is a common feature of prokaryotes and organelles (Slomovic et al., 2006) and contrasts with the stabilizing effect of poly(A) tails in the cytosol (Dreyfus and Regnier, 2002). Furthermore, as a phosphorylytic enzyme, PNPase differs from most cytosolic ribonucleases, which are hydrolytic and often activated under P deficiency (Green, 1994).
Because the twin anabolic and catabolic activities of PNPase either generate or consume Pi, the enzyme has the potential to affect P pools in addition to, or by way of, regulating RNA levels. Indeed, the Arabidopsis mutant rif10 overcomes the inhibition of a key enzyme in the plastid isoprenoid pathway through the inactivation of PNPase, which in turn affects metabolic flux (Sauret-Gueto et al., 2006). Using Chlamydomonas, we explored whether strains defective in PNPase expression or regulation would also be defective in acclimation to P limitation. Here, we present evidence that chloroplast DNA (cpDNA) and RNA (cpRNA) levels are both adjusted to optimize cell acclimation and that correct regulation of PNPase may be required for cell survival under P limitation.
RESULTS
Creation of PNPase-Deficient Strains
A Chlamydomonas PNP cDNA was identified through an EST project and extended using RT-PCR (see Methods). Based on available genomic sequence (Joint Genome Institute version 3.0), Chlamydomonas appears to have a single gene encoding PNPase, in contrast with the duplicated genes found in higher plants, which specify plastid (Hayes et al., 1996) and mitochondrial (Perrin et al., 2004) isozymes. The Chlamydomonas cDNA sequence (GenBank accession number DQ492261) predicts a protein with the catalytic core domains I and II and the KH and S1 RNA binding domains, which are conserved in bacterial, mitochondrial, and chloroplast PNPases (Yehudai-Resheff et al., 2003).
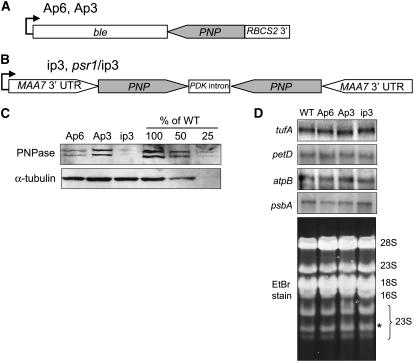
We designed constructs for antisense RNA or RNA interference (RNAi) inhibition of PNP expression (Figures 1A and 1B, respectively) and obtained multiple colonies for each construct. The effects of both antisense expression and RNAi were highly variable, as judged by RT-PCR and immunoblot analysis of PNP expression. For reasons discussed below, three strains were selected for detailed analysis: Ap6, Ap3, and ip3. As shown in Figure 1C, an antibody raised against a portion of the Chlamydomonas protein revealed that PNPase accumulation in Ap6 was ∼25% of that in the wild type, whereas the other antisense RNA–derived strain, Ap3, was only slightly reduced relative to the wild type, retaining ∼75% of the wild-type level based on multiple immunoblots. By contrast, the RNAi strain was strongly affected, with residual PNPase being estimated at 10 to 20% of the wild-type level, also based on multiple experiments. PNPase generally appeared as a doublet, probably reflecting two slightly different forms of the protein, which accumulate in parallel and, for this study, were considered in total. The low overall abundance of PNPase (even in the wild type) precludes precise quantification, but in general, Ap3, Ap6, and ip3 constitute an allelic series with successively decreasing amounts of PNPase.
Figure 1.
Generation and Characterization of PNPase-Deficient Strains.
(A) Diagram of the antisense transformation construct used to generate Ap3 and Ap6. The antisense PNP moiety is cotranscribed with the Ble selectable marker.
(B) Diagram of the RNAi transformation construct used to generate ip3 and psr1/ip3. Transcription of the tandem MAA7/PNP hairpin activates the RNAi pathway to downregulate both genes. UTR, untranslated region.
(C) Immunoblot analysis of PNPase in the wild-type and PNP-deficient strains, with tubulin as a loading control. Chlamydomonas PNPase migrates at ∼95 kD, similar to other chloroplast PNPase enzymes. A dilution series of wild-type protein was used to estimate the residual PNPase amount in Ap6, Ap3, and ip3. The reason for the doublet is not known; however, the bands coaccumulate and are not seen with preimmune serum.
(D) Ethidium bromide–stained gel and filter hybridization comparing the accumulation of chloroplast mRNAs in wild-type and PNPase-deficient cells grown under nutrient-replete conditions. The 23S rRNA band that is shifted in Arabidopsis PNPase mutants is marked with an asterisk.
In Arabidopsis, a cosuppressed line was studied that was largely depleted of the chloroplast PNPase (Walter et al., 2002). This line failed to fully process the 23S rRNA, as revealed by a mobility shift on an ethidium bromide–stained gel. In addition, novel psbA and rbcL transcripts accumulated with 3′ end extensions. In contrast with those results, none of the Chlamydomonas strains showed visible differences when total RNA was analyzed by gel electrophoresis and ethidium bromide staining, nor were migration differences observed for the four chloroplast mRNAs tested (Figure 1D). This suggests either that residual PNPase is sufficient for RNA maturation functions or that it is redundant, for example with RNase II, as is the case in Escherichia coli (Donovan and Kushner, 1986), when these Chlamydomonas strains are grown under standard, nutrient-replete conditions.
Chloroplast RNA Accumulates in Response to Phosphate Limitation
The catalytic activity of chloroplast PNPase is reversible in vitro, in which a relatively high NDP:Pi ratio favors polymerization and a low ratio favors degradation (Yehudai-Resheff et al., 2001). As discussed in the Introduction, given that PNPase uses and consumes major P-containing metabolites, its activity might be relevant to the P limitation response. A simple way of conceptualizing this potential integration is to hypothesize that under P limitation, PNPase-catalyzed RNA degradation and thus the consumption of P would slow, whereas polymerization would be favored, assuming that the NDP concentration remained relatively high. This same polymerization reaction would release Pi as a form of P mobilization.
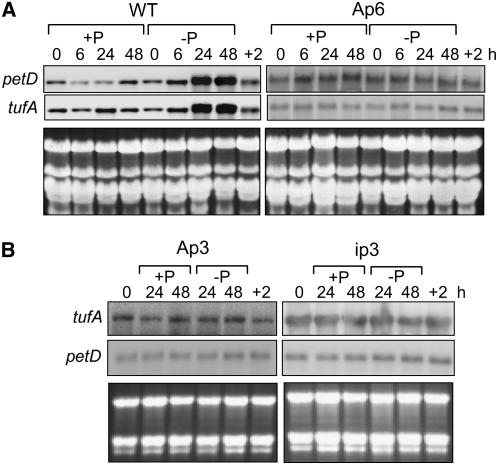
As a first step, cpRNA levels were measured after P depletion. Wild-type or PNPase-deficient strains were first grown in the presence of P, then the culture was divided into two portions, one with P and one without. Samples were taken over a 48-h time course, and at the end, the starved culture was transferred back to P-containing medium for 2 h. Figure 2A shows a representative gel blot that reveals that in wild-type cells, accumulation of the petD and tufA chloroplast transcripts increased substantially under P limitation. This increase is most evident at the 24- and 48-h time points and was fully reversed 2 h after P was restored (Figure 2A, lane +2). This accumulation is consistent with diminished PNPase catabolic activity along with continued transcription, and the 24 h delay in the manifestation of the phenomenon could be explained as the time taken for the Pi concentration in the chloroplast to change under P starvation.
Figure 2.
Chloroplast RNA Accumulation under P Limitation.
(A) Wild-type and Ap6 cells were grown to early log phase in TAP medium, then collected and resuspended in TAP (+P) or TA (−P) medium. Total RNA was isolated immediately (0) or after 6, 24, or 48 h. An additional sample was obtained from cells in TA, after they were transferred to TAP medium for an additional 2 h (+2). A total of 3 μg of RNA was loaded in each lane, and blots were hybridized with the experimental probes listed at left. To demonstrate the equality of loading, gels were stained with ethidium bromide.
(B) Ap3 and ip3 cells were analyzed as described for (A), except that the 0 time point was taken before division of the culture and the 6-h time point was omitted.
Figure 2A also shows that this phenomenon was not observed in Ap6 cells, which under +P conditions have similar cpRNA accumulation as the wild type (Figure 1D). In −P conditions, Ap6 mRNA levels remained constant, reacting to neither P depletion nor the readdition of P to the culture. This finding implicates decreased PNPase exonucleolytic activity as being responsible for cpRNA accumulation in wild-type cells acclimating to P deficiency; in Ap6, the initial PNPase level is low, so minimal activity changes might be expected under P limitation (Figure 3B). Two additional strains, Ap3 and ip3, were also examined (Figure 2B). Ap3 showed slight abundance increases under P limitation, with a subsequent reduction when P was restored. This phenotype was intermediate between those of the wild type and Ap6, consistent with the relative residual levels of PNPase. Strain ip3, however, showed no variation in RNA accumulation. These results confirm that PNPase deficiency is associated with a lack of cpRNA accumulation in response to P limitation.
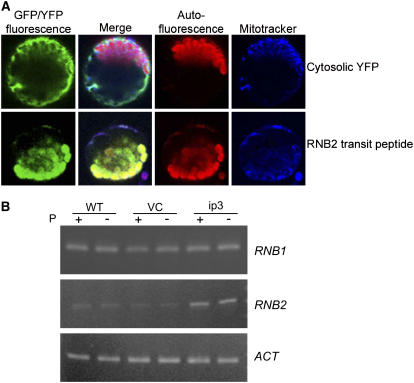
Figure 3.
Subcellular Localization and Expression of RNB2.
(A) YFP (cytosolic control) and GFP (RNB2) fusions were expressed in tomato protoplasts as described in Methods. The protoplasts were stained with Mitotracker for mitochondrial localization, and fluorescence images were collected as shown across the top. The GFP/YFP and chlorophyll autofluoresence channels were merged to show the relative localization of the two signals.
(B) RT-PCR analysis for the strains shown across the top (WT, recipient strain CC-406; VC, vector control in CC-406) from cells grown under +P or −P conditions. The vector control was transformed with the RNAi vector targeting only MAA7. The genes are listed at right, with RT-PCR performed as described in Methods. Actin (ACT) was used as a loading control.
Expression of a Putative Chloroplast RNase II Homolog Is Increased in ip3
This apparent ability of PNPase to limit mRNA accumulation under P stress in the wild type raises the question of why ip3, in particular, does not overaccumulate cpRNAs under +P conditions (Figure 1D). We speculated that other ribonucleases might mask the depletion of PNPase under those circumstances. As an initial test of this hypothesis, we focused on nuclear genes that might encode other chloroplast exoribonucleases, in particular RNase II homologs. PNPase and RNase II are redundant in E. coli, as mentioned above, and the RNase II homolog RNR1 is known to have a key role in chloroplast rRNA maturation in Arabidopsis (Kishine et al., 2004; Bollenbach et al., 2005). Furthermore, the E. coli pnp7 mutant exhibits a 2- to 2.5-fold increase in RNB activity and a similar increase in rnb mRNA (Zilhao et al., 1996), suggesting that rnb is regulated by PNPase in wild-type cells. Therefore, we explored whether any Chlamydomonas RNB genes might be similarly affected in ip3.
Examination of the most current version of the Chlamydomonas nuclear genome assembly (version 3) revealed two gene models that might encode chloroplast-targeted RNase II–like proteins, based on TargetP (Emanuelsson et al., 2000) predictions. These loci were named RNB1 (scaffold 19:799951-806626) and RNB2 (scaffold 11:108345-114817). To gain evidence for subcellular localization, cDNAs encoding the N-terminal 189 amino acids (RNB1) or 115 amino acids (RNB2) were fused to a green fluorescent protein (GFP)–encoding gene, and the proteins were expressed transiently in tomato (Solanum lycopersicum) protoplasts. As shown in Figure 3A, a cytosolic version of yellow fluorescent protein (YFP) generated fluorescence throughout the cell, except the large vacuole. RNB2 fusions were clearly targeted to the chloroplasts, because chlorophyll autofluorescence colocalized with the GFP signal but not with the Mitotracker signal, which labels mitochondria. RNB1 also appeared to target GFP to the chloroplast, although the data were ambiguous because of low expression of the fusion protein (data not shown). We conclude that RNB2 very likely encodes a chloroplast RNase II homolog and that RNB1 may encode a second plastid homolog.
We next examined the expression of RNB1 and RNB2 in wild-type, ip3, or vector control cells grown under +P or −P conditions, as shown in Figure 3B. In multiple experiments, little or no variation was seen for the RNB1 transcript level between strains or growth conditions. On the other hand, RNB2 mRNA was reproducibly found at an increased level in ip3 compared with the wild type or the vector control in both +P and −P conditions. This increase was not quantified, but examination of the gels in Figure 3B and in other experiments was consistent with a twofold to threefold difference compared with the actin control. If this reflects an increase of RNase II–like activity in ip3 chloroplasts, this might account for the lack of increased cpRNA when cells are grown under nutrient-replete conditions (Figure 1D) and may help to explain the failure of cpRNA levels to change under P deficiency even in partially PNPase-deficient strains (Figure 2).
PNP Expression Is Controlled by the Phosphate Response Regulator PSR1
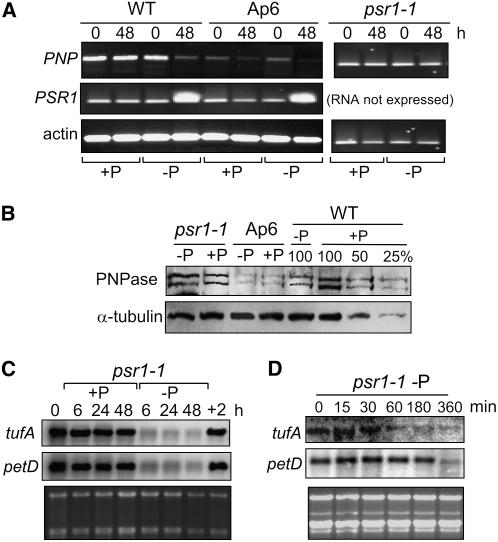
As discussed in the Introduction, the psr1 mutant is defective in the P limitation response as a result of the absence of a probable transcription factor. Known PSR1 targets include genes encoding chloroplast-localized proteins involved in photosynthesis, gene expression, and other processes (Moseley et al., 2006), but PNPase had not been tested. Given that PNPase was associated with a chloroplast gene expression response to P limitation, it was of interest to determine whether the PNP gene is influenced by PSR1. To answer this question, we initially compared transcript accumulation for the PNP gene under +P and −P conditions in the wild-type, Ap6, and psr1 mutant backgrounds using RT-PCR (Figure 4A). This experiment revealed that PNP transcript accumulation decreases in response to 48 h of P starvation in both wild-type and Ap6 cells, although the initial level is much reduced in the antisense strain Ap6. This is opposite to the behavior of the PSR1 transcript, which increases under P limitation compared with the actin mRNA control. However, −P conditions did not result in a decrease of PNP mRNA in the psr1 mutant. This is consistent with PSR1 functioning to repress PNP transcription, directly or indirectly, in response to P limitation.
Figure 4.
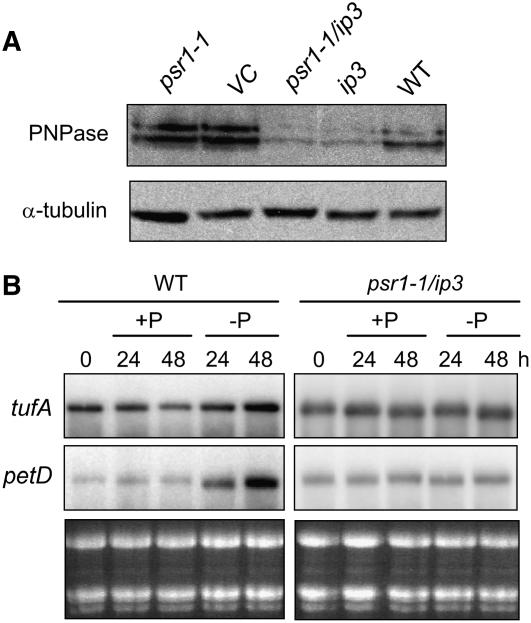
Effects of the psr1-1 Mutation on PNPase and cpRNA Accumulation under P Limitation.
(A) Approximate transcript levels for the PNP and PSR1 genes were measured by RT-PCR with 40 cycles of PCR for the strains, growth conditions, and times of P deprivation shown. PSR1 cDNA cannot be amplified from psr1-1. Actin RNA was used as an internal standard, using 20 PCR cycles.
(B) Immunoblot analysis of a dilution series of wild-type total protein from cells grown under +P conditions, compared with total protein from wild-type, psr1, or Ap6 cells starved for phosphate for 48 h, using an anti-PNPase antibody. A tubulin antibody was used as a loading control. A similar comparison was made for psr1-1 cells, and no decrease in PNPase was observed.
(C) Total RNA was isolated from psr1 cells grown under +P conditions, then transferred to +P or −P conditions for the number of hours shown. Starved cells were returned to TAP medium for 2 h (+2) for an additional sample. Loading was estimated by staining the gel with ethidium bromide.
(D) An experiment was performed for psr1-1 as described for (C), except that the time points were shorter and no restoration of P was performed.
We used immunoblot analysis to determine whether the PNPase protein level mirrors PNP transcript accumulation. Figure 4B shows that after 48 h of P starvation, the protein level declined, based on multiple blots, to between 25 and 50% of the initial level in the wild type when tubulin was used as a loading control. The psr1 mutant, however, retained 100% or possibly a slightly increased level of PNPase even when starved for P. Thus, both PNP mRNA and protein expression fail to respond to P limitation in the psr1 mutant.
Given this result, it was of interest to determine whether PSR1 function was also linked to cpRNA accumulation under P limitation. To do so, RNA gel blot analysis was performed, as shown in Figure 4C. We found that in psr1 cells, tufA and petD transcript accumulation declined under P limitation, in contrast with the increase observed in wild-type cells and the constant amount measured in PNPase-deficient cells (Figure 2). The same phenomenon was observed for a second mutant allele, psr1-2 (data not shown). Because transcript levels had fully declined at 6 h, the earliest time point measured, still earlier time points were examined in a follow-up experiment (Figure 4D). The results show a decrease in tufA mRNA within 60 min, whereas petD mRNA abundance began to decrease between 3 and 6 h. The short half-life of tufA mRNA (Hwang et al., 1996) may explain this difference. Together, the results in Figure 4 connect PSR1 with the repression of PNP expression, in turn allowing cpRNA accumulation. In the psr1 mutant, PNPase activity does not decrease and cpRNA accumulation declines. This finding implies that excess PNPase causes additional RNA degradation, even under P limitation.
Because this evidence is to some extent circumstantial, we created a double mutant by transforming psr1 with the RNAi construct shown in Figure 1B (ip3), yielding the new strain psr1 ip3. If psr1 and PNPase are not related as hypothesized above, then the double mutant should still degrade cpRNA under P limitation. Figure 5A shows immunoblot analysis of the single and double mutants, along with a vector control. The results show that the double mutant, when grown under +P conditions, has approximately the same PNPase abundance as the original RNAi mutant discussed above (Figures 1 and 2). Figure 5B shows cpRNA accumulation under P limitation in the wild type and the double mutant. Whereas cpRNA abundance increased in the wild type after 24 to 48 h under −P conditions, no change was seen in the double mutant. This supports the contention that a major reason why the psr1 mutant fails to increase cpRNA abundance under P limitation is its failure to repress PNPase expression.
Figure 5.
The psr1-1 ip3 Double Mutant Resembles Strain ip3.
(A) PNPase protein accumulation in the strains shown across the top, with tubulin as a loading control. Strain vector control (VC) was transformed with the RNAi vector targeting only MAA7.
(B) Chloroplast RNA accumulation under P limitation in wild-type or double mutant cells. A 0 time point was taken before division of the cultures.
RNA Stability after P Limitation
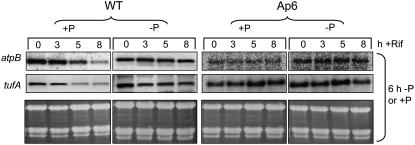
These data support the concept that a posttranscriptional mechanism involving PNPase contributes to cpRNA accumulation under P limitation. To gain direct evidence for changes in RNA stability, we followed RNA accumulation after transcription inhibition by rifampicin, a global chloroplast transcription inhibitor in Chlamydomonas (Hwang et al., 1996; Eberhard et al., 2002). Rifampicin was introduced 6 h after division of the initial culture into +P and −P subcultures, to coincide with the period during which cpRNA accumulation increases in wild-type cells under −P conditions (Figure 2). Figure 6 shows that after the addition of rifampicin to wild-type cells, RNA levels decreased substantially in 5 to 8 h when cells remained in +P medium, whereas little change was seen when cells were kept in −P medium. This finding indicates lesser RNA stability in +P versus −P conditions; it is thus likely that the observed RNA accumulation under −P conditions is a posttranscriptional effect. Unlike the wild type, Ap6 showed little decay of RNA after the addition of rifampicin, regardless of P availability. This suggests that under either +P or −P conditions, PNPase is required for rapid adjustments of cpRNA abundance.
Figure 6.
Analysis of cpRNA Stability after the Addition of Rifampicin.
Cells were grown to early log phase in TAP medium, resuspended, and grown in +P or −P medium for 6 h, and then rifampicin (Rif) was added to a concentration of 350 μg/mL. RNA was isolated and analyzed at the time points shown across the top.
PNPase Regulation Contributes to Viability under P Limitation
These data link PNP expression to the response regulator PSR1, raising the possibility that cells acclimate to P limitation in part by repressing PNPase activity. Ultimately, cell survival is at stake during P limitation; thus, viability is one measure of the ability to acclimate. We measured viability in two different ways, comparing several strains and stress conditions. The strains chosen were the wild type, Ap6, psr1, and sulfur acclimation1 (sac1), the latter being a mutant unable to acclimate to sulfur deprivation (Davies et al., 1994). Using them, we examined the growth of colonies and also viability as indicated by the dye Evans blue, which only stains nonviable Chlamydomonas cells (Crutchfield et al., 1999).
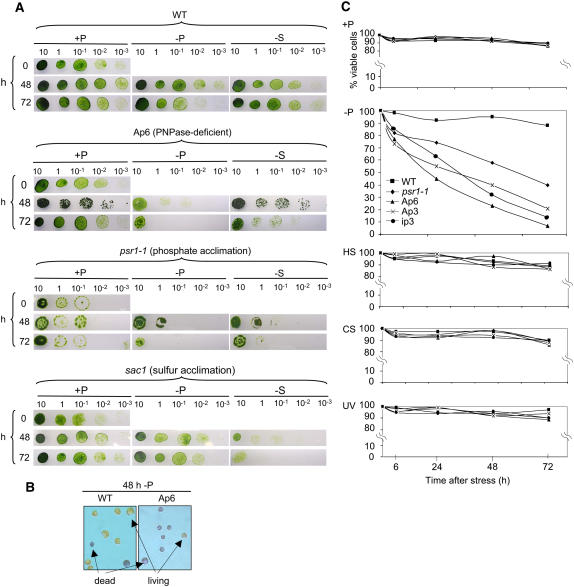
We initially compared plating efficiencies of cells after growth in +P, −P, and −S conditions, as shown in Figure 7A. Cultures were initiated in rich medium and then divided into three portions, in which they remained in rich medium (+P), were deprived for P (−P), or were deprived for sulfur (−S). The psr1 and sac1 mutants were used as controls for the expected loss of viability on −P and −S medium, respectively. The results show that all strains retained viability under +P conditions, as expected, although all of the mutants and particularly psr1 achieved lower cell densities than the wild type. Under −P conditions, however, Ap6 and psr1 cells progressively lost viability, whereas wild-type and sac1 cells remained viable. This finding suggests that, like psr1, Ap6 cannot acclimate to −P conditions. A complementary picture was observed under −S conditions. In this case, the wild type and Ap6 remained viable, whereas sac1 lost viability. This result suggested that Ap6 was defective in its P limitation, but not its S limitation, response.
Figure 7.
PNPase-Deficient and psr1-1 Cells Lose Viability under −P Conditions.
(A) Cells were grown in TAP medium (+P) and then placed on replete (+P) or nutrient-deprived (−P or −S) medium for the number of hours indicated at left. At those time points, cells were spotted onto TAP medium at the indicated dilutions (10 represents a 10-fold concentration of the culture). The images show growth after 4 to 7 d (depending on the strain) in continuous light at 25°C.
(B) An example showing that Evans blue stain differentiates viable and nonviable cells, because it cannot cross the intact plasma membrane. At 48 h under −P conditions, most Ap6 cells stain positively and are thus nonviable.
(C) Percentage of viable cells (y axes) after the indicated times of growth (x axes) under the following stress conditions (see Methods for details): −P, phosphate-deprived; HS, heat-stressed; CS, cold-stressed; UV, treated with short-wave UV light. The data points shown are based on Evans blue staining and are averages of at least three experiments.
Because P and S limitation are both nutrient stresses, and because of possible crosstalk between the response pathways (Moseley et al., 2006), we tested PNPase-deficient strains under other types of stress, in case the strains lost viability under P limitation as a result of a general stress intolerance, the S limitation results notwithstanding. For a more direct measure of cell viability, we used Evans blue, whose staining of wild-type versus Ap6 cells subjected to 48 h of P limitation is shown as an example in Figure 7B (the dye only stains dead cells). The experiment for which results are shown in Figure 7C included the three PNPase-deficient strains studied here as well as the wild type and psr1. The top two graphs show that all strains retained high viability for 72 h under +P conditions but that psr1 and the PNPase-deficient strains lost viability under −P conditions, as expected. The same strains were then subjected to heat, cold, and UV light stress, using conditions established previously for Chlamydomonas. Under all of these conditions, no appreciable difference was observed when comparing the wild type with other strains. Thus, neither Ap6 nor other PNPase-deficient strains have a general sensitivity to abiotic stress.
Phosphite Substitution for Phosphate Prevents PSR1 Induction and PNPase Repression
The results described above demonstrate that P limitation leads to altered expression of PNPase, with consequences for cpRNA accumulation and cell viability. Furthermore, the response regulator PSR1 appears to play a role in this process. An additional way to gauge whether the chloroplast effects we have observed result from P limitation signaling, as opposed to a purely metabolic response, is to substitute phosphite (Phi) for phosphate. Phi is a chemical mimic of P and represses many P limitation responses, for example in Arabidopsis (reviewed in Poirier and Bucher, 2002), although it cannot be as readily metabolized and in fact inhibits plant growth to some degree (Ticconi et al., 2001; Varadarajan et al., 2002).
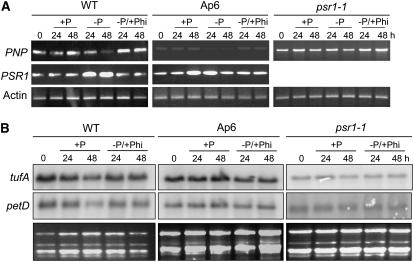
To test the effects of Phi, wild-type, Ap6, and psr1 cells were grown under +P conditions and then transferred to medium lacking P, supplemented or not with 1.0 mM Phi, a much lower concentration than the >2.5 mM that inhibits the growth of vascular plants and the chlorophyte Ulva lactuca (Lee et al., 2005). As shown in Figure 8A, Phi prevents the induction of PSR1 gene expression that normally occurs under −P conditions and concomitantly prevents the reduction of PNP transcript levels in either a psr1 or PSR1 background. Given the results shown in Figure 8A and the lack of a growth phenotype, we expected that cpRNA levels would also mimic +P levels when Phi was substituted for P. Figure 8B shows that this is indeed the case, because cells grown under −P/+Phi conditions did not exhibit cpRNA abundance changes for any of the three strains examined. In conclusion, the effects studied here appear to be related to the perception of Pi limitation (see Discussion).
Figure 8.
Response of Cells to Phi under −P Conditions.
(A) Cells were grown in TAP medium (+P) and then divided into thirds and resuspended in P-replete (+P), nutrient-deprived (−P), or Phi-substituted (−P/+Phi) medium for the number of hours indicated. Transcript abundance for the genes indicated at left was observed using RT-PCR, with actin as a loading control.
(B) Cells were grown as described for (A), and RNA gel blotting was performed using the probes shown at left. Loading can be estimated by the ethidium bromide–stained gels.
Chloroplast DNA May Be Mobilized upon P Starvation
Given that cpRNA is very stable in the wild type under P limitation, we considered that DNA might be a source of mobilizable P in the chloroplast. Indeed, plants can use DNA as a sole source of P (Chen et al., 2000). The chloroplast genome is polyploid, although only a few of the ∼80 copies of cpDNA in Chlamydomonas are required to maintain gene expression (Eberhard et al., 2002). At the same time, it is known that Chlamydomonas cells are capable of rapidly degrading cpDNA after the fusion of opposite mating types (Nishimura et al., 1999, 2002).
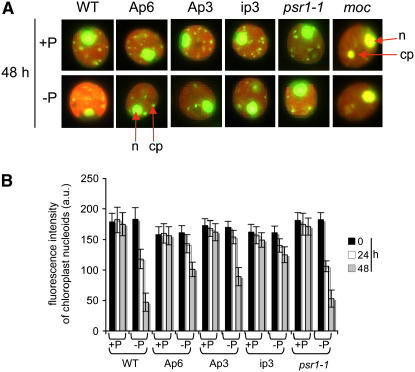
Chloroplast DNA is organized into nucleoprotein complexes called nucleoids (Ris and Plaut, 1962). These can be visualized by staining cells with SYBR Green I, as shown in Figure 9A. In these images, the nucleus appears as a large fluorescent area, whereas nucleoids are dispersed in the single large chloroplast; fainter mitochondrial nucleoids are generally not visible at this resolution. We additionally examined an otherwise phenotypically normal mutant called moc (for monokaryotic chloroplast), which possesses a single, larger, and thus easily visible nucleoid (Misumi et al., 1999). The representative results shown in Figure 9A reveal that in the wild type, the number and intensity of nucleoids (or the intensity of the moc nucleoid) appear to diminish within 48 h of transfer to −P medium. This was confirmed by quantitative treatment of the image data, as shown for nucleoid intensity in Figure 9B. This summation of total cpDNA fluorescence shows that wild-type cpDNA decreases by ∼35% after 24 h and by 75% after 48 h. This decrease can be largely explained by reduced numbers of nucleoids (data not shown), although the measurements are somewhat subjective. psr1 mutant cells behaved similarly to wild-type cells, suggesting that cpDNA copy number reduction does not require PSR1. In PNPase-deficient cells, cpDNA fluorescence and nucleoid numbers also declined, but to a lesser extent than in wild-type cells, particularly in ip3. This finding is most likely attributable at least in part to slower cell (and therefore chloroplast) division in these strains under −P conditions.
Figure 9.
Reduction in Chloroplast Genome Copy Number during Phosphate Starvation.
(A) Cells of the indicated strains were stained with SYBR Green at 48 h after division of early log phase cultures into TAP (+P) or TA (−P) medium. Representative images of individual cells are shown. cp, chloroplast; n, nucleus.
(B) Cumulative intensities of clearly visible nucleoids (see Methods) were measured for 10 cells for each strain and growth condition. Values shown are means ± se. The intensity of nuclear fluorescence was also measured but did not vary appreciably (data not shown). a.u., arbitrary units.
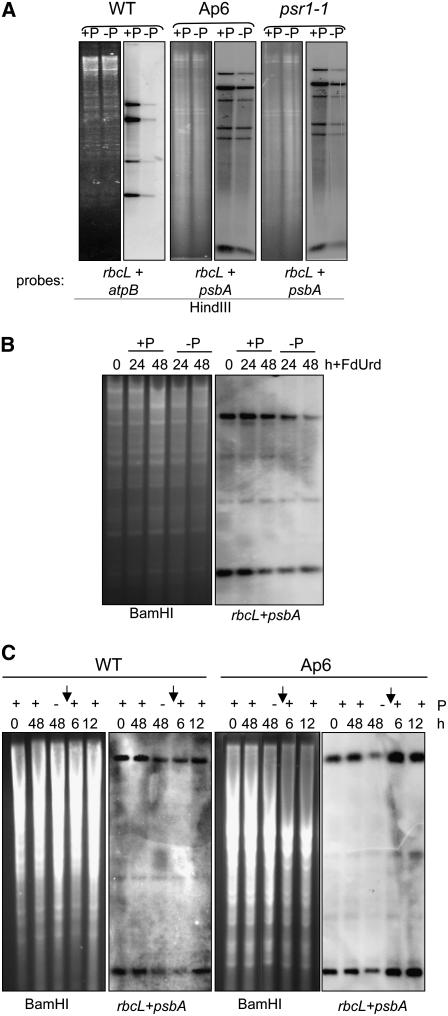
To measure cpDNA using an independent method, we performed DNA gel blot analysis, as shown in Figure 10A. The gels were loaded equally with total DNA, based on ethidium bromide staining, and hybridized with combined probes for two chloroplast genes. A substantial reduction of all fragments was observed in −P samples from the wild type and psr1, with a lesser effect in Ap6 cells. These results support the microscopy data. Thus, copy number reduction of cpDNA accompanies the P stress response in Chlamydomonas, perhaps as a way of regenerating free P through nucleotide scavenging and/or conserving P through reduced replication of cpDNA.
Figure 10.
Dynamics of Chloroplast Genome Copy Number during Phosphate Starvation and Recovery.
(A) Total DNA was isolated from cells grown under the same conditions described for Figure 9A, and 10 μg/lane was digested with HindIII and hybridized with mixed probes corresponding to the genes listed at the bottom of each gel. Loading was estimated by staining gels before blotting with ethidium bromide.
(B) Total DNA was isolated from cells grown under the conditions shown across the top, with 5-fluoro-2′-deoxyuridine (FdUrd) being added as the cultures were divided into +P and −P portions. The DNAs were digested and probed as shown across the bottom.
(C) Total DNA was isolated from cells grown under the conditions shown across the top. At time 0, cultures were divided into +P and −P portions and grown for an additional 48 h. The −P culture was subsequently returned to +P conditions for an additional 6 or 12 h. The DNAs were digested and probed as shown across the bottom.
The cpDNA signal detected by gel blot analysis, as a function of total DNA amount, can be influenced by cpDNA degradation, failure to replicate, and/or dilution through cell division. To eliminate any effects of DNA replication, cpDNA dynamics were reexamined with cells grown in the presence of 5-fluorodeoxyuridine, which inhibits cpDNA replication in Chlamydomonas (Wurtz et al., 1977). Figure 10B shows that under +P conditions, cpDNA copy number slowly declines in the wild type, as reported previously. Under −P conditions, however, there is a greater copy number decline over the same time period, even though cells divide more slowly when faced with nutrient limitation. This result strongly suggests that cpDNA is actively digested under −P conditions.
If digestion of cpDNA is a P mobilization response, then copy number should recover when cells are returned to +P conditions, even as the division rate accelerates. To test this hypothesis, cpDNA was examined after the restoration of P to starved cultures. Figure 10C shows that within 12 h in the wild type, and within 6 h in Ap6, cpDNA returned to the level found in cells grown in P-containing medium. This rapid recovery may indicate an increased replication rate upon restoration of P, although this was not tested directly.
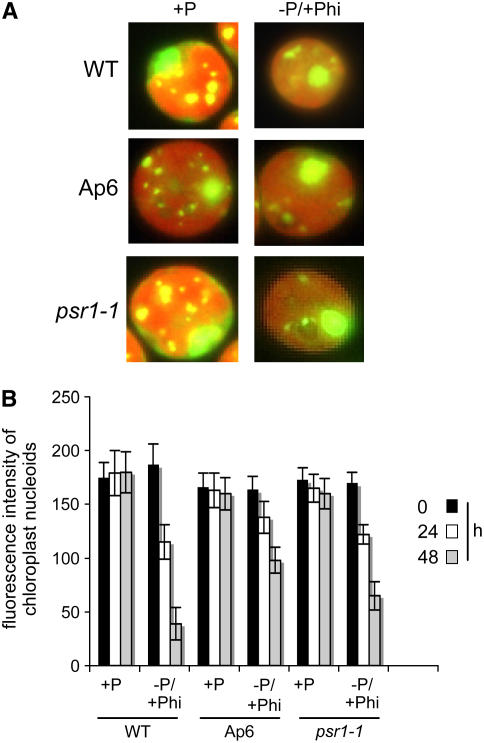
Finally, we tested whether Phi would prevent decreased cpDNA abundance under P limitation by interfering with P limitation signaling. Because Phi prevented PSR1 induction and associated RNA responses (Figure 8), this seemed a reasonable hypothesis. Figure 11, however, shows that cpDNA still decreased when cells were starved for P, even when Phi was present. The dynamics were very similar to those seen under −P conditions without Phi (Figure 9), suggesting that cpDNA copy number adjustments are Phi-insensitive. Because these adjustments are also PSR1-insensitive, this finding argues for an independent response system to P limitation.
Figure 11.
Substitution of Phi Does Not Prevent the Reduction in Chloroplast Genome Copy Number during Phosphate Starvation.
(A) Cells of the indicated strains were stained with SYBR Green at 48 h after division of early log phase cultures into TAP (+P) or Phi (−P/+Phi) medium. Representative images of individual cells are shown.
(B) Cumulative intensities of clearly visible nucleoids (see Methods) were measured for 10 cells for each strain and growth condition. Values shown are means ± se. The intensity of nuclear fluorescence was also measured but did not vary appreciably (data not shown).
DISCUSSION
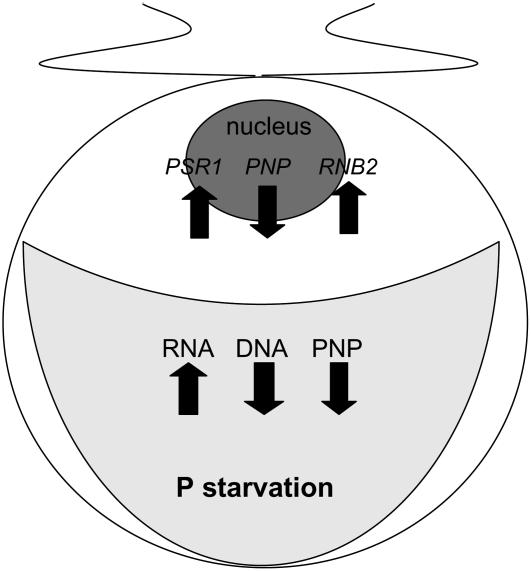
Although acclimation to P limitation has been studied intensively, the role of the chloroplast as a possible P reservoir has been largely unexplored. In this study, we have described opposite responses of chloroplast RNA and DNA pools to P limitation in Chlamydomonas, as summarized in Figure 12. Our data suggest that cpRNA abundance increases after the repression of PNPase expression by PSR1, a phosphate response regulator. Because PNPase is phosphorylytic, reducing its activity might preserve limiting Pi. Although this in turn would reduce P mobilization from RNA, the chloroplast appears to compensate by degrading or failing to replicate a significant proportion of its polyploid genome. That PNPase has a role in the P limitation response is also suggested by the P starvation–specific inviability of PNPase-deficient strains, which is consistent with a severe metabolic defect under these growth conditions. Although we have not measured P metabolites directly, our data support the notion that chloroplast nucleic acid metabolism is integral to the P budget of the organelle and, in turn, the cell.
Figure 12.
Scheme of Chlamydomonas P Starvation Responses in the Wild Type Relevant to This Study.
Both the nuclear PNP gene and chloroplast PNPase protein accumulation are shown as decreasing under P starvation; RNB2 protein levels are unknown. Changes in activates involved in extracellular scavenging, transport, and cytosolic metabolism are omitted; these have been reviewed recently (Grossman, 2000).
PNPase Is among Chloroplast Functions Whose Expression Is Repressed under P Limitation
Our results show that during P starvation, cpRNA hyperaccumulates, whereas both transcript and protein levels decrease for PNPase (Figure 4). As suggested by these results, cpRNA is more stable under −P conditions (Figures 2 and 5). The decrease in PNPase expression under P stress is PSR1-dependent (Figure 4), although it remains to be tested whether PSR1, which encodes a probable transcription factor (Wykoff et al., 1999), regulates the PNP gene directly or acts through an intermediary such as a microRNA, as was discussed previously for PHR1 in Arabidopsis (Bari et al., 2006). PSR1 also regulates P uptake and extracellular mobilization (Shimogawara et al., 1999); thus, it appears to coordinate at least some P limitation responses outside and within the cell, including the chloroplast. Integrating intrachloroplast and extrachloroplast stress responses is perhaps a necessary outcome of endosymbiosis and is also a property of SAC3, a Chlamydomonas sulfur limitation response factor that regulates sulfur acquisition as well as chloroplast transcription (Davies et al., 1999; Irihimovitch and Stern, 2006).
A potentially broad role for PSR1 is supported by a recent array analysis that compared gene expression in the wild type and psr1 under +P and −P conditions (Moseley et al., 2006). This study measured transcript abundance for ∼3000 genes and found 152 that exhibited PSR1-dependent changes under P limitation, whereas 83 changed in a PSR1-independent manner. PNP, studied here, falls into the former category, because its mRNA abundance declined under P limitation, but only in the wild type. Interestingly, several other transcripts exhibited similar behavior when measured using microarrays. Most prominent among these were mRNAs encoding chloroplast ribosomal proteins and the translation initiation factor IF-3. Therefore, PSR1 appears to repress both mRNA degradation and translation initiation in the chloroplast. Translation being an energy-intensive process, its decrease under P limitation is consistent with the well-known general metabolic slowdown. Transcriptome analysis in Arabidopsis also revealed repression under P limitation of several genes encoding translational functions in the chloroplast (Misson et al., 2005). These included ribosomal proteins, a peptide release factor, and DAL, a gene required for ribosome assembly (Bisanz et al., 2003). Whether these are regulated by the PSR1 ortholog PHR1 was not tested.
Phi Represses Certain Chloroplast P Limitation Responses
We also studied how P limitation sensing was related to chloroplast RNA and DNA dynamics by providing Phi in the absence of P. Phi represses many P limitation responses because it is chemically sensed as P, both in plants and in cultured plant cells, although the effect varies between genes and as a function of Phi concentration (Varadarajan et al., 2002). Phi is also less readily metabolized than P, and at the concentration used in this study (1 mM), it can inhibit growth. This effect is not uniform, however. For example, weight gain in tomato is strongly affected but plant height is only slightly affected (Varadarajan et al., 2002). In a study of Arabidopsis seedlings, fresh weight was only affected at Phi concentrations of >2.5 mM when P was omitted from the medium (Ticconi et al., 2001). We found that Chlamydomonas grows as robustly when provided with Phi as when provided with P, which was surprising. This raises the possibility that Phi is easily metabolized in Chlamydomonas. Indeed, microorganisms have several pathways for reducing Phi or related compounds to P, with a recent discovery being a catalysis of Phi by alkaline phosphatase, which also yields H2 (Yang and Metcalf, 2004).
Given that Chlamydomonas cells appear to grow well for 1 week when provided with either P or Phi as a source of P, it is not entirely surprising that neither PNP nor PSR1 gene expression, nor cpRNA abundance, could be differentiated between cells grown under +P versus −P/+Phi conditions within this time period. This, in turn, makes it somewhat ambiguous whether −P/+Phi cells are in fact sensing P limitation. One clue relevant to this issue is the fact that cpDNA abundance declined when Phi was the sole external P source (Figure 11), as it did under −P conditions. If cells were fully metabolizing Phi into P, then cpDNA abundance should have resembled what is observed under +P conditions. This suggests that Phi represses P limitation signaling, leading to cpRNA metabolic adjustments and other responses controlled by PSR1, but that the pathway leading to cpDNA degradation operates independently. Indeed, Phi also represses P starvation signaling, leading to changes in chloroplast photosynthetic membrane composition (Kobayashi et al., 2006).
RNA Metabolism under P Limitation
PNPase activity (Yehudai-Resheff et al., 2001) and cpRNA degradation rates (Schuster et al., 1999) respond to the relative concentrations of Pi and nucleotides in vitro, but how its directionality is regulated in vivo is poorly understood. Our results with wild-type cells show that cpRNA stability increases under −P conditions (Figure 6), although RNA accumulation is not seen until the 24-h time point (Figure 2). The delayed response likely reflects the time required for the chloroplast Pi concentration to decrease sufficiently so that PNPase nucleolytic activity is disfavored. Whether the decrease in Pi concentration leads to increased mRNA polyadenylation via the reverse PNPase reaction is unknown, and quantitative measurements of either chloroplast polyadenylation or NDP:Pi ratio are highly problematic. In any event, although polyadenylation normally destabilizes cpRNA (Lisitsky et al., 1997), in a low P situation PNPase would not be prone to degrade these polyadenylated species, thus allowing RNA levels to increase. Together with the reduction of PNPase itself (Figure 4B), our results indicate a general slowing of cpRNA metabolism under P limitation.
In the PNPase-deficient ip3 line, the putative RNase II encoded by RNB2 appears to be upregulated (Figure 3) under both +P and −P conditions. Although in E. coli the reciprocal relationship between pnp and rnb expression can be explained by the activity of each respective enzyme on the other's mRNA (Zilhao et al., 1996), this cannot be the case in Chlamydomonas, in which the mRNAs are cytosolic and the enzymes are in the chloroplast. This finding, therefore, suggests some kind of metabolic or chemical signal resulting from PNPase deficiency that ultimately results in a transcription increase for RNB2.
The situation we have described for ribonucleases in the wild-type chloroplast contrasts sharply with the upregulation of cytosolic ribonucleases that often occurs during P limitation. For example, RNS1 and RNS2 are induced under P stress in Arabidopsis (Bariola et al., 1994). Cultured plant cells, bacteria, and fungi also induce and sometimes secrete RNases under P stress (Nürnberger et al., 1989; Hahnen et al., 2000; Tasaki et al., 2004). These RNases, however, are almost certainly hydrolytic, rather than phosphorylytic, allowing them to serve as scavengers under these circumstances. Why the presumably chloroplastic RNB2 would not be induced as a scavenger in Chlamydomonas is unclear.
Requirement of PNPase for Acclimation to P Limitation
A striking result was the progressive loss of viability for PNPase-deficient strains, which was manifested between 6 and 72 h in −P conditions (Figure 7). The psr1 mutant also dies under −P conditions, ultimately because of an inability to control oxidative damage (Moseley et al., 2006). PNPase-deficient cells do not lose viability under +P conditions or a variety of other stress situations, suggesting that they are specifically unable to cope with P limitation. Indeed, Arabidopsis plants lacking detectable PNPase were reported to grow normally when planted in soil (Walter et al., 2002), and PNPase is also not essential in E. coli (Donovan and Kushner, 1986).
On the other hand, prokaryotic PNPase has been shown to participate in other abiotic and biotic stress responses. In bacteria, PNPase induction is integral to cold shock adaptation through its regulation of mRNA stability (Clarke and Dowds, 1994; Goverde et al., 1998; Yamanaka and Inouye, 2001). PNPase is also required for the complete degradation of glucose transporter mRNA in response to blockage of the glycolytic pathway in E. coli (Morita et al., 2004) and is required for the full development of competence in Bacillus subtilis (Luttinger et al., 1996), which is a pathway induced by nutrient deficiency. PNPase has further been linked to the ability of Yersinia to infect animal cells via induction of the bacterial type III secretion system (Rosenzweig et al., 2005). These results, along with the finding that ∼100 mRNAs are differentially expressed in a Salmonella PNPase mutant (Clements et al., 2002) and a more recent link to apoptosis (Sarkar and Fisher, 2006), suggest that the regulatory properties of PNPase have only begun to be revealed.
DNA Mobilization in the Chloroplast
The reduction of DNA copy number under P limitation was unexpected, given the predominance of mobilization mechanisms harnessing RNA, polyphosphate, or other metabolites. However, the chloroplast is polyploid, leaving it in a situation of genic excess. The evolutionary basis for chloroplast polyploidy has been much debated (Bendich, 1987; Race et al., 1999; Allen, 2003; Koumandou et al., 2004), but our data suggest that at least in Chlamydomonas it can serve as a repository of P, as was suggested previously (Sears and VanWinkle-Swift, 1994). If one assumes 80 copies of a 203-kb genome per chloroplast, a cell volume of 10 μm3, and that the chloroplast occupies 60% of the cell volume, the release of all P in cpDNA would lead to a concentration of 1 mM. Although cpDNA did not disappear completely in the wild type under our experimental conditions (Figures 9 and 10), the difference in cpDNA amount between +P and −P conditions could be metabolically meaningful. Reduction of cpDNA copy number has also been associated with leaf maturation or senescence in several species (Shaver et al., 2006), which also may contribute to P mobilization. These observations, however, have been challenged for some species (Li et al., 2006).
In summary, our data suggest that, apart from the known role of PNPase in polyadenylation-mediated RNA degradation, there is an additional and perhaps even more important role under P limitation. We hypothesize that this role ultimately can be described as altering metabolic flux, which not only determines the fate of RNA and DNA but also influences photosynthesis and the P economy of the cell as a whole.
METHODS
Culture Conditions and Treatments
Cultures were initiated in Tris-acetate-phosphate (TAP) medium (Harris, 1989) under continuous light at 25°C, and after 3 to 4 d cell densities had reached 1 to 2 × 106 cells/mL. Phosphate was depleted by washing and then resuspending cells in TA medium (TAP − P). Repletion was achieved by washing cells and resuspending them in TAP medium, followed by additional growth. For Phi substitution experiments, the P in TAP medium was replaced by 1.0 mM H3PO3. To observe RNA stability, cells were grown as described above, and either 0 or 6 h after establishing subcultures, rifampicin was added to a final concentration of 250 μg/mL.
Stress conditions were as follows: for heat shock, cells in log phase were grown in TAP medium for 72 h under continuous light at 32°C; cold shock was as for heat shock but at 16°C; for UV stress, cells at log phase were exposed to 60 mJ·m−2.s−1 254-nm UV light for 24 h, then grown for 72 h in TAP medium under continuous light at 25°C. For 5-fluoro-2′-deoxyuridine treatment, cells were incubated with a 0.5 mM 5-fluoro-2′-deoxyuridine concentration, which is known to reduce chloroplast genome copy number (Wurtz et al., 1977). Evans blue staining used a 10% solution of the dye at a final concentration of 1%.
Generation of PNPase-Deficient Strains and Anti-PNPase Antibodies
The overlapping EST clones AV396679 and AV644809 were obtained from the Kazusa DNA Research Institute (Asamizu et al., 1999). Degenerate primers were designed for RT-PCR, leading to the complete cDNA sequence mentioned in Results (GenBank accession number DQ492261). The antisense construct was based on plasmid pCB797 (Schroda et al., 2002). PCR was used to amplify base pairs 158 to 522 of the coding region, with the addition of ClaI and EcoRI sites, which were ligated into the same sites of pCB797. Nuclear transformants were generated by electroporation (Shimogawara et al., 1998) with the linearized construct and selected on TAP medium containing 2 μg/mL zeocin (Invitrogen).
The RNAi construct was based on the vector containing the Maa7/X inverted repeat transgene from Rohr et al. (2004). To facilitate the insertion of an inverted repeat into this vector, modifications were performed to make it Gateway (Invitrogen) compatible, similar to pHELLSGATE (Helliwell and Waterhouse, 2003). The new vector was named pGwyRNAi and contains the Arabidopsis thaliana PDK intron in the spacer region. PCR was used to amplify base pairs 277 to 522 of the PNP coding region and insert it into pENTR (Invitrogen). A double Gateway LR recombination was then used to insert this fragment in inverted orientations into pGwyRNAi. Chlamydomonas reinhardtii transformants of the recipient strain CC-406 were selected on TAP medium containing 8 μM 5-fluoroindole, 10 μg/mL paromomycin, and 1.5 mM Trp on plates covered with a single layer of paper towels and under light.
To generate the anti-PNPase antibody, a cDNA fragment extending from position 172 to 522 of the coding region was inserted into pENTR. Gateway LR Clonase facilitated recombination of the fragment into the pDEST17 expression vector with an N-terminal His tag. BL21-AI cells carrying the vector grown to an OD of 0.44 were induced with 0.2% arabinose for 4.5 h at 25°C. The cells were pelleted and resuspended in lysis buffer (300 mM NaCl, 10 mM imidazole, and 50 mM Tris, pH 8). After the cells were passed two times through a French press, it was determined that the overexpressed antigen was in the insoluble fraction. The insoluble pellet was resuspended in urea buffer (10 mM Tris, 100 mM sodium phosphate, 50 mM NaCl, and 8 M urea) and clarified before mixing with nickel–nitrilotriacetic acid agarose slurry (Qiagen) for 1.5 h at room temperature and transferring to a column. After washing, the column was eluted with 100 mM NaH2PO4, 10 mM Tris HCl, and 8 M urea, pH 4.4. A total of 2.5 mg of the eluted polypeptide was excised from a polyacrylamide gel and sent to Lampire Biological Laboratories for antibody production in rabbits using their Express-line protocol.
Targeting Analysis of RNB1 and RNB2
The RNB1 fragment encoding amino acids 1 to 189 was amplified from cDNA, inserted into the Gateway entry vector pENTR, and recombined into pMDC83 (Curtis and Grossniklaus, 2003), a vector designed for N-terminal fusions with GFP. The intronless RNB2 fragment encoding amino acids 1 to 115 was amplified from genomic DNA and cloned in the same manner. The predicted RNB2 amino acid sequence differs from the gene model at the N terminus, based on our cDNA sequencing (GenBank accession number EF431887). The expression cassette for the YFP cytosolic control was described previously (Bollenbach et al., 2005). A total of 20 μg of the vectors was used in polyethylene glycol–mediated transformation of tomato (Solanum lycopersicum) protoplasts as described (Xing et al., 2001). Protoplasts were visualized at 24 h after transformation by confocal microscopy. The dye Mitotracker CMXRos (Molecular Probes) was used at ∼200 nM to visualize mitochondria.
RNA and DNA Analysis
Total RNA was isolated and analyzed as described by Drapier et al. (1998). Radiolabeled probes were derived from intragenic DNA fragments. RT-PCR was performed using the Access PCR system (Promega) to generate products corresponding to PNP, PSR1, and actin, with 0.1 μg of DNase-treated total RNA as starting material. PCR conditions were 95°C for 10 s followed by cycles of 95, 62, and 75°C for 1 min each. The reactions were stopped after 20 cycles for actin and after 40 cycles for PNP and PSR1. The primers used were as follows: for PNPase, 5′-CCGCTTGCGTGACTGCAAATGC-3′ and 5′-CATCCGACGCAGCGATGTCC-3′; for PSR1, 5′-CAGAAGTACCGCCTCAACATCC-3′ and 5′-CCTCCAAGCTCAGCTGCAGTTG-3′; and for actin, 5′-AATCGTGCGCGACATCAAGGAGAA-3′ and 5′-TTGGCGATCCACATTTGCTGGAAGGT-3′. For the experiment shown in Figure 3, cDNA was generated from 0.5 μg of DNase-treated RNA (TAP-grown cells; ip3 and vector control cultures contained 8 μM 5-fluoroindole) and random hexamers using the SuperScript III kit (Invitrogen). One-tenth of the reaction was used in a 50-μL PCR with Promega GoTaq polymerase, 1 M betaine, and intron-spanning primers (available upon request) for RNB1 (37 cycles), RNB2 (35 cycles), or actin (30 cycles).
For DNA gel blot analysis, total DNA was prepared as described by Rochaix (1980), digested with restriction enzymes, and, after gel electrophoresis and transfer, probed with radiolabeled gene-specific fragments.
SYBR Green I Staining and Quantification of Nucleoids
To stain the nuclear and chloroplast DNAs in live cells, SYBR Green I (Molecular Probes) was added to a final dilution of 1:1000. Images were recorded under blue excitation with a Zeiss Axioscope fluorescence microscope. Nucleoid numbers were estimated by counting clearly visible fluorescent spots within the outlines of the chloroplast. Questionable or indistinct spots were excluded. To quantify fluorescence, images were imported into ImageQuant version 1.2 (Molecular Dynamics), and areas were selected corresponding to nucleoids as described above. The volumes (i.e., densities) of these were determined and summed, leading to an estimation of total cpDNA fluorescence per cell.
Accession Numbers
GenBank/EMBL accession numbers and Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: Chlamydomonas RNB2, EF431887; Chlamydomonas PNP, DQ492261; Arabidopsis PHR1, At4g28610; and Chlamydomonas PSR1, AF174480.
Acknowledgments
We thank Osumi Misumi (Rikkyo University) for providing the moc strain, Michel Schroda (University of Freiburg) for pCB797, Arthur Grossman (Carnegie Institution) for psr1 and sac1, Steve MacKinnon for completion of the PNP and RNB2 cDNA sequences, and Katia Wostrikoff, Tom Bollenbach, and especially Gadi Schuster for helpful suggestions and critical readings of the manuscript. Work at the Boyce Thompson Institute was supported by Postdoctoral Fellowship Award FI-346-2003 from the Binational Agriculture Research and Development Fund (BARD) to S.Y.-R. and by BARD Award IS-3605-04, Binational Science Foundation Award 2001090, and National Science Foundation Grant MCB-0091020 to D.B.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: David B. Stern (ds28@cornell.edu).
Open Access articles can be viewed online without a subscription.
References
- Abel, S., Ticconi, C.A., and Delatorre, C.A. (2002). Phosphate sensing in higher plants. Physiol. Plant. 115 1–8. [DOI] [PubMed] [Google Scholar]
- Allen, J.F. (2003). The function of genomes in bioenergetic organelles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 19–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu, E., Nakamura, Y., Sato, S., Fukuzawa, H., and Tabata, S. (1999). A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. DNA Res. 6 369–373. [DOI] [PubMed] [Google Scholar]
- Aung, K., Lin, S.I., Wu, C.C., Huang, Y.T., Su, C.L., and Chiou, T.J. (2006). pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari, R., Datt Pant, B., Stitt, M., and Scheible, W.R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola, P.A., Howard, C.J., Taylor, C.B., Verburg, M.T., Jaglan, V.D., and Green, P.J. (1994). The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 6 673–685. [DOI] [PubMed] [Google Scholar]
- Bendich, A.J. (1987). Why do chloroplasts and mitochondria contain so many copies of their genome? Bioessays 6 279–282. [DOI] [PubMed] [Google Scholar]
- Bisanz, C., Begot, L., Carol, P., Perez, P., Bligny, M., Pesey, H., Gallois, J.L., Lerbs-Mache, S., and Mache, R. (2003). The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol. Biol. 51 651–663. [DOI] [PubMed] [Google Scholar]
- Bollenbach, T.J., Lange, H., Gutierrez, R., Erhardt, M., Stern, D.B., and Gagliardi, D. (2005). RNR1, a 3′-5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res. 33 2751–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D.L., Delatorre, C.A., Bakker, A., and Abel, S. (2000). Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta 211 13–22. [DOI] [PubMed] [Google Scholar]
- Chiou, T.J., Aung, K., Lin, S.I., Wu, C.C., Chiang, S.F., and Su, C.L. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciereszko, I., Johansson, H., Hurry, V., and Kleczkowski, L.A. (2001). Phosphate status affects the gene expression, protein content and enzymatic activity of UDP-glucose pyrophosphorylase in wild-type and pho mutants of Arabidopsis. Planta 212 598–605. [DOI] [PubMed] [Google Scholar]
- Clarke, D.J., and Dowds, B.C. (1994). The gene coding for polynucleotide phosphorylase in Photorhabdus sp. strain K122 is induced at low temperatures. J. Bacteriol. 176 3775–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements, M.O., Eriksson, S., Thompson, A., Lucchini, S., Hinton, J.C., Normark, S., and Rhen, M. (2002). Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. USA 99 8784–8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crutchfield, A.L.M., Diller, K.R., and Brand, J.J. (1999). Cryopreservation of Chlamydomonas reinhardtii (Chlorophyta). Eur. J. Phycol. 34 43–52. [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, J.P., Yildiz, F., and Grossman, A.R. (1994). Mutants of Chlamydomonas with aberrant responses to sulfur deprivation. Plant Cell 6 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, J.P., Yildiz, F.H., and Grossman, A.R. (1999). Sac3, an Snf1-like serine/threonine kinase that positively and negatively regulates the responses of Chlamydomonas to sulfur limitation. Plant Cell 11 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize, E., and Randall, P.J. (1995). Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 107 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan, W.P., and Kushner, S.R. (1986). Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 83 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapier, D., Suzuki, H., Levy, H., Rimbault, B., Kindle, K.L., Stern, D.B., and Wollman, F.-A. (1998). The chloroplast atpA gene cluster in Chlamydomonas reinhardtii: Functional analysis of a polycistronic transcription unit. Plant Physiol. 117 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus, M., and Regnier, P. (2002). The poly(A) tail of mRNAs. Bodyguard in eukaryotes, scavenger in bacteria. Cell 111 611–613. [DOI] [PubMed] [Google Scholar]
- Eberhard, S., Drapier, D., and Wollman, F.A. (2002). Searching limiting steps in the expression of chloroplast-encoded proteins: Relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 31 149–160. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Fujii, H., Chiou, T.J., Lin, S.I., Aung, K., and Zhu, J.K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15 2038–2043. [DOI] [PubMed] [Google Scholar]
- Goverde, R.L., Huis in't Veld, J.H., Kusters, J.G., and Mooi, F.R. (1998). The psychrotrophic bacterium Yersinia enterocolitica requires expression of pnp, the gene for polynucleotide phosphorylase, for growth at low temperature (5°C). Mol. Microbiol. 28 555–569. [DOI] [PubMed] [Google Scholar]
- Green, P.J. (1994). The ribonucleases of higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45 421–445. [Google Scholar]
- Grossman, A. (2000). Acclimation of Chlamydomonas reinhardtii to its nutrient environment. Protist 151 201–224. [DOI] [PubMed] [Google Scholar]
- Hahnen, E., Znamenskaya, L., Koczan, D., and Leshchinskaya, I. (2000). A novel secreted ribonuclease from Bacillus intermedius: Gene structure and regulatory control. Mol. Gen. Genet. 263 571–580. [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press). [DOI] [PubMed]
- Hausler, R.E., Schlieben, N.H., Schulz, B., and Flugge, U.I. (1998). Compensation of decreased triose phosphate/phosphate translocator activity by accelerated starch turnover and glucose transport in transgenic tobacco. Planta 204 366–376. [DOI] [PubMed] [Google Scholar]
- Hayes, R., Kudla, J., Schuster, G., Gabay, L., Maliga, P., and Gruissem, W. (1996). Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J. 15 1132–1141. [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C., and Waterhouse, P. (2003). Constructs and methods for high-throughput gene silencing in plants. Methods 30 289–295. [DOI] [PubMed] [Google Scholar]
- Hwang, S., Kawazoe, R., and Herrin, D.L. (1996). Transcription of tufA and other chloroplast-encoded genes is controlled by a circadian clock in Chlamydomonas. Proc. Natl. Acad. Sci. USA 93 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irihimovitch, V., and Stern, D.B. (2006). The SAC3 kinase is required for chloroplast transcriptional repression under sulfur limitation in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 103 7911–7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishine, M., Takabayashi, A., Munekage, Y., Shikanai, T., Endo, T., and Sato, F. (2004). Ribosomal RNA processing and an RNase R family member in chloroplasts of Arabidopsis. Plant Mol. Biol. 55 595–606. [DOI] [PubMed] [Google Scholar]
- Kobayashi, K., Masuda, T., Takamiya, K.-i., and Ohta, H. (2006). Membrane lipid alteration during phosphate starvation is regulated by phosphate signaling and auxin/cytokinin cross-talk. Plant J. 47 238–248. [DOI] [PubMed] [Google Scholar]
- Koumandou, V.L., Nisbet, R.E.R., Barbrook, A.C., and Howe, C.J. (2004). Dinoflagellate chloroplasts—Where have all the genes gone? Trends Genet. 20 261–267. [DOI] [PubMed] [Google Scholar]
- Kudla, J., Hayes, R., and Gruissem, W. (1996). Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J. 15 7137–7146. [PMC free article] [PubMed] [Google Scholar]
- Lee, T.-M., Tsai, P.-F., Shyu, Y.-T., and Sheu, F. (2005). The effects of phosphite on phosphate starvation responses of Ulva lactuca (Ulvales, Chlorophyta). J. Phycol. 41 975–982. [Google Scholar]
- Li, W., Ruf, S., and Bock, R. (2006). Constancy of organellar genome copy numbers during leaf development and senescence in higher plants. Mol. Genet. Genomics 275 185–192. [DOI] [PubMed] [Google Scholar]
- Lisitsky, I., Kotler, A., and Schuster, G. (1997). The mechanism of preferential degradation of polyadenylated RNA in the chloroplast: The exoribonuclease 100RNP-polynucleotide phosphorylase displays high binding affinity for poly(A) sequences. J. Biol. Chem. 272 17648–17653. [DOI] [PubMed] [Google Scholar]
- Luttinger, A., Hahn, J., and Dubnau, D. (1996). Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 19 343–356. [DOI] [PubMed] [Google Scholar]
- Misson, J., et al. (2005). A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. USA 102 11934–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misumi, O., Suzuki, L., Nishimura, Y., Sakai, A., Kawano, S., Kuroiwa, H., and Kuroiwa, T. (1999). Isolation and phenotypic characterization of Chlamydomonas reinhardtii mutants defective in chloroplast DNA segregation. Protoplasma 209 273–282. [Google Scholar]
- Miura, K., Rus, A., Sharkhuu, A., Yokoi, S., Karthikeyan, A.S., Raghothama, K.G., Baek, D., Koo, Y.D., Jin, J.B., Bressan, R.A., Yun, D.-J., and Hasegawa, P.M. (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA 102 7760–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, T., Kawamoto, H., Mizota, T., Inada, T., and Aiba, H. (2004). Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 54 1063–1075. [DOI] [PubMed] [Google Scholar]
- Moseley, J.L., Chang, C.-W., and Grossman, A.R. (2006). Genome-based approaches to understanding phosphorus deprivation responses and PSR1 control in Chlamydomonas reinhardtii. Eukaryot. Cell 5 26–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, Y., Kikis, E.A., Zimmer, S.L., Komine, Y., and Stern, D.B. (2004). Antisense transcript and RNA processing alterations suppress instability of polyadenylated mRNA in Chlamydomonas chloroplasts. Plant Cell 16 2849–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, Y., Misumi, O., Kato, K., Inada, N., Higashiyama, T., Momoyama, Y., and Kuroiwa, T. (2002). An mt(+) gamete-specific nuclease that targets mt(−) chloroplasts during sexual reproduction in C. reinhardtii. Genes Dev. 16 1116–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, Y., Misumi, O., Matsunaga, S., Higashiyama, T., Yokota, A., and Kuroiwa, T. (1999). The active digestion of uniparental chloroplast DNA in a single zygote of Chlamydomonas reinhardtii is revealed by using the optical tweezer. Proc. Natl. Acad. Sci. USA 96 12577–12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger, T., Abel, S., Jost, W., and Glund, K. (1989). Induction of an extracellular ribonuclease in cultured tomato cells upon phosphate starvation. Plant Physiol. 92 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin, R., Lange, H., Grienenberger, J.M., and Gagliardi, D. (2004). AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res. 32 5174–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, B.L., Lagerstedt, J.O., Pratt, J.R., Pattison-Granberg, J., Lundh, K., Shokrollahzadeh, S., and Lundh, F. (2003). Regulation of phosphate acquisition in Saccharomyces cerevisiae. Curr. Genet. 43 225–244. [DOI] [PubMed] [Google Scholar]
- Poirier, Y., and Bucher, M. (September 30, 2002). Phosphate transport and homeostasis in Arabidopsis. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0099, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Race, H.L., Herrmann, R.G., and Martin, W. (1999). Why have organelles retained genomes? Trends Genet. 15 364–370. [DOI] [PubMed] [Google Scholar]
- Ris, H., and Plaut, W. (1962). Ultrastructure of DNA-containing areas in the chloroplast of Chlamydomonas. J. Cell Biol. 13 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix, J.-D. (1980). Restriction fragments from Chlamydomonas chloroplast DNA. Methods Enzymol. 65 785–795. [DOI] [PubMed] [Google Scholar]
- Rohr, J., Sarkar, N., Balenger, S., Jeong, B.R., and Cerutti, H. (2004). Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 40 611–621. [DOI] [PubMed] [Google Scholar]
- Rosenzweig, J.A., Weltman, G., Plano, G.V., and Schesser, K. (2005). Modulation of Yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J. Biol. Chem. 280 156–163. [DOI] [PubMed] [Google Scholar]
- Rubio, V., Linhares, F., Solano, R., Martin, A.C., Iglesias, J., Leyva, A., and Paz-Ares, J. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, D., and Fisher, P.B. (2006). Human polynucleotide phosphorylase (hPNPase old-35): An RNA degradation enzyme with pleiotrophic biological effects. Cell Cycle 5 1080–1084. [DOI] [PubMed] [Google Scholar]
- Sauret-Gueto, S., Botella-Pavia, P., Flores-Perez, U., Martinez-Garcia, J.F., San Roman, C., Leon, P., Boronat, A., and Rodriguez-Concepcion, M. (2006). Plastid cues posttranscriptionally regulate the accumulation of key enzymes of the methylerythritol phosphate pathway in Arabidopsis. Plant Physiol. 141 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda, M., Beck, C.F., and Vallon, O. (2002). Sequence elements within an HSP70 promoter counteract transcriptional transgene silencing in Chlamydomonas. Plant J. 31 445–455. [DOI] [PubMed] [Google Scholar]
- Schuster, G., Lisitsky, I., and Klaff, P. (1999). Polyadenylation and degradation of mRNA in the chloroplast. Plant Physiol. 120 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears, B.B., and VanWinkle-Swift, K. (1994). The salvage/turnover/repair (STOR) model for uniparental inheritance in Chlamydomonas: DNA as a source of sustenance. J. Hered. 85 366–376. [DOI] [PubMed] [Google Scholar]
- Shaver, J.M., Oldenburg, D.J., and Bendich, A.J. (2006). Changes in chloroplast DNA during development in tobacco, Medicago truncatula, pea, and maize. Planta 224 72–82. [DOI] [PubMed] [Google Scholar]
- Shimogawara, K., Fujiwara, S., Grossman, A., and Usuda, H. (1998). High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawara, K., Wykoff, D.D., Usuda, H., and Grossman, A.R. (1999). Chlamydomonas reinhardtii mutants abnormal in their responses to phosphorus deprivation. Plant Physiol. Biochem. 120 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic, S., Portnoy, V., Liveanu, V., and Schuster, G. (2006). RNA polyadenylation in prokaryotes and organelles: Different tails tell different tales. CRC Crit. Rev. Plant Sci. 25 65–77. [Google Scholar]
- Tasaki, Y., Azwan, A., Hara, T., and Joh, T. (2004). Structure and expression of a phosphate deficiency-inducible ribonuclease gene in Pholiota nameko. Curr. Genet. 45 28–36. [DOI] [PubMed] [Google Scholar]
- Ticconi, C.A., Delatorre, C.A., and Abel, S. (2001). Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol. 127 963–972. [PMC free article] [PubMed] [Google Scholar]
- Varadarajan, D.K., Karthikeyan, A.S., Matilda, P.D., and Raghothama, K.G. (2002). Phosphite, an analog of phosphate, suppresses the coordinated expression of genes under phosphate starvation. Plant Physiol. 129 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaw, W.K., and Harrison, M.J. (2002). A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14 1751–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, M., Kilian, J., and Kudla, J. (2002). PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. EMBO J. 21 6905–6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, A.P. (2004). Solute transporters as connecting elements between cytosol and plastid stroma. Curr. Opin. Plant Biol. 7 247–253. [DOI] [PubMed] [Google Scholar]
- Wu, P., Ma, L., Hou, X., Wang, M., Wu, Y., Liu, F., and Deng, X.W. (2003). Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 132 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz, E.A., Boynton, J.E., and Gillham, N.W. (1977). Perturbation of chloroplast DNA amounts and chloroplast gene transmission in Chlamydomonas reinhardtii by 5-fluorodeoxyuridine. Proc. Natl. Acad. Sci. USA 74 4552–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff, D.D., Grossman, A.R., Weeks, D.P., Usuda, H., and Shimogawara, K. (1999). Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl. Acad. Sci. USA 96 15336–15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, T., Malik, K., Martin, T., and Miki, B.L. (2001). Activation of tomato PR and wound-related genes by a mutagenized tomato MAP kinase kinase through divergent pathways. Plant Mol. Biol. 46 109–120. [DOI] [PubMed] [Google Scholar]
- Yamanaka, K., and Inouye, M. (2001). Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J. Bacteriol. 183 2808–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, K., and Metcalf, W.W. (2004). A new activity for an old enzyme: Escherichia coli bacterial alkaline phosphatase is a phosphite-dependent hydrogenase. Proc. Natl. Acad. Sci. USA 101 7919–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., Hirsh, M., and Schuster, G. (2001). Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 21 5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., Portnoy, V., Yogev, S., Adir, N., and Schuster, G. (2003). Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with exosome proteins. Plant Cell 15 2003–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilhao, R., Cairrao, F., Regnier, P., and Arraiano, C.M. (1996). PNPase modulates RNase II expression in Escherichia coli: Implications for mRNA decay and cell metabolism. Mol. Microbiol. 20 1033–1042. [DOI] [PubMed] [Google Scholar]