Abstract
Chlorophyll is a central player in harvesting light energy for photosynthesis, yet the rate-limiting steps of chlorophyll catabolism and the regulation of the catabolic enzymes remain unresolved. To study the role and regulation of chlorophyllase (Chlase), the first enzyme of the chlorophyll catabolic pathway, we expressed precursor and mature versions of citrus (Citrus sinensis) Chlase in two heterologous plant systems: (1) squash (Cucurbita pepo) plants using a viral vector expression system; and (2) transiently transformed tobacco (Nicotiana tabacum) protoplasts. Expression of full-length citrus Chlase resulted in limited chlorophyll breakdown in protoplasts and no visible leaf phenotype in whole plants, whereas expression of a Chlase version lacking the N-terminal 21 amino acids (ChlaseΔN), which corresponds to the mature protein, led to extensive chlorophyll breakdown in both tobacco protoplasts and squash leaves. ChlaseΔN-expressing squash leaves displayed a dramatic chlorotic phenotype in plants grown under low-intensity light, whereas under natural light a lesion-mimic phenotype occurred, which was demonstrated to follow the accumulation of chlorophyllide, a photodynamic chlorophyll breakdown product. Full-length and mature citrus Chlase versions were localized to the chloroplast membrane fraction in expressing tobacco protoplasts, where processing of the N-terminal 21 amino acids appears to occur. Results obtained in both plant systems suggest that Chlase functions as a rate-limiting enzyme in chlorophyll catabolism controlled via posttranslational regulation.
INTRODUCTION
The chlorophyll molecule is a central player in harvesting light energy channeled for photosynthesis. In its functional context as part of the multiple protein–pigment photosynthetic complex, the capture of a light photon by chlorophyll is the first step in converting light energy into chemical energy and carbon assimilation. Outside of the photosynthetic complex, the very same photodynamic properties of chlorophyll result in phototoxicity of its anabolic and catabolic metabolites. Because both the biosynthesis and breakdown of chlorophyll occur throughout plant development, both pathways need to be tightly regulated in a manner that prevents the accumulation of intermediate compounds that can potentially damage plant tissue. However, knowledge of the regulation of chlorophyll catabolism in plant tissue is incomplete; in particular, the mechanisms regulating the catabolic enzymes remain largely unknown (Kräutler and Matile, 1999; Hörtensteiner, 2006).
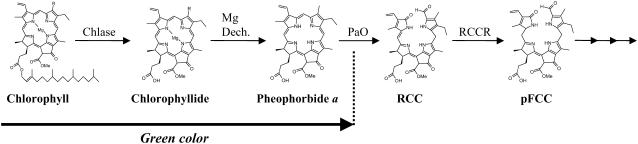
Chlorophyll catabolism is dramatically visualized during leaf senescence and fruit ripening but also occurs at a basal level during natural turnover (Goldschmidt, 2001), which is affected by environmental conditions such as excessive light, leading to photoinhibition (Prasil et al., 1992; Andersson and Barber, 1996). Insight into the chlorophyll catabolic pathway was obtained fairly recently after the identification and characterization of chlorophyll breakdown products in plant tissue (Amir-Shapira et al., 1987; Engel et al., 1991; Matile et al., 1996; Hörtensteiner, 1999). The breakdown of chlorophyll into phytol, magnesium, and the primary cleavage product of the porphyrin ring occurs in four consecutive steps catalyzed by chlorophyllase (Chlase), Mg-dechelatase, pheophorbide a oxygenase, and red chlorophyll catabolite reductase (Figure 1). Loss of the typical chlorophyll green color occurs only after cleavage of the porphyrin ring catalyzed by pheophorbide a oxygenase. Further downstream, the porphyrin breakdown products are modified and exported to the vacuole, where they are stored indefinitely (Hörtensteiner, 1999, 2004; Matile et al., 1999; Takamiya et al., 2000). Thus, chlorophyll breakdown is a multistep enzymatic process, and although the majority of the enzymes involved were identified several years ago (Terpstra, 1981; Trebitsh et al., 1993; Rodoni et al., 1997; Tsuchiya et al., 1997; Hörtensteiner et al., 1998), the delay in gene cloning presented a major obstacle to the study of chlorophyll catabolism (Takamiya et al., 2000). The isolation of genes encoding key enzymes of the catabolic pathway in recent years (Chlase [Jacob-Wilk et al., 1999; Tsuchiya et al., 1999]; red chlorophyll catabolite reductase [Wuthrich et al., 2000]; pheophorbide a oxygenase [Pružinská et al., 2003]) provided tools for an in-depth study of their role and regulation during chlorophyll breakdown.
Figure 1.
Chlorophyll Catabolic Pathway.
The first four enzymatic steps of the chlorophyll catabolic pathway leading to loss of the typical green color are outlined. Structures of chlorophyll and the intermediate breakdown products are shown. Enzyme-catalyzed chlorophyll breakdown steps are as follows. (1) Chlase, an esterase, catalyzes the cleavage of the hydrophobic thylakoid-anchoring phytol chain of chlorophyll from the porphyrin ring, resulting in the product chlorophyllide, which retains the typical green color. (2) Mg-dechelatase is responsible for removing the Mg ion from chlorophyllide to yield pheophorbide a, which retains the green color. (3) Pheophorbide a oxygenase (PaO) catalyzes the cleavage of the porphyrin ring, resulting in the red chlorophyll catabolite (RCC), which is further reduced by the enzyme red chlorophyll catabolite reductase (RCCR) to a primary fluorescent chlorophyll catabolite (pFCC). pFCCs are further metabolized by additional enzymes downstream and exported to the vacuole, where they are nonenzymatically converted to nonfluorescent chlorophyll catabolites (NCCs) and stored indefinitely.
The first genes encoding Chlase were isolated concomitantly from citrus (Citrus sinensis) and Chenopodium based on protein sequence data (Jacob-Wilk et al., 1999; Tsuchiya et al., 1999) and revealed an N-terminally encoded sequence absent from the mature protein. In both cases, the N-terminal sequence is short (21 amino acids in citrus and 30 amino acids in Chenopodium) and not characteristic of chloroplast transit peptides (the Chenopodium sequence is more reminiscent of an endoplasmic reticulum signal peptide [Takamiya et al., 2000]), a finding that questioned whether all Chlases are located in the chloroplast and raised the possibility that some chlorophyll catabolism could occur in the vacuole after export of whole chlorophyll molecules (Tsuchiya et al., 1999; Takamiya et al., 2000). The gene encoding Chlase from citrus (Jacob-Wilk et al., 1999) is the only Chlase gene cloned based on an enzyme purified from chloroplasts. Therefore, despite the short (21 amino acid) and uncharacteristic N-terminal sequence, the experimental evidence places the encoded protein within the chloroplast (Trebitsh et al., 1993; Jacob-Wilk et al., 1999).
The regulation of chlorophyll catabolism and the mechanism regulating the catabolic enzymes have been elusive research targets and the subject of much debate. Chlase, the first enzyme in the catabolic pathway, is an obvious candidate for being a rate-limiting enzyme in the pathway (Tsuchiya et al., 1999). Indeed, some data, such as the correlation between degreening and Chlase expression induction in ethylene-treated citrus fruit (Jacob-Wilk et al., 1999), support this possibility. Yet, other data reported in the literature are inconsistent with the possibility that Chlase is a rate-limiting enzyme regulated at the level of transcription. (1) Chlase was found to be expressed in low constitutive levels throughout natural fruit development (in citrus), and it is not clear how chlorophyll breakdown is set into motion toward senescence or fruit ripening (Jacob-Wilk et al., 1999). (2) Chlase activity in plant tissues, as analyzed in vitro, does not always correlate well with degreening during natural senescence and fruit ripening (Minguez-Mosquera and Gallardo-Guerrero, 1996; Fang et al., 1998). (3) Chlase enzyme has been localized to the inner envelope membrane of the chloroplast (Brandis et al., 1996; Matile et al., 1997), and it is not clear how the enzyme and its chlorophyll substrate come into contact. Thus, integration of the data implies two main possibilities regarding the regulation of Chlase and its role during natural turnover, senescence, and degreening processes in the chlorophyll catabolic pathway. One possibility is that Chlase comprises a constitutive, nonregulated step in chlorophyll catabolism. Alternatively, the data are also consistent with Chlase constituting a rate-limiting step in the case of regulation at the posttranslational level. Recent work by Benedetti and Arruda (2002) and Kariola et al. (2005) showed that overexpression of the Chlase-encoding gene in Arabidopsis thaliana affected the chlorophyll:chlorophyllide ratio in leaf tissue to some extent, but no visible phenotype was obtained. These results provided an important demonstration of the activity of the Arabidopsis Chlase gene product in vivo but did not shed light on the regulation of Chlase and its role in chlorophyll catabolism.
The open questions surrounding the role and regulation of Chlase in chlorophyll catabolism encouraged us to conduct a physiological study in whole plants and plant cells expressing several versions of the citrus Chlase gene. We provide evidence that overexpression of a Chlase version lacking the N-terminal 21 amino acids, which corresponds to the mature protein, results in extensive chlorophyll breakdown and chlorosis, suggesting that it enters the chloroplast by a noncanonical mechanism and is devoid of the regulatory constraints of the full-length protein. Both full-length and mature citrus Chlase expressed in protoplasts are localized to the chloroplast membrane fraction, where processing of the N-terminal 21 amino acids occurs. We suggest that Chlase functions as a rate-limiting enzyme in chlorophyll catabolism controlled via posttranslational regulation.
RESULTS
Ser-147 Is Essential for Citrus Chlase Activity
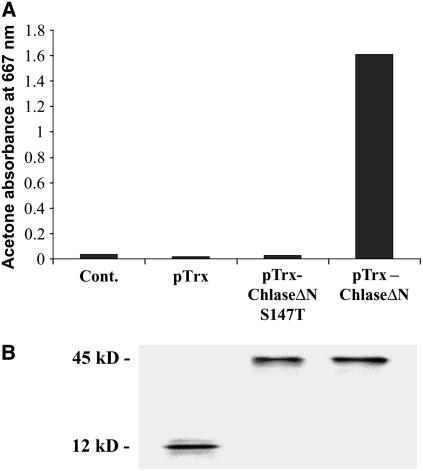
Sequence analysis of functionally characterized Chlase genes, using motif detection software, indicated the presence of a putative Ser lipase domain (Jacob-Wilk et al., 1999; Tsuchiya et al., 1999; Takamiya et al., 2000). The Ser lipase domain constitutes the core of the catalytic site of Ser lipases, including an entirely conserved and essential Ser residue (Brady et al., 1990; Winkler et al., 1990; Blow, 1991; Lowe, 1992). Recently, Tsuchiya et al. (2003) showed that the corresponding Ser residue in Chlase from Chenopodium album was essential for enzyme function in vitro, providing experimental support for the role of the Ser lipase domain in Chlase catalytic activity. To establish the general relevance of this result to Chlase enzymes from other species, and to develop and establish a negative-control tool for in planta experiments, we introduced a Ser-to-Thr mutation into the corresponding Ser residue (codon 147) of the citrus Chlase gene and studied the function of the recombinant enzyme expressed in Escherichia coli by in vitro assay. Both wild-type and S147T mutant forms of citrus Chlase were expressed in E. coli in the mature form (ChlaseΔN, lacking the first 21 amino acids encoded by the cDNA) as fusion proteins with thioredoxin, as a result of the superior solubility, stability, and function reported for recombinant thioredoxin fusion proteins (Holmgren, 1985; LaVallie et al., 1992). Soluble protein extracts were analyzed in vitro for Chlase activity as described previously (Jacob-Wilk et al., 1999). These results establish that although the wild-type recombinant enzyme (pTrx-ChlaseΔN) was functionally active, the conservative S147T mutation entirely abolished Chlase activity in vitro (Figure 2A). Chlorophyllide accumulation was not detected in the recombinant S147T mutant or in the thioredoxin (pTrx) control reaction. The uniformity of soluble thioredoxin-Chlase fusion protein expression and the thioredoxin control (pTrx control) in the relevant samples was confirmed by protein gel blot analysis using anti-thioredoxin monoclonal antibodies (Figure 2B).
Figure 2.
In Vitro Enzyme Activity of Recombinant Chlase Versions.
Citrus Chlase versions were expressed as recombinant proteins in E. coli. Total soluble proteins were extracted from E. coli harboring pTrxFus plasmid expressing thioredoxin (pTrx; 12 kD), pTrxFus-Chlase plasmid containing the mature form of Chlase (lacking the N-terminal 21 amino acids) fused to thioredoxin (pTrx-ChlaseΔN; 45 kD), or pTrxFus-Chlase-S147T plasmid containing a version of ChlaseΔN bearing a Ser-to-Thr point mutation at position 147 (pTrx-ChlaseΔN-S147T; 45 kD). Protein extracts, and a protein-free reaction control, were examined for Chlase in vitro activity (A) as described in Methods and for Chlase-thioredoxin recombinant fusion protein level by protein gel blot analysis (B) using thioredoxin-specific antibodies.
Overexpression of Citrus Chlase in Cucurbits Using a Viral Vector System
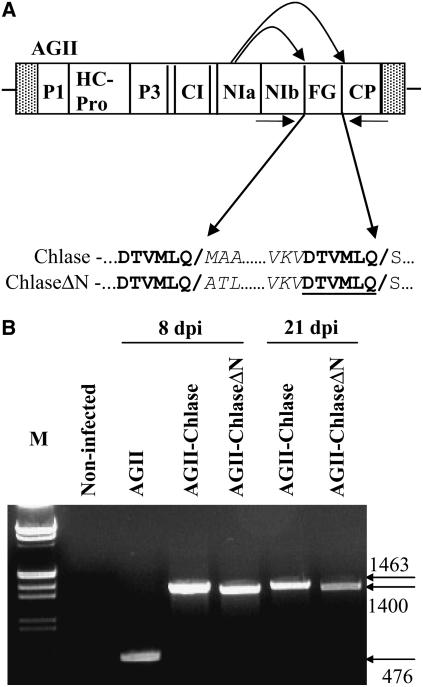
We opted to analyze Chlase gene product function in planta using a ZYMV-based viral vector infective clone system (AGII) designed for the expression of genes of interest in cucurbits (Arazi et al., 2001, 2002; Shoresh et al., 2006). The viral genes (including the gene of interest) are translated as a polyprotein that is proteolytically cleaved to obtain equal amounts of each of the viral proteins. Two versions of the citrus Chlase gene were cloned into the AGII viral vector polylinker sequence localized between the NIb and coat protein genes (Figure 3A): (1) full-length Chlase consisting of the entire coding sequence of the cDNA (AGII-Chlase); and (2) a Chlase version corresponding to the mature protein (determined by protein sequence [Trebitsh et al., 1993]) that lacks the first 21 amino acids encoded by the cDNA (AGII-ChlaseΔN). The polylinkers are flanked by NIa proteolysis sites, ensuring that the product of the foreign gene of interest would be released from the polyprotein by NIa protease–catalyzed proteolytic cleavage (Arazi et al., 2001). Both clones were designed to be processed in planta to their precise original sequence on the N terminus but retain an extension of six amino acids (after processing), essential for NIa protease recognition, at the C terminus. This extension is identical for both clones and therefore should affect them equally.
Figure 3.
Construction and Expression of a Viral Vector Containing the Citrus Chlase1 Gene.
(A) Scheme of the ZYMV-based AGII viral vector. AGII noncoding (hatched boxes) and coding (open boxes) regions, including the foreign gene (FG) cloning site, are shown. The sequence blow-up indicates the NIa protease cleavage sites involved in proteolysis of the foreign gene product from the viral polyprotein. Amino acid sequences corresponding to the NIa protease recognition motif are indicated in boldface, and cleavage sites are indicated by slashes interrupting the sequence and by the arrows. An extension of six amino acids added to the C terminus of Chlase versions (after processing) is underlined. P1, first protein; HC-Pro, helper component-proteinase; P3, third protein; CI, cytoplasmic inclusion; NIa, nuclear inclusion a; NIb, nuclear inclusion b; CP, coat protein; FG, foreign gene.
(B) RT-PCR analysis of viral RNA from infected plants. Total RNA was extracted from noninfected squash leaves (virus free) or from squash leaves systemically infected with empty viral vector (AGII), viral vector harboring full-length Chlase (AGII-Chlase), or N-terminally deleted Chlase (AGII-ChlaseΔN) at 8 and 21 d after inoculation (dpi). RNA was subjected to RT-PCR with primers flanking the foreign gene (FG) insertion site (see arrows in [A]). Amplified products were separated by gel electrophoresis, together with molecular weight markers (M), and stained with ethidium bromide.
To test expression in planta of the Chlase gene versions using the viral system, squash (Cucurbita pepo cv Ma'ayan) cotyledons were inoculated by particle bombardment with infectious AGII-Chlase and AGII-ChlaseΔN clones. The third true leaf of each infected plant was harvested, and the stability of the Chlase gene within the AGII genome was tested at 8 and 21 d after inoculation by RT-PCR (Figure 3B). RT-PCR products of the expected sizes were obtained for plants infected with AGII viral vector control (476 bp), AGII-Chlase (1463 bp), and AGII-ChlaseΔN (1400 bp).
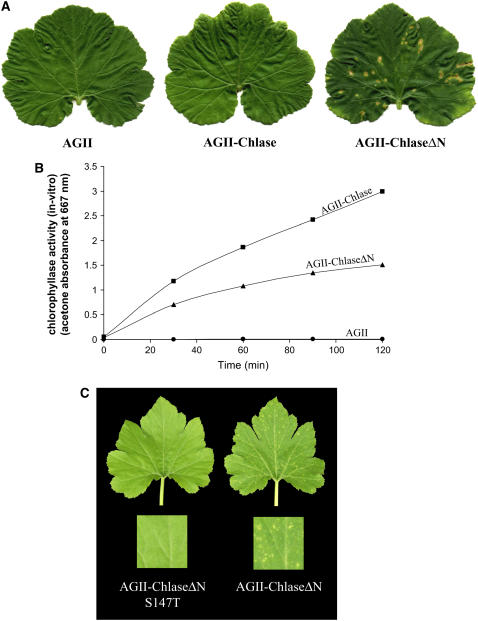
Overexpression of ChlaseΔN in Squash Grown under Low-Intensity Light Results in a Chlorotic Phenotype
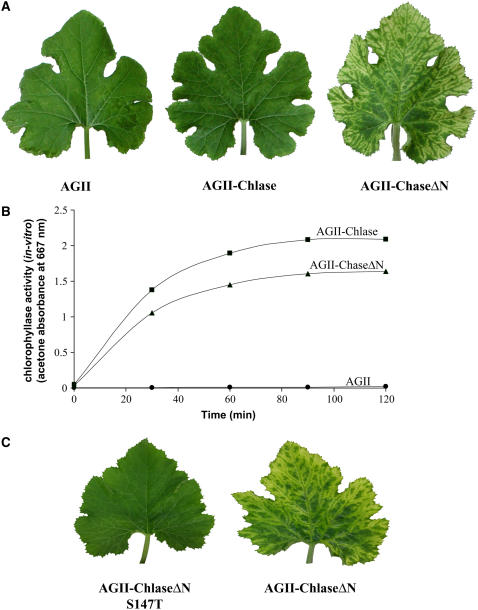
The physiological effect of citrus Chlase overexpression in squash plants was studied by inoculating squash cotyledons with AGII viral vectors harboring citrus Chlase versions. After inoculation, plants were grown in a growth room under continuous low-intensity fluorescent light (15 μE) for 10 d, and the third true leaf (fully expanded at 10 d after inoculation) was monitored for phenotype. These experiments were repeated in at least 30 plants for each construct, and representative plants are shown in the figures. Plants infected with the empty control viral vector (AGII) and with the vector encoding full-length Chlase (AGII-Chlase) displayed a normal phenotype, which included the very minor symptoms of viral infection (Figure 4A). By contrast, plants infected with vector encoding a version of Chlase corresponding to the mature protein (AGII-ChlaseΔN) developed a distinct blotchy, chlorotic phenotype (Figure 4A). Despite the different phenotypes observed, Chlase in vitro activity in crude leaf extracts of plants expressing both forms of Chlase (full length and ChlaseΔN) was comparable and was measured to be >400-fold the Chlase activity in the AGII control and noninoculated plants (Figure 4B; data not shown). Chlase in vitro activity was higher (∼25%) in plants infected with AGII-Chlase than in plants infected with AGII-ChlaseΔN, in contrast with the phenotype observed.
Figure 4.
Phenotypes of Plants Overexpressing Chlase Versions under Low-Intensity Light.
(A) Squash plants were infected with empty viral vector (AGII), viral vector harboring the full-length Chlase gene (AGII-Chlase), or viral vector harboring the N-terminally deleted form of the Chlase gene (AGII-ChlaseΔN). Plants were grown for 10 d after inoculation under low-intensity fluorescent light (15 μE), and leaf 3 of each plant was documented.
(B) Kinetics of in vitro Chlase activity was measured from crude extracts of leaf 4 of the plants used for (A), as described in Methods. Data points represent AGII-Chlase (squares), AGII-ChlaseΔN (triangles), and AGII (circles).
(C) Squash plants were infected with viral vector harboring the N-terminally deleted form of the Chlase gene in the wild-type form (AGII-ChlaseΔN) or mutated at Ser-147 to encode Thr (AGII-ChlaseΔN S147T). Plants were grown for 10 d after inoculation under low-intensity fluorescent light (15 μE), and leaf 3 of each plant was documented.
The Chlorotic Phenotype of ChlaseΔN-Overexpressing Plants Is Associated with Chlase Activity in Vivo
To establish the correlation between the chlorotic phenotype observed in plants overexpressing ChlaseΔN and Chlase enzyme activity in vivo, we tested the physiological effect of expressing the citrus ChlaseΔN S147T null-activity mutation in squash plants. The chlorotic leaf phenotype was repeatedly observed in plants infected with AGII-ChlaseΔN, whereas no visible phenotype was observed in plants expressing the S147T mutant version (AGII-ChlaseΔN S147T) (Figure 4C). Viral replication and polyprotein translation were confirmed for plants infected with both Chlase version viral vector constructs by RT-PCR and protein gel blot analysis using anti-ZYMV coat protein antibodies (data not shown).
The chlorotic phenotype was further studied during leaf development and was found to follow a distinct kinetic pattern. The chlorotic phenotype was observed in the third true leaf upon emergence and continued to develop during leaf maturation until ∼10 d after infection (Figure 5). In the days after day 10, the phenotype was reversed and the leaves gradually regreened.
Figure 5.
Dynamics of the Chlorotic Phenotype in ChlaseΔN-Expressing Plants.
Phenotype was monitored and documented once every 2 d for leaf 4 of squash plants infected with the viral vector harboring the N-terminally deleted Chlase gene (AGII-ChlaseΔN). Phenotypes shown were documented for the same leaf on days 8 to 16 after viral vector inoculation (dpi).
Overexpression of ChlaseΔN in Squash Grown under Natural Light Results in a Lesion-Mimic Phenotype
The physiological effect of overexpressing full-length Chlase and ChlaseΔN was also examined in plants exposed to natural light in a greenhouse (∼500 μE). As described above, these experiments were repeated in at least 30 plants for each construct, and representative plants are shown in the figures. Although plants infected with AGII-Chlase or AGII control showed, again, no visible phenotype, plants infected with AGII-ChlaseΔN displayed a distinct lesion-mimic phenotype (Figure 6A). The necrotic lesions, resembling those observed in the pathogen-induced hypersensitive response (Dangl et al., 1996), were observed all over the leaf surface (Figure 6A). Chlase in vitro activity was monitored in leaf crude extracts and showed that overexpression of both full-length Chlase and ChlaseΔN resulted in up to 200-fold higher Chlase activity than in the AGII-infected control (Figure 6B). Chlase in vitro activity for the phenotype-less full-length Chlase-expressing leaves was almost twofold higher than in plants expressing ChlaseΔN, which displayed the lesion-mimic phenotype.
Figure 6.
Phenotypes of Plants Overexpressing Chlase Versions under Natural Light.
(A) Squash plants were infected with empty viral vector (AGII), viral vector harboring the full-length Chlase gene (AGII-Chlase), or viral vector harboring the N-terminally deleted form of the Chlase gene (AGII-ChlaseΔN). Plants were grown for 10 d after inoculation under natural light in a greenhouse (500 μE), and leaf 3 of each plant was documented.
(B) Kinetics of in vitro Chlase activity was measured in crude extracts of leaf 4 of the plants used for (A), as described in Methods. Data points represent full-length AGII-Chlase (squares), N-terminally deleted AGII-ChlaseΔN (triangles), and vector control AGII (circles).
(C) Squash plants were infected with viral vector harboring the N-terminally deleted form of the Chlase gene in the wild-type form (AGII-ChlaseΔN) or mutated at Ser-147 to encode Thr (AGII-ChlaseΔN S147T). Plants were grown for 10 d after inoculation under natural light in a greenhouse (500 μE), and leaf 3 of each plant was documented. Blow-ups of the leaves (insets) enable a more detailed perspective of the leaf surface and phenotype.
The Lesion-Mimic Phenotype of ChlaseΔN-Overexpressing Plants Is Associated with Chlase Activity and Is Light-Dependent
The correlation between the lesion-mimic phenotype and Chlase activity was tested by overexpression and analysis of the null-activity S147T mutant in squash plants exposed to natural light. The lesion-mimic phenotype typical of ChlaseΔN-expressing plants (AGII-ChlaseΔN) was absent in the null-activity mutant–expressing plants (AGII-ChlaseΔN S147T), showing direct correlation between the phenotype and Chlase enzyme activity (Figure 6C). Viral accumulation and polyprotein translation were confirmed for plants infected with both Chlase version viral vector constructs by RT-PCR as well as by protein gel blot analysis using anti-ZYMV coat protein antibodies (data not shown).
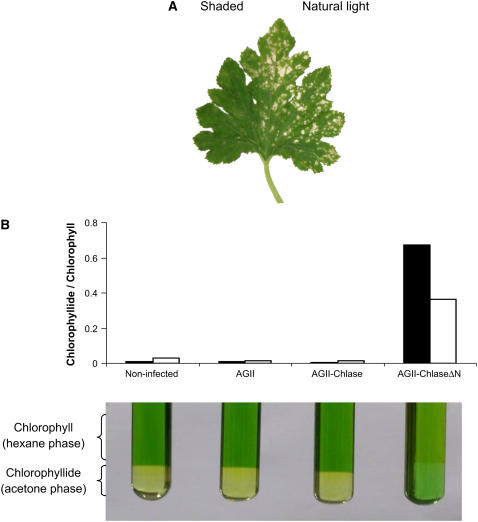
Because lesion-mimic phenotypes have been known to arise from a variety of causes (Molina et al., 1999; Mittler and Rizhsky, 2000; Pružinská et al., 2007), including photooxidative stress in chlorophyll biosynthesis and catabolism mutants (Hu et al., 1998; Mock et al., 1999; Ishikawa et al., 2001; Pružinská et al., 2007), it was of interest to directly test the involvement of light in the development of the lesion-mimic phenotype in plants expressing ChlaseΔN. Plants infected with AGII-ChlaseΔN were grown in the greenhouse under natural light conditions, and in each case half of the third leaf was shaded to block >90% of the light. The exposed part of the leaf developed the characteristic lesion-mimic phenotype, whereas the shaded part was virtually lesion-free (Figure 7A).
Figure 7.
The Lesion-Mimic Phenotype Caused by ChlaseΔN Overexpression Is Light-Dependent and Results from the Accumulation of Chlorophyll Breakdown Product.
(A) Plants infected with viral vector harboring the N-terminally deleted Chlase gene (AGII-ChlaseΔN) were grown under natural light conditions so that half of leaf 3 was exposed and half was shaded by black netting, blocking ∼90% of the light. The displayed leaf was documented at 10 d after inoculation.
(B) Leaves (leaf 3) weighing ∼100 mg were collected from noninfected plants and from plants infected with empty viral vector (AGII), viral vector harboring the full-length Chlase gene (AGII-Chlase), or viral vector harboring the N-terminally deleted form of the Chlase gene (AGII-ChlaseΔN). Chlorophylls were extracted from pools of five leaves each, and chlorophyll and chlorophyllide were separated by an acetone/hexane phase separation assay as described in Methods. The ratio of chlorophyllide to chlorophyll is displayed visually (phase separations at bottom) and as a histogram (top) showing the spectral quantitation results separately for chlorophyllide a/chlorophyll a (black columns) and chlorophyllide b/chlorophyll b (white columns).
Appearance of the Lesion-Mimic Phenotype Is Preceded by the Accumulation of a Photodynamic Chlorophyll Breakdown Product
Previous work suggested that lesion-mimic phenotypes may occur as a result of the oxidative stress generated from the accumulation of photodynamic chlorophyll synthesis or breakdown products (Hu et al., 1998; Mock et al., 1999; Ishikawa et al., 2001; Pružinská et al., 2003, 2007). Therefore, we hypothesized that similarly, the lesion-mimic phenotype generated in squash plants expressing ChlaseΔN results from the accumulation of chlorophyll breakdown products attributable to enhanced Chlase function, resulting in an enzymatic bottleneck farther downstream. Indeed, a dramatic accumulation of chlorophyllide was observed in ChlaseΔN-expressing squash leaves at a developmental stage that precedes the lesion phenotype by 2 to 3 d (Figure 7B). The chlorophyllide a:chlorophyll a ratio in these leaves was ∼90-fold that of leaves expressing full-length Chlase or that of AGII virus–infected or noninfected leaves. The chlorophyllide b:chlorophyll b ratio in these leaves was ∼20-fold that of leaves expressing full-length Chlase or the control leaves. Chlorophyllide accumulation in plants expressing ChlaseΔN, however, was transient, and levels decreased dramatically after the appearance of the lesion-mimic phenotype (data not shown). We note that it was not possible to conduct this experiment as a kinetic study of chlorophyllide levels, because the variability between plants attributable to the developmental stage of the leaf (leaf 3) was high. However, the results shown are representative and were qualitatively repeated in three independent experiments.
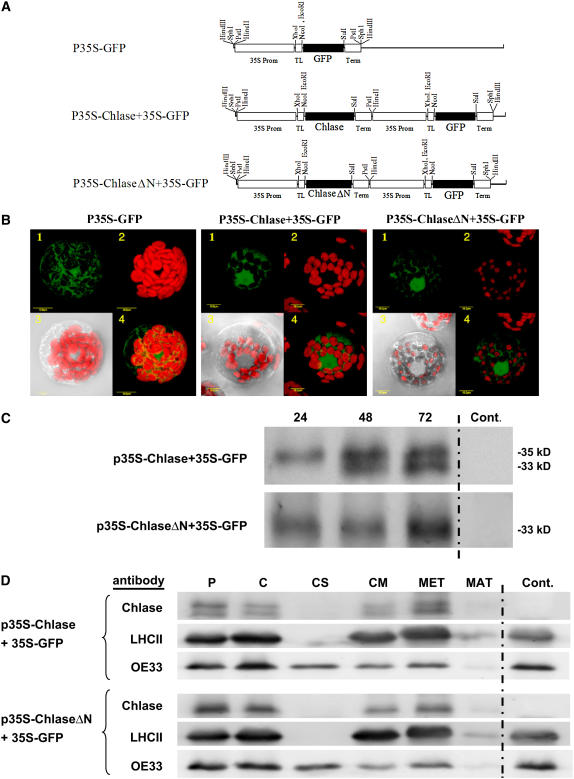
Overexpression of ChlaseΔN in Tobacco Protoplasts Results in Enhanced Chlorophyll Breakdown
To complement the physiological studies conducted in squash plants expressing citrus Chlase versions, we also analyzed the effect of citrus Chlase transient expression in tobacco (Nicotiana tabacum) protoplasts (Figure 8). Thus, the same citrus Chlase versions (full-length and ChlaseΔN) were analyzed in an additional plant system at the cellular level. Two plasmid constructs were designed to contain the gene encoding green fluorescent protein (GFP) driven by a 35S promoter and either full-length Chlase (p35S-Chlase+35S-GFP) or ChlaseΔN (p35S-ChlaseΔN+35S-GFP) separately driven by an additional 35S promoter (Figure 8A). An additional control construct consisted only of the GFP-encoding gene driven by a 35S promoter (p35S-GFP). The three constructs were electroporated into tobacco protoplasts for transient expression, and cell fluorescence was visualized by confocal microscopy (Figure 8B). Transformed cells were detected by the fluorescence of GFP that typically accumulates in the cytoplasm and nucleus. Chlorophyll levels in the chloroplasts of transformed cells were monitored by chlorophyll red autofluorescence. Figure 8B provides a semiquantitative analysis of Chlase in vivo activity by summarizing seven independent experiments. Representative cells displaying similar GFP fluorescence intensities from each of the three constructs were monitored for chlorophyll levels measured by red fluorescence. Chlorophyll levels of the control cells expressing GFP alone were measured to have chlorophyll area intensity (CAI) = 15.55 ± 1.8. Cells overexpressing full-length Chlase displayed reduced levels of chlorophyll in their chloroplasts relative to the GFP-expressing control (∼23%; CAI = 3.6 ± 0.5), demonstrating some chlorophyll breakdown activity in vivo, but all chloroplasts still retained significant amounts of chlorophyll (Figure 8B). Cells overexpressing ChlaseΔN displayed chloroplasts devoid of chlorophyll or containing very low levels of chlorophyll (average of 4.5% of wild-type levels; CAI = 0.71 ± 0.64), demonstrating enhanced chlorophyll breakdown activity in vivo.
Figure 8.
Relative Chlorophyll Levels, Chlase Processing, and Localization in Tobacco Protoplasts Transiently Expressing Citrus Chlase Versions.
(A) Tobacco protoplasts were transiently transformed with plasmids harboring the gene encoding GFP alone (p35S-GFP) or in combination with either full-length Chlase (p35S-Chlase+35S-GFP) or N-terminally deleted Chlase (p35S-ChlaseΔN+35S-GFP).
(B) After 72 h of transient expression, fluorescence was visualized using a laser scanning confocal microscope as described in Methods. One representative protoplast for each construct is visually presented as follows: panel 1, green fluorescence corresponds to GFP; panel 2, red fluorescence corresponds to chlorophyll; panel 3, confocal image recorded simultaneously in transmitted and red fluorescence mode (i.e., chlorophyll fluorescence overlaid on the bright-field image); and panel 4, confocal image recorded simultaneously for red and green fluorescence (i.e., GFP and chlorophyll fluorescence overlaid). Protoplasts displaying similar levels of GFP fluorescence were selected for semiquantitative analysis of chlorophyll relative levels by fluorescence. Average fluorescence measured for protoplasts expressing GFP alone (CAI = 15.55 ± 1.8; n = 3) was designated as 100%. Average fluorescence for protoplasts expressing Chlase with GFP (CAI = 3.6 ± 0.5; n = 3) and protoplasts expressing ChlaseΔN with GFP (CAI = 0.71 ± 0.64; n = 5) are depicted as percentages relative to protoplasts expressing GFP alone.
(C) Analysis of expression and processing kinetics of citrus Chlase versions in protoplasts. Tobacco protoplasts were transiently transformed with plasmids directing coexpression of Chlase and GFP (p35S-Chlase+35S-GFP), coexpression of ChlaseΔN and GFP (p35S-ChlaseΔN+35S-GFP), or GFP alone as a control. Protoplasts were harvested at 24, 48, and 72 h after transformation, and total proteins extracted with USB protein extraction buffer were separated by SDS-PAGE (30 μg per sample), blotted, and dressed with anti-citrus Chlase antibodies for the detection of citrus Chlase versions. Protoplasts expressing GFP alone served as a negative control for citrus Chlase–specific detection.
(D) Intracellular localization of Chlase versions in protoplasts. Tobacco protoplasts transiently transformed with the constructs mentioned above were harvested at 72 h after transformation. Intact chloroplasts were isolated from lysed protoplasts by Percoll gradient centrifugation and were in turn lysed and fractionated to membrane and soluble fractions. Membrane-associated proteins were washed with buffer containing 200 mM NaCl (data not shown) followed by extraction with buffer containing 0.5% Triton X-100. Proteins were precipitated with acetone and resuspended in USB protein extraction buffer from intact protoplasts, intact chloroplasts, chloroplast soluble and membrane fractions, 0.5% Triton X-100 wash of membrane fractions, and membrane pellets after Triton wash. Proteins were separated by SDS-PAGE (loading based on equal chlorophyll: 6 μg per lane), blotted, and dressed with anti-citrus Chlase antibodies for the detection of expressed Chlase versions, anti-LHCII (thylakoid integral membrane protein) antibodies, and anti-OE33 (lumen protein) antibodies for the detection of the endogenous proteins that serve as controls. The GFP control lane contains total protein extract from transformed protoplasts expressing GFP but not expressing any of the citrus Chlase versions.
Chlase and ChlaseΔN Expressed in Tobacco Protoplasts Are Localized in Chloroplast Membranes
The differential phenotypes obtained by expression of the Chlase versus ChlaseΔN versions prompted us to study their intracellular localization. Initial studies consisted of visualization of Chlase- and ChlaseΔN-GFP fusions (N-terminal, C-terminal, and internal fusions) expressed transiently in tobacco protoplasts. However, protoplasts expressing all GFP fusion versions retained normal chlorophyll levels, suggesting that mislocalization or inactivation had occurred. Therefore, we opted for an alternative localization approach involving immunodetection of Chlase versions in fractionated chloroplasts from protoplasts expressing Chlase or ChlaseΔN (Figures 8C and 8D). Full-length Chlase expressed in tobacco protoplasts is initially detected as an ∼35-kD precursor form that is consequently processed to a mature form of ∼33 kD, whereas expression of ChlaseΔN gives rise to the mature form (∼33 kD) (Figure 8C). Transiently transformed tobacco protoplasts expressing Chlase or ChlaseΔN for 72 h were chosen for further studies on the localization of the expressed Chlase forms (Figure 8D). Immunoblots using citrus Chlase–specific antibodies show that comparable levels of full-length and mature Chlase are present in Percoll gradient–purified intact chloroplasts versus protoplasts when proteins are loaded based on equal chlorophyll (chlorophyll degradation occurring in Chlase-expressing protoplasts was not a concern, because they constitute only 5 to 10% of the protoplast population). Similarly, ChlaseΔN was detected at comparable levels in chloroplasts and protoplasts, demonstrating that both Chlase versions are localized in the chloroplast.
For further localization, intact chloroplasts were aggressively lysed by freeze/thaw cycles (see Methods) to ensure complete chloroplast lysis and subjected to fractionation into soluble and membrane fractions by ultracentrifugation. In full-length Chlase-expressing protoplasts, both precursor and mature forms were detected in the chloroplast membrane fraction but not in the soluble fraction. Extraction of the membrane fraction with buffer containing 200 mM NaCl did not release any of the membrane-associated Chlase versions (data not shown), whereas extraction with buffer containing 0.5% Triton X-100 resulted in complete release of both Chlase forms. Protoplasts expressing ChlaseΔN yielded similar results; ChlaseΔN was detected in the membrane fraction but not in the soluble fraction and was released by extraction with buffer containing 0.5% Triton X-100. Probing the membranes with antibodies against the endogenous chloroplast proteins LHCII (for light-harvesting complex of photosystem II) and OE33 (for 33-kD subunit of oxygen-evolving complex) confirmed the fractionation to soluble and membrane fractions. The integral membrane protein LHCII was detected only in the membrane fraction and was extracted in 0.5% Triton X-100–containing buffer. By contrast, the lumen protein OE33, which is present in the lumen both in unassembled soluble form and bound to thylakoid-embedded photosystem II core proteins (Ettinger and Theg, 1991), was found in both membrane and soluble fractions as a result of the aggressive freeze/thaw lysis protocol we used that was previously shown to cause significant thylakoid rupture and release of lumen proteins (Hincha et al., 1987; Grouneva et al., 2006).
DISCUSSION
In this study, we exploited two different expression systems in two distant plant species to address pivotal questions concerning the role and regulation of Chlase in chlorophyll catabolism. Although the EST database suggests that most plants contain more than one Chlase homolog, we chose to conduct this study using the Chlase1 gene from citrus (Jacob-Wilk et al., 1999) because (1) it is the only Chlase gene encoding an enzyme experimentally shown to be localized to the chloroplast (Trebitsh et al., 1993; Jacob-Wilk et al., 1999) and (2) it is one of only two Chlases for which the processing site of the mature protein was experimentally determined (Jacob-Wilk et al., 1999; Tsuchiya et al., 1999). The citrus Chlase1 gene was overexpressed in two systems: (1) a ZYMV-based viral vector expression system that efficiently expressed citrus Chlase versions in squash plants for at least 21 d, which was more than sufficient time to study the physiological effects of Chlase expression in plants; and (2) a transient expression system in tobacco cells that allowed monitoring of the physiological effects of citrus Chlase expression at the cellular level. The results obtained in both plant systems were consistent and support the major conclusions we discuss below.
Perhaps the most provocative and intriguing result we present is that the expression of ChlaseΔN, which corresponds to the mature citrus Chlase, resulted in dramatic chlorosis in plants grown under low light as well as in protoplasts, whereas expression of the full-length enzyme resulted in only moderate chlorophyll breakdown. The apparent loss of natural regulatory constraints in ChlaseΔN is reminiscent of other enzymes that are activated by deletion of either (1) regulatory domains (e.g., deletion of the calmodulin binding domain of the enzyme Glu decarboxylase [Baum et al., 1996]) or (2) domains responsible for enzyme localization within the cell (e.g., deletion of the membrane binding domain of the enzyme HMGR [Ayora-Talavera et al., 2002]). Thus, the moderate phenotype obtained under overexpression of the full-length Chlase suggests that, similar to full-length HMGR and Glu decarboxylase, Chlase is posttranslationally regulated.
Although overexpression of ChlaseΔN in plants grown under low light resulted in chlorosis, the very same construct expressed in plants grown under natural light intensities caused a lesion-mimic phenotype, suggesting that they were subject to oxidative stress. We provide evidence that this phenotype results from the photoexcitation of transiently accumulating chlorophyllide, a photoreactive chlorophyll intermediate breakdown product. The light energy, absorbed by chlorophyllide outside of the context of the photosynthetic apparatus, is apparently converted into singlet oxygen and oxygen free radicals, triggering a genetically programmed apoptotic response (Wagner et al., 2004). Under low light intensities, the stress generated is expected to be much lower and appears to be dealt with successfully by the relevant enzyme machinery of the plant. Indeed, under low light conditions, no damage beyond chlorophyll breakdown is apparent and the chlorotic leaves eventually regreen (Figure 5), presumably after the reduction in viral vector replication in fully expanded leaves, as noted previously for other potyviruses (Hull, 2002). The relevance of light and photodynamic chlorophyll intermediate breakdown products to the lesion-mimic phenotype is further substantiated by the lack of phenotype in the shaded half of leaves (Figure 7A) and by the fact that similar lesion-mimic phenotypes have been documented in mutant plants arrested in either biosynthesis or breakdown of chlorophyll (Hu et al., 1998; Mock et al., 1999; Molina et al., 1999; Ishikawa et al., 2001; Mach et al., 2001; Pružinská et al., 2003). These results are consistent with the notion that the chlorophyll breakdown pathway is a detoxification process necessary for the tightly controlled degradation of the photodynamically active pigments (Vicentini et al., 1995; Matile et al., 1996; Hörtensteiner, 2004; Yang et al., 2004). In this context, it was also recently suggested that the chlorophyll catabolic pathway, and specifically the enzyme Chlase, may be involved in modulating the plant defense response by affecting damage-derived photodynamic free chlorophyll levels leading to reactive oxygen species (Kariola et al., 2005).
The vast majority of nucleus-encoded chloroplast proteins are directed to the chloroplast via a mechanism involving an N-terminal transit peptide, which typically undergoes cleavage upon entry into the chloroplast (reviewed in Fuks and Schnell, 1997). In the case of Chlase enzymes, which are nucleus-encoded, N-terminal protein sequencing of the mature protein conducted on citrus and Chenopodium Chlase revealed that these proteins undergo processing at amino acid positions 21 and 30, respectively (Trebitsh et al., 1993; Jacob-Wilk et al., 1999; Tsuchiya et al., 1999). However, the role of this cleaved N-terminal domain is something of an enigma, because the sequences do not conform to any known criteria for chloroplast transit peptides. Based on the data presented here, we conclude that the N-terminal 21–amino acid domain of citrus Chlase does not function as a classical N-terminal chloroplast-directing transit peptide, because both the full-length and ChlaseΔN versions enter the chloroplast and catalyze the breakdown of chlorophyll. This notion is further supported by experiments demonstrating that the N-terminal domain of citrus Chlase does not direct a GFP fusion protein into the chloroplast (data not shown). Therefore, it is likely that a different protein domain is responsible for Chlase chloroplast import, as has been noted for other chloroplast inner envelope membrane proteins (Miras et al., 2002). In this context, it is also worth noting that Chlase may be a glycoprotein (Terpstra, 1981; Tsuchiya et al., 1997, 1999); therefore, its route to the chloroplast may be more complex and may be directed via organelles involved in glycosylation, as described recently for carbonic anhydrase (Villarejo et al., 2005).
We can only speculate at this stage on the nature of the mechanism regulating Chlase activity and on whether or not processing of the N-terminal 21 amino acids is the trigger that sets chlorophyll degradation into motion. One possible explanation for the enhanced chlorophyll breakdown activity of Chlase ΔN is that removal of a regulatory domain present in the N-terminal 21 amino acids results in a much more active enzyme in vivo. This possibility is reminiscent of the N-terminal flap mechanism that regulates the catalytic site of Ser lipases (Hermoso et al., 1996; Pignol et al., 2000), although the flap domain in lipases is shifted by substrate threshold levels and is not removed by proteolysis. A second possibility is that the N-terminal 21 amino acids are directly or indirectly involved in the compartmentalization of Chlase and that their removal enables Chlase to come into direct contact with the substrate chlorophyll. This possibility is in agreement with work that localized Brassica and barley (Hordeum vulgare) Chlase to the chloroplast inner envelope membrane, spatially separated from the chlorophyll substrate (Matile et al., 1997). However, in those experiments, only 6% of the total cell Chlase activity was accounted for in the envelope fraction, perhaps partly as a result of the technical difficulty of separating chloroplast inner envelope membranes from stromal and granal lamellae.
In this context, it is important to note that recent work using electron microscopy suggests that a physical connection may exist between the inner envelope and the stromal lamellae (Shimoni et al., 2005). Thus, localization of Chlase to the inner envelope membrane of the plastid appears to be definite (also shown in chromoplast envelopes by Hirschfeld and Goldschmidt [1983] and Brandis et al. [1996]), but other chloroplast membrane locations cannot be ruled out based on the data presented here. The only consistent finding regarding the localization of Chlase from various plant species is the association of Chlase activity with chloroplast membranes (Ardao and Vennesland, 1960; Terpstra, 1974, 1976; Tarasenko et al., 1986; Trebitsh et al., 1993; Schellenberg and Matile, 1995).
To obtain further insights into Chlase localization and the Chlase posttranslational regulatory mechanism, we addressed the localization of citrus Chlase versions in our experimental system by visualization of expressed Chlase-GFP fusions and by immunodetection of Chlase versions in purified chloroplast soluble and membrane fractions. Chlase-GFP fusion localizations proved to be of no value, likely because of mislocalization, which is commonly encountered using GFP fusions (Tian et al., 2004). Chloroplast fractionation studies conducted on Percoll gradient–purified chloroplasts from transiently expressing tobacco protoplasts show that both full-length and mature Chlase versions localize to the chloroplast membrane fraction, where the processing of the N-terminal 21 amino acids apparently occurs. This result is not surprising, considering that Chlase activity in young, senescing, or ripening tissue is consistently associated with the membrane fraction (Trebitsh et al., 1993). Although these results do not provide a mechanistic explanation for the posttranslational regulation of Chlase in vivo activity, the processing kinetics suggest that processing of the full-length enzyme to its mature form may be a rate-limiting step. Further work, perhaps using electron microscopy, will be required to determine whether the different versions are located in different membrane compartments in the chloroplast and whether processing of the N terminus is correlated with a change in membrane compartment localization. In this context, we note that spatial relocation between membrane compartments and processing within chloroplast membranes have been documented previously for the D1 protein (Mattoo and Edelman, 1987).
The possible role of Chlase as a rate-limiting enzyme in chlorophyll catabolism has been a topic of much controversy, and the data in the literature are inconclusive. Chlase gene expression and in vitro activity were often found to be constitutive or noncorrelated with natural degreening of leaves and fruit (Minguez-Mosquera and Gallardo-Guerrero, 1996; Fang et al., 1998; Jacob-Wilk et al., 1999), suggesting that either Chlase is not a rate-limiting enzyme in chlorophyll catabolism or that Chlase function is regulated posttranslationally, resulting in latent function (Matile et al., 1997, 1999; Jacob-Wilk et al., 1999). In this work, we show that a Chlase version devoid of regulatory constraints is sufficient to trigger complete chlorosis, suggesting that Chlase functions as a rate-limiting enzyme in chlorophyll catabolism, controlled by posttranslational regulation. Because chlorophyll levels are continuously modulated throughout the plant life cycle and the breakdown pathway must be tightly controlled to avoid the accumulation of photodynamically active pigments, a posttranslational regulatory mechanism (as opposed to transcriptional regulation) makes physiological sense, allowing the plant much more flexibility in the response to environmental cues. This does not rule out the parallel existence of transcriptional regulation; indeed, in citrus fruit, Chlase transcript is responsive to ethylene (Jacob-Wilk et al., 1999).
The mechanism by which Chlase and its substrate chlorophyll come into contact to initiate chlorophyll breakdown has been a controversial issue, and several suggestions were raised in the literature concerning possible mechanisms for the transfer of chlorophyll destined for breakdown to Chlase, which was suggested to be located in the inner envelope membrane (Matile et al., 1997; Hörtensteiner, 2006). Although we have not presented any data to favor or rule out such a mechanism, our results show that ChlaseΔN catalyzes massive chlorosis, thus supporting the possibility that Chlase reaches chlorophyll assembled within the photosynthetic complex, and not vice versa, as was suggested previously.
Our results regarding the correlation between in vitro and in vivo activity raise questions regarding the interpretation of the role of Chlase in previous physiological studies based on in vitro activity. Expression of full-length citrus Chlase and ChlaseΔN in squash leaves resulted in similar levels of Chlase activity measured by in vitro assay but in dramatically different phenotypes. Thus, the results suggest that great care must be taken when interpreting in vitro Chlase activity results. Although in vitro activity can attest to the general level of Chlase enzyme in the tissue, the data suggest that it does not attest to the actual function of the enzyme in vivo. This is most likely explained by the notion that Chlase enzyme function is regulated posttranslationally; the in vitro activity assay abolishes the natural functional regulation either by destruction of suborganellar compartment barriers or by directly affecting enzyme activity (i.e., buffer and detergent components that affect enzyme function). This important lesson may be applicable to the study of the correlation between Chlase function and the regulation of chlorophyll breakdown during the developmental stages of ripening and senescence, in which previously in vitro activity was the scale by which Chlase function was measured. Similarly, Chlase in vitro activity was previously used to rule out the involvement of Chlase in stay-green mutants (Akhtar et al., 1999). Future knowledge of the posttranslational regulatory mechanism of Chlase may facilitate a more accurate assessment of the potential involvement of Chlase in stay-green mutants and the processes of chlorophyll catabolism during senescence and ripening.
METHODS
Expression of Recombinant Wild-Type and Mutant Chlase Versions in Bacteria
The Chlase cDNA sequence excluding the codons for the first 21 amino acids was cloned into the thioredoxin fusion expression vector pTrxFus (Invitrogen) as described previously (Jacob-Wilk et al., 1999). A mutant version of Chlase (Ser-147→Thr) was created by site-directed mutagenesis (Kunkel et al., 1987) using the primer 5′-GCTGTTATGGGGCACACCCGCGGAGGACAAACG-3′. Chlase versions were expressed in Escherichia coli as thioredoxin fusion proteins essentially as described previously (Jacob-Wilk et al., 1999). Soluble protein extracts were obtained by centrifuging 10 mL of bacterial cells at 4000g for 10 min. The pellet was resuspended in 2 mL of grinding buffer (20 mM Na-phosphate, pH 8, 500 mM NaCl, and 0.1% Triton X-100). Lysosyme was added to a concentration of 1 mg/mL, and the suspension was incubated for 1 h at 4°C and then subjected to three cycles of freeze/thaw using liquid nitrogen. Samples were centrifuged for 30 min at 15,000g at 4°C, and the soluble and pellet fractions were separated. In vitro activity assays for the soluble Chlase recombinant protein were performed as described (Jacob-Wilk et al., 1999).
Construction of AGII-Chlase Plasmids for Viral Vector–Based Expression in Squash
To facilitate the cloning of Chlase into the AGII viral vector (Arazi et al., 2001), two PstI restriction sites present within the original citrus (Citrus sinensis) Chlase cDNA (at positions 76 and 426) were cancelled by site-directed mutagenesis (Kunkel et al., 1987) without altering the encoded amino acids using the mutagenic primers 5′-GTGGACGCCAAGCCGGCAGCTTCAGTGC-3′ and 5′-GCTACCCCAAGGCCTTCAGCAAAATCTCCCAG-3′. Full-length (990-bp open reading frame) Chlase sequence versions (wild type and S147T-mutated) were amplified by PCR using primers containing PstI and SalI restriction sites necessary for subcloning into the AGII viral vector (5′-GAACTGCAGATGGCAGCAATGGTGGACG-3′ and 5′-GATGTCGACTACTTTTACATGTGTTGTGGTAAGCATGCTTGATGC-3′). N-terminally deleted (924-bp open reading frame) Chlase sequence versions (wild type and S147T-mutated) were amplified by PCR using primers containing PstI and SalI restriction sites necessary for subcloning into the AGII viral vector (5′-CATTATCTGCAGGCCACACTTCCGGTATTTAC-3′ and 5′-GATGTCGACTACTTTTACATGTGTTGTGGTAAGCATGCTTGATGC-3′). Primers were designed to include NIa proteolysis sites (Arazi et al., 2001) and a reading frame matching the AGII genes without any stop codon at the 3′ terminus, allowing translation of Chlase in planta as part of the virus-encoded polyprotein and its release after NIa proteolysis. Chlase gene PCR-amplified products were initially cloned into pGEM-T Easy plasmid (Promega), sequenced, and then subcloned into the AGII viral vector. Positive clones were used to inoculate squash (Cucurbita pepo) plants as described below.
Squash Plant Growth, Inoculation, and Symptom Evaluation
Squash seeds of a commercial cultivar (cv Ma'ayan) were used for all whole-plant experiments, and seedlings were selected for experimental use when the cotyledons were fully expanded. Particle bombardment inoculation with the various plasmid–virus constructs was performed using a handgun device (Gal-On et al., 1997). After inoculation, squash seedlings were grown either in a growth chamber under continuous light (∼11 μE) at 23°C or in a whitewashed greenhouse under natural light conditions (∼500 μE). Symptom development was examined daily, and the first appearance was recorded. Confirmation of the systemic spread of intact virus was performed by RT-PCR according to Arazi et al. (2001).
Chlase in Vitro Activity Assay
Chlase enzyme activity assay in squash leaf crude extracts was analyzed essentially as described (Jacob-Wilk et al., 1999) but with the following slight modifications: no preincubation in reaction buffer was done, and reaction buffer consisted of a mixture of 0.05 mL of extraction buffer (50 mM Na-phosphate, pH 7.4, 50 mM KCl, 5 mM MgCl2, 5 mM DTT, and 1% Triton X-100), 0.45 mL of reaction buffer A (50 mM Na-phosphate, pH 7.4, and 50 mM KCl), and 2.5 mL of reaction buffer B (50 mM Na-phosphate, pH 7.4, 50 mM KCl, and 0.5% Triton X-100). Chlorophyll a substrate was prepared from parsley (Petroselinum sativum) leaves according to Bazzaz and Rebeiz (1978), stored dry at −20°C, and redissolved in acetone before use.
Determination of Chlorophyll:Chlorophyllide Ratios in Squash Leaves
Chlorophylls were extracted from squash leaves by incubating 200 mg of sliced leaves in 6 mL of 100% acetone for 72 h at 4°C in the dark. Four milliliters from each sample was added to a Corex tube containing 6 mL of hexane and 1 mL of KOH (10 mM). The mixture was vortexed briefly and centrifuged for 2 min at 12,000g to obtain phase separation. Chlorophyll levels were determined in the hexane phase, and chlorophyllide levels were determined in the acetone phase spectrophotometrically using the formulae Chla (μg/mL) = 12.7 × A663 – 2.69 × A645 and Chlb (μg/mL) = 22.9 × A645 – 4.68 × A663, as described by Arnon (1949).
Protein Extraction from Plants, Protoplasts, Chloroplasts, and Fractionated Chloroplasts
Samples of squash leaf tissue (150 mg) were homogenized in extraction buffer containing 50 mm Tris-HCl, 5% β-mercaptoethanol, 10% glycerin, and 2% SDS, pH 6.8, and denatured by boiling for 5 min. Extracts were centrifuged for 10 min at 21,000g, the supernatant was transferred to a new tube, and glycerol was added to a concentration of 10%. Equal amounts of the supernatants were separated by SDS-PAGE (12%) and subjected to protein gel blot analysis as described below.
Tobacco (Nicotiana tabacum) protoplast total proteins were extracted with USB protein extraction buffer (20 mM Tris-HCl, pH 7.5, 8 M urea, 4.5% SDS, and 1 M β-mercaptoethanol), separated by SDS-PAGE, and subjected to protein gel blot analysis as described below. For preparation of chloroplasts for fractionation, 3.5 × 106 protoplasts were pooled for transformation of each construct. Protoplasts were lysed for 30 min on ice in protoplast lysis buffer (50 mM HEPES, pH 8, 2 mM EDTA, 1 mM MgCl2, 1 mM MnCl2, 0.33 M sorbitol, 1 mM ascorbic acid, and 20 μg/mL BSA). Intact chloroplasts were isolated on a continuous Percoll gradient according to Adam and Hoffman (1993) and Levy and Adam (1995). Isolated intact chloroplasts were aggressively lysed in chloroplast lysis buffer (20 mM Tris-HCl, pH 7.5) by three freeze/thaw cycles in liquid nitrogen, each followed by vigorous vortex on the partly thawed samples. Membrane and soluble fractions were separated by centrifugation at 100,000g (rotor SW-50; Beckman) for 1 h. The membrane pellet was extracted with 20 mM Tris-HCl, pH 7.5, and 200 mM NaCl for 10 min and centrifuged at 100,000g for 1 h. The pellet was then extracted with 20 mM Tris-HCl, pH 7.5, and 0.5% Triton X-100 for 10 min and centrifuged at 100,000g for 1 h. The washed pellet was resuspended in chloroplast lysis buffer. Samples were brought to a final volume of 100 μL, and 5 volumes of cold acetone was added. Precipitated proteins were extracted in USB protein extraction buffer. Chlorophyll concentration was determined according to Arnon (1949) and used to calculate the protein equivalent to 6 μg of chlorophyll from each fraction for SDS-PAGE.
Protein Gel Blot Analysis
Proteins separated by SDS-PAGE were transferred onto nitrocellulose membranes (Schleicher and Schuell) as described (Page and Thorpe, 1996). Membranes were blocked and incubated overnight with the following antibodies: mouse anti-E. coli thioredoxin monoclonal antibodies (BioVendor) diluted 1:80,000; rabbit anti-ZYMV coat protein polyclonal antibody diluted 1:40,000 (Arazi et al., 2001); rabbit anti-LHCII polyclonal antibodies diluted 1:80,000 (a gift from Oren Ostersetzer); rabbit anti-OE33 antibodies diluted 1:20,000 (a gift from Zach Adam); and mouse anti-citrus Chlase antibodies diluted 1:1000 (see below). Immunoblots were developed using the SuperSignal West Dura Substrate (Pierce) after incubation for 1 h with goat anti-mouse or goat anti-rabbit horseradish peroxidase–conjugated secondary antibody (Jackson ImmunoResearch Laboratories) diluted 1:10,000. Dressed immunoblots were visualized in a FluorChem 8800 imaging system (Alpha Innotech). Between application of different antibodies, membranes were stripped using Restore protein gel blot stripping buffer (Pierce). Specific anti-citrus Chlase polyclonal ascites antibodies were raised in mice against a recombinant StrepTag-thioredoxin-ChlaseΔN fusion protein expressed in bacteria using the pTrxFus system (LaVallie et al., 1992) and purified using a StrepTactin affinity column as described by the manufacturer (IBA).
Transient Expression of Chlase Versions in Tobacco Protoplasts and Confocal Microscopy
Constructs for transient protoplast transformation were prepared in the cloning vector pUC-19 and are presented schematically in Figure 8A. Plasmid p35S-GFP was created by cloning 900 bp of the 35S promoter upstream of the tobacco etch virus (TEV) translation enhancer (Gallie et al., 1995), the gene encoding enhanced green fluorescent protein (GFP-S65T) (Heim et al., 1995), and the 35S terminator. Plasmid p35S-Chlase+35S-GFP was created by cloning a Chlase expression cassette (i.e., 35S promoter, TEV translational enhancer, full Chlase cDNA coding sequence, 35S terminator) into the p35S-GFP plasmid. Plasmid p35S-ChlaseΔN+35S-GFP was created by cloning a ChlaseΔN expression cassette (i.e., 35S promoter, TEV translational enhancer, Chlase cDNA coding sequence lacking the first 21 codons but supplemented with an ATG initiator codon, 35S terminator) into the p35S-GFP plasmid. Relevant restriction sites for all constructs are shown in Figure 8A.
Mesophyll protoplasts were isolated from tobacco (Nicotiana tabacum cv Samsun NN) plants grown under sterile conditions (Draper et al., 1988). Electroporation of 2.5 to 5 × 105 protoplasts per construct was performed in prechilled electroporation medium (Fromm et al., 1985) using 5 to 20 μg of plasmid DNA, 15 μg of calf thymus DNA, and 0.5 mL of electroporation solution. After electroporation, the protoplasts were transferred into growth medium and incubated in darkness at 27°C before visual analysis or harvesting.
Images were obtained using an IX 81 fully automated Olympus microscope equipped with a 488-nm argon laser for GFP excitation and a narrow band emission filter (515 to 525 nm) to filter GFP from chlorophyll green autofluorescence, a BA 660 IF emission filter for chlorophyll red autofluorescence, and a PLAPO60XWLSM (numerical aperture 1.00) objective. Transmitted light images were acquired using Nomarski differential interference contrast.
Accession Number
The accession number for the citrus Chlase gene used throughout this work (Chlase1 [Jacob-Wilk et al., 1999]) is AF160869.
Acknowledgments
We thank Hillel Fromm and Gozal Ben-Hayyim for critical reading of the manuscript, Avihai Danon, Alex Levitan (of blessed memory), Zach Adam, Einat Sadot, Victor Gaba, and Oren Ostersetzer for helpful discussions and advice, and Edi Belausov for assistance with confocal microscopy. We thank Zach Adam and Oren Ostersetzer for generous contributions of antibodies. This research was supported by grants from the Chief Scientist of the Israeli Ministry of Agriculture, the Israel Science Foundation (Grant 1291/04), and the Swiss National Science Foundation. S.H.-S. was the recipient of a European Molecular Biology Organization short-term scholarship. T.A. was the recipient of a Levi Eshkol Ph.D. scholarship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yoram Eyal (eyalab@volcani.agri.gov.il).
References
- Adam, Z., and Hoffman, N.E. (1993). Biogenesis of a photosystem I light-harvesting complex (evidence for a membrane intermediate). Plant Physiol. 102 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar, M.S., Goldschmidt, E.E., John, I., Rodoni, S., Matile, P., and Grierson, D. (1999). Altered patterns of senescence and ripening in gf, a stay-green mutant of tomato. J. Exp. Bot. 50 1115–1122. [Google Scholar]
- Amir-Shapira, D., Goldschmidt, E.E., and Altman, A. (1987). Chlorophyll catabolism in senescing plant tissues: In vivo breakdown intermediates suggest different degradative pathways for citrus fruit and parsley leaves. Proc. Natl. Acad. Sci. USA 84 1901–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, B., and Barber, J. (1996). Mechanisms of photodamage and protein degradation during photoinhibition of photosystem II. In Photosynthesis and the Environment, N.R. Baker, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 101–121.
- Arazi, T., Lee Huang, P., Huang, P.L., Zhang, L., Shiboleth, Y.M., Gal-On, A., and Lee-Huang, S. (2002). Production of antiviral and antitumor proteins MAP30 and GAP31 in cucurbits using the plant virus vector ZYMV-AGII. Biochem. Biophys. Res. Commun. 292 441–448. [DOI] [PubMed] [Google Scholar]
- Arazi, T., Slutski, S.G., Shiboleth, Y.M., Wang, Y., Rubinstein, M., Barak, S., Yang, J., and Gal-On, A. (2001). Engineering zucchini yellow mosaic potyvirus as a non-pathogenic vector for expression of heterologous proteins in cucurbits. J. Biotechnol. 87 67–82. [DOI] [PubMed] [Google Scholar]
- Ardao, C., and Vennesland, B. (1960). Chlorophyllase activity of spinach chloroplastin. Plant Physiol. 35 368–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon, D.I. (1949). Copper enzymes in isolated chloroplasts: Polyphenol-oxidase in Beta vulgaris. Plant Physiol. 24 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayora-Talavera, T., Chappell, J., Lozoya-Gloria, E., and Loyola-Vargas, V.M. (2002). Overexpression in Catharanthus roseus hairy roots of a truncated hamster 3-hydroxy-3-methylglutaryl-CoA reductase gene. Appl. Biochem. Biotechnol. 97 135–145. [DOI] [PubMed] [Google Scholar]
- Baum, G., Lev-Yadun, S., Fridmann, Y., Arazi, T., Katsnelson, H., Zik, M., and Fromm, H. (1996). Calmodulin binding to glutamate decarboxylase is required for regulation of glutamate and GABA metabolism and normal development in plants. EMBO J. 15 2988–2996. [PMC free article] [PubMed] [Google Scholar]
- Bazzaz, M.B., and Rebeiz, C.A. (1978). Chloroplast culture: The chlorophyll repair potential of mature chloroplast incubated in a simple medium. Biochim. Biophys. Acta 504 310–323. [DOI] [PubMed] [Google Scholar]
- Benedetti, C.E., and Arruda, P. (2002). Altering the expression of the chlorophyllase gene ATHCOR1 in transgenic Arabidopsis caused changes in the chlorophyll-to-chlorophyllide ratio. Plant Physiol. 128 1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow, D. (1991). Lipases reach the surface. Nature 351 444–445. [DOI] [PubMed] [Google Scholar]
- Brady, L., Brzozowski, A.M., Derewenda, Z.S., Dodson, E., Tolley, S., Turkenburg, J.P., Christiansen, L., Huge-Jensen, B., Norskov, L., Thim, L., and Menge, U. (1990). A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature 343 767–770. [DOI] [PubMed] [Google Scholar]
- Brandis, A., Vainstein, A., and Goldschmidt, E.E. (1996). Distribution of chlorophyllase among components of chloroplast membranes in orange (Citrus sinensis) leaves. Plant Physiol. Biochem. 34 49–54. [Google Scholar]
- Dangl, J.L., Dietrich, R.A., and Richberg, M.H. (1996). Death don't have no mercy: Cell death programs in plant-microbe interactions. Plant Cell 8 1793–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper, J., Scott, R., and Hamil, J. (1988). Transformation of dicotyledonous plant cells using the Ti plasmid of A. tumefaciens and the Ri plasmid of A. rhizogenes. In Plant Genetic Transformation and Gene Expression: A Laboratory Manual, J. Draper, R. Scott, P. Armitage, and R. Walden eds (Oxford, UK: Blackwell Scientific Publications), pp. 71–160.
- Engel, N., Jenny, T.A., Mooser, V., and Gossauer, A. (1991). Chlorophyll catabolism in Chlorella protothecoides. Isolation and structure elucidation of a red bilin derivative. FEBS Lett. 293 131–133. [DOI] [PubMed] [Google Scholar]
- Ettinger, W.F., and Theg, S.M. (1991). Physiologically active chloroplasts contain pools of unassembled extrinsic proteins of the photosynthetic oxygen-evolving enzyme complex in the thylakoid lumen. J. Cell Biol. 115 321–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Z., Bouwkamp, J.C., and Solomos, T. (1998). Chlorophyllase activities and chlorophyll degradation during leaf senescence in non-yellowing mutant and wild type of Phaseolus vulgaris L. J. Exp. Bot. 49 503–510. [Google Scholar]
- Fromm, M., Taylor, L.P., and Walbot, V. (1985). Expression of genes transferred into monocot and dicot plant cells by electroporation. Proc. Natl. Acad. Sci. USA 82 5824–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks, B., and Schnell, D.J. (1997). Mechanism of protein transport across the chloroplast envelope. Plant Physiol. 114 405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie, D.R., Tanguay, R.L., and Leathers, V. (1995). The tobacco etch viral 5′ leader and poly(A) tail are functionally synergistic regulators of translation. Gene 165 233–238. [DOI] [PubMed] [Google Scholar]
- Gal-On, A., Meiri, E., Elman, C., Gray, D.J., and Gaba, V. (1997). Simple hand-held devices for the efficient infection of plants with viral-encoding constructs by particle bombardment. J. Virol. Methods 64 103–110. [DOI] [PubMed] [Google Scholar]
- Goldschmidt, E.E. (2001). Chlorophyll decomposition in senescing leaves and ripening fruits: Functional and evolutionary perspectives. Acta Hortic. 553 331–335. [Google Scholar]
- Grouneva, I., Jacob, T., Wilhelm, C., and Goss, R. (2006). Influence of ascorbate and pH on the activity of the diatom xanthophyll cycle-enzyme diadinoxanthin de-epoxidase. Physiol. Plant. 126 205–211. [Google Scholar]
- Heim, R., Cubitt, A.B., and Tsien, R.Y. (1995). Improved green fluorescence. Nature 373 663–664. [DOI] [PubMed] [Google Scholar]
- Hermoso, J., Pignol, D., Kerfelc, B., Crenon, I., Chapus, C., and Fontecilla-Camps, J.C. (1996). Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex. J. Biol. Chem. 271 18007–18016. [DOI] [PubMed] [Google Scholar]
- Hincha, D.K., Höfner, R., Schwab, K.B., Heber, U., and Schmitt, J.M. (1987). Membrane rupture is the common cause of damage to chloroplast membranes in leaves injured by freezing or excessive wilting. Plant Physiol. 83 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld, K.R., and Goldschmidt, E.E. (1983). Chlorophyllase activity in chlorophyll-free citrus chromoplasts. Plant Cell Rep. 2 117–118. [DOI] [PubMed] [Google Scholar]
- Holmgren, A. (1985). Thioredoxin. Annu. Rev. Biochem. 54 237–271. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner, S. (1999). Chlorophyll breakdown in higher plants and algae. Cell. Mol. Life Sci. 56 330–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner, S. (2004). The loss of green color during chlorophyll degradation—A prerequisite to prevent cell death? Planta 219 191–194. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner, S. (2006). Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 57 55–77. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner, S., Wuthrich, K.L., Matile, P., Ongania, K.H., and Kräutler, B. (1998). The key step in chlorophyll breakdown in higher plants. Cleavage of pheophorbide A macrocycle by a monooxygenase. J. Biol. Chem. 273 15335–15339. [DOI] [PubMed] [Google Scholar]
- Hu, G., Yalpani, N., Briggs, S.P., and Johal, G.S. (1998). A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant in maize. Plant Cell 10 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, R. (2002). Matthews' Plant Virology. (London: Academic Press).
- Ishikawa, A., Okamoto, H., Iwasaki, Y., and Asahi, T. (2001). A deficiency of coproporphyrinogen III oxidase causes lesion formation in Arabidopsis. Plant J. 27 89–99. [DOI] [PubMed] [Google Scholar]
- Jacob-Wilk, D., Holland, D., Goldschmidt, E.E., Riov, J., and Eyal, Y. (1999). Chlorophyll breakdown by chlorophyllase: Isolation and functional expression of the Chlase1 gene from ethylene-treated citrus fruit and its regulation during development. Plant J. 20 653–661. [DOI] [PubMed] [Google Scholar]
- Kariola, T., Brader, G., Li, J., and Tapio Palva, E. (2005). Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 17 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräutler, B., and Matile, P. (1999). Solving the riddle of chlorophyll breakdown. Acc. Chem. Res. 32 35–43. [Google Scholar]
- Kunkel, T.A., Robert, J.D., and Zakour, R.A. (1987). Rapid and efficient site-specific mutagenesis without phenotype selection. Methods Enzymol. 154 367–382. [DOI] [PubMed] [Google Scholar]
- LaVallie, E.R., DiBlasio, E.A., Kovacic, S., Grant, K.L., Schendel, P.F., and McCoy, J.M. (1992). A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology 11 187–193. [DOI] [PubMed] [Google Scholar]
- Levy, M., and Adam, Z. (1995). Mutations in the processing site of the precursor of ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit: Effects on import, processing, assembly and stability. Plant Mol. Biol. 29 53–61. [DOI] [PubMed] [Google Scholar]
- Lowe, M.E. (1992). The catalytic site residues and interfacial binding of human pancreatic lipase. J. Biol. Chem. 267 17069–17073. [PubMed] [Google Scholar]
- Mach, J.M., Castillo, A.R., Hoogstraten, R., and Greenberg, J.T. (2001). The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl. Acad. Sci. USA 98 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile, P., Hörtensteiner, S., and Thomas, H. (1999). Chlorophyll degradation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 67–95. [DOI] [PubMed] [Google Scholar]
- Matile, P., Hörtensteiner, S., Thomas, H., and Kräutler, B. (1996). Chlorophyll breakdown in senescent leaves. Plant Physiol. 112 1403–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile, P., Schellenberg, M., and Vicentini, F. (1997). Localization of chlorophyllase in the chloroplast envelope. Planta 201 96–99. [Google Scholar]
- Mattoo, A.K., and Edelman, M. (1987). Intramembrane translocation and posttranslational palmitoylation of the chloroplast 32-kDa herbicide-binding protein. Proc. Natl. Acad. Sci. USA 84 1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguez-Mosquera, M.I., and Gallardo-Guerrero, L.G. (1996). Role of chlorophyllase in chlorophyll metabolism in olives cv. Gordal. Phytochemistry 41 691–697. [Google Scholar]
- Miras, S., Salvi, D., Ferro, M., Grunwald, D., Garin, J., Joyard, J., and Rolland, N. (2002). Non-canonical transit peptide for import into the chloroplast. J. Biol. Chem. 277 47770–47778. [DOI] [PubMed] [Google Scholar]
- Mittler, R., and Rizhsky, L. (2000). Transgene-induced lesion mimic. Plant Mol. Biol. 44 335–344. [DOI] [PubMed] [Google Scholar]
- Mock, H.P., Heller, W., Molina, A., Neubohn, B., Sandermann, H., and Grimm, B. (1999). Expression of uroporphyrinogen decarboxylase or coproporphyrinogen oxidase RNA in tobacco induces pathogen defense responses conferring increased resistance to tobacco mosaic virus. J. Biol. Chem. 274 4231–4238. [DOI] [PubMed] [Google Scholar]
- Molina, A., Volrath, S., Guyer, D., Maleck, K., Ryals, J., and Ward, E. (1999). Inhibition of protoporphyrinogen oxidase expression in Arabidopsis causes a lesion-mimic phenotype that induces systemic acquired resistance. Plant J. 17 667–678. [DOI] [PubMed] [Google Scholar]
- Page, M., and Thorpe, R. (1996). Protein blotting by electroblotting. In The Protein Protocols Handbook, J.M. Walker, ed (Totowa, NJ: Humana Press), pp. 245–247.
- Pignol, D., Ayvazian, L., Kerfelec, B., Timmins, P., Crenon, I., Hermoso, J., Fontecilla-Camps, J.C., and Chapus, C. (2000). Critical role of micelles in pancreatic lipase activation revealed by small angle neutron scattering. J. Biol. Chem. 275 4220–4224. [DOI] [PubMed] [Google Scholar]
- Prasil, O., Adir, N., and Ohad, I. (1992). Dynamics of photosystem II: Mechanism of photoinhibition and recovery process. In Topics in Photosynthesis: The Photosystem Structure, Function and Molecular Biology, Vol. 11, J. Barber, ed (Amsterdam: Elsevier), pp. 295–384.
- Pružinská, A., Anders, I., Aubry, S., Schenk, N., Tapernoux-Lüthi, E., Müller, T., Kräutler, B., and Hörtensteiner, S. (2007). In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19 369–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pružinská, A., Tanner, G., Anders, I., Roca, M., and Hörtensteiner, S. (2003). Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 100 15259–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodoni, S., Muhlecker, W., Anderl, M., Kraütler, B., Moser, D., Thomas, H., Matile, P., and Hörtensteiner, S. (1997). Chlorophyll breakdown in senescent chloroplasts. Plant Physiol. 115 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg, M., and Matile, P. (1995). Association of components of the chlorophyll catabolic system with pigment-protein complexes from solubilized chloroplast membranes. J. Plant Physiol. 146 604–608. [Google Scholar]
- Shimoni, E., Rav-Hon, O., Ohad, I., Brumfeld, V., and Reich, Z. (2005). Three-dimensional organization of higher-plant chloroplast thylakoid membranes revealed by electron tomography. Plant Cell 17 2580–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoresh, M., Gal-On, A., Leibman, D., and Chet, I. (2006). Characterization of a MAPK gene from cucumber required for Trichoderma-conferred plant resistance. Plant Physiol. 142 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamiya, K.I., Tsuchiya, T., and Ohta, H. (2000). Degradation pathway(s) of chlorophyll: What has gene cloning revealed? Trends Plant Sci. 5 426–431. [DOI] [PubMed] [Google Scholar]
- Tarasenko, L.G., Khodasevich, E.V., and Orlovskaya, K.I. (1986). Location of chlorophyllase in chloroplast membranes. Photobiochem. Photobiophys. 12 119–121. [Google Scholar]
- Terpstra, W. (1974). Properties of chloroplasts and chloroplast fragments as deduced from internal chlorophyll-chlorophyllide conversion. Z. Pflanzenphysiol. 71 129–143. [Google Scholar]
- Terpstra, W. (1976). Chlorophyllase and lemellar structure in Phaeodactylum triconutum. Situation of chlorophyllase in pigment membranes. Z. Pflanzenphysiol. 80 177–188. [Google Scholar]
- Terpstra, W. (1981). Identification of chlorophyllase as a glycoprotein. FEBS Lett. 126 231–235. [Google Scholar]
- Tian, G.-W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh, T., Goldschmidt, E.E., and Riov, J. (1993). Ethylene induces de novo synthesis of chlorophyllase, a chlorophyll degrading enzyme, in citrus fruit peel. Proc. Natl. Acad. Sci. USA 90 9441–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, T., Ohta, H., Masuda, T., Mikami, B., Kita, N., Shioi, Y., and Takamiya, K.I. (1997). Purification and characterization of two isozymes of chlorophyllase from mature leaves of Chenopodium album. Plant Cell Physiol. 38 1026–1031. [Google Scholar]
- Tsuchiya, T., Ohta, H., Okawa, K., Iwamatsu, A., Shimaba, H., Masuda, T., and Takamiya, K.I. (1999). Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: Finding of a lipase motif and the induction by methyl jasmonate. Proc. Natl. Acad. Sci. USA 96 15362–15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, T., Suzuki, T., Yamada, T., Shimada, H., Masuda, T., Ohta, H., and Takamiya, K.I. (2003). Chlorophyllase as a serine hydrolase: Identification of a putative catalytic triad. Plant Cell Physiol. 44 96–101. [DOI] [PubMed] [Google Scholar]
- Vicentini, F., Hörtensteiner, S., Schellenberg, M., Thomas, H., and Matile, P. (1995). Chlorophyll breakdown in senescent leaves: Identification of the lesion in a stay-green genotype of Festuca pratensis. New Phytol. 129 247–252. [DOI] [PubMed] [Google Scholar]
- Villarejo, A., et al. (2005). Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7 1224–1231. [DOI] [PubMed] [Google Scholar]
- Wagner, D., Przybyla, D., Op den Camp, R., Kim, C., Landgraf, F., Lee, K.P., Wursch, M., Laloi, C., Nater, M., Hideg, E., and Apel, K. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306 1183–1185. [DOI] [PubMed] [Google Scholar]
- Winkler, F.K., D'Arcy, A., and Hunziker, W. (1990). Structure of human pancreatic lipase. Nature 343 771–774. [DOI] [PubMed] [Google Scholar]
- Wuthrich, K.L., Bovet, L., Hunziker, P.E., Donnison, I.S., and Hörtensteiner, S. (2000). Molecular cloning, functional expression and characterization of RCC reductase involved in chlorophyll catabolism. Plant J. 21 189–198. [DOI] [PubMed] [Google Scholar]
- Yang, M., Wardzala, E., Johal, G.S., and Gray, J. (2004). The wound-inducible Lls1 gene from maize is an orthologue of the Arabidopsis Acd1 gene, and the LLS1 protein is present in non-photosynthetic tissues. Plant Mol. Biol. 54 175–191. [DOI] [PubMed] [Google Scholar]