Abstract
As in other eukaryotes, progression through the cell cycle in plants is governed by cyclin-dependent kinases. Phosphorylation of a canonical Thr residue in the T-loop of the kinases is required for high enzyme activity in animals and yeast. We show that the Arabidopsis thaliana Cdc2+/Cdc28 homolog CDKA;1 is also phosphorylated in the T-loop and that phosphorylation at the conserved Thr-161 residue is essential for its function. A phospho-mimicry T161D substitution restored the primary defect of cdka;1 mutants, and although the T161D substitution displayed a dramatically reduced kinase activity with a compromised ability to bind substrates, homozygous mutant plants were recovered. The rescue by the T161D substitution, however, was not complete, and the resulting plants displayed various developmental abnormalities. For instance, even though flowers were formed, these plants were completely sterile as a result of a failure of the meiotic program, indicating that different requirements for CDKA;1 function are needed during plant development.
INTRODUCTION
The decision to divide or not is one of the most crucial choices made by a cell. Extrinsic cues, such as nutrient availability, need to be sensed, evaluated, and integrated with intrinsic cues, such as the developmental program. The central convergence point of these regulatory pathways are cyclin-dependent kinases (CDKs), and only if a certain threshold of CDK activity is reached will the entry into the next cell cycle phase be promoted. The activity of CDKs can be modulated at several levels, and it is this parallelism of regulatory inputs that makes CDKs function as integrating units (Morgan, 1997). The four major regulatory inputs affecting CDK activity are the binding of positive cofactors (i.e., cyclins), negative regulators (i.e., CDK inhibitors), and positive and negative phosphorylation events (i.e., at the Thr and/or Tyr residues of the T- and P-loops, respectively) (Pines, 1995).
Analysis of the Arabidopsis thaliana and rice (Oryza sativa) genome sequences has revealed that most of the core cell cycle regulators known from yeast and animals are also present in plants (Vandepoele et al., 2002; Wang et al., 2004a). For instance, Arabidopsis contains one Cdc2+/Cdc28 homolog, designated CDKA;1, that appears to be the main kinase involved at both major transition points: entry into S- and M-phases (Reichheld et al., 1999; Menges and Murray, 2002). Even though the theme of CDK–cyclin-regulated cell cycle progression appears to be conserved, there are also pronounced differences between animal and plant cell cycle controls. Plants possess an additional type of CDK, B-type CDKs, that do not occur in other species (Joubes et al., 2000). In particular, B-type CDKs have been found to play a role in asymmetric cell division during stomata development (Boudolf et al., 2004). Also, on the cyclin level, functional differences between plants and animals have been identified. For example, while D-type cyclins function solely at the G1-S transition in animals, D-type cyclins in plants can regulate the G2–M transition as well (Schnittger et al., 2002; Koroleva et al., 2004). Although plants possess two classes of CDK inhibitors (CKIs; the ICK/KRP and the SIM/EL2 classes), they show very little similarity with animal CDK inhibitors (Verkest et al., 2005b; Churchman et al., 2006; Wang et al., 2006). Furthermore, for the ICK/KRP class, it was found that at least one member can function in a non-cell-autonomous manner (Weinl et al., 2005).
Although the importance of regulatory cofactors (i.e., cyclins and CDK inhibitors) has been functionally addressed in plants, little is known about CDK regulation by phosphorylation. In particular, the importance of the phosphorylation of a conserved Thr residue (Thr-161) in the T-loop (also called the activation loop) of CDKA;1 is not understood.
Based on a substantial body of work, the function of the T-loop of animal and yeast CDKs is understood at atomic resolution (for review, see Morgan, 2007, and references therein). In the monomeric CDK, the T-loop is folded over the active site. This may ensure that the inactivated kinase indeed has no kinase activity. After cyclin binding, a global change in conformation is induced in the CDK structure that also causes the T-loop to swing over, liberating the catalytic core. However, kinase activity is still very low, and only upon phosphorylation of a canonical Thr residue in the T-loop can full activity be reached. The phosphorylation in the T-loop has been found to stabilize the structure of the CDK–cyclin complex; in particular, the opening of the T-loop by phosphorylation is crucial for substrate recognition and binding during the catalytic reaction of the CDK.
The phosphorylation of the T-loop is catalyzed by CDK-activating kinases (CAKs) that are also cyclin-dependent kinases (Draetta, 1997; Kaldis, 1999). In the Arabidopsis genome, four putative CAKs are present—CDKD;1, CDKD;2, CDKD;3 and CDKF;1—that have been found to have at least partially overlapping functions (Vandepoele et al., 2002; Umeda et al., 2005). The situation is even more complex in that D-type CDKs, similar to CAKs from other species, also phosphorylate the C-terminal domain of RNA polymerase II and participate in both RNA polymerase II–dependent transcription and nucleotide excision repair of DNA (Shimotohno et al., 2006).
Here, we have analyzed the function of T-loop phosphorylation in plants by starting from the CDKA;1 protein. We show that Arabidopsis CDKA;1 is phosphorylated in the T-loop region in vivo and demonstrate that phosphorylation is essential for kinase function. In contrast with other eukaryotes analyzed to date, we recovered homozygous cdka;1 mutants by expressing the phospho-mimicry T161D substitution. This substitution compromised the kinase activity as a result of reduced binding to substrates. Thus, the T161D CDK variant represents a weak cdka;1 allele that allows the analysis of postembryonic requirements of CDKA;1 activity in vivo.
RESULTS
CDKA;1 Is Phosphorylated in the T-Loop Region in Vivo
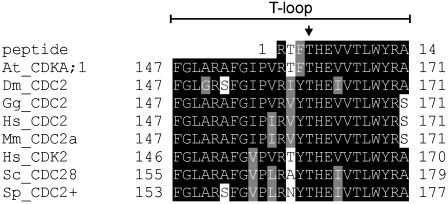
The Arabidopsis CDKA;1 shares 65, 67, and 62% of its amino acid residues with the human CDK1 (CDC2), the human CDK2, and the fission yeast Cdc2+, respectively. Moreover, we modeled CDKA;1 onto the known crystal structures of human CDK2 and found that all of the important structural elements of CDK2 are conserved in the Arabidopsis kinase, among them the T-loop (Figure 1; data not shown).
Figure 1.
Sequence Similarities in the T-Loop Regions between Different Cdc2+/Cdc28 Proteins.
Alignment of the T-loop regions of different CDKs (At, Arabidopsis thaliana; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Dm, Drosophila melanogaster; Mm, Mus musculus; Gg, Gallus gallus), including the phosphopeptide identified by mass spectrometric analysis of the Arabidopsis CDKA;1. The Thr in the T-loop of Arabidopsis corresponding to the canonical Thr residue phosphorylated in yeast or animals (Thr-160 for human CDK2, Thr-169 for S. cerevisiae, and Thr-167 for S. pombe) is indicated with an arrow.
To address whether plant CDKs are also regulated by the T-loop, we first analyzed its potential phosphorylation. Both A- and B-type CDK complexes from crude Arabidopsis cell suspension extracts were separated and purified using a four-step purification procedure.
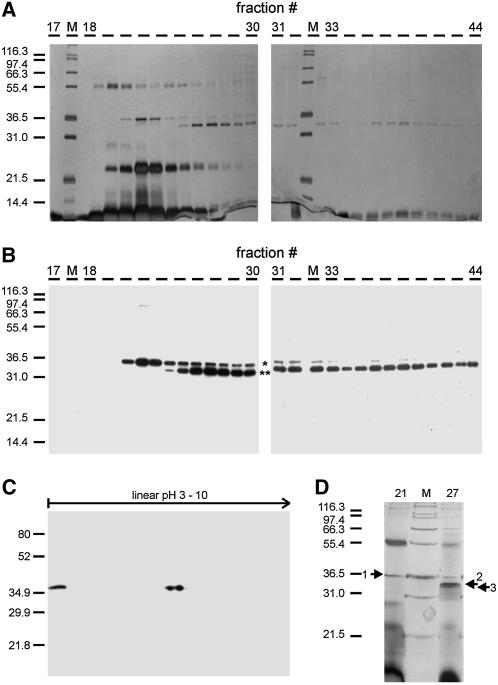
In short, after first preclearing the crude extract on an anion-exchange column, CDK-related complexes were bound to an affinity column containing the Arabidopsis CDK-SUBUNIT1 (CKS1, also called SUC1) covalently bound to Sepharose. More than 90% of the CDK-dependent histone H1 kinase activity was eluted from the affinity matrix using an excess of free CKS1. After a size-exclusion chromatography step, A- and B-type CDK complexes were separated on an anion-exchange column and eluted into two fractions, as shown by silver staining and immunoblotting with CDKA;1 and CDKB1;1 antisera after SDS-PAGE (Figures 2A and 2B). This final anion-exchange step led to an efficient purification of both CDKs, because only a small fraction of proteins coeluted with the CDKs (Figures 2A and 2D).
Figure 2.
Purification of CDKA;1 Complexes.
(A) and (B) Native A- and B-type CDK complexes were purified using a four-step purification scheme. One-tenth of each fraction eluting from the Source15Q column was analyzed via silver staining after SDS-PAGE (A) and probed with a mixture of CDKA;1 and CDKB1;1 antisera (B). The asterisk marks CDKB1;1, and the double asterisk indicates CDKA;1.
(C) A fraction of the CKS1-Sepharose-CDK complexes was separated by two-dimensional gel electrophoresis, blotted and probed with CDKA;1 antiserum.
(D) Eighty percent of fractions 21 and 27 eluted from the Source15Q column was resolved by 12% SDS-PAGE and stained with CBB-G. Protein bands 1, 2, and 3 were excised and further analyzed via MALDI-TOF MS.
A first hint that CDKA;1 is phosphorylated came from the separation of the CKS1-eluted fraction by isoelectric focusing followed by SDS-PAGE and protein gel blotting. Using a CDKA;1 antiserum, we detected two CDKA;1 isoforms migrating with the same molecular weight but with a shift in pI (Figure 2C). This finding suggested that one of the spots that we detected by two-dimensional gel electrophoresis is a CDKA;1 phospho-form, as phosphorylation can cause a shift in the pI of proteins.
Because the use of a phospho-Thr antibody gave variable results in our hands, we analyzed the peak fractions of the anion-exchange column via matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF MS). Proteins coeluting with the CDKs were resolved using SDS-PAGE and visualized by Coomassie Brilliant Blue G 250 (CBB-G) staining (Figure 2D). Several protein bands were excised. These gel slices were digested with trypsin, followed by MALDI-TOF peptide mass fingerprinting (PMF) of the fragments. Using the SEQUEST algorithm, acquired fragmentation spectra of the peptides were correlated with predicted amino acid sequences from the standard Arabidopsis open reading frames. In this way, we identified the protein present in band 1 in Figure 2D as CDKB1;1 (see Supplemental Figure 1 online). The proteins present in bands 2 and 3 were both identified as CDKA;1 (see Supplemental Figures 2A and 2B online). The only difference between the spectra derived from both CDKA;1 bands was a peptide with a mass of 1631.74 D (see Supplemental Figures 2A and 2B online). This peptide was identified as the phosphorylated form of (R)TFTHEVVTLWYR(A), corresponding to amino acid residues 159 to 170 (Figure 1), which have a calculated mass of 1551.7954 D. Upon phosphorylation, this mass shifts by 80 D, resulting in a peptide peak with a calculated mass of 1631.795 D.
These results show that the Cdc2+/Cdc28 homolog from Arabidopsis is also phosphorylated at a single residue in the T-loop region in vivo. Although the identified phospho-peptide contains three Thr residues as potential phosphorylation sites, given the sequence homology, these data suggest that CDKA;1 is phosphorylated at the highly conserved residue Thr-161 (Figure 1).
The De-Phospho Mutation Val-161 Cannot Rescue cdka;1 Mutants
The previous experiments raised the hypothesis that phosphorylation of the conserved Thr-161 is as crucial in plants as it is in animals and yeast. To test this importance, we replaced Thr-161 with a nonphosphorylatable Val residue (referred to as T161V).
To first address whether such a mutation might unspecifically interfere with the CDKA;1 structure or stability, we investigated the lifetime of this mutant version in transient expression assays. Analysis of a yellow fluorescent protein (YFP) fusion with wild-type CDKA;1 and CDKA;1T161V did not reveal an obvious difference in the stability of the two proteins (see Supplemental Figures 3A, 3B, 3D, and 3E online). In addition, CDKA;1T161V was found to bind to typical interaction partners such as CYCLIN B1;2 (CYCB1;2) in bimolecular fluorescent complementation (BiFC) assays (see Supplemental Figures 3D and 3E online).
Next, this mutated CDK variant was cloned behind the endogenous promoter (ProCDKA;1:CDKA;1T161V) for a functional test in planta. Because cdka;1 mutants are homozygous lethal, this construct was transformed into heterozygous cdka;1 mutant plants that we and others have described previously (Iwakawa et al., 2006; Nowack et al., 2006). More than 1300 descendents of 14 independent primary transformants that were heterozygous for the cdka;1 mutation were genotyped, but no homozygous cdka;1 mutant plants were identified. In comparison, we transformed a CDKA;1 rescue construct with the nonmutated CDK version under the control of the same promoter fragment. For this wild-type rescue construct, we tested >500 individual plants from the progeny of eight independent T1 plants that were heterozygous for the cdka;1 mutation. All eight lines segregated homozygous cdka;1 mutant plants, with ratios between 19 and 29%, consistent with our previous experiments (Nowack et al., 2006).
The Phospho-Mimicry Asp-161 Can Rescue cdka;1 Mutants
Next, we conducted the converse experiment and substituted Thr-161 with an Asp residue to mimic a phosphorylated Thr (from now on referred to as T161D). In transient expression assays, no deviation in stability from wild-type CDKA;1 was observed (see Supplemental Figures 3A and 3C online). Also, this CDK variant was found to interact with CYCB1;2 in BiFC assays (see Supplemental Figures 3D and 3F online). The CDKA;1T161D construct driven by the previously used endogenous promoter was then introduced into heterozygous cdka;1 mutant plants. The progeny of 10 heterozygous cdka;1 mutant T1 plants (with >700 individuals) carrying the transgenic construct were analyzed. All 10 lines segregated for homozygous cdka;1 mutants (referred to as T161D plants here), with ratios between 6 and 32%.
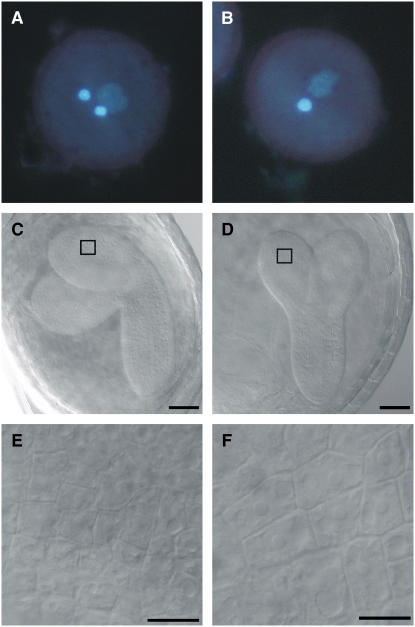
To corroborate the finding that CDKA;1T161D can rescue the cdka;1 mutation, the pollen of two lines heterozygous for cdka;1 and homozygous for the ProCDKA;1:CDKA;1T161D construct was analyzed. Heterozygous cdka;1 mutant plants display a defect during gametogenesis: the mutant microspores fail to undergo the second mitotic division, leading to mature pollen with only one instead of two gametes (Figures 3A and 3B). In contrast with the heterozygous mutants, we found in T161D plants pollen resembling the wild-type morphology, with two sperm cells and one vegetative cell (Figures 3A and 3B, Table 1).
Figure 3.
Morphological Analysis of Pollen and Progeny from Heterozygous cdka;1 Mutants Carrying a CDKA;1T161D Transgene.
(A) Pollen in wild-type and T161D plants is three-celled, with two brightly 4′,6-diamidino-2-phenylindole (DAPI)–stained sperm cell nuclei.
(B) cdka;1 mutant pollen is two-celled, with only one sperm cell–like cell. The expression of CDKA;1T161V cannot restore the wild-type morphology.
(C) Wild-type embryo at 10 d after pollination. The cotyledons start to bend toward the chalazal pole of the seed. The box indicates the magnified area in (E).
(D) A T161D mutant embryo at 10 d after pollination is smaller than the wild-type embryo, and the cotyledons are not bent. The box indicates the magnified area in (F).
(E) Detail of (C). Epidermal cells of the wild-type embryonic cotyledon.
(F) Detail of (D). Epidermal cells of the T161D mutant embryonic cotyledon are considerably larger than the corresponding wild-type cells.
Bars = 50 μm in (C) and (D) and 5 μm in (E) and (F).
Table 1.
Pollen Phenotypes of T161D and T161V
| Line | Three-Celled (%) | Two-Celled (%) | n |
|---|---|---|---|
| Columbia | 97 | 3 | 287 |
| cdka;1+/− | 54 | 46 | 393 |
| Wild-type rescue | 97 | 3 | 279 |
| T161D #1 | 97 | 3 | 464 |
| T161D #2 | 98 | 2 | 485 |
n, number of pollen scored.
T161D Plants Show a Drastically Reduced Kinase Activity
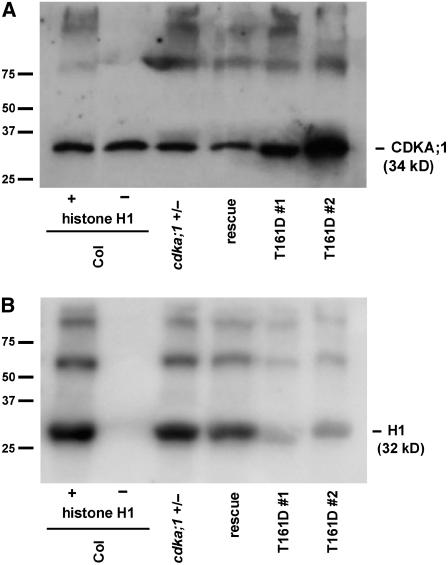
Phosphorylation of the T-loop has been found to tremendously stimulate catalytic efficiency and overall turnover rate in animals and yeast (Brown et al., 1999b; Hagopian et al., 2001). Thus, a phospho-mimicry of the canonical Thr residue could theoretically result in increased kinase activity levels. However, a phospho-mimicry substitution in the T-loop of Cdc2+ was found to mimic very inefficiently a phosphate group in S. pombe, and the resulting kinase displayed very low activity levels (Gould et al., 1998). To test the kinase activity of T161D plants, we first performed kinase assays with p13Suc1-bound CDKs pulled down from crude extracts of flower buds. Extracts of T161D plants showed a strong reduction compared with p13Suc1-bound kinase activity from wild-type plants or homozygous cdka;1 mutant plants expressing a wild-type CDKA;1 variant (Figures 4A and 4B).
Figure 4.
In Vitro Histone H1 Kinase Assays of T161D Plants.
(A) Of a crude plant extract derived from flower buds, 150 μg of total protein was subjected to a p13Suc1 pull-down and a subsequent kinase assay, separated by SDS-PAGE, blotted, and probed with a CDK antibody (α-PSTAIRE) to ensure equal loading of the beads. As controls, Columbia (Col), heterozygous cdka;1 mutants, and homozygous mutant plants carrying the rescue construct ProCDKA;1:CDKA;1 were used.
(B) Two independent homozygous mutant lines rescued by ProCDKA;1:CDKA;1T161D (lines #1 and #2) were investigated for their in vitro kinase activity toward their substrate histone H1. The homozygous mutant T161D lines show a strong decline in kinase activity.
Similar results were observed in at least three independent repetitions; the left two lanes show loading (A) and phosphorylation (B) in control reactions of CDK pull-downs from wild-type material with (+) and without (−) histone H1.
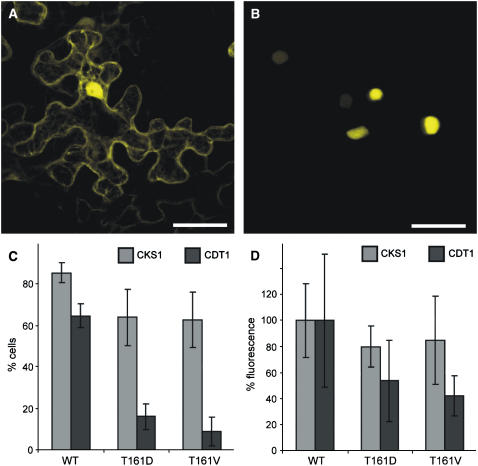
Based on the crystal structure of human CDK–cyclin complexes, a function of the T-loop in substrate binding has been realized (Russo et al., 1996; Brown et al., 1999a). Therefore, we sought for a system to monitor in vivo CDK–substrate interaction. Because the BiFC assay is known to stabilize weak or transient interactions, we wanted to know whether this interaction system would detect CDKA;1 binding to known bona fide substrates such as CDT1 and CDC6. Indeed, a strong fluorescence signal was obtained, demonstrating that CDKA;1 interacts with both proteins in a BiFC assay (Figure 5B; data not shown). The interaction of CDKA;1 with CDT1 or CDC6 was preferentially nuclear, although in some cells in addition to a nuclear fluorescence, interaction in the cytoplasm was also detected. These localization and interaction patterns are consistent with previous reports of the CDT1 and CDC6 localization in animals (Lei and Tye, 2001; Prasanth et al., 2004).
Figure 5.
Enzyme–Substrate Interaction of CDKA;1 Variants.
(A) CDKA;1 interacts with the nonsubstrate CKS1 in both the cytoplasm and the nucleus. The same pattern was observed for CDKA;1T161D and CDKA;1T161V. Bar = 40 μm.
(B) CDKA;1 binds to the bona fide substrate CDT1 predominantly in the nucleus. Although much weaker, CDKA;1T161D and CDKA;1T161V interaction with CDT1 also was observed. Bar = 40 μm.
(C) and (D) Quantification of the interaction between different CDKA;1 variants and CKS and CDT1. Quantifications were done with at least 25 cells each, and error bars represent sd from at least three independent experiments.
(C) The number of fluorescent cells in a given area of 25 cells is reduced slightly in interactions of the nonsubstrate CKS1 with the T-loop mutants CDKA;1T161D and CDKA;1T161V compared with wild-type CDKA;1. By contrast, the number of YFP-positive cells is strongly reduced in interactions of the substrate CDT1 with the T-loop mutants CDKA;1T161D and CDKA;1T161V compared with wild-type CDKA;1. The number of positive cells is most reduced in interaction assays with CDKA;1T161V.
(D) Fluorescence intensities are reduced slightly in interactions of the nonsubstrate CKS1 with the T-loop mutants CDKA;1T161D and CDKA;1T161V compared with wild-type CDKA;1, which was arbitrarily set to 100%. By contrast, the YFP intensities are strongly reduced in interactions of the substrate CDT1 with the T-loop mutants CDKA;1T161D and CDKA;1T161V compared with wild-type CDKA;1; the fluorescence reduction is most pronounced in interaction assays with CDKA;1T161V.
Next, we compared the binding of the wild type and T-loop mutants to CDT1 compared with CKS1 as a nonsubstrate interaction partner. CKS1 interacted with wild-type CDKA;1 equally strongly in both the nucleus and the cytoplasm (Figure 5A). For the two T-loop mutants, T161D and T161V, a slight reduction in the binding to CKS1 was observed (Figures 5C and 5D). By contrast, we found that the T161D substitution strongly reduced the binding of CDKA;1 to its substrates (Figures 5C and 5D). Interaction between CDKA;1 and CDC6 or CDT1 was even more reduced through the T161V substitution. This became obvious by analyzing the fluorescence intensities of the observed interactions and was even more pronounced by the reduction of YFP-positive cells expressing the T161D and T161V variants (Figures 5C and 5D; data not shown).
We conclude that the T161D mutant represents a weak cdka;1 allele, most likely attributable to a reduced ability to bind substrates during the phosphorylation reaction.
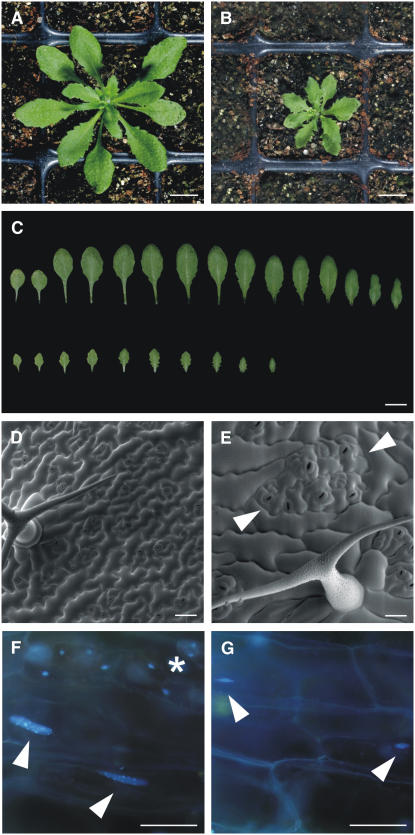
T161D Plants Are Strongly Reduced in Growth
Consistent with a strongly decreased kinase activity, T161D plants showed various developmental defects; most prominently, these plants were strongly reduced in growth (Figures 6A and 6B). The reduction of growth was already obvious during embryogenesis at 10 d after pollination. Approximately one-quarter of the seeds in the siliques of heterozygous cdka;1 mutants and those homozygous for the CDKA;1T161D construct contained embryos that were small and displayed cotyledons that did not, or only partially, fold to one side (Figures 3C and 3D). These embryos exhibited fewer but enlarged cells (Figures 3E and 3F).
Figure 6.
Morphological Analyses of T161D Plants.
(A) and (B) Wild-type (A) and T161D (B) plants at 3 weeks after germination. T161D plants are reduced in growth.
(C) Comparison of wild-type (top row) and T161D (bottom row) leaves from 3-week-old plants. T161D plants have fewer leaves that are smaller and strongly serrated.
(D) and (E) Scanning electron micrographs.
(D) Typical leaf surface of wild-type plants with equally distributed stomata cells and a three-branched trichome.
(E) Epidermis of T161D leaves display larger epidermal pavement cells that surround islands of stomata lineage cells (marked by arrowheads). Trichomes are reduced in branching, here showing a two-branched trichome.
(F) and (G) Fluorescence micrographs of DAPI-stained leaves.
(F) Wild-type leaf of the same age as in (E) stained with DAPI. Two nuclei of the large cells overlying the midvein are indicated by arrowheads. For comparison, a stomata cell with a DNA content of 2C is marked by an asterisk.
(G) The DNA amount of nuclei in T161D plants is reduced dramatically, and midvein overlying cells contain very small nuclei, highlighted with arrowheads.
Bars = 1 cm in (A) to (C) and 50 μm in (D) to (G).
After germination, fewer and smaller leaves were generated by the T161D plants compared with the wild type (Figures 6A to 6C). Leaves from T161D plants were strongly serrated and had a rough appearance. Closer examination at the cellular level by scanning electron microscopy revealed that these leaves comprised fewer yet in general much larger cells (Figure 6D). In the wild type, it has been estimated that >75% of leaf epidermal cells are generated by the activity of transiently amplifying cells of the stomata lineage (Geisler et al., 2000). Interestingly, cell divisions in the stomata lineage appeared to be less affected than in other epidermal cells in the T161D plants, and we found islands of stomata lineage cells that were separated by highly enlarged pavement cells (Figure 6E).
Finally, we analyzed the DNA content of these leaves by flow cytometry. In Arabidopsis as in many other species, differentiating cells in maturating organs, for instance the leaf, enter an endoreplication cycle in which the DNA continues to be replicated but without subsequent mitosis and cytokinesis; this leads to polyploid cells, and on average, the degree of polyploidization in Arabidopsis ranges between 8C and 16C (Galbraith et al., 1991). Whereas leaves from wild-type plants or cdka;1 mutants rescued by a wild-type CDK variant displayed the typical DNA profile with increasing DNA levels over time, the cells of leaves from T161D plants largely resided in G1-phase with a DNA content of 2C (Table 2). Endoreplication was almost completely blocked in these plants, and the endocycle levels were also not found to have increased over time (Table 2). Staining of leaves with the DNA dye DAPI confirmed a dramatic reduction of endoreplication levels in T161D plants. For instance, the epidermal cells overlying the midvein are among the largest cells found in an Arabidopsis leaf, with the highest endoreplication levels. However, in T161D plants, the nuclei of these cells were dramatically reduced in size (Figures 6F and 6G). Consistent with a previously reported upper limit for cell expansion without concomitant endoreplication, we found that these cells were smaller than those in the wild type (Schnittger et al., 2003). This effect was even more pronounced in trichomes, which are usually three- to four-branched in the wild type. In T161D plants, the branch number was reduced to two to three (Table 3).
Table 2.
DNA Profiles of T161D Plants
| Line | Leaf | 2C | 4C | 8C | 16C | n |
|---|---|---|---|---|---|---|
| Columbia | 1 + 2 | 13 ± 13 | 23 ± 11 | 46 ± 11 | 18 ± 16 | 13,980 |
| 3 + 4 | 19 ± 1 | 49 ± 3 | 31 ± 3 | 1 ± 0 | 8,678 | |
| cdka;1+/− | 1 + 2 | 13 ± 6 | 21 ± 7 | 40 ± 5 | 27 ± 12 | 7,898 |
| 3 + 4 | 11 ± 3 | 39 ± 9 | 45 ± 8 | 6 ± 2 | 5,237 | |
| Wild-type rescue | 1 + 2 | 20 ± 9 | 21 ± 10 | 26 ± 6 | 33 ± 19 | 3,035 |
| 3 + 4 | 11 ± 3 | 33 ± 9 | 46 ± 5 | 11 ± 8 | 1,532 | |
| T161D #1 | 1 + 2 | 72 ± 7 | 26 ± 7 | 2 ± 0 | 0 ± 0 | 4,327 |
| 3 + 4 | 66 ± 7 | 32 ± 8 | 2 ± 1 | 0 ± 0 | 4,577 | |
| T161D #3 | 1 + 2 | 71 ± 7 | 26 ± 6 | 2 ± 2 | 0 ± 0 | 537 |
| 3 + 4 | 80 ± 3 | 18 ± 4 | 2 ± 1 | 0 ± 0 | 1,486 |
n, number of cells scored. Mean DNA content ± sd is given.
Table 3.
Trichome Branch Number
| Number of Branches in Percentage per Leaf
|
||||||
|---|---|---|---|---|---|---|
| Line | 1 | 2 | 3 | 4 | 5 | n |
| Columbia | 0.0 ± 0.0 | 0.6 ± 0.3 | 78.0 ± 4.2 | 20.8 ± 4.1 | 0.6 ± 0.9 | 813 |
| T161D #1 | 1.6 ± 2.3 | 25.6 ± 11.2 | 71.8 ± 11.6 | 0.9 ± 1.6 | 0.0 ± 0.0 | 330 |
| T161D #2 | 1.0 ± 2.2 | 32.5 ± 23.4 | 64.7 ± 23.1 | 1.7 ± 3.1 | 0.0 ± 0.0 | 226 |
All trichomes on rosette leaves 3 and 4 were counted from at least two plants per line. Average values ± sd are given.
Thus, in contrast with all other higher eukaryotes analyzed to date, a phospho-mimicry substitution in the T-loop can rescue cdc2/cdc28 mutants in Arabidopsis. Yet, this rescue is partial, and in T161D plants, cell cycle progression is strongly affected, with entry into S-phase most affected.
T161D Plants Are Sterile as a Result of Failures in the Meiotic Program
One of the most striking features of T161D plants was the sterile phenotype. Although all floral organs were present, the plants were entirely female- and male-sterile, as revealed by reciprocal crosses with wild-type plants (data not shown). No pollen was found in mature anthers, and clearing preparations revealed that although the development of the ovule integuments and overall ovule morphology appeared normal, female gametophyte development was arrested right after or at meiosis (data not shown). Both defects suggested a requirement of CDKA;1 in the early progamic phase or during meiosis. Therefore, we investigated in detail the development of the germ line lineage by confocal laser scanning microscopy, focusing on the development of the male gametes.
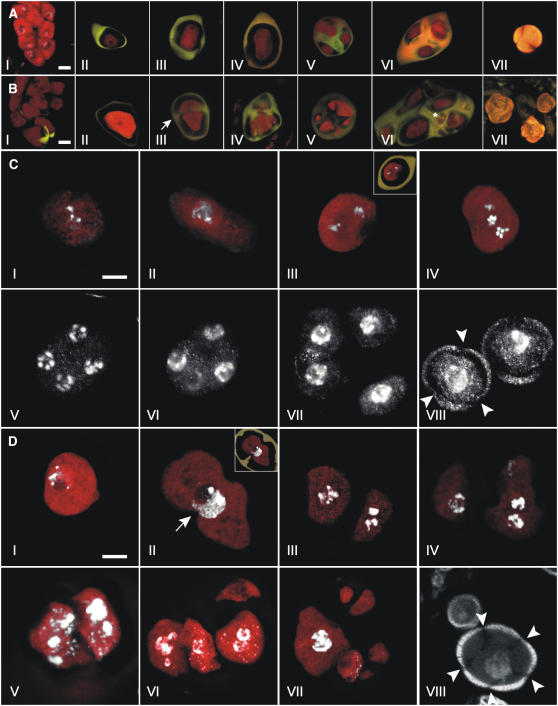
In Arabidopsis anthers, meiosis is initiated in a group of archesporial cells termed pollen mother cells (Figure 7A, I). Meiosis results in a tetrad of four haploid microspores encapsulated in a layer of callose (Figures 7A, II to VI, and 7C, I to VII) (Hamant et al., 2006; Ma, 2006). After release from the tetrad (Figures 7A, VII, and 7C, VIII), the male haploid microspores develop through two mitotic divisions to mature pollen, consisting of two sperm cells residing inside the cytoplasm of a vegetative cell (Figure 3A).
Figure 7.
Male Meiosis in Wild-Type and T161D Plants.
Confocal laser scanning microscopy of developmental stages of male meiosis in wild-type ([A] and [C]) and T161D ([B] and [D]) plants.
(A) and (B) Propidium iodide (PI; red) and aniline blue (AB; yellow) stains were used to paint meiotic cells and callose walls, respectively.
(A) I, wild-type pollen mother cells before callose deposition (triple stain with PI/AB/DAPI). II, premetaphase I. III, telophase I. IV, telophase II. V, early tetrad stage. VI, late tetrad stage. VII, released haploid uninucleate microspore. Bar = 5 μm.
(B) I, T161D pollen mother cells. Callose deposition has started in the bottom part of the micrograph. II, premetaphase I. III, dyad undergoing cytokinesis and deposition of callose between cells (arrow), presumably after telophase I. IV, continued callose wall deposition. V, heptad. Note different compartment sizes. VI, late-stage polyad with 11 nonuniform cellular compartments. Note division in the small compartment (asterisk). VII, released T161D microspores. Bar = 5 μm.
(C) and (D) PI (red) and DAPI (white) stains were used to visualize meiotic cells and DNA, respectively.
(C) Male meiosis in the wild type. I, pachytene. II, early diplotene. III, metaphase II. The inset depicts the same micrograph in overlay with callose deposition visualized by AB staining. IV, anaphase II. Two spindle poles reside in different focal planes in the center of the cell. V, early telophase II showing five chromatids at each spindle pole. VI, cytokinesis divides the nuclei at each pole of the second meiotic spindle. VII, tetrad stage. Callose is deposited between the four haploid meiotic products. VIII, released haploid uninucleate microspores with three pores (arrowheads). Bar = 5 μm.
(D) Male meiosis in T161D. I, meiocyte from T161D in pachytene/early diplotene. II, premetaphase I meiocyte. Note invaginations in the meiotic cell (arrow). The inset depicts the same micrograph in overlay with callose deposition visualized by AB staining. Note that callose projects into the meiotic cell invaginations. III to V, T161D dyads after cytokinesis and callose deposition. III, the left nuclei appear to be in anaphase/metaphase. IV, in the right cell, nuclear division appears to have taken place. V, cytokinesis appears to have taken place between the two nuclei in the left cellular compartment. VI, callose deposition has taken place between cells in a T161D tetrad. Note two prominent nuclei in the bottom left cellular compartment and no nuclei in the top left cell. VII, heptad with nonuniform cells embedded in callose. Note the large cell at left with large nuclei. The other cellular compartments appear to be enucleate. VIII, released T161D microspores with multiple pores (arrowheads). A large and a small microspore are shown. The exine layer appears normal in all sizes. Bar = 5 μm.
The differentiation of archesporial cells into pollen mother cells in T161D plants appeared normal compared with that in the wild type (Figures 7A, I, and 7B, I), and the meiocytes were found to enter meiosis (Figures 7A, II, 7B, II, 7C, I, and 7D, I). However, already during the first meiotic division of the pollen mother cell, differences compared with the wild type was observed. We found that the first meiotic division was followed by cytokinesis, as seen by the deposition of callose (Figures 7B, III, and 7D, II). This led to the development of dyads. In the wild type, cytokinesis is initiated in anaphase II of the second meiotic division (Figure 7C, VII). Remarkably, we found that cytokinesis started even though the genetic material was not completely distributed to the two daughter cells (Figure 7D, II).
Although in the wild type cytokinesis marks the end of the meiotic program (Figure 7C, IV to VII), the two cells of the dyad in T161D plants appeared to initiate a second division that was in many cases asynchronous (Figure 7D, III and IV). Also, the amount of DNA in the two products of the first division sometimes appeared to be unequal, arguing for problems in DNA distribution and/or replication (Figure 7D, IV). After this second division in T161D plants, a second cytokinesis was initiated that took place in an uncoordinated manner between the two cellular compartments (Figure 7D, V), as did callose deposition between cells or cytoplasmic pockets (Figure 7D, VI and VII). At late stages, the number of callose-encaged compartments in the normally four-celled tetrad ranged from 5 to 11 (Figures 7B, V and VI, and 7D, VI and VII). The cell or compartment size was variable and seemed to be in proportion with the nuclear material present within each compartment. This relationship, however, was not absolute, as some small compartments appeared to be enucleate (Figure 7D, VI and VII). After release from the callose cage, young microspores of different sizes were found in T161D anthers. Compared with wild-type microspores, more than the typical three pores were observed in mutant pollen (Figures 7B, VII, and 7D, VIII). No mutant pollen was observed to develop past this stage or to initiate the first pollen mitosis.
Together, these data demonstrated that CDKA;1 is also a major regulator of plant meiosis. Notably, there are different requirements for CDKA;1 function during development; in particular, meiosis seems to necessitate high levels of CDK activity.
Expression of CDKA;1T161D Causes a cdc Phenotype in S. pombe
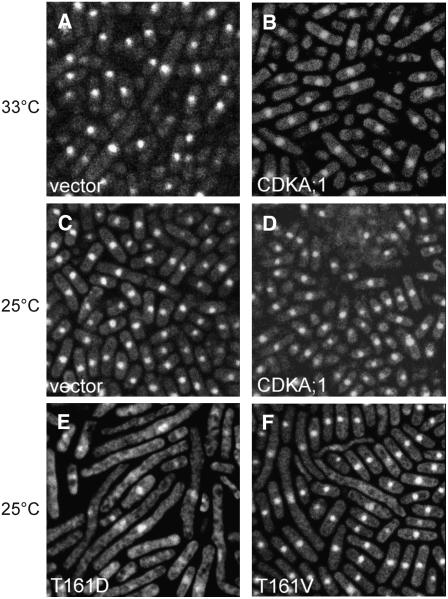
Although the T161D substitution reduced the kinase activity of CDKA;1, viable cdka;1 mutant plants were recovered by expression of this kinase variant. To test whether T161D is a weak allele not only in Arabidopsis but also in other model organisms, we expressed this mutant kinase in the temperature-sensitive cdc2 mutant of the fission yeast S. pombe. Previously, it was reported that plant kinases can at least partially rescue a temperature-sensitive cdc2 mutant (Hirayama et al., 1991; Hirt et al., 1991; Porceddu et al., 1999). Consistent with these reports, we found that CDKA;1 could partially suppress a cdc2 phenotype in temperature-sensitive mutants (Figures 8A and 8B).
Figure 8.
Heterologous Complementation Assays in S. pombe.
Cytological analysis of S. pombe cdc2-M63 cells expressing different variants of Arabidopsis CDKA;1 in pREP81X. Cells were fixed by heat and stained with DAPI. Image width = 50 μm.
(A) and (B) Complementation of the S. pombe mutation by wild-type CDKA;1 when incubated at semirestrictive 33°C. Cells transformed with empty vector show elongated cells at 33°C (A) but grow normally when carrying CDKA;1 (B).
(C) to (F) cdc2-M63 cells at 25°C. Transformants carrying either empty vector (C) or CDKA;1 (D). Cells expressing CDKA;1T161D (E) and CDKA;1T161V (F) both display a strong dominant negative effect with elongated cells.
Conversely, it has been reported that expression of the cdc2T167E phospho-mimicry substitution could not suppress temperature-sensitive cdc2 mutants, and the respective yeast cells were found to be arrested in metaphase (Gould et al., 1998). Notably, we found that expression of CDKA;1T161D caused a strong cdc phenotype at temperatures already below the restrictive temperature (Figures 8C to 8E). This phenotype mimics that of cdc2 mutants, suggesting that T161D functions as a dominant negative kinase variant. In this, the Arabidopsis phospho-mimicry kinase behaves similar to other heterologous phospho-mimicry CDK variants expressed in yeast (Ducommun et al., 1991; Krek et al., 1992; Atherton-Fessler et al., 1993). Moreover, although T161D and T161V behaved differently in plants (i.e., T161D could complement a cdka;1 mutant, whereas T161V could not), both kinase versions displayed a similar dominant negative effect in yeast (Figures 8E and 8F).
Together, these data suggest that the Arabidopsis Cdc2+/Cdc28 homolog has similar enzymatic properties as other Cdc2+/Cdc28 proteins, as judged by the effect of its expression in a heterologous system. Nonetheless, the reduced kinase activity of a T161D variant is sufficient to drive a mitotic cycle in plants. Thus, these data suggest that plants can progress through the cell cycle with very low Cdc2+/Cdc28-like kinase activity.
DISCUSSION
Plants Only Require Low Levels of CDK Activity to Progress through Mitosis
Here, we have shown that the Arabidopsis homolog of Cdc2+/Cdc28 is phosphorylated in vivo in the T-loop region and that a phosphorylation at the canonical Thr-161 residue is essential for CDK function. Phosphorylation in the T-loop has been found to be required for CDK function in organisms from three eukaryotic kingdoms—animals, yeast, and now plants—and thus appears to be a unifying principle of CDK regulation and action in eukaryotes. However, a phospho-mimicry substitution can complement a cdc2/cdc28 mutant only in plants, whereas similar phospho-mimicry substitutions did not rescue the respective mutants in budding or fission yeast (Lim et al., 1996; Gould et al., 1998). This difference between Arabidopsis and yeast becomes most obvious in the complete rescue of the gametophytic program by CDKA;1T161D in heterozygous cdka;1 mutants. In this situation, CDKA;1 activity diminishes, yet the expression of CDKA;1T161D promoted progression through the second mitosis and restored the generation of three-celled pollen. By contrast, expression of CDKA;1T161D in wild-type S. pombe resulted in a cdc phenotype. This effect was even more pronounced in a temperature-sensitive cdc2 mutant background grown just below the restrictive temperature.
Consistent with previous data, we found that Thr-mutated CDKA;1 also has a dramatically reduced kinase activity in plants, most likely attributable to a reduced substrate binding, as visualized in BiFC assays with CDT1 and other bona fide substrates. In fission yeast, a reduction of CDK activity results first in the failure to enter mitosis (MacNeill et al., 1991). Thus, it has been suggested that especially the entry and progression through mitosis requires higher levels of Cdc2+/Cdc28 activity (Stern and Nurse, 1996). By contrast, we found that cells of T161D plants reside predominantly in G1-phase. One possible explanation that could resolve these apparent discrepancies is that CDKA;1 function might be supported by B-type CDKs in plants. B-type CDKs have been found to be present in the cell cycle, especially from S-phase throughout mitosis (Porceddu et al., 2001); thus, we speculate that an assistance of CDKA;1 by B-type CDKs is the reason why plants can tolerate lower levels of Cdc2+/Cdc28 activity. Currently, we are testing the relationship between B-type CDKs and A-type CDKs. However, because B-type CDKs consist of a small family with four members that appear to act in a highly redundant manner, further work is required to untangle this relationship.
Low CDK Activity and Plant Development
The phenotype of T161D plants resembled the effects seen in plants misexpressing plant CDK inhibitors of the ICK/KRP-type, which share a limited sequence similarity to the animal Cip/Kip inhibitors (Verkest et al., 2005b; Wang et al., 2006). Misexpression of ICK1/KRP1 or ICK2/KRP2 in Arabidopsis resulted in dwarfed plants with serrated leaves composed of fewer and less endoreplicated cells (Wang et al., 2000; De Veylder et al., 2001).
However, the similarity of T161D plants is restricted to strong CKI misexpression plants. In contrast with the findings described above, weak misexpression of CKIs has been found to result in a specific block to entering mitosis while allowing progression through S-phase; consequently, weak misexpression plants displayed enhanced levels of endoreplication (Verkest et al., 2005a; Weinl et al., 2005). ICK/KRPs are largely believed to block CDKA;1 function, because in yeast two-hybrid interaction assays they strongly bind to A-type CDKs but no interaction of any of the ICK/KRPs to B-type CDKs has been reported (De Veylder et al., 2001; Zhou et al., 2002). In light of the findings discussed above that plants require little CDKA;1 activity, this preferential binding of ICK/KRPs leads to an apparent discrepancy. Based on our hypothesis that CDKA;1 is assisted by B-type CDKs during mitosis, one would expect weak ICK/KRP misexpression plants to have reduced rather than increased levels of endoreplication. However, the preference of ICK/KRPs to A-type CDKs is restricted to yeast two-hybrid data, and strikingly, a recent report using purified CDK complexes and adding ICK/KRP to them found that a B-type CDK complex was most susceptible to CKIs (Pettko-Szandtner et al., 2006). Thus, it will be an important task in the future to decipher the full spectrum of ICK/KRP targets.
Control of the Meiotic Program by CDKA;1
The similarity between the strong ICK1/KRP1 misexpression and T161D plants also extends to the sterile phenotype (Wang et al., 2000). Even though the reason for the sterility of ICK/KRP-misexpressing plants was not reported, it is plausible in the light of our data that ICK/KRP misexpression interfered with meiosis as well.
A large number of genes essential for meiosis have been identified in a variety of organisms (for review, see Dresser, 2000), and several homologs of these genes can be identified in Arabidopsis, predominantly genes involved in homologous recombination (Ma, 2006). However, little is known about the cell cycle machinery that controls the meiotic program in plants. Here, we have identified CDKA;1 as an important regulator of the meiotic program, and the observed defects in T161D plants fall into the class of true meiotic mutants, because sporogenic cells appear to be initiated correctly.
For proper progression through meiosis, it seems that higher levels of CDKA;1 activity are required than for mitosis. One reason may be that immediately before meiosis, the S-phase is very sophisticated and already includes preparations for the interaction of the homologous chromosomes. Also, meiosis might be under a tight developmental regime to ensure the timely and synchronous development of the female and male gametes required for fertilization. In this context, it is remarkable that there is apparently no spindle checkpoint during Arabidopsis meiosis. In T161D plants, the meiotic program continues, although the genetic material is unequally distributed. This observation is consistent with previously described mutants that displayed defects in meiosis yet continued with the separation and distribution program (Caryl et al., 2003).
Currently, only two other cell cycle regulators have been identified that affect plant meiosis. The first is SOLO DANCERS (SDS), which encodes a putative plant-specific cyclin that is phylogenetically isolated from all other cyclins in Arabidopsis (Azumi et al., 2002; Wang et al., 2004a). sds mutants display defects in the pairing and/or synapsis formation of homologous chromosomes during prophase I, leading to greatly reduced levels of meiotic recombination. The SDS protein has been found to interact with both CDKA;1 and CDKB1;1 in yeast two-hybrid interaction assays (Azumi et al., 2002). Furthermore, an interesting finding in this context is that the Ph1 locus from wheat (Triticum aestivum) has been mapped to a region also containing a cluster of cdc2+/CDC28-related genes (Griffiths et al., 2006). Ph1 is a major trait of modern wheat because it prevents the pairing of related chromosomes, allowing for the fecund propagation of the polyploid genome.
The second cell cycle mutant described to date is TARDY ASYNCHRONOUS MEIOSIS, which encodes CYCLIN A1;2 (CYCA1;2) (Wang et al., 2004b). CYCA1;2 is required for fast progression through the male meiosis I and II. Similar to T161D plants, cyca1;2 mutants form dyads instead of tetrads and meiosis progresses asynchronously. However, as with CYCA1;2, a detailed molecular understanding of its function during cell cycle regulation is pending. It is likely that the defects seen in T161D plants are compound, because CDKA;1 might pair with different cyclin partners to fulfill specific tasks at defined stages during meiosis. Consequently, the meiotic defects seen in T161D plants are much more severe than those in cyca1;2 mutants, which are fertile despite displaying a delayed and asynchronous progression through meiosis.
The reason why very few cell cycle regulators of the plant meiotic program have been identified to date might be the high overlap between the components for a mitotic and a meiotic cell cycle. Consequently, mutants in cell cycle regulators that affect plant growth would preclude analysis of the meiotic phase. Indeed, homozygous cdka;1 mutants appeared to be early embryonically lethal, whereas heterozygous mutant plants did not display developmental defects during the development of the sporophyte (Iwakawa et al., 2006; Nowack et al., 2006). Thus, a weak cdka;1 allele, as presented here, is a helpful tool to dissect cell cycle control and development in general and to explore the regulation of the meiotic program in particular.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana plants were grown under standard greenhouse conditions (16/8 h of light/dark) between 18 and 25°C. Arabidopsis cell suspension cultures (ecotype Columbia; a gift of Michèle Axelos) were refreshed weekly in Gamborg's B5 medium with minimal organics, 3% sucrose, 1 μM naphthalene acetic acid, and 0.5 μM kinetin at pH 5.7 and grown at 22°C and 110 rpm in a 16/8-h light/dark regime. Cells were harvested at 2 d after subculturing by filtration, frozen in liquid N2, and stored at −70°C until use. Mutant plants heterozygous for a T-DNA insertion in the cdka;1 locus were transformed using the Agrobacterium tumefaciens–mediated floral dip method.
Yeast Strains, Vectors, and Transformation
Schizosaccharomyces pombe strains (all strains were a kind gift of Ursula N. Fleig) were grown in rich medium or minimal medium (MM) with appropriate supplements according to standard protocols; amino acids were supplied as required. The mutant variants of Arabidopsis CDKA;1 were cloned into pREP81X using the primers P183 and P184 (see Supplemental Table 1 online). The nmt1+ promoter was repressed in medium containing 5 μg/mL thiamine. The constructs, including empty vector and cdc2+ controls, were transformed into either wild-type S. pombe (leu1-32, h-) or the temperature-sensitive strain cdc2-M63 (carrying the mutation G227C; restrictive temperature of 35°C; leu1-32, h+) (Nurse et al., 1976; Carr et al., 1989). The strains were transformed and selected for Leu prototrophy at 25°C on MM plates for 2 d; colonies were transferred to MM without thiamine and kept at 25°C for 24 h, then shifted to 28, 30, 32, 33, 34, or 35°C for 12 h, and growth phenotypes were examined. DNA staining was done with 10 μg/mL DAPI.
DNA Work
All primer sequences are listed in Supplemental Table 1 online. The phosphorylation site mutant variants of CDKA;1 were generated by PCR-mediated site-directed mutagenesis using the primers ND01/ND02 and ND03/ND04. The variants were sequenced and, via a Gateway LR reaction, cloned into pAM-PATGWProCDKA;1 (Nowack et al., 2006) and transformed into cdka;1 heterozygous mutant plants. Transformed plants were genotyped using the T-DNA left border primer J504 together with the CDKA;1 gene-specific primer N034 or the CDKA;1-specific pair N048 and N049. For BiFC assays, CDKA;1 variants were introduced into a previously described Gateway-based expression system after PCR using the primers P182 and SP22 (Jakoby et al., 2006).
Purification of Native CDKA;1 and CDKB1;1 Complexes
Frozen cell material of exponentially grown Arabidopsis cells was suspended in extraction buffer (25 mM Tris-Cl, pH 7.6, 85 mM NaCl, 15 mM MgCl2, 5 mM NaF, 15 mM EGTA, 0.1 mM Na3VO4, 15 mM p-NO2PhePO4, 60 mM β-glycerophosphate, 1 mM DTT, 0.1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM benzamidine, 50 μg/mL N-tosyl-l-Phe chloromethyl ketone, 10 μg/mL leupeptin, 10 μg/mL aprotinin, 5 μg/mL antipain, 5 μg/mL chymostatin, 10 μg/mL pepstatin A, and 10 μg/mL soybean trypsin inhibitor) and homogenized at 4°C using an UltraTurrax T25 disperser (Jahnke and Kunkel). The lysate was clarified by centrifugation at 40,000g for 40 min and at 200,000g for 1 h.
The clear extract was loaded onto a DEAE SepharoseFF (GE Healthcare Life Sciences) column (Omnifit) preequilibrated in extraction buffer (diluted 1:5 in 25 mM Tris-Cl, pH 7.6, and 85 mM NaCl). The flow-through of the DEAE SepharoseFF column was then bound to a CKS1-Sepharose affinity column (HR5/5; GE Healthcare Life Sciences). Expression and purification of recombinant CKS1 protein were performed as described (Landrieu et al., 1999). After washing the CKS1-Sepharose with bead buffer (see below), the affinity column was eluted with an excess of free CKS1 and the CDK-containing fractions were separated from the excess of free ligand via size exclusion (Superdex, 75 pg [GE Healthcare Life Sciences]; column, 25/325 mm [Omnifit]). All proteins >44 kD were pooled and further resolved on an ion-exchange chromatography column (Source15Q HR5/5; GE Healthcare Life Sciences) by a pH and NaCl gradient (buffer A, 25 mM Tris-Cl, pH 9.2; buffer B, 25 mM Tris-Cl, pH 7.8, 1 M NaCl, and 0.25 mM phenylmethylsulfonyl fluoride). The separated proteins (500 μL/fraction) were concentrated through binding to StrataClean resin (Stratagene), followed by elution with SDS sample buffer. Denatured CDK complexes were further resolved on a 12% SDS-PAGE gel, probed with CDKA;1 and CDKB1;1 antibodies, and stained with silver or CBB-G.
MALDI-TOF MS Protein Identification
MALDI-TOF PMF was used to identify gel-separated CDK complexes according to previously published methods (Gevaert et al., 2001).
In Vitro Histone H1 Kinase Assays
Flower buds were harvested and either snap-frozen in liquid N2 and stored at −70°C or immediately ground in ice-cold extraction buffer (2 mL/g fresh weight; 25 mM Tris-Cl, pH 7.6, 85 mM NaCl, 15 mM MgCl2, 2.5 mM NaF, 15 mM EGTA, 1 mM Na3VO4, 15 mM p-NO2PhePO4, 60 mM β-glycerophosphate, 5 mM DTT, 0.1% Tween 20, 0.1% Nonidet P-40, and a protease inhibitor cocktail composed of 1 mM phenylmethylsulfonyl fluoride, 0.1 mM benzamidine, leupeptin, aprotinin, pepstatin A, 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, and soybean trypsin inhibitor [10 μg/mL each]). The protein content of the cleared supernatant was measured (protein assay kit; Bio-Rad Laboratories) and adjusted to 1 mg/mL. Based on the desired kinase reaction, equal amounts of total protein (∼150 μg) were incubated with 30 μL of p13Suc1-Sepharose beads (50% [v/v] slurry; Upstate Biotechnology). The beads were washed three times with 500 μL of ice-cold bead buffer (50 mM Tris-Cl, pH 7.6, 250 mM NaCl, 5 mM NaF, 5 mM EDTA, 5 mM EGTA, 1 mM Na3VO4, 10 mM β-glycerophosphate, 1 mM DTT, 0.1% Tween 20, 0.1% Nonidet P-40, and the inhibitor mix). The protein extract was added to the aspirated beads and tumbled for 2 h at 4°C. The beads were washed three times with 500 μL of ice-cold bead buffer and once with 500 μL of prekinase buffer (50 mM Tris-Cl, pH 7.8, 15 mM MgCl2, 5 mM EGTA, 2 mM DTT, and the inhibitor mix). The assay reaction was prepared by adding 11.75 μL of water, 3.5 μL each of 500 mM Tris-Cl, pH 7.8, 150 mM MgCl2, 50 mM EGTA, and 10 mM DTT and was complemented with 1.75 μL of histone H1 (10 mg/mL; Millipore). The reactions were started by adding 4.1 μL of water, 0.3 μL of 1 mM Li-ATP, and 6 μCi of [γ-32P]ATP (220 TBq/mmol; Hartmann Analytic) and incubating for 45 min at room temperature. Reactions were terminated by the addition of 7.5 μL of 6× SDS sample buffer and boiling for 5 min. The samples were analyzed by 12% SDS-PAGE, blotted to nitrocellulose, probed with Cdc2 antibody (α-PSTAIRE; Santa Cruz Biotechnology), and detected with enhanced chemiluminescence (Pierce); phosphorylated histone was detected via PhosphorImager scanning (Molecular Dynamics).
BiFC Assays
The BiFC assays with leaves of Nicotiana benthamiana were performed as described previously (Jakoby et al., 2006).
Histology and Histochemistry
Pistils and siliques of different developmental stages were prepared as described previously (Nowack et al., 2006). For confocal laser scanning microscopy of male meiotic stages, whole inflorescences were fixed in 4% paraformaldehyde in microtubule-stabilizing buffer (MTSB; 50 mM PIPES, 5 mM EGTA, and 5 mM MgSO4, pH 6.9) at room temperature and 10 mmHg for 1 h, washed twice for 10 min in MTSB, dehydrated in a series of 10, 25, 40, and 55% ethanol in MTSB, and stored in 70% ethanol/MTSB. After rehydration, male meiosis was inspected by mounting single anthers of different stages in staining buffer (50 mM NaPO4 buffer, pH 7.2, 5% DMSO, and 0.01% Tween 20) containing combinations of 0.1 μg/mL propidium iodide, 2 μg/mL DAPI, or 0.01% (w/v) aniline blue.
Flow Cytometry
For flow cytometry analysis, rosette leaves were chopped with a razor blade in nuclear extraction buffer (CyStain UV-precise kit; Partec). All preparations were subsequently filtered through a 30-μm nylon mesh and stained with nuclear staining solution (CyStain) containing DAPI. Flow cytometry was performed on a three-laser LSRII analytical flow cytometer (BD Biosciences) using the 405-nm solid state laser for excitation and a 440/40 band-pass filter. The ploidy levels of three individual plants per set were averaged, and doublets were excluded from the analysis by gating on single nuclei in a DAPI-width versus DAPI-area display according to Wersto et al. (2001). Data were presented with Flowjo analysis software (TreeStar).
Microscopy
Light microscopy was performed with an Axiophot microscope (Zeiss) or a DM RA2 microscope (Leica) equipped with differential interference contrast (Nomarski) and epifluorescence optics. Confocal laser scanning microscopy was performed with a Leica TCS SP2 AOBS microscope or an Olympus META system microscope. Images were processed using Photoshop and Illustrator (Adobe) and ImageJ (rsb.info.nih.gov/ij).
Accession Numbers
The sequence data from this article are as follows. CDKA;1 has the Arabidopsis Genome Initiative code At3g48750 and GenBank accession number AY090353. The T-DNA insertion sites for the cdka;1 mutant allele used (cdka;1-1) have the GenBank accession numbers DQ156166 and DQ156167.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Primer Sequences.
Supplemental Figure 1. MALDI-TOF PMF Spectrum of CDKB1;1.
Supplemental Figure 2. MALDI-TOF MS PMF Spectra of CDKA;1 Isoforms.
Supplemental Figure 3. Localization and Stability of CDKA;1 Mutant Proteins.
Acknowledgments
We thank Jennifer Chiang, Reza Shirzadi, and Ulrich Dissmeyer for critical reading and helpful comments on the manuscript. We thank Annika K. Weimer, Janina Albert, and Samson Simon for their help in genotyping. We kindly acknowledge Andreas Dolf and Elmar Endl from the University of Bonn for their help with the flow cytometric analysis. Elmon Schmelzer, Maret Kalda, and Rolf-Dieter Hirtz are acknowledged for excellent technical assistance in microscopy and documentation. We are grateful to Ursula N. Fleig and Jennifer Poehlmann from the University of Düsseldorf for their assistance in yeast transformation and sharing of reagents. N.D. and M.K.N. are fellows of the International Max Planck Research School. P.E.G. is supported by a grant from the Norwegian Research Council. This work was supported by grants from the Deutsche Forschungsgemeinschaft (Grant SFB572) and the Volkswagen-Stiftung to A.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Arp Schnittger (schnitt@mpiz-koeln.mpg.de).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Atherton-Fessler, S., Parker, L.L., Geahlen, R.L., and Piwnica-Worms, H. (1993). Mechanisms of p34cdc2 regulation. Mol. Cell. Biol. 13 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azumi, Y., Liu, D., Zhao, D., Li, W., Wang, G., Hu, Y., and Ma, H. (2002). Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J. 21 3081–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf, V., Barroco, R., de Almeida Engler, J., Verkest, A., Beeckman, T., Naudts, M., Inze, D., and De Veylder, L. (2004). B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, N.R., Noble, M.E., Endicott, J.A., and Johnson, L.N. (1999. a). The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1 438–443. [DOI] [PubMed] [Google Scholar]
- Brown, N.R., Noble, M.E., Lawrie, A.M., Morris, M.C., Tunnah, P., Divita, G., Johnson, L.N., and Endicott, J.A. (1999. b). Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity. J. Biol. Chem. 274 8746–8756. [DOI] [PubMed] [Google Scholar]
- Carr, A.M., MacNeill, S.A., Hayles, J., and Nurse, P. (1989). Molecular cloning and sequence analysis of mutant alleles of the fission yeast cdc2 protein kinase gene: Implications for cdc2+ protein structure and function. Mol. Gen. Genet. 218 41–49. [DOI] [PubMed] [Google Scholar]
- Caryl, A.P., Jones, G.H., and Franklin, F.C. (2003). Dissecting plant meiosis using Arabidopsis thaliana mutants. J. Exp. Bot. 54 25–38. [DOI] [PubMed] [Google Scholar]
- Churchman, M.L., et al. (2006). SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18 3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., Krols, L., Terras, F., Landrieu, I., van der Schueren, E., Maes, S., Naudts, M., and Inze, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draetta, G.F. (1997). Cell cycle: Will the real Cdk-activating kinase please stand up? Curr. Biol. 7 R50–R52. [DOI] [PubMed] [Google Scholar]
- Dresser, M.E. (2000). Meiotic chromosome behavior in Saccharomyces cerevisiae and (mostly) mammals. Mutat. Res. 451 107–127. [DOI] [PubMed] [Google Scholar]
- Ducommun, B., Brambilla, P., Felix, M.A., Franza, B.R., Jr., Karsenti, E., and Draetta, G. (1991). cdc2 phosphorylation is required for its interaction with cyclin. EMBO J. 10 3311–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith, D.W., Harkins, K.R., and Knapp, S. (1991). Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 96 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler, M.D., Nadeau, J.A., and Sack, F.D. (2000). Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert, K., Demol, H., Martens, L., Hoorelbeke, B., Puype, M., Goethals, M., Van Damme, J., De Boeck, S., and Vandekerckhove, J. (2001). Protein identification based on matrix assisted laser desorption/ionization-post source decay-mass spectrometry. Electrophoresis 22 1645–1651. [DOI] [PubMed] [Google Scholar]
- Gould, K.L., Feoktistova, A., and Fleig, U. (1998). A phosphorylation site mutant of Schizosaccharomyces pombe cdc2p fails to promote the metaphase to anaphase transition. Mol. Gen. Genet. 259 437–448. [DOI] [PubMed] [Google Scholar]
- Griffiths, S., Sharp, R., Foote, T.N., Bertin, I., Wanous, M., Reader, S., Colas, I., and Moore, G. (2006). Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439 749–752. [DOI] [PubMed] [Google Scholar]
- Hagopian, J.C., Kirtley, M.P., Stevenson, L.M., Gergis, R.M., Russo, A.A., Pavletich, N.P., Parsons, S.M., and Lew, J. (2001). Kinetic basis for activation of CDK2/cyclin A by phosphorylation. J. Biol. Chem. 276 275–280. [DOI] [PubMed] [Google Scholar]
- Hamant, O., Ma, H., and Cande, W.Z. (2006). Genetics of meiotic prophase I in plants. Annu. Rev. Plant Biol. 57 267–302. [DOI] [PubMed] [Google Scholar]
- Hirayama, T., Imajuku, Y., Anai, T., Matsui, M., and Oka, A. (1991). Identification of two cell-cycle-controlling cdc2 gene homologs in Arabidopsis thaliana. Gene 105 159–165. [DOI] [PubMed] [Google Scholar]
- Hirt, H., Pay, A., Gyorgyey, J., Bako, L., Nemeth, K., Bogre, L., Schweyen, R.J., Heberle-Bors, E., and Dudits, D. (1991). Complementation of a yeast cell cycle mutant by an alfalfa cDNA encoding a protein kinase homologous to p34cdc2. Proc. Natl. Acad. Sci. USA 88 1636–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa, H., Shinmyo, A., and Sekine, M. (2006). Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45 819–831. [DOI] [PubMed] [Google Scholar]
- Jakoby, M.J., Weinl, C., Pusch, S., Kuijt, S.J., Merkle, T., Dissmeyer, N., and Schnittger, A. (2006). Analysis of the subcellular localization, function and proteolytic control of the Arabidopsis CDK inhibitor ICK1/KRP1. Plant Physiol. 141 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubes, J., Chevalier, C., Dudits, D., Heberle-Bors, E., Inze, D., Umeda, M., and Renaudi, J.P. (2000). CDK-related protein kinases in plants. Plant Mol. Biol. 43 607–620. [DOI] [PubMed] [Google Scholar]
- Kaldis, P. (1999). The Cdk-activating kinase (CAK): From yeast to mammals. Cell. Mol. Life Sci. 55 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva, O.A., Tomlinson, M., Parinyapong, P., Sakvarelidze, L., Leader, D., Shaw, P., and Doonan, J.H. (2004). CycD1, a putative G1 cyclin from Antirrhinum majus, accelerates the cell cycle in cultured tobacco BY-2 cells by enhancing both G1/S entry and progression through S and G2 phases. Plant Cell 16 2364–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek, W., Marks, J., Schmitz, N., Nigg, E.A., and Simanis, V. (1992). Vertebrate p34cdc2 phosphorylation site mutants: Effects upon cell cycle progression in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 102 43–53. [DOI] [PubMed] [Google Scholar]
- Landrieu, I., Casteels, P., Odaert, B., De Veylder, L., Portetelle, D., Lippens, G., Van Montagu, M., and Inze, D. (1999). Recombinant production of the p10CKS1At protein from Arabidopsis thaliana and 13C and 15N double-isotopic enrichment for NMR studies. Protein Expr. Purif. 16 144–151. [DOI] [PubMed] [Google Scholar]
- Lei, M., and Tye, B.K. (2001). Initiating DNA synthesis: From recruiting to activating the MCM complex. J. Cell Sci. 114 1447–1454. [DOI] [PubMed] [Google Scholar]
- Lim, H.H., Loy, C.J., Zaman, S., and Surana, U. (1996). Dephosphorylation of threonine 169 of Cdc28 is not required for exit from mitosis but may be necessary for start in Saccharomyces cerevisiae. Mol. Cell. Biol. 16 4573–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H. (June 6, 2006). A molecular portrait of Arabidopsis meiosis. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0095, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- MacNeill, S.A., Creanor, J., and Nurse, P. (1991). Isolation, characterisation and molecular cloning of new mutant alleles of the fission yeast p34cdc2+ protein kinase gene: Identification of temperature-sensitive G2-arresting alleles. Mol. Gen. Genet. 229 109–118. [DOI] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30 203–212. [DOI] [PubMed] [Google Scholar]
- Morgan, D.O. (1997). Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13 261–291. [DOI] [PubMed] [Google Scholar]
- Morgan, D.O. (2007). The Cell Cycle. (London: New Science Press).
- Nowack, M.K., Grini, P.E., Jakoby, M.J., Lafos, M., Koncz, C., and Schnittger, A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38 63–67. [DOI] [PubMed] [Google Scholar]
- Nurse, P., Thuriaux, P., and Nasmyth, K. (1976). Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146 167–178. [DOI] [PubMed] [Google Scholar]
- Pettko-Szandtner, A., Meszaros, T., Horvath, G.V., Bako, L., Csordas-Toth, E., Blastyak, A., Zhiponova, M., Miskolczi, P., and Dudits, D. (2006). Activation of an alfalfa cyclin-dependent kinase inhibitor by calmodulin-like domain protein kinase. Plant J. 46 111–123. [DOI] [PubMed] [Google Scholar]
- Pines, J. (1995). Cyclins and cyclin-dependent kinases: A biochemical view. Biochem. J. 308 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porceddu, A., De Veylder, L., Hayles, J., Van Montagu, M., Inze, D., and Mironov, V. (1999). Mutational analysis of two Arabidopsis thaliana cyclin-dependent kinases in fission yeast. FEBS Lett. 446 182–188. [DOI] [PubMed] [Google Scholar]
- Porceddu, A., Stals, H., Reichheld, J.P., Segers, G., De Veylder, L., Barroco, R.P., Casteels, P., Van Montagu, M., Inze, D., and Mironov, V. (2001). A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J. Biol. Chem. 276 36354–36360. [DOI] [PubMed] [Google Scholar]
- Prasanth, S.G., Mendez, J., Prasanth, K.V., and Stillman, B. (2004). Dynamics of pre-replication complex proteins during the cell division cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld, J., Vernoux, T., Lardon, F., Van Montagu, M., and Inze, D. (1999). Specific checkpoints regulate plant cell cycle progression in response to oxidative stress. Plant J. 17 647–656. [Google Scholar]
- Russo, A.A., Jeffrey, P.D., and Pavletich, N.P. (1996). Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat. Struct. Biol. 3 696–700. [DOI] [PubMed] [Google Scholar]
- Schnittger, A., Schobinger, U., Bouyer, D., Weinl, C., Stierhof, Y.D., and Hulskamp, M. (2002). Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc. Natl. Acad. Sci. USA 99 6410–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., Weinl, C., Bouyer, D., Schobinger, U., and Hulskamp, M. (2003). Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno, A., Ohno, R., Bisova, K., Sakaguchi, N., Huang, J., Koncz, C., Uchimiya, H., and Umeda, M. (2006). Diverse phosphoregulatory mechanisms controlling cyclin-dependent kinase-activating kinases in Arabidopsis. Plant J. 47 701–710. [DOI] [PubMed] [Google Scholar]
- Stern, B., and Nurse, P. (1996). A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 12 345–350. [PubMed] [Google Scholar]
- Umeda, M., Shimotohno, A., and Yamaguchi, M. (2005). Control of cell division and transcription by cyclin-dependent kinase-activating kinases in plants. Plant Cell Physiol. 46 1437–1442. [DOI] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouze, P., Rombauts, S., and Inze, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest, A., Manes, C.L., Vercruysse, S., Maes, S., Van Der Schueren, E., Beeckman, T., Genschik, P., Kuiper, M., Inze, D., and De Veylder, L. (2005. a). The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17 1723–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest, A., Weinl, C., Inze, D., De Veylder, L., and Schnittger, A. (2005. b). Switching the cell cycle. Kip-related proteins in plant cell cycle control. Plant Physiol. 139 1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., Kong, H., Sun, Y., Zhang, X., Zhang, W., Altman, N., DePamphilis, C.W., and Ma, H. (2004. a). Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 135 1084–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Zhou, Y., and Fowke, L.C. (2006). The emerging importance of cyclin-dependent kinase inhibitors in the regulation of the plant cell cycle and related processes. Can. J. Bot. 84 640–650. [Google Scholar]
- Wang, H., Zhou, Y., Gilmer, S., Whitwill, S., and Fowke, L.C. (2000). Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 24 613–623. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Magnard, J.L., McCormick, S., and Yang, M. (2004. b). Progression through meiosis I and meiosis II in Arabidopsis anthers is regulated by an A-type cyclin predominately expressed in prophase I. Plant Physiol. 136 4127–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl, C., Marquardt, S., Kuijt, S.J., Nowack, M.K., Jakoby, M.J., Hulskamp, M., and Schnittger, A. (2005). Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell 17 1704–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wersto, R.P., Chrest, F.J., Leary, J.F., Morris, C., Stetler-Stevenson, M.A., and Gabrielson, E. (2001). Doublet discrimination in DNA cell-cycle analysis. Cytometry 46 296–306. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Fowke, L.C., and Wang, H. (2002). Plant CDK inhibitors: Studies of interactions with cell cycle regulators in the yeast two-hybrid system and functional comparison in transgenic Arabidopsis plants. Plant Cell Rep. 20 967–975. [Google Scholar]