Abstract
RNA silencing can be induced by highly transcribed transgenes through a pathway dependent on RNA-DEPENDENT RNA POLYMERASE6 (RDR6) and may function as a genome protection mechanism against excessively expressed genes. Whether all transcripts or just aberrant transcripts activate this protection mechanism is unclear. Consistent RNA silencing induced by a transgene with three direct repeats of the β-glucuronidase (GUS) open reading frame (ORF) is associated with high levels of truncated, unpolyadenylated transcripts, probably from abortive transcription elongation. Truncated, unpolyadenylated transcripts from triple GUS ORF repeats were degraded in the wild type but accumulated in an rdr6 mutant, suggesting targeting for degradation by RDR6-mediated RNA silencing. A GUS transgene without a 3′ transcription terminator produced unpolyadenylated readthrough mRNA and consistent RDR6-dependent RNA silencing. Both GUS triple repeats and terminator-less GUS transgenes silenced an expressed GUS transgene in trans in the wild type but not in the rdr6 mutant. Placing two 3′ terminators in the GUS transgene 3′ reduced mRNA 3′ readthrough, decreased GUS-specific small interfering RNA accumulation, and enhanced GUS gene expression. Moreover, RDR6 was localized in the nucleus. We propose that improperly terminated, unpolyadenylated mRNA from transgene transcription is subject to RDR6-mediated RNA silencing, probably by acting as templates for the RNA polymerase, in Arabidopsis thaliana.
INTRODUCTION
In plants, RNA silencing can be induced efficiently by expressing transgenes with inverted repeats (Chuang and Meyerowitz, 2000; Smith et al., 2000). In Arabidopsis thaliana, RNA silencing induced by transgenes with inverted repeats does not require the putative RNA-dependent RNA polymerase RDR6 (also known as SDE1 or SGS2), most likely because the transcripts from such transgenes can directly fold back to form double-stranded (ds)RNA (Beclin et al., 2002). RNA silencing can also be induced frequently by sense transgenes that are designed for overexpression (Napoli et al., 1990; van der Krol et al., 1990). Sense transgene-induced RNA silencing requires RDR6 in Arabidopsis (Beclin et al., 2002). Since transcription of sense transgenes generally does not produce dsRNA, RDR6 may recognize, directly or indirectly, certain transcripts of silenced transgenes as templates for synthesis of dsRNA to trigger RNA silencing. How RDR6 distinguishes between the transcripts of silenced sense transgenes and the far greater amounts of transcripts from expressing endogenous genes is not fully understood.
Sense transgene-induced RNA silencing often occurs in a portion of a transgenic plant population and may be associated with certain specific events, such as high transgene copy number, the use of strong promoters, and special arrangements and/or insertion locations of transgenes occurring during integration (Jorgensen et al., 1996; Que et al., 1997; Muskens et al., 2000). While more recent studies indicated that position effects, inverted repeat T-DNA configurations, and arrangements of tandemly repeated transgenes may not be sufficient to trigger transgene silencing (Lechtenberg et al., 2003), there is strong evidence that expression levels of transgenes affect the frequencies of transgene silencing. It has been observed that highly expressing transgenes are often associated with high frequencies of transgene silencing (Lindbo et al., 1993; Vaucheret et al., 1998; Schubert et al., 2004). By comparing the frequency and degree of cosuppression by sense chalcone synthase transgenes driven by strong and weak promoters, it has been demonstrated that a strong transgene promoter is required for high frequency cosuppression of chalcone synthase genes and for the production of the full range of cosuppression phenotypes (Que et al., 1997). The major effect of transgene expression levels on transgene silencing may also account for the positive correlation between the transgene copy number and silencing frequency, since gene copy number and expression are often positively correlated (Schubert et al., 2004). Based on these observations, it appears that sense transgene-induced RNA silencing is a genome surveillance system that detects and eliminates transcripts from excessively expressed genes, including transgenes (Schubert et al., 2004).
While the transcript threshold model accounts for a probable cause of sense transgene-induced RNA silencing in transgenic plants, it does not necessarily reveal the specific trigger directly responsible for the activation of silencing. In a transgenic plant harboring a highly expressing transgene, the genome surveillance mechanism may sense the high levels of the transgene transcripts and activate silencing when transcripts of the transgene reach a gene-specific threshold level. Expression of transgenes may also generate certain aberrant RNAs with unusual structures that could be recognized by the cellular RNA silencing mechanism. It is possible, for a variety of reasons, that highly transcribed transgenes may generate more aberrant RNAs as collateral products than poorly transcribed transgenes and are, therefore, more prone to RNA silencing. The distinction between these two models for the activation of sense transgene-induced RNA silencing is important when considering the mechanistic aspects of the cellular RNA surveillance system. It also has important implications in transgene expression used extensively for basic research and for many applications in plant biotechnology. If the cellular RNA surveillance mechanism triggers the silencing of a transgene in direct response to the high transcript levels of the transgene, it will be difficult to achieve expression of the transgene to very high levels. On the other hand, if the RNA surveillance mechanism triggers the silencing of a transgene primarily because of aberrant RNAs generated from transcription of the transgene, it should be possible to achieve very high expression levels of the transgene if ways are found to reduce or eliminate the generation of aberrant RNAs.
There is indirect evidence that expressing transgenes generate unusual transcripts that are not produced from expressing endogenous genes. For example, when transgenic plants harboring an expressing Nia2 transgene encoding a nitrate reductase were used as scions and grafted onto transgenic tobacco (Nicotiana tabacum) harboring a silenced Nia2 transgene, systemic acquired silencing of the Nia2 transgene in the scion was observed (Palauqui et al., 1997). By contrast, when nontransgenic wild-type tobacco plants were grafted onto the transgenic plants harboring a silenced Nia2 transgene, the endogenous Nia2 gene in the wild-type scion was not silenced (Palauqui et al., 1997). Furthermore, virus vectors carrying parts of a GREEN FLUORESCENT PROTEIN (GFP) transgene can target RNA silencing in Nicotiana benthamiana and Arabidopsis harboring an expressing GFP transgene (Vaistij et al., 2002). The silencing can spread from the initiator region into the adjacent 5′ and 3′ regions of the target gene. This spread of RNA silencing, however, was not found with endogenous genes, including highly expressed endogenous genes such as RbcS (Vaistij et al., 2002). It appears that the expression of transgenes, but not endogenous genes, generates certain signals that allow for sensitive responses to systemic and spreading silencing stimuli. Because of the sequence specificity of the process, these signals are probably nucleic acids.
A number of studies have shown that unpolyadenylated or unproductive RNAs accumulate in plants showing RNA silencing. These aberrant RNAs have been reported in transgenic tomato (Solanum lycopersicum) lines containing a truncated ripening-specific polygalacturonase (PG) gene, in which the endogenous PG gene and the transgene were both silenced (Han and Grierson, 2002). In these plants, RNA molecules distinct from and smaller than the endogenous mRNA and the transgene transcript accumulated at high levels (Han and Grierson, 2002). In transgenic Petunia plants in which the chalcone synthase genes were silenced, a variety of polyadenylated and unpolyadenylated aberrant chalcone synthase RNAs have also been detected (Metzlaff et al., 2000). Posttranscriptional silencing of basic β-1,3-glucanase genes in tobacco was also associated with the generation of mainly 3′ truncated, unpolyadenylated RNAs for the silenced genes (van Eldik et al., 1998). Unpolyadenylated RNA was also associated with high-efficiency silencing of a β-glucuronidase (GUS) gene in transgenic rice (Oryza sativa) lines supertransformed with a set of constructs designed to silence the resident GUS gene (Wang and Waterhouse, 2000). However, these observations were made from populations of transgenic plants harboring the same transgene constructs, and the correlative nature of these data make it difficult to determine whether these aberrant RNA species were the triggers or the products of RNA silencing. To provide direct evidence for or against aberrant RNAs as a trigger of silencing, it may be necessary to design transgene constructs with enhanced or reduced production of aberrant RNAs and then to test them for corresponding promotion or suppression of silencing in transgenic plants. A similar approach with promoters of different strengths has been used to provide direct evidence that high expression levels promote transgene silencing (Que et al., 1997).
However, since the specific structures of aberrant RNAs important for activating RNA silencing, if any, are unknown, it is difficult to design transgene constructs with predictable enhancement or reduction in aberrant RNA generation in transgenic plants. A recent study has reported high efficiency of RNA silencing by transgene constructs containing three or four transgene direct repeats (Ma and Mitra, 2002). The resulting silencing is associated with the accumulation of gene-specific small interfering RNA (siRNA), indicating the involvement of a posttranscriptional RNA silencing mechanism (Ma and Mitra, 2002). As with sense transgenes, transcription of transgene direct repeats is unlikely to generate dsRNA directly and, therefore, may require RDR6 for the activation of dsRNA-mediated posttranscriptional gene silencing. If certain aberrant RNAs are primary triggers for RDR6-mediated silencing, constructs containing transgene direct repeats, which generate consistent RNA silencing, must be unusually efficient in generating aberrant RNAs and could serve as a good system to study the structures and biogenesis of the silencing-inducing aberrant RNAs.
Indeed, we found that RNA silencing induced by three direct repeats of the GUS open reading frame (ORF) was RDR6-dependent in Arabidopsis and correlated with the accumulation of high levels of truncated, unpolyadenylated mRNAs, apparently due to abortive transcription elongation and premature termination of transcription of the long transgene direct repeats. These truncated, unpolyadenylated mRNAs accumulated in a silencing-deficient rdr6 mutant and not in the wild-type plants; therefore, they were not the degradation products of RNA silencing. Furthermore, a GUS transgene driven by the cauliflower mosaic virus (CaMV) 35S promoter with no transcription terminator at its 3′ end also led to the generation of improperly terminated, unpolyadenylated readthrough mRNA and consistent RDR6-dependent RNA silencing. Both the GUS triple repeats and terminator-less GUS transgenes could silence an expressed GUS transgene in trans in the wild type but not in the sde1-1 mutant. By contrast, a GUS transgene with two terminators at its 3′ end had a significant decrease in mRNA 3′ readthrough and RNA silencing. Using GFP fusion proteins, we found that Arabidopsis RDR6 was localized in the nucleus, a cellular compartment where unpolyadenylated RNAs are known to accumulate. These results provide strong evidence that improperly terminated, unpolyadenylated mRNAs generated from abortive elongation or readthrough of transgene transcription can trigger RDR6-mediated RNA silencing, probably by acting as templates for the cellular RNA polymerase.
RESULTS
Effects of Transgene Copy Number on Transgene Expression and RDR6-Mediated Silencing
A number of studies have shown a positive correlation among transgene copy number, transgene expression, and silencing. In these studies, transgenic lines were generated in the wild-type background with active RNA silencing; therefore, the effects of transgene copy number on transgene expression and silencing are difficult to separate. In this study, we have generated and comparatively analyzed a large number of transgenic plants harboring a transgene construct (pGts) in both the wild-type and silencing-deficient sde1-1 mutant backgrounds. pGts contains a single copy of the GUS reporter gene flanked by the constitutive CaMV 35S promoter with duplicated enhancers and the 35S transcription terminator (Figure 1A). The copy numbers of T-DNA insertion in these transgenic lines were determined by DNA gel blot analysis. From ∼180 transgenic plants analyzed, 30 to 40% were single-copy transgenic plants, whereas the remaining 60 to 70% had multiple transgenes in the genome. DNA gel blot analysis also revealed that a majority of single-copy transgenic lines (>90%) contained intact GUS transgene constructs, while >30% of lines with multiple T-DNA insertions contained both truncated and intact GUS transgenes.
Figure 1.
Scheme of the GUS Constructs in the Binary Vector pOCA30.
(A) The T-DNA region of the binary vector pOCA30 contains the CaMV 35S promoter with duplicated enhancers. GUS constructs were inserted behind the 35S promoter. LB, left border; RB, right border; E35S promoter, CaMV 35S promoter with duplicated enhancers; GUS, β-glucuronidase; 35S terminator, the CaMV 35S transcription terminator; nos terminator, the transcription terminator from the nos gene of A. tumefaciens; EcoRI/ClaI fragment, the ∼0.8-kb EcoRI-ClaI DNA fragment from pOCA30 located 3′ to the insertion site of the GUS constructs.
(B) DNA sequences of the CaMV 35S terminator and the Agrobacterium nos gene terminator used in the GUS gene constructs.
To determine the effects of transgene copy number on transgene expression and silencing, we compared GUS activities in both wild-type and sde1-1 transformants with one to five copies of the T-DNA insertion in the genome; transformants with more than five copies were not included due to very limited numbers. As shown in Figure 2A, the average GUS activities in the wild-type transformants decreased by 2.5-fold as the copy number increased from one to five. In the sde1-1 mutant transformants, on the other hand, the GUS activities increased steadily with the transgene copy number, increasing from ∼630 units in single-copy plants to ∼2000 units in plants containing five copies (Figure 2A). As a result of the opposite effects, the difference in GUS activities between the wild type and the sde1-1 transformants increased markedly with transgene copy number (Figure 2B). These results support the positive correlation among transgene copy number, expression, and RDR6-mediated RNA silencing. Because of the major effects of the transgene copy number on transgene expression and silencing, we used only transgenic plants with a single copy of the T-DNA insertion in the genome for the comparative analysis described below.
Figure 2.
Effects of Transgene Copy Number on Transgene Expression.
(A) GUS activities in the wild-type and sde1-1 transformants with one to five copies of a GUS transgene driven by the CaMV 35S promoter. The means and se of GUS activities were calculated from 8 to 30 T1 transformants for each copy number. GUS activities are expressed in units (nanomoles of 4-methylumbelliferone per minute per milligram of total soluble protein).
(B) Ratios of GUS activities in the sde1-1 transformants over those in the wild-type transformants harboring the same copy numbers of the pGts transgene construct.
RNA Silencing Induced by Transgene Direct Repeats Is RDR6-Dependent
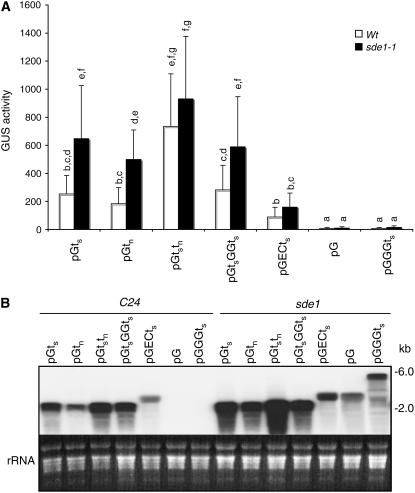
To determine whether RDR6 is required for efficient RNA silencing induced by transgene direct repeats, we compared two constructs (pGts and pGGGts) in both the wild type and an sde1-1 mutant (Dalmay et al., 2000). As described above, the pGts construct contains a single copy of the GUS reporter gene flanked by the constitutive CaMV 35S promoter with duplicated enhancers and the 35S transcription terminator (Figure 1). pGGGts contains three GUS ORF direct repeats between the enhanced 35S promoter and the 35S terminator (Figure 1A). Thirty independent single-copy wild-type transformants or sde1-1 transformants were assayed for GUS activities for each construct. The wild-type transformants harboring the pGts construct had a wide range of GUS activities, with ∼30% of plants containing little or no GUS activities (see Supplemental Figure 1 online). Because of the substantial percentage of transformants with little or no GUS activities, the average GUS activity of the wild-type plants harboring the pGts construct was only ∼250 units (Figure 3A). In the sde1-1 mutant background, all of the primary transformants harboring the pGts construct contained GUS activities (see Supplemental Figure 1 online); as a result, the average GUS activity for the population increased by >2.5 fold (Figure 3A). It is also worthy to note that the GUS activities in a substantial number of the sde1-1 transformants harboring the pGts construct were higher than the highest ones found in the wild-type pGts transformants (see Supplemental Figure 1 online), suggesting that partial silencing by RDR6-mediated pathways occurred even in those wild-type transformants with relatively high GUS activities. Enhanced transgene expression in the silencing-deficient mutant background has been reported previously (Butaye et al., 2004).
Figure 3.
GUS Activities and GUS mRNA Accumulation in Arabidopsis Wild-Type and sde1-1 Transformants.
(A) GUS activities. Average GUS activity and sd for each construct calculated from 30 T1 wild-type or sde1-1 transformants containing a single-copy T-DNA insertion. GUS activities are expressed in units (nanomoles of 4-methylumbelliferone per minute per milligram of total soluble protein). According to Duncan's multiple range test (P = 0.05), means of GUS activities do not differ significantly if they are indicated with the same letters.
(B) Accumulation of GUS transcripts. Total RNA was pooled from 9 to 10 randomly selected T1 transformants with a single-copy T-DNA insertion for each construct in the wild-type or sde1-1 background and probed with the full-length GUS gene fragment. The ethidium bromide stain of rRNA is shown for each lane to allow assessment of equal loading.
When the pGGGts construct was transformed into the wild-type plants, all of the transformants tested had little or no GUS activities (see Supplemental Figure 1 online), supporting the previous study, which found that RNA silencing induced by transgene direct repeats is highly efficient (Ma and Mitra, 2002). When the same construct was introduced into the sde1-1 mutant, the GUS activities were higher than those in the wild-type transformants but amounted to only ∼3% of the activities detected in the pGts transformants in the sde1-1 mutant background (Figure 3A; see Supplemental Figure 1 online).
It was possible that the GUS activities reflected the GUS protein levels but not the levels of GUS transcripts. To test this possibility, we analyzed the levels of GUS transcript in the pGts and pGGGts transformants. About 60 to 70% of the wild-type transformants but 100% of the sde1-1 transformants harboring the pGts construct contained high levels of GUS transcripts (Figure 3B; see Supplemental Figure 2 online). In the wild-type pGGGts transformants, there was little accumulation of GUS transcripts (Figure 3B; see Supplemental Figure 2 online), as reported previously (Ma and Mitra, 2002). The lack of accumulation of GUS transcripts in the wild-type transformants was consistent with little or no GUS activities in these plants (Figure 3A). In the sde1-1 transformants harboring the pGGGts construct, however, we observed an accumulation of substantial levels of GUS transcripts (Figure 3B; see Supplemental Figure 2 online). The accumulation of GUS transcripts in the sde1-1 transformants harboring the pGGGts construct but not in the wild-type transformants indicated that RNA silencing induced by transgene direct repeats is indeed RDR6-dependent. However, the accumulated mRNAs in the sde1-1 mutant plants transformed with pGGGts were apparently not functional for the production of active GUS proteins, resulting in low GUS activities in these plants (Figure 3A). One possible reason could be the poor translation of the polycistronic transcripts. However, in eukaryotes, distal ORFs should not affect the translation of the first ORF. Thus, there should have been ample GUS activity from the polycistronic GUS transcripts in the sde1-1 transformants, given the abundant GUS mRNA detected there. This raised the possibility that other structural features of the transcripts caused the low GUS activities associated with the pGGGts construct.
Premature Transcription Termination of Transgene Direct Repeats Generated Partial, Unpolyadenylated mRNA
From the RNA gel blots (Figure 3B; see Supplemental Figure 2 online) it was apparent that the accumulated GUS transcripts isolated from the sde1-1 mutant transformants differed in size between the two constructs. Unlike the sde1-1 transformants containing the pGts construct that accumulated GUS transcripts with expected ∼2.0-kb size (Figure 3B), the sde1-1 transformants harboring the pGGGts construct accumulated GUS transcripts of various high molecular sizes, as exhibited by the slow migrating bands on the RNA gel blot (Figure 3B). Most of these GUS transcripts had sizes larger than that of a single copy of GUS transcript (∼2 kb) but smaller than the size of three direct GUS repeats (∼6 kb) (Figure 3B; see Supplemental Figure 2 online). This observation suggested that transcription of the three GUS ORF repeats continued beyond the first GUS ORF repeat but did not go through the third repeat to produce full-length GUS triple repeat transcripts. To test this possibility, we probed the total RNA from the sde1-1 transformants harboring either the pGts or the pGGGts construct with a DNA fragment corresponding to the 35S terminator sequence placed behind the GUS gene in the two constructs (Figure 1A). If transcription of the GUS gene or GUS ORF repeats generated full-length transcripts, we expected that these transcripts would contain most of the 35S terminator sequence at their 3′ ends that should be detected through RNA gel blotting using the terminator sequence as probe. As shown in Figure 4, when the total RNA isolated from the sde1-1 transformants harboring the pGts construct was probed with the terminator sequence, a major RNA species of ∼2 kb was detected and the intensities of the bands from individual transformants were generally correlated with those from the blot probed with a GUS gene fragment (Figure 4). When the total RNA isolated from the sde1-1 transformants harboring the pGGGts construct was probed with the terminator sequence, little signal was detected despite the fact that some of these transformants produced intensive bands when probed with the GUS gene fragment (Figure 4). These results indicated that a majority of transcripts in the sde1-1 transformants harboring the pGGGts construct did not contain the 35S terminator sequence at their 3′ ends.
Figure 4.
Analysis of Total and Full-Length GUS Transcripts in T1 pGts and pGGGts Transformants in the sde1-1 Mutant Background.
Total RNA was isolated from four randomly selected T1 transformants with a single-copy T-DNA insertion for each construct in the sde1-1 background and probed first with the GUS gene fragment (A). After stripping the GUS probe, the blot was rehybridized with the CaMV 35S terminator sequence (B). GUS activities for individual transformants analyzed are shown at the bottom of the blot. The ethidium bromide stain of rRNA is shown for each lane to allow assessment of equal loading.
The polyadenylation status of the transcripts was also examined in these transgenic lines harboring the pGts or pGGGts construct. Total RNA isolated from these lines and polyadenylated RNAs were isolated with oligo(dT)-cellulose and analyzed by RNA gel blotting using the GUS gene fragment as probe. As shown in Figure 5, in the poly(A)+ fraction, we detected high GUS RNA levels from the transgenic sde1-1 mutant transformants harboring the pGts constructs, consistent with the high GUS activities in these plants. In the sde1-1 transformants harboring the pGGGts construct, however, we detected little polyadenylated GUS RNA even though those transformants accumulated substantial levels of total GUS transcripts (Figure 5). Thus, a majority of the GUS transcripts in the sde1-1 mutant transformants harboring the pGGGts construct were not polyadenylated. As polyadenylation of mRNA is important both for export into cytoplasm and for translation (Eckner et al., 1991; Huang and Carmichael, 1996, 2001; Zhao et al., 1999), the unpolyadenylated nature of the majority of the GUS transcripts in the sde1-1 mutant transformants harboring the pGGGts construct accounts, at least in part, for the low GUS activities in these plants.
Figure 5.
Analysis of Total and Polyadenylated GUS mRNA in T1 pGts, pGGGts, and pG Transformants.
(A) Total GUS transcripts. Total RNA was pooled from 10 T1 transformants with a single-copy T-DNA insertion for each construct in the sde1-1 background and probed with the GUS gene fragment to determine the total GUS transcripts in the transformants. The ethidium bromide stain of rRNA is shown for each lane to allow assessment of equal loading.
(B) Polyadenylated GUS transcripts. The total RNA used for analysis of total GUS transcripts shown in (A) was mixed with oligo(dT) cellulose. After washing, polyadenylated mRNA was eluted and probed with the GUS gene fragment. The polyadenylated mRNA was reprobed with a β-tubulin gene fragment (TUB8; At5g23860) after stripping of the first probe.
Insertion of a Transcription Termination Sequence Decreases Silencing of Transgene Direct Repeats
To study consistent silencing induced by transgene direct repeats, we analyzed a third construct (pGtsGGts) that contains three GUS ORF repeats but with a 35S terminator inserted between the first and second GUS ORF repeats (Figure 1A). In both the wild-type and sde1-1 mutant backgrounds, the pGtsGGts construct gave much higher GUS activities than the pGGGts construct (Figure 3A; see Supplemental Figure 1 online). The enhanced GUS activities in the pGtsGGts transformants were correlated with increased GUS transcripts (Figure 3B; see Supplemental Figure 2 online). Thus, insertion of a transcription termination sequence after the first ORF repeat effectively abrogated efficient RNA silencing induced by transgene direct repeats. It appears that if transgene repeats are cotranscribed as a single polycistronic transcript, as in the pGGGts construct, they are effective at inducing silencing. However, if transcription is terminated after the first transgene repeat, as in pGtsGGts, they become ineffective at inducing gene silencing.
Consistent and RDR6-Dependent RNA Silencing of a GUS Transgene with No Transcription Terminator
A majority of the GUS transcripts accumulated in sde1-1 mutant transformants harboring the pGGGts construct had two abnormal structural features: three tandem repeats of the GUS ORF, and improperly terminated, unpolyadenylated 3′ ends. To determine which of these two features is more important for efficient, RDR6-dependent RNA silencing, we analyzed another construct named pG that contains a single GUS gene driven by the same enhanced CaMV 35S promoter without a transcription terminator sequence at its 3′ end (Figure 1A). Because of the lack of a terminator, we expected that transcription of the transgene would continue beyond the GUS gene and generate the transcripts without appropriately processed or polyadenylated 3′ ends. As shown in Figure 3A and Supplemental Figure 1 online, in the wild-type background, little or no GUS activities were detected in the pG transformants. In the sde1-1 mutant background, the GUS activities in all of the transformants were also very low (Figure 3A; see Supplemental Figure 1 online). Thus, the GUS activities in both the wild-type and sde1-1 mutant plants harboring the pG constructs were similar to those in the pGGGts transformants.
We again examined GUS transcripts in both the wild-type and sde1-1 mutant transformants harboring the pG construct. Wild-type plants transformed with the pG construct accumulated little of the GUS transcript, while substantial amounts were found in the sde1-1 mutant background (Figure 3B; see Supplemental Figure 2 online). Direct comparison indicated that the levels of GUS transcripts in the sde1-1 mutant transformants harboring the pG construct were lower than those in the sde1-1 mutant transformants harboring the pGts construct (Figure 3B). This observation indicated that GUS transcripts generated from the pG construct were in part degraded by an RDR6-independent mechanism. Nevertheless, the difference in the accumulation of GUS transcripts between the wild-type and the sde1-1 mutant transformants harboring the pG construct indicated that RDR6 played a significant role in the degradation of transcripts from the terminator-less GUS transgene.
In the sde1-1 mutant transformants containing the pG construct, a majority of GUS transcripts had a size of ∼2.8 kb, longer than the expected ∼2.0 kb size for the GUS gene. When polyadenylated RNA was isolated and probed with the GUS gene fragment (Figure 5), little signal was detected in the sde1-1 transformants harboring the terminator-less GUS gene. Thus, a majority of the GUS transcripts in these transformants were unpolyadenylated, as expected.
As observed with pGGGts, efficient, RDR6-dependent silencing of the pG construct is associated with the accumulation of improperly terminated, unpolyadenylated transcripts. These aberrant transcripts are also longer than those produced by pGTs, raising the possibility that the additional sequences present in these transcripts make them susceptible to silencing. To test this possibility, we analyzed a modified pGts construct (pGECts) in which the ∼0.8-kb EcoRI-ClaI DNA fragment downstream of the terminator sequences on the binary vector (Figure 1A) had been inserted between the GUS reporter gene and the 35S terminator. Because of the insertion of the DNA fragment, we expected that transcription of pGECts would generate transcripts with size and sequence similar to those of the readthrough transcripts from the pG construct. Unlike transcripts from pG, however, transcripts from pGECts are expected to be properly terminated and polyadenylated due to the 35S terminator at the 3′ end. As shown in Figure 3A, the wild-type and sde1-1 transformants harboring the pGECts construct had average GUS activities of 89 and 159 units, respectively. Although these GUS activities were lower than those from the pGts transformants, they were ∼10 fold higher than those from the pG transformants. Unlike the wild-type transformants harboring the pG construct, a large percentage of the wild-type transformants harboring the pGECts construct accumulated GUS transcripts (Figure 3B; see Supplemental Figure 2 online). These results suggest that proper termination of transgene transcripts reduced their susceptibility to RDR6-mediated RNA silencing.
Silencing of an Expressing GUS Transgene by Triple Repeat and Terminator-Less GUS Transgenes
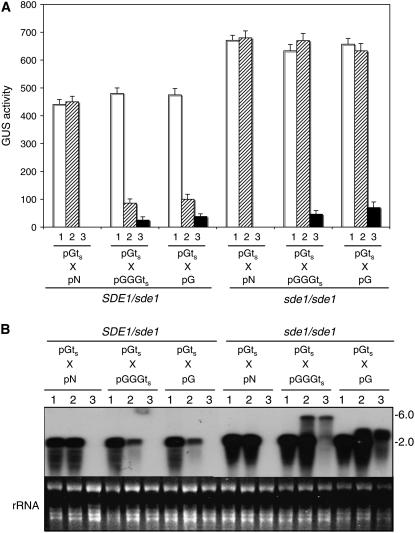
The accumulation of GUS transcripts from the pGGGts and pG constructs in the sde1-1 mutant but not in the wild-type background indicates that unpolyadenylated mRNA is targeted by RDR6-mediated silencing mechanisms. Previously, it was shown that silencing caused by CAT transgene direct repeats can inactivate a nonsilenced CAT gene in trans in the same plants generated through genetic crossing or double transformation (Ma and Mitra, 2002). To determine whether the triple repeat and terminator-less GUS transgenes can also inactivate a homologous nonsilenced GUS transgene, we crossed a nonsilenced wild-type pGts line with an sde1 pGGGts or pG line. As a control, the same nonsilenced wild-type pGts line was also crossed with an sde11 line harboring a terminator-less bacterial nahG gene (pN) (Figure 1). The resulting F1 progeny were all in the SDE1/sde1 genetic background, and those harboring pGts alone or both pGts and a silencing construct (pGGGts, pG, or pN) were identified by PCR genotyping and analyzed for both GUS activities and GUS transcripts. As shown in Figure 6, transgenic pGts plants with or without pN had very similar levels of GUS activities and GUS transcripts. On the other hand, the GUS activities in the transgenic pGts plants were reduced by >80% by the presence of the pGGGts or pG construct (Figure 6A). The reduction in GUS activities in these plants was correlated with reduced levels of GUS transcripts (Figure 6B). Thus, the triple repeats and terminator-less GUS transgenes inactivated a nonsilenced GUS reporter gene in trans.
Figure 6.
GUS Activities and GUS mRNA Accumulation in Arabidopsis pGts Transformants with or without the pGGGts, pG, or pN Construct.
F1 progeny in the silencing-competent SDE1/sde1 or silencing-deficient sde1/sde1 background were generated from crosses between a single-copy paternal wild-type or sde1 pGts transformant and a single-copy maternal sde1 pGGGts, pG, or pN transformant. F1 progeny were genotyped by PCR, and plants containing one or both constructs from their parental lines were identified. Means and se of GUS activities (A) and levels of GUS transcripts (B) were determined from 10 F1 progeny for each genotype containing only the paternal pGts construct (column 1 and lane 1), only the maternal pGGGts, pG, or pN construct (column 3 and lane 3), or both the paternal pGts construct and a corresponding maternal construct (column 2 and lane 2). GUS activities are expressed in units (nanomoles of 4-methylumbelliferone per minute per milligram of total soluble protein).
To determine whether the silencing of an expressing GUS gene by the triple repeats and terminator-less GUS transgenes is SDE1-dependent, we crossed the same sde1 pGGGts and pG lines with an sde1 pGts line. The F1 progeny from the crosses were still homozygous for the sde1 mutant gene and, therefore, deficient in SDE1-dependent RNA silencing. As shown in Figure 6, the resulting transgenic pGts lines with or without the pGGGts, pG, or pN construct had very similar levels of GUS activities and GUS transcripts. Thus, inactivation of a nonsilenced GUS gene by the GUS triple repeats or terminator-less GUS gene is SDE1-dependent.
Double Transcription Terminators Enhance Transgene Expression
To complement the studies with the terminator-less GUS transgene, we examined the expression of a GUS transgene that contains two terminators at its 3′ end. It has been reported that the commonly used CaMV 35S terminator was leaky when used with a transgene driven by a strong promoter (Rose and Last, 1997). For this purpose, we examined two additional constructs: pGtn and pGtstn. pGtn contains a single GUS transgene flanked by the enhanced 35S promoter at its 5′ end and a transcriptional terminator from the nos gene of Agrobacterium tumefaciens at its 3′ end (Figure 1). pGtstn contains the same enhanced 35S promoter and the GUS gene but has both the 35S and nos terminators at its 3′ end (Figure 1). As shown in Figure 3A, in pGtn-transformed wild-type plants, GUS activities were slightly lower (∼15%) than those found in the transformants harboring the pGts construct. In the sde1-1 mutant background, the pGts construct also appeared to be slightly superior over the pGtn construct (Figure 3A).
In the sde1-1 mutant transformants harboring the pGtstn construct, we observed a 1.5-fold increase in the GUS activities over those of the sde1-1 mutant transformants harboring the pGts construct (Figure 3A). Thus, adding the second nos terminator had a positive effect on the GUS activities in the sde1-1 mutant plants that are defective in RNA silencing. In the wild-type background, we observed an approximately threefold to fourfold increase in GUS activities with the double terminators when compared with the 35S or nos single terminator (Figure 3A; see Supplemental Figure 1 online). In fact, the average GUS activity in the wild-type transformants harboring the pGtstn construct was even higher than the GUS activity in the sde1-1 transformants harboring the pGts or pGtn construct (Figure 3A). We compared the PGts and PGtstn lines at very young (3 weeks old) and old (7 weeks old) stages and found similar difference between the constructs. When all of the transformants (∼300) regardless of their transgene copy number were compared, a similar threefold to fivefold increase in GUS activities was observed with the double terminators over the 35S or nos single terminator (data not shown). RNA gel blot analysis revealed that increased GUS activities in the wild-type transformants harboring the pGtstn construct were generally correlated with an increased number of transformants with high levels of GUS transcripts (Figure 3B; see Supplemental Figure 2 online). Thus, strengthening termination capacities by incorporating double terminators significantly improves transgene expression.
Readthrough of the 35S and nos 3′ Terminators in Transgenic Plants
To examine the possibility that a single terminator might not be efficient in terminating the transcription of a transgene driven by the strong 35S promoter, leading to RNA 3′ readthrough, we isolated total RNA from both wild-type and sde1-1 lines transformed with various GUS constructs and probed them with a DNA fragment corresponding to the sequence downstream of the terminator sequences on the plant transformation vector (the EcoRI-ClaI fragment in the vector shown in Figure 1A). As shown in Figure 6A, we detected little hybridization signal in the wild-type transformants harboring the pGts construct. However, significant levels of hybridization signals were detected in the sde1-1 mutant plants transformed with this construct (Figure 6A). Even greater amounts of hybridization signals were observed in the sde1-1 mutant plants transformed with pGtn (Figure 7A). These results indicated that both the 35S and nos terminators were leaky for terminating transcription of a transgene driven by the strong CaMV 35S promoter. These readthrough mRNAs were apparently targeted by RDR6-mediated silencing mechanisms as they accumulated at higher levels in the sde1-1 mutant transformants than in the wild-type plants (Figure 7A). When the total RNA isolated from the sde1-1 mutant transformants harboring the pGtstn construct was probed with the same DNA fragment, the signals were lower than those detected for the pGts and pGtn constructs (Figure 7A). This result demonstrated that the use of two terminators at the 3′ end of the GUS gene driven by an enhanced 35S promoter substantially reduced mRNA 3′ readthrough.
Figure 7.
Analysis of mRNA 3′ Readthrough in T1 Transformants Transformed with pGts, pGtn, and pGtstn.
(A) Detection of readthrough transcripts. Total RNA was isolated from T1 transformants for each construct in the wild-type (W) or sde1-1 (s) background and probed with the EcoRI-ClaI fragment corresponding to the region downstream of the GUS construct in the T-DNA part of the binary vector pOCA30 (see Figure 1A).
(B) Polyadenylation status of the readthrough transcripts. Total RNA (10 μg for pGts and pGtn and 1 μg for pGECts) from T1 sde1-1 transformants was separated and probed with the pOCA30 EcoRI-ClaI DNA fragment (top panel). Poly(A)+ mRNA was isolated from 20 μg of total RNA from the transformants. All isolated polyadenylated mRNA for pGts and pGtn and one-tenth for pGECts was fractionated and probed with the pOCA30 EcoRI-ClaI fragment and reprobed with the β-tubulin gene fragment after stripping of the first probe (bottom panel).
To determine whether these readthrough transcripts were properly terminated, we examined their polyadenylation status. As a control, we included the sde1-1 transformants harboring the pGECts construct. Transcripts from pGECts, which are expected to be largely polyadenylated based on relatively high GUS activities in the transformants, contain the same sequence at the 3′ end as readthrough transcripts from pGts and pGtn. Both total RNA and poly(A)+ RNAs were analyzed by RNA gel blotting using the EcoRI-ClaI vector DNA fragment as a probe. As shown in Figure 7B, in the poly(A)+ fraction, we detected high GUS RNA levels from the sde1-1 transformants harboring the pGECts constructs, consistent with the relatively high GUS activities in these plants. In the sde1-1 transformants harboring the pGts or pGtn construct, however, the same probe detected little polyadenylated RNA (Figure 7B). Thus, the readthrough GUS transcripts from pGts or pGtn were largely unpolyadenylated.
siRNA Accumulation
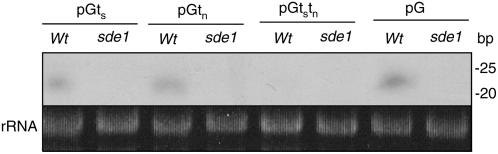
In order to determine whether the variation of GUS activities and GUS transcripts in the GUS transgene constructs differing in terminators at their 3′ end was associated with posttranscriptional gene silencing, we determined GUS-specific siRNA in these transgenic plants. Total RNA was isolated from independent transformants for each construct and separated on denatured polyacrylamide gels after enrichment for siRNA. The separated RNA was then probed with the GUS gene fragment. As shown in Figure 8, in the wild-type transformants harboring the pGts or pGtn construct, a species of RNA with an approximate molecular size of 21 to 23 bp was detected. In the wild-type transformants harboring the pGtstn construct, the level of the GUS-specific siRNA was substantially reduced (Figure 8). Little GUS-specific siRNA was detected in the sde1-1 mutants harboring these constructs (Figure 8). Thus, the enhanced levels of GUS activities and GUS transcripts in the pGtstn transformants were associated with reduced accumulation of GUS-specific siRNA, which most likely resulted from a reduction in RDR6-dependent RNA silencing of the transgene.
Figure 8.
Accumulation of siRNA in T1 Transformants Transformed with the pGts, pGtn, pGtstn, and pG Constructs.
Total RNA was isolated from 10 randomly selected T1 transformants for each construct in the wild-type or the sde1-1 background and enriched for low molecular weight RNA. After fractionation on a polyacrylamide gel, the RNA was probed with the full-length GUS gene fragment. Equivalent loading of the samples was shown by detection of 5S rRNA in the lanes prior to blot transfer.
Nuclear Localization of RDR6
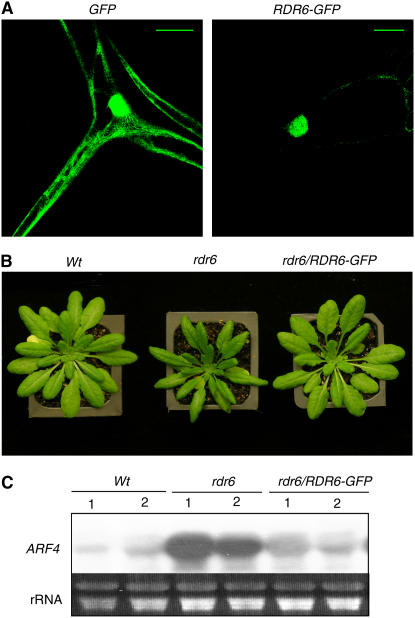
Two different GUS constructs, pGGGts and pG, produced improperly terminated, unpolyadenylated GUS transcripts that were degraded in the wild-type transformants but accumulated in the sde1-1 mutant transformants (Figure 3B; see Supplemental Figure 2 online). Since unpolyadenylated RNAs are poor substrates for export into the cytoplasm and usually accumulate in the nucleus (Eckner et al., 1991; Huang and Carmichael, 1996, 2001; Zhao et al., 1999), it was of interest to determine whether RDR6 is also localized to this subcellular compartment. For this purpose, we generated a construct of an RDR6-eGFP fusion gene driven by the CaMV 35S promoter and transformed it into the wild-type plants. However, little GFP signal was detected in these transformants, probably because of silencing as a result of overexpression of the RDR6 fusion gene. We then transformed the same construct into the sde1-1 and rdr6-11 (Peragine et al., 2004) mutant plants and found GFP signals in trichome cells of a substantial percentage of the rdr6-11 transformants. As shown in Figure 9A, the stably expressed RDR6-GFP fusion protein was localized predominantly to the nucleus of the Arabidopsis cells. By contrast, the native GFP protein, which is small enough to enter the nucleus without a targeting sequence (Keminer and Peters, 1999), was found in both the nucleus and the cytoplasm (Figure 9A).
Figure 9.
Nuclear Localization of RDR6 in Arabidopsis Seedlings.
(A) Detection of the RDR6-GFP in the nucleus of Arabidopsis trichome cells. The Arabidopsis rdr6-11 mutant plants were transformed with a 35S:RDR6-GFP or a 35S:GFP construct, and the seedlings of positive transformants were visualized by a confocal microscope. The RDR6-GFP fusion protein is predominantly localized in the nuclei of trichome cells. The low green fluorescence signals around the edges of cells containing the RDR6-GFP fusion protein may result from autofluorescence, since similar levels of signals were also observed in nontransgenic wild-type seedlings. The native GFP is localized in both the cytoplasm and the nuclei due to its small size. More than 10 T1 plants were investigated for each construct, and all showed the same fluorescence pattern. Bars = 100 μm.
(B) Leaf morphology of wild-type, sde1-1, and sde1-1 transformants harboring the 35S:RDR6-GFP construct. The photograph was taken 5 weeks after germination.
(C) Transcript levels of ARF4 in wild-type, sde1-1, and sde1-1 transformants harboring the 35S:RDR6-GFP construct.
To determine whether the tagged RDR6 is functional in Arabidopsis, we analyzed the phenotypes of the Arabidopsis rdr6-11 mutant transformants expressing the tagged RDR6. RDR6 is required for juvenile development, and the first few leaves of the rdr6 mutants are elongated and curl downward. In addition, RDR6 is required for the production of trans-acting siRNAs that target the cleavage of a number of protein-coding transcripts, including those for the AUXIN RESPONSE FACTOR gene ARF4. As a result, the transcript levels for ARF4 are increased in the rdr6 mutants. In the rdr6-11 mutant transformants expressing the tagged RDR6, the elongated and curled leaf morphology observed in the original rdr6-11 mutant mostly disappeared (Figure 9B). Furthermore, while the transcript levels of ARF4 are increased substantially in the rdr6 mutant, its transformants expressing the tagged RDR6 had ARF4 transcript levels similar to those in the wild-type plants (Figure 9C). These results indicate that the tagged RDR6 can rescue the rdr6 mutant phenotype and, therefore, is functional in the transformants.
DISCUSSION
Improperly Terminated, Unpolyadenylated Transcripts Are Targeted by RDR6-Mediated Silencing
In this study, we tested seven different GUS transgene constructs to assess whether the levels of total, productive, or unproductive transcripts of the GUS transgenes determine the frequency and magnitude of their silencing in transgenic plants. Unlike previously reported studies, we tested these constructs in both the wild-type and sde1-1 mutant backgrounds so that the extents of RDR6 dependence of the silencing of the transgenes could be assessed. In addition, for those constructs such as pGGGts and pG that generate consistent silencing, it was difficult to analyze their transcripts in the wild-type background, as they were degraded by RNA silencing (Figure 3B; see Supplemental Figure 2 online). In the silencing-deficient sde1-1 mutant background, we were able to determine the levels of the transgene transcripts and examined their unique structures associated with consistent silencing of these constructs. These experiments have generated new insights into differential susceptibilities of various types of transgene transcripts to RDR6-mediated, transgene-induced RNA silencing.
By analyzing the GUS activities in silencing-deficient sde1-1 mutant transformants harboring the pGts construct, we confirmed the positive correlation between the copy number and expression level of the transgene (Figure 2). The enhanced transgene expression observed in the sde1-1 transformants with increased copy numbers was associated with decreased transgene expression in the wild-type transformants (Figure 2), consistent with the transcript threshold model that highly expressed genes are more prone to RNA silencing than poorly expressed genes. However, as discussed earlier, highly expressed transgenes may generate not only high levels of normal transcripts but also aberrant transcripts as collateral products. The absolute levels of such aberrant RNAs will rise as the total transcript levels increase whenever genes are highly expressed, even if the probability of making aberrant transcripts remains constant as transgene expression levels increase. The levels of aberrant RNAs would increase even further if the probability of making aberrant transcripts rises because one or more error-reduction processes operate inefficiently when transcription rates are high. Therefore, the positive correlation between the expression level and the silencing of transgenes did not necessarily reveal whether it was the normal transcripts or structurally aberrant RNAs that acted as primary triggers for RDR6-mediated RNA silencing. To address this question, we analyzed additional constructs, including pGGGts and pG, and obtained important new insights.
Although the total GUS transgene transcripts from the pGGGts construct accumulated in the absence of silencing (i.e., in the sde1-1 mutant background), their levels were similar to those produced from other constructs (e.g., pGts, pGtsGGts, pGtn, and pGtstn) that showed lower degrees of silencing (Figures 3 and 8). Thus, the levels of total transcripts of the GUS gene were not correlated with the magnitude of RNA silencing, suggesting that high levels of total transcripts of a transgene are not the primary trigger for silencing. Further analysis indicated that normal, translationally competent transcripts were unlikely to function as effective triggers for silencing, since the pGGGts construct generated little polyadenylated GUS transcripts (Figure 5) but was extremely efficient in silencing (Figure 3). Instead, we found that the high efficiency of gene silencing with the triple GUS ORF repeats was associated with high levels of truncated, unpolyadenylated GUS mRNA in the transformants (Figures 3 and 5). This result suggested that it is these inappropriately generated mRNAs that might function as effective triggers for RDR6-mediated silencing.
Two additional lines of evidence from this study support this conclusion. First, when a terminator-less GUS transgene driven by the strong 35S promoter was introduced into wild-type plants, there was little accumulation of GUS transcripts in the wild-type transformants (Figure 3B; see Supplemental Figure 2 online). Transcription of a terminator-less transgene is expected to generate improperly terminated, unpolyadenylated mRNAs that are known to be unstable (Drummond et al., 1985). However, we found that in the sde1-1 mutant background, substantial levels of mostly unpolyadenylated GUS transcripts were detected in the transformants harboring the terminator-less GUS construct (Figures 3B and 5). Thus, RDR6-mediated RNA silencing appeared to play an important role in the degradation of these GUS readthrough transcripts. To determine whether it was the improper termination or the extra readthrough sequence that renders the transcripts highly susceptible to RDR6-mediated silencing, we tested a construct (pGECts) that produces properly terminated transcripts containing the same extra sequence as the readthrough transcripts from the pG construct. The wild-type transformants harboring the pGECts construct had much higher levels of GUS activities and GUS transcripts than the pG transformants (Figure 2; see Supplemental Figures 1 and 2 online). These results indicated that improperly terminated transgene transcripts were highly susceptible to RDR6-mediated RNA silencing.
Second, when the 35S and nos double terminators were placed 3′ to the 35S promoter–driven GUS transgene (Figure 1), transcriptional readthrough was reduced (Figure 7A) and GUS activities were increased by threefold to fourfold over the construct with a single terminator (Figure 3A). This increase of GUS activity was most likely caused by reduced RNA silencing, as shown by its RDR6 dependence, increased levels of GUS transcripts, and reduced accumulation of GUS-specific siRNA (Figures 3 and 8). These studies have again helped us to assess the possible association of various types of transcripts with RNA silencing. We noted that in the sde1-1 mutant background, the GUS activities for the GUS transgene with double terminators were significantly higher than those for the GUS transgene with a 35S or nos single terminator (Figure 3A). If RNA silencing was triggered whenever total or normal transcript levels surpassed a gene-specific threshold, we would expect that the construct with the double terminators would be more prone to silencing than the construct with the 35S or nos single terminator. Instead, we observed that substantially higher GUS activities (Figure 3A) and GUS transcripts (Figure 3B) from a construct with a double terminator correlated with reduced 3′ mRNA readthrough (Figure 7), leading to less production of unpolyadenylated RNAs. Thus, for a number of transgene constructs designed to enhance or reduce the production of improperly terminated, unpolyadenylated RNA, we observed a corresponding increase or decrease in RNA silencing. These results provide direct evidence that improperly terminated, unpolyadenylated RNA of the GUS transgene acts as an aberrant RNA targeted by RDR6-mediated gene silencing.
By crossing a nonsilenced pGts line with pGGGts and pG lines, we have demonstrated that direct repeats and terminator-less GUS transgenes can substantially inactivate an expressing GUS transgene in trans (Figure 6). Thus, unpolyadenylated RNAs can act as both targets and triggers of RDR6-mediated RNA silencing. It was shown recently that GUS gene–derived sequences placed between two convergent promoters produce unpolyadenylated transcripts that are highly effective at silencing an expressed GUS transgene in trans, supporting the notion that unpolyadenylated transcripts may represent highly accessible substrates for RNA silencing (Yan et al., 2006). The same study also tested other constructs that were expected to generate unpolyadenylated transcripts with varying efficiencies in silencing an expressing GUS transgene. However, the GUS sequence used in these constructs is a 300-bp GUS fragment, and we did not examine whether these constructs generated high levels of unpolyadenylated GUS transcripts, which would be required for efficient silencing of an expressing GUS gene. More recently, it was reported that mutations of genes encoding homologs of the mRNA 3′ end formation proteins CstF64, sympletin PTA1, and CPSF100 result in an enhanced silencing phenotype in Arabidopsis esp mutants (Herr et al., 2006). This study further supports the idea that defective mRNA 3′ end formation enhances RNA silencing.
While these studies have provided strong evidence that improperly terminated, unpolyadenylated transcripts are highly susceptible to RDR6-mediated silencing, they do not rule out the possibility that normal transcripts at high levels may also directly trigger silencing. It is even possible that highly expressed transgenes are prone to silencing due to the combination of aberrant RNAs and the high levels of normal transcripts generated in the transgenic plants. In this study, we tested a number of constructs efficient in producing improperly terminated, unpolyadenylated mRNA and found these constructs to be highly susceptible to RDR6-mediated silencing. To test whether high levels of normal transcripts can directly trigger silencing, one would need to take an opposite approach of designing constructs that generate no or little aberrant RNA even when the transgene is expressed at excessively high levels. This would require a good knowledge of the mechanisms by which aberrant transcripts are generated from highly expressed transgenes.
Biogenesis of Improperly Terminated, Unpolyadenylated RNAs
In the sde1-1 transformants containing the triple GUS ORF repeats, we observed an accumulation of GUS transcripts that lacked the 35S terminator sequence (Figure 4) and were unpolyadenylated (Figure 5). It is likely that these unpolyadenylated RNAs resulted from abortive elongation and premature termination of transcription caused either by the unusual structure or the length of the transgene. Abortive elongation of transcription may also occur when there is a single ORF in the transgene. Recent studies in eukaryotic model systems have shown that transcription elongation is a highly regulated process and plays an important role in the regulation of transcription (Sims et al., 2004). Once transcription has been initiated, further elongation is blocked by negative transcription elongation factors that pause RNA polymerase II (RNAP II), resulting in arrested transcription (Sims et al., 2004). The positive transcription elongation factor b (P-TEFb), which consists of a CDK9 kinase and a regulatory cyclin T subunit, can overcome the blocking and release RNAP II from the arrest through phosphorylation of the C-terminal domain of the largest subunit of RNAP II (Sims et al., 2004). When the genes encoding P-TEFb were overexpressed in human HeLa cells, transcription of a reporter gene driven by the strong cytomegavirus promoter was stimulated by ∼40-fold (Peng et al., 1998). Thus, in vivo expression of a transgene driven by a strong promoter can be greatly limited by transcription elongation factors. If similar mechanisms exist in plants, abortive transcription elongations may occur whenever the availability of transcription elongation factors becomes limiting, for example, when transgenes are driven by the strong CaMV 35S promoter.
Improperly terminated, unpolyadenylated RNA may also be produced from mRNA 3′ readthrough that was detected when the strong 35S promoter was used in conjunction with the 35S or nos terminator (Figure 7) (Rose and Last, 1997). Transcription termination is also a highly regulated process that involves extensive interactions between cis-acting nucleic acid elements and trans-acting protein factors. In addition, processing of eukaryotic mRNA, including 5′ capping, 3′ cleavage, and polyadenylation and splicing, starts throughout transcription elongation and functionally influences or may even be coupled with each other (Neugebauer, 2002; Proudfoot et al., 2002). Biochemical errors leading to the production of unpolyadenylated mRNA could occur at any one of these steps. In human, for example, efficient cleavage of the 3′ end of the mRNA, which is required for polyadenylation, requires 5′ capping (Adamson et al., 2005), while polyadenylation, in turn, is highly coordinately or coupled with splicing (Cooke and Alwine, 2002). In mammalian cells, the coordination between these two steps in RNA processing can involve sequences found in the last exon of pre-mRNA (Cooke and Alwine, 1996, 2002). Mutations in the 3′ splicing site of the last exon, which eliminated splicing, can inhibit polyadenylation (Niwa et al., 1990). For RNA synthesized without introns, the 5′ cap can positively affect the efficiency of polyadenylation (Cooke and Alwine, 1996). Decapped transgene mRNA has been shown to be produced in transgenic plants and functionally linked with RDR6-mediated silencing of transgenes, probably by acting directly as a template for the RNA-dependent RNA polymerase (RdRP) (Gazzani et al., 2004).
In the sde1-1 transformants harboring the pGGGts or pG construct, the accumulated transcripts were detected as distinct bands of various sizes (Figure 3B; see Supplemental Figure 2 online). The levels of the transcripts, particularly in the transformants harboring the pG construct, were also markedly lower than those harboring the pGts construct (Figure 3B; see Supplemental Figure 2 online). The reduced levels of these multiple-sized transcripts may have resulted from the action of cryptic termination sites in the constructs and/or the existence of RDR6-independent RNA decay mechanisms. Both of these mechanisms could provide additional pathways for the generation of aberrant RNAs capable of triggering RDR6-mediated silencing. Because of the polycistronic nature of pGGGts and the readthrough transcription from pG, long transcripts produced from the two constructs contain stop codons distant from their 3′ ends and, therefore, may be subject to nonsense-mediated mRNA decay (NMD). However, studies from other eukaryotic systems have shown that NMD is elicited upon alteration of the translation termination process and, therefore, is translation-dependent (Baker and Parker, 2004). Since pGGGts and pG produce mostly unpolyadenylated mRNAs that are poor substrates for translation, translation-independent mRNA decay mechanisms other than NMD are more likely to be responsible for the observed reduction of the transcripts from the constructs. These mRNA decay mechanisms could function in concert with RDR6 in the genome surveillance mechanisms. For example, the intermediate degradation produced from the mRNA decay mechanisms could be recognized by RDR6 for the initiation of silencing.
Targeting of Improperly Terminated, Unpolyadenylated RNA by RDR6-Mediated Silencing
Improperly terminated mRNAs are subject to degradation in an RDR6-dependent manner, probably by functioning as templates for the RNA polymerase. An important distinction between the properly and improperly terminated transcripts is the 3′ poly(A) tail. The ability of RDR6 to distinguish between those RNAs with normal 3′ ends and those without might be determined simply by the colocalization of the enzyme and its potential templates. Unlike polyadenylated RNA that is rapidly exported to cytoplasm for translation, unpolyadenylated RNA is known to accumulate in the nucleus (Eckner et al., 1991; Huang and Carmichael, 1996, 2001; Zhao et al., 1999). Likewise, localization studies using GFP fusion proteins have demonstrated that RDR6 localizes in the nucleus (Figure 9).
Cellular RdRPs are known to catalyze both primer-dependent and primer-independent synthesis of complementary RNAs (cRNAs) using single-stranded (ss)RNA molecules as templates (Schiebel et al., 1993; Makeyev and Bamford, 2002). Primer-independent synthesis of cRNA using aberrant RNAs as templates may provide a mechanism for triggering sense RNA-induced gene silencing (Baulcombe, 2004). An RdRP activity in wheat germ extract has been shown to catalyze primer-independent generation of cRNA products with the same or similar sizes as the ssRNA templates provided (Tang et al., 2003). In order to synthesize cRNA products with the same or similar sizes as the ssRNA templates, the initiation of polymerization would have to start from the 3′ ends of the templates, raising the possibility that in the absence of primers, the RdRP activity may recognize the 3′ end of ssRNA molecules (Allen et al., 2005). The 3′ end of polyadenylated mRNA is well bound by a variety of proteins, including poly(A) binding proteins, and therefore may not be accessed by RDR6 as the recognition/binding site for dsRNA synthesis. On the other hand, unpolyadenylated RNAs may not be as well protected at their 3′ ends due to the lack of poly(A) tail and become accessible for RDR6 (Baulcombe, 2004).
MicroRNA (miRNA)-mediated cleavage of a target RNA generates a truncated unpolyadenylated 5′ fragment and a truncated uncapped 3′ fragment (Allen et al., 2005). If uncapped and unpolyadenylated transcripts are targeted by RDR6-mediated RNA silencing, one would expect miRNA-mediated cleavage products of a target RNA to enter an RDR6-mediated silencing pathway. It has been reported that siRNAs corresponding to highly upregulated miRNA target genes accumulate in wild-type plants but not in the rdr6 mutant (Ronemus et al., 2006). In addition, the 0.44-kb nonpolyadenylated 5′ cleavage fragment of the trans-acting siRNA TAS2 transcript accumulates to higher levels in the rdr6 mutant than in the wild-type plants (Yoshikawa et al., 2005). Thus, miRNA-mediated RNA cleavage products, including the 5′ fragments from the TAS loci, do enter an RDR6 pathway after miRNA-mediated cleavage, but probably at a low efficiency, based on the levels of intact and cleaved target transcripts and corresponding siRNA. The low efficiency may be caused by differential subcellular localization of miRNA-mediated cleavage products in the cytoplasm and RDR6 in the nucleus (Figure 9). In addition, if uncapped and unpolyadenylated ends of these miRNA-mediated cleavage products of target transcripts are bound and, therefore, protected by miRNA and its associated protein factors, they would be poorly accessible to RDR6-mediated silencing mechanisms.
METHODS
T-DNA Construction
An EcoRI-HindIII fragment that contains the CaMV 35S promoter with double enhancers, multiple cloning sites, and a 35S terminator was excised from pFF19 (Timmermans et al., 1990) and cloned into the same sites of the Arabidopsis thaliana transformation vector pOCA28 to generate pOCA30 (Chen and Chen, 2002). A DNA fragment containing the full GUS ORF was then subcloned into the XbaI site of pOCA30 to generate pGts. A second GUS ORF with a SpeI site at its 5′ end was ligated into the BamHI and SpeI sites upstream of the first GUS ORF in pGts, resulting in a construct of double GUS ORF repeats with BamHI and SpeI sites at its 5′ end. A third GUS ORF was then ligated again into the BamHI and SpeI site of the double GUS ORF construct to generate the triple GUS ORF construct pGGGts. To generate pGtsGGts, the most upstream GUS ORF repeat in pGGGts was replaced with a GUS gene fragment containing a 35S terminator at its 3′ end. The pG plasmid was generated by deleting the 35S terminator from pGts through restriction digestion and religation. pGtn was generated by replacing the 35S terminator in pGts with the nos terminator from pBlue-GFP-TYG-nos SK (provided by J. Sheen, Harvard University). pGtstn was generated by adding the nos terminator behind the 35S terminator in pGts. pGECts was generated by inserting the EcoRI-ClaI fragment from pOCA28 between the GUS ORF and the 35S terminator in pGts.
Plant Transformation, Crossing, and Growth Conditions
All of the constructs were introduced into Agrobacterium tumefaciens strain GV3301. The stability of the constructs was verified through isolation of plasmid DNA from the Agrobacterium transformants, transfer back to Escherichia coli, and diagnostic restriction digestion of reisolated plasmid DNA. The transformed Agrobacterium cells were then used to transform both the wild-type and sde1-1 mutant plants (both in the C24 ecotype) through Agrobacterium-mediated transformation using the floral dip procedure (Clough and Bent, 1998). Seeds were screened on Murashige and Skoog medium containing kanamycin (50 mg/L) for selection of positive transformants. For each construct, 80 to 100 independent transformants in the wild-type or sde1-1 mutant background were transferred into soil. All of the transformants for the seven constructs analyzed were grown in a large walk-in growth chamber at the same time and in identical conditions (at 22°C and 120 mE·m−2·s−1 light on a photoperiod of 12 h of light and 12 h of dark). Four fully expanded leaves were harvested from each 4.5-week-old transformant, quickly frozen in liquid nitrogen, and stored at −80°C for analysis of GUS activities and GUS transcripts. GUS activities and GUS transcripts are stable in Arabidopsis leaves stored at −80°C for at least 1 month. Three additional leaves were harvested from each transformant 1 week later for the isolation of genomic DNA and DNA gel blot analysis of the copy number of T-DNA insertion in the genome.
A single-copy pGGGts, pG, or pN transformant in the sde1 background was crossed to a single-copy pGts transformant in either the wild-type or the sde1 background. The F1 progeny were genotyped by PCR to identify plants containing pGts with or without the pGGGts, pG, or pN construct. Locations and sequences of the PCR primers used in genotyping of F1 progeny are provided in Supplemental Figure 3 online.
Isolation of Arabidopsis Genomic DNA and DNA Gel Blot Analysis
For genomic DNA isolation, frozen leaves were ground to fine powder in liquid nitrogen and mixed with 500 μL of an extraction buffer consisting of 800 mM NaCl, 1% sarkosyl, 140 mM sorbitol, 22 mM EDTA, 220 mM Tris, pH 8.0, and 0.8% cetyltrimethylammonium bromide. The mixture was incubated at 65°C for 20 min and homogenized with 200 μL of chloroform. After centrifugation for 5 min, the aqueous phase was mixed with 340 μL of isopropanol and centrifuged again for 5 min. The precipitate was resuspended in 80 μL of TE buffer (10 mM Tris-HCl, pH 7.5, and 1 mM EDTA), mixed with 80 μL of 4 M lithium acetate, and incubated on ice for 20 min. After centrifugation for 10 min, the supernatant was mixed with 320 μL of ethanol and incubated on ice for 20 min. The DNA was collected after centrifugation, resuspended in TE, and extracted with phenol/chloroform three times before ethanol precipitation. To determine the T-DNA copy number, the isolated DNA (5 μg) from each transformant was digested with EcoRI-BamHI, separated on a 0.7% agarose gel, and probed with a PstI-KpnI APHII DNA fragment within the T-DNA region of pOCA28 (Figure 1A). To assess the intactness of the GUS transgene constructs in transformants, genomic DNA from single-copy transformants and plasmid DNA from corresponding GUS constructs (Figure 1) were digested with HindIII-EcoRI and separated on a 0.7% agarose gel. The blots were probed with a GUS gene fragment, and the sizes of detected GUS constructs from the transformants were directly compared with those of the GUS constructs from corresponding GUS plasmids.
GUS Protein Assay
GUS activity was measured through a 4-methylumbelliferyl-d-glucuronide substrate assay (Jefferson et al., 1987) and was expressed in units defined as nanomoles of 4-methylumbelliferon per minute per milligram of total soluble proteins.
RNA Gel Blot Analysis
For RNA isolation, leaf tissues were ground to a fine powder in liquid nitrogen, mixed with 500 μL of hot (80°C) extraction buffer (phenol: 0.1 M LiCl/100 mM Tris-HCl, pH 8.0/10 mM EDTA/1% SDS, 1:1), and homogenized with 250 μL of chloroform:isoamyl alcohol (24:1). After 5 min of centrifugation, the upper phase was mixed with 1 volume of 4 M LiCl. The mixture was stored at 4°C overnight and centrifuged for 10 min. The RNA pellets were dissolved in water and then precipitated with ethanol. Hybridization conditions were as described previously (Yu et al., 2001).
Small RNA Detection
Small RNAs were extracted as described previously (Goto et al., 2003) with minor modifications. In brief, total RNA was isolated with TRIzol (Invitrogen) following the manufacture's instructions. Polyethylene glycol was added to a final concentration of 20%, and NaCl was added to a final concentration of 1 M. After a 30-min incubation on ice, the RNA was centrifuged at 10,000g for 10 min. Low molecular weight RNAs in the supernatant were precipitated with ethanol. Small RNAs were separated through a 15% polyacrylamide/8 M urea/0.5× Tris-borate EDTA gel and transferred electrophoretically onto a nylon membrane at 250 mA for 30 min. Prehybridization was performed in PerfectHyb plus hybridization buffer (Sigma-Aldrich) at 50°C for 2 h. Random-primed 32P-labeled full-length GUS cDNA was used for overnight hybridization at 50°C. The membrane was washed three times at 50°C with 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.2% SDS for 10 min, one time with 2× SSC for 5 min, and then three times with 2× SSC and 1% SDS for 20 min.
Poly(A)+ RNA Selection
Total RNA was isolated with LiCl as described above and mixed with oligo(dT) cellulose (Sigma-Aldrich) (1.6 mg cellulose/20 μg RNA). The resin was washed two times with a binding buffer (0.5 M NaCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 0.05% SDS) and two times with a wash buffer (0.2 M NaCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 0.05% SDS). The bound RNA was then eluted with an elution buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 0.05% SDS) twice. Both elutions were combined as poly(A)+ RNA.
Subcellular Localization
To generate the RDR6-GFP fusion, a XhoI site was generated by PCR at the end of an RDR6 cDNA fragment, and the GFP gene from pBlue-GFP-TYG-nos SK was added. The fusion construct was then subcloned into pOCA30. Arabidopsis plants were transformed with the vector using the floral dip procedure, and positive transformants were identified through selection for antibiotic resistance as described for other T-DNA constructs. The T1 seedlings (9 d old) were visualized by confocal microscopy using a MRC Bio-Rad 2100 confocal microscope, and confocal images were analyzed with ImageJ 1.34 software (Le et al., 2003).
Accession Numbers
Arabidopsis Genome Initiative numbers for the genes discussed in this article are as follows: RDR6, At3g49500; TUB8, At5g23860; ARF4, At5g60450.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. GUS Activities in Arabidopsis Wild-Type and sde1-1 Background Transformed with Various GUS Transgene Constructs.
Supplemental Figure 2. GUS mRNA Accumulation in Arabidopsis Wild-Type and sde1-1 Background Transformed with Various GUS Transgene Constructs.
Supplemental Figure 3. Locations and Sequences of PCR Primers for Genotyping of F1 Progeny from Genetic Crosses between pGts Transformants and the pGGGts, pG, and pN Transformants.
Supplementary Material
Acknowledgments
We thank David Baulcombe for the sde1-1 mutant, Scott Poethig for the rdr6-11 mutant, and Jen Sheen for the GFP plasmid. We thank Dan Szymanski and Jie Le for access to and help with the confocal microscope. We also thank Sribah Roy for his help with the construction of the RDR6-GFP plasmid, Allan Caplan for critically reading the manuscript, and other members of our laboratory for help and discussion. This project was supported in part by the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service (Grant 2002-04590). This is Journal Paper 2007-18096 of the Purdue University Agricultural Research Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zhixiang Chen (zhixiang@purdue.edu).
Online version contains Web-only data.
References
- Adamson, T.E., Shutt, D.C., and Price, D.H. (2005). Functional coupling of cleavage and polyadenylation with transcription of mRNA. J. Biol. Chem. 280 32262–32271. [DOI] [PubMed] [Google Scholar]
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. [DOI] [PubMed] [Google Scholar]
- Baker, K.E., and Parker, R. (2004). Nonsense-mediated mRNA decay: Terminating erroneous gene expression. Curr. Opin. Cell Biol. 16 293–299. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Beclin, C., Boutet, S., Waterhouse, P., and Vaucheret, H. (2002). A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12 684–688. [DOI] [PubMed] [Google Scholar]
- Butaye, K.M., Goderis, I.J., Wouters, P.F., Pues, J.M., Delaure, S.L., Broekaert, W.F., Depicker, A., Cammue, B.P., and De Bolle, M.F. (2004). Stable high-level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. Plant J. 39 440–449. [DOI] [PubMed] [Google Scholar]
- Chen, C., and Chen, Z. (2002). Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, C.F., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cooke, C., and Alwine, J.C. (1996). The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol. Cell. Biol. 16 2579–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke, C., and Alwine, J.C. (2002). Characterization of specific protein-RNA complexes associated with the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 22 4579–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101 543–553. [DOI] [PubMed] [Google Scholar]
- Drummond, D.R., McCrae, M.A., and Colman, A. (1985). Stability and movement of mRNAs and their encoded proteins in Xenopus oocytes. J. Cell Biol. 100 1148–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner, R., Ellmeier, W., and Birnstiel, M.L. (1991). Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 10 3513–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani, S., Lawrenson, T., Woodward, C., Headon, D., and Sablowski, R. (2004). A link between mRNA turnover and RNA interference in Arabidopsis. Science 306 1046–1048. [DOI] [PubMed] [Google Scholar]
- Goto, K., Kanazawa, A., Kusaba, M., and Masuta, C. (2003). A simple and rapid method to detect plant siRNAs using nonradioactive probes. Plant Mol. Biol. Rep. 21 51–58. [Google Scholar]
- Han, Y., and Grierson, D. (2002). Relationship between small antisense RNAs and aberrant RNAs associated with sense transgene mediated gene silencing in tomato. Plant J. 29 509–519. [DOI] [PubMed] [Google Scholar]
- Herr, A.J., Molnar, A., Jones, A., and Baulcombe, D.C. (2006). Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. USA 103 14994–15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., and Carmichael, G.C. (1996). Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., and Carmichael, G.G. (2001). Nucleocytoplasmic mRNA transport. Results Probl. Cell Differ. 34 139–155. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, R.A., Cluster, P.D., English, J., Que, Q., and Napoli, C.A. (1996). Chalcone synthase cosuppression phenotypes in petunia flowers: Comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol. Biol. 31 957–973. [DOI] [PubMed] [Google Scholar]
- Keminer, O., and Peters, R. (1999). Permeability of single nuclear pores. Biophys. J. 77 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, J., El-Assal Sel, D., Basu, D., Saad, M.E., and Szymanski, D.B. (2003). Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr. Biol. 13 1341–1347. [DOI] [PubMed] [Google Scholar]
- Lechtenberg, B., Schubert, D., Forsbach, A., Gils, M., and Schmidt, R. (2003). Neither inverted repeat T-DNA configurations nor arrangements of tandemly repeated transgenes are sufficient to trigger transgene silencing. Plant J. 34 507–517. [DOI] [PubMed] [Google Scholar]
- Lindbo, J.A., Silva-Rosales, L., Proebsting, W.M., and Dougherty, W.G. (1993). Induction of a highly specific antiviral state in transgenic plants: Implications for regulation of gene expression and virus resistance. Plant Cell 5 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C., and Mitra, A. (2002). Intrinsic direct repeats generate consistent post-transcriptional gene silencing in tobacco. Plant J. 31 37–49. [DOI] [PubMed] [Google Scholar]
- Makeyev, E.V., and Bamford, D.H. (2002). Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol. Cell 10 1417–1427. [DOI] [PubMed] [Google Scholar]
- Metzlaff, M., O'Dell, M., Hellens, R., and Flavell, R.B. (2000). Developmentally and transgene regulated nuclear processing of primary transcripts of chalcone synthase A in petunia. Plant J. 23 63–72. [DOI] [PubMed] [Google Scholar]
- Muskens, M.W., Vissers, A.P., Mol, J.N., and Kooter, J.M. (2000). Role of inverted DNA repeats in transcriptional and post-transcriptional gene silencing. Plant Mol. Biol. 43 243–260. [DOI] [PubMed] [Google Scholar]
- Napoli, C., Lemieux, C., and Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer, K.M. (2002). On the importance of being co-transcriptional. J. Cell Sci. 115 3865–3871. [DOI] [PubMed] [Google Scholar]
- Niwa, M., Rose, S.D., and Berget, S.M. (1990). In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 4 1552–1559. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.C., Elmayan, T., Pollien, J.M., and Vaucheret, H. (1997). Systemic acquired silencing: Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Zhu, Y., Milton, J.T., and Price, D.H. (1998). Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12 755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. (2002). Integrating mRNA processing with transcription. Cell 108 501–512. [DOI] [PubMed] [Google Scholar]
- Que, Q., Wang, H.Y., English, J.J., and Jorgensen, R.A. (1997). The frequency and degree of cosuppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell 9 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus, M., Vaughn, M.W., and Martienssen, R.A. (2006). MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell 18 1559–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, A.B., and Last, R.L. (1997). Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J. 11 455–464. [DOI] [PubMed] [Google Scholar]
- Schiebel, W., Haas, B., Marinkovic, S., Klanner, A., and Sanger, H.L. (1993). RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J. Biol. Chem. 268 11858–11867. [PubMed] [Google Scholar]
- Schubert, D., Lechtenberg, B., Forsbach, A., Gils, M., Bahadur, S., and Schmidt, R. (2004). Silencing in Arabidopsis T-DNA transformants: The predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 16 2561–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims, R.J., III, Belotserkovskaya, R., and Reinberg, D. (2004). Elongation by RNA polymerase II: The short and long of it. Genes Dev. 18 2437–2468. [DOI] [PubMed] [Google Scholar]
- Smith, N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407 319–320. [DOI] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans, M.C., Maliga, P., Vieira, J., and Messing, J. (1990). The pFF plasmids: Cassettes utilising CaMV sequences for expression of foreign genes in plants. J. Biotechnol. 14 333–344. [DOI] [PubMed] [Google Scholar]
- Vaistij, F.E., Jones, L., and Baulcombe, D.C. (2002). Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krol, A.R., Mur, L.A., Beld, M., Mol, J.N., and Stuitje, A.R. (1990). Flavonoid genes in petunia: Addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eldik, G.J., Litiere, K., Jacobs, J.J., Van Montagu, M., and Cornelissen, M. (1998). Silencing of beta-1,3-glucanase genes in tobacco correlates with an increased abundance of RNA degradation intermediates. Nucleic Acids Res. 26 5176–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.B., Mourrain, P., Palauqui, J.C., and Vernhettes, S. (1998). Transgene-induced gene silencing in plants. Plant J. 16 651–659. [DOI] [PubMed] [Google Scholar]
- Wang, M.B., and Waterhouse, P.M. (2000). High-efficiency silencing of a beta-glucuronidase gene in rice is correlated with repetitive transgene structure but is independent of DNA methylation. Plant Mol. Biol. 43 67–82. [DOI] [PubMed] [Google Scholar]
- Yan, H., Chretien, R., Ye, J., and Rommens, C.M. (2006). New construct approaches for efficient gene silencing in plants. Plant Physiol. 141 1508–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, M., Peragine, A., Park, M.Y., and Poethig, R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D., Chen, C., and Chen, Z. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13 1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Hyman, L., and Moore, C. (1999). Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.