Abstract
Cellulose is synthesized by cellulose synthases (CESAs) contained in plasma membrane–localized complexes. In Arabidopsis thaliana, three types of CESA subunits (CESA4/IRREGULAR XYLEM5 [IRX5], CESA7/IRX3, and CESA8/IRX1) are required for secondary cell wall formation. We report that mutations in these proteins conferred enhanced resistance to the soil-borne bacterium Ralstonia solanacearum and the necrotrophic fungus Plectosphaerella cucumerina. By contrast, susceptibility to these pathogens was not altered in cell wall mutants of primary wall CESA subunits (CESA1, CESA3/ISOXABEN RESISTANT1 [IXR1], and CESA6/IXR2) or POWDERY MILDEW–RESISTANT5 (PMR5) and PMR6 genes. Double mutants indicated that irx-mediated resistance was independent of salicylic acid, ethylene, and jasmonate signaling. Comparative transcriptomic analyses identified a set of common irx upregulated genes, including a number of abscisic acid (ABA)–responsive, defense-related genes encoding antibiotic peptides and enzymes involved in the synthesis and activation of antimicrobial secondary metabolites. These data as well as the increased susceptibility of ABA mutants (abi1-1, abi2-1, and aba1-6) to R. solanacearum support a direct role of ABA in resistance to this pathogen. Our results also indicate that alteration of secondary cell wall integrity by inhibiting cellulose synthesis leads to specific activation of novel defense pathways that contribute to the generation of an antimicrobial-enriched environment hostile to pathogens.
INTRODUCTION
The presence of a cell wall confers many of the gross morphological characteristics of plants and is one of the features that distinguish them from animals. The cell wall is a complex composite of cellulose, high molecular weight polysaccharides, proteins, and aromatic substances that undergoes dynamic changes. In addition to providing structural support and a passive barrier against invading pathogens and pests, the cell wall controls cell expansion and is involved in the exchange of water and substances throughout plant development (Carpita and McCann, 2000). It also constitutes a reservoir of antimicrobial compounds and is a source of signaling molecules (Carpita and McCann, 2000).
Cellulose is a linear polymer of β-(1-4)–linked glucose, which is synthesized at the plasma membrane by a large membrane-bound complex whose only identified components are several structurally similar cellulose synthase (CESA) subunits (Somerville et al., 2004). Arabidopsis thaliana contains 10 CESA genes, six of which encode proteins with known functions (Somerville et al., 2004). The CESA1 (RADIAL SWELLING1 [RSW1]), CESA3 (ISOXABEN RESISTANT1 [IXR1]), and CESA6 (PRC1/IXR2) subunits are largely responsible for cellulose production during the formation of the primary cell wall in most tissues (Fagard et al., 2000; Scheible et al., 2001; Desprez et al., 2002; Somerville et al., 2004), whereas cellulose synthesis for the secondary cell wall, which takes place after the arrest of cell expansion, requires CESA4 (IRX5), CESA7 (IRX3), and CESA8 (IRX1) and a diverse set of recently identified proteins (Taylor et al., 2003; Somerville et al., 2004; Brown et al., 2005; Persson et al., 2005). Mutations in any of these six CESA genes lead to a reduced level of cellulose synthesis and consequently to modifications in the composition and structure of either the primary or the secondary cell wall (Fagard et al., 2000; Scheible et al., 2001; Desprez et al., 2002; Taylor et al., 2003; Somerville et al., 2004; Brown et al., 2005; Persson et al., 2005).
Initial evidence for a connection between modifications of the cell wall structure and stress responses came from the finding that alteration of the primary cell wall of the Arabidopsis ixr1/cev1 (for constitutive expression of VSP1) mutant, caused by partial loss of CESA3 function, leads to the constitutive activation of the jasmonate (JA) and ethylene (ET) signaling pathways and to enhanced resistance to some pathogens (Ellis et al., 2002). Also, disruption of the Arabidopsis secondary cell wall by inactivation of the CESA8/IRX1 gene in the leaf wilting2 (lew2)/irx1 mutant causes an increase of the endogenous abscisic acid (ABA) levels and an enhanced tolerance to drought and osmotic stress (Chen et al., 2005). Cell wall polysaccharide composition is also a determinant of disease development (Vogel et al., 2002, 2004; Somerville et al., 2004). For example, a search for Arabidopsis mutants with enhanced resistance to virulent strains of powdery mildew (Erysiphe cichoracearum) led to the identification of powdery mildew–resistant5 (pmr5) and pmr6 mutants, which exhibit an altered cell wall composition (Vogel et al., 2002, 2004).
Our knowledge of the plant molecular mechanisms controlling infections caused by the necrotrophic fungus Plectosphaerella cucumerina and the vascular bacterial pathogen Ralstonia solanacearum is limited. Arabidopsis resistance to the fungus depends on the ET, JA, and salicylic acid (SA) signaling pathways, whereas tolerance to the bacterium is just ET-dependent (Berrocal-Lobo et al., 2002a; Hirsch et al., 2002). Arabidopsis resistance to various strains of R. solanacearum is monogenic and depends on RRS1-R, a resistance gene present in the Niederzenz (Nd-1)–resistant ecotype but truncated in the susceptible Columbia (Col-0) ecotype (Deslandes et al., 2003). By contrast, Arabidopsis resistance to P. cucumerina is multigenic (Llorente et al., 2005). We have shown previously that the receptor-like kinase ERECTA was required for Arabidopsis resistance to both P. cucumerina and strain 14-25 of R. solanacearum (Godiard et al., 2004; Llorente et al., 2005).
To identify additional Arabidopsis genes that control the development of the infections caused by these pathogens, we searched for mutants unable to develop disease symptoms upon infection. Here, we show that mutations (irx) in any of the three Arabidopsis CESAs required for secondary cell wall formation (IRX1/CESA8, IRX3/CESA7, and IRX5/CESA4) conferred enhanced resistance to these pathogens as well as to Botrytis cinerea and powdery mildew fungi. Genetic and transcriptomic analyses indicated that irx/cesa-mediated resistance was independent of SA, ET, and JA signaling and allowed the identification in irx plants of a group of constitutively upregulated genes, which included ABA-responsive, defense-related genes. These results suggest that modifications of plant cell wall integrity could activate specific defensive pathways, which could lead to plant enhanced resistance to different type of pathogens.
RESULTS
Isolation and Characterization of ern1/irx1 and nws2/irx5 Mutants
To identify Arabidopsis genes that affect the development of the diseases caused by the necrotroph P. cucumerina and the soil-borne pathogen R. solanacearum GMI1000, we inoculated an ethyl methanesulfonate–mutagenized population with the fungus and T-DNA–tagged mutants with the bacterium, and we searched for mutants unable to develop disease symptoms. In these two independent screens, we isolated the ern1 (enhanced resistance to necrotrophs1) and nws2 (no wilt symptoms2) mutants, which showed reduced disease symptoms or remained symptomless upon infection with the fungus and the bacterium, respectively (Figures 1A and 1D). The ern1 and nws2 mutations were identified by map-based cloning and inverse PCR of T-DNA flanking sequences (see Supplemental Figure 1 online), and they were found to be impaired in the CESA8/IRX1 and CESA4/IRX5 proteins, respectively, required for secondary cell wall synthesis (Taylor et al., 2003; Brown et al., 2005; Chen et al., 2005; Persson et al., 2005). The ern1 mutant was identical to the lew2 mutant (Chen et al., 2005), in which a point mutation leads to the production of a truncated CESA8/IRX1 protein lacking the catalytic domain. The ern1/lew2 mutant was renamed irx1-6, because it was allelic to the previously described irx1-1 to irx1-5 mutants (Taylor et al., 2003; Brown et al., 2005). In the nws2 mutant, the T-DNA was inserted in the second exon of the CESA4/IRX5 gene, and no transcript could be detected (see Supplemental Figure 1 online; data not shown). The nws2 mutant was renamed irx5-5, because it was allelic to the loss-of-function mutants irx5-1 to irx5-4 (Taylor et al., 2003; Brown et al., 2005).
Figure 1.
Alteration of Secondary Cell Walls by Loss of Function of IRX/CESA Genes Reduced Arabidopsis Susceptibility to Pathogens.
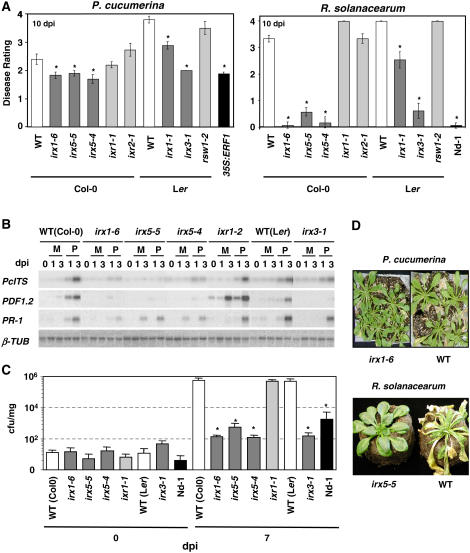
(A) Average disease rating (±sd) of wild-type plants (white bars) and mutants in secondary cell walls (irx1-6/ern1, irx5-5/nws2, irx5-4, irx1-1, and irx3-1 [dark gray bars]) or primary cell walls (ixr1-2, ixr2-1, and rsw2-1 [light gray bars]) at 10 d after inoculation (dpi) with P. cucumerina or R. solanacearum. The disease rating varies between 0 (no symptoms) and 4 (dead plants). The alleles used were in either the Col-0 or Ler genetic background. The transgenic 35S:ERF1 plants (Col-0) and the Nd-1 ecotype that showed enhanced or full resistance to P. cucumerina and R. solanacearum, respectively, were included as controls (black bars) (Berrocal-Lobo et al., 2002a; Deslandes et al., 2003). Asterisks indicate values significantly different (P < 0.01, t test) from those of wild-type plants.
(B) RNA gel blot analysis of the expression of the P. cucumerina Interspace Transcribed Sequence (Pc ITS) and of the defense genes PR1 and PDF1.2 in wild-type (Col-0 and Ler), irx, and ixr plants. Total RNA (2.5 μg/lane) was extracted from plants collected at the indicated times after inoculation with the fungus (P) or mock-inoculation (M). β-Tubulin (β-TUB) was included as a loading control.
(C) R. solanacearum GMI1000 growth (log cfu [colony-forming units]/mg fresh tissue) in wild-type (Col-0 and Ler), irx, and ixr plants at 0 and 7 d after inoculation. The resistant Nd-1 ecotype (Deslandes et al., 2003) was included for comparison. Error bars represent sd. At least 10 plants per genotype were tested. Asterisks indicate values significantly different (P < 0.01, t test) from those of wild-type plants.
(D) Disease symptoms of ixr1-6, irx5-5, and wild-type plants inoculated with P. cucumerina (12 d after inoculation) or R. solanacearum GMI1000 (10 d after inoculation).
To further demonstrate the significance of the impairment of secondary cell wall CESAs in the reduced susceptibility to both pathogens, we examined the disease responses of additional alleles of irx1 (irx1-1) and irx5 (irx5-4) as well as that of irx3-1, a null mutant of CESA7/IRX3, the third subunit required for the synthesis of secondary cell wall cellulose (Taylor et al., 2003; Somerville et al., 2004; Brown et al., 2005). Consistently, all of these mutants showed reduced susceptibility to P. cucumerina comparable to that of transgenic 35S:ERF1 plants, in which the ET and JA defensive pathways are constitutively activated by the overexpression of the ERF1 transcriptional factor, an integrator of both pathways (Figure 1A) (Berrocal-Lobo et al., 2002a). The resistance of these irx mutants to R. solanacearum was similar to that of the resistant Nd-1 ecotype (Figure 1A) (Deslandes et al., 2003). Interestingly, mutants of the three CESAs required for primary cell wall synthesis (ixr1-1/cev1, ixr2-1, and rws1-1) (Fagard et al., 2000; Scheible et al., 2001; Desprez et al., 2002; Ellis et al., 2002) were as susceptible to these pathogens as wild-type plants (Figure 1A). To verify whether the disease symptoms observed in these mutants correlated with a concomitant diminution of pathogen multiplication in planta, internal growth curves of R. solanacearum were plotted and determination of P. cucumerina biomass was performed in the inoculated plants. These analyses revealed that the reduced disease symptoms observed in the irx secondary cell wall mutants correlated with a concomitant decrease in the proliferation of these pathogens in planta (Figures 1B and 1C). By contrast, in the ixr primary cell wall mutants, the two pathogens grew to similar levels than in wild-type plants (Figures 1B and 1C). These results proved that reduction of cellulose content in secondary cell walls specifically activates Arabidopsis resistance to P. cucumerina and R. solanacearum.
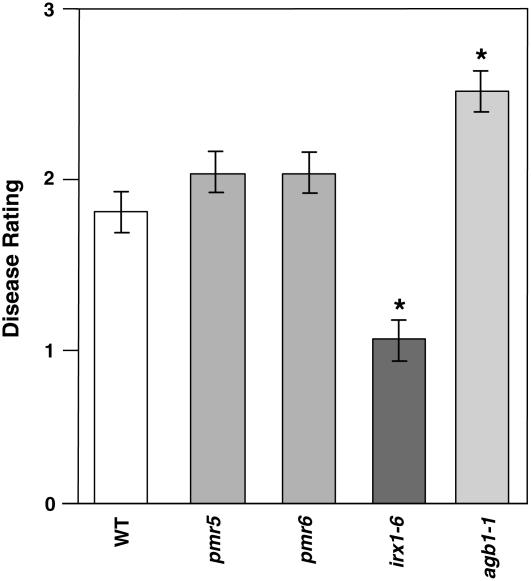
Besides P. cucumerina and R. solanacearum, we also tested the responses of the irx1-6 and irx5-5 mutant alleles to a broad range of pathogens, including oomycetes, fungi, and bacteria. These mutants were as susceptible as wild-type plants to most of these pathogens, except for the necrotrophic fungus B. cinerea and a virulent isolate of powdery mildew (Figure 2, Table 1). However, the reduced susceptibility to powdery mildew and B. cinerea may not be specific to secondary cell wall mutants, because the primary cell wall mutants ixr1/cev1 and ixr2 were also more resistant to these pathogens (Figure 2) (Ellis et al., 2002). To further determine the specificity of the disease resistance activated by the impairment of the CESA required for secondary cell wall formation, we analyzed the susceptibility to P. cucumerina and R. solanacearum of additional cell wall mutants, such as pmr5 and pmr6, that showed, like the irx mutants, powdery mildew resistance (Vogel et al., 2002, 2004). Interestingly, the susceptibility of these cell wall mutants to P. cucumerina, R. solanacearum, and B. cinerea was similar to that of wild-type plants (Figure 3; data not shown). These results illustrate the specificity of the resistance of irx/cesa mutants toward R. solanacearum and P. cucumerina and also show that alterations of different components of primary or secondary cell walls lead to enhanced resistance to different subsets of pathogens.
Figure 2.
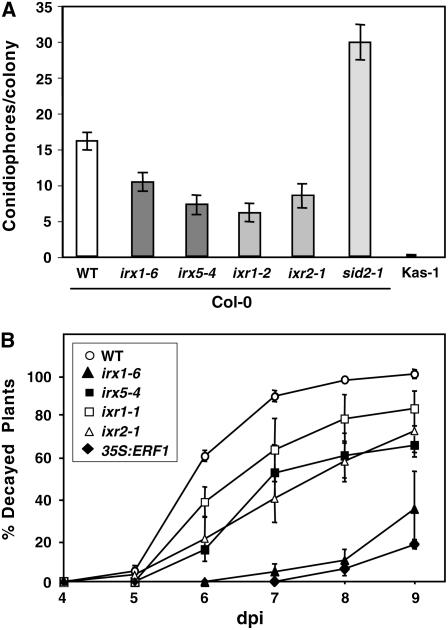
Susceptibility of irx and ixr Mutants to E. cichoracearum USC1 and B. cinerea.
(A) Quantification of E. cichoracearum growth (conidiophores per colony at 6 d after inoculation) in wild-type plants (Col-0) and irx and ixr mutants. Mean values ± sd based on 15 colonies are represented. The resistant Kashmir (Kas-1) ecotype and the susceptible sid2-1 mutant were included for comparison (Vogel et al., 2002).
(B) Percentage of wild-type (Col-0), irx, and ixr decayed plants at several days after inoculation (dpi) with 5 × 104 spores/mL B. cinerea. Data values represent averages ± sd of three independent experiments. Partially resistant, transgenic 35S:ERF1 plants were included for comparison (Berrocal-Lobo et al., 2002a).
Table 1.
Response of irx Mutants to Various Pathogens
| Pathogen | Wild Type | irx5-5 | irx1-6 |
|---|---|---|---|
| Botrytis cinerea | S | WR | WR |
| Erysiphe cichoracearum USC1 | S | WR | WR |
| Hyaloperonospora parasitica Noco2 | S | S | S |
| H. parasitica Emwa1 | HR | HR | HR |
| Pseudomonas syringae DC3000 | S | S | S |
| P. syringae DC3000 (AvrRPM1) | HR | HR | HR |
| Xanthomonas campestris pv campestris 8004 | S | S | ND |
| X. campestris pv campestris 147 | HR | HR | ND |
HR, hypersensitive response; ND, not determined; S, susceptible; WR, weak enhanced resistance.
Figure 3.
Susceptibility of pmr5 and pmr6 Cell Wall Mutants to P. cucumerina.
Average disease rating (±sd) of wild-type plants and mutants at 10 d after inoculation with P. cucumerina. The disease rating varies between 0 (no symptoms) and 4 (dead plants). At least 10 plants per genotype were tested. The agb1-1 mutant, which shows enhanced susceptibility to P. cucumerina, was included as a control (Llorente et al., 2005). Asterisks indicate values significantly different (P < 0.01, t test) from those of wild-type plants.
irx-Mediated Resistance Is SA-, ET-, and JA-Independent
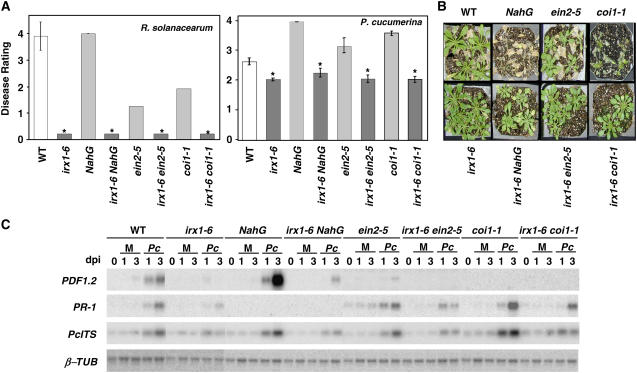
Distinct mechanisms control infection caused by the necrotroph P. cucumerina and the vascular pathogen R. solanacearum, two pathogens whose mode of colonization of host plants differ widely. Indeed, resistance to the fungus depends on the ET, JA, and SA signaling pathways, as revealed by the enhanced susceptibility to the fungus of NahG plants and sid2 mutants, which are impaired in SA signaling, of the ein2-5 mutant, which is blocked in the ET pathway, and of the coi1-1 and jar1-1 mutants, which are defective in JA signaling (Figures 4A and 4B) (Berrocal-Lobo et al., 2002a; Stein et al., 2006). By contrast, Arabidopsis tolerance to the bacterium is just ET- and JA-dependent, as disease development was reduced in ein2-5 and coi1-1 mutants compared with wild-type plants (Figure 4A) (Hirsch et al., 2002). To determine the relevance of these defense signaling pathways (Glazebrook, 2001) in irx-mediated resistance, we disabled each of them in the irx1-6 mutant by generating irx1-6 NahG lines and the irx1-6 sid2-1, irx1-6 coi1-1, irx1-6 jar1-1, and irx1-6 ein2-5 double mutants. However, the inactivation of these defense pathways did not affect the response to R. solanacearum and P. cucumerina, demonstrating that they did not contribute to the observed reduced susceptibility of irx plants (Figures 4A and 4B; data not shown). Similarly, inactivation of the defense regulator protein NPR1 (Cao et al., 1994; Glazebrook, 2001) in the double irx1-6 npr1-1 mutant did not affect irx-mediated resistance (data not shown).
Figure 4.
irx-Mediated Resistance Is SA-, ET-, and JA-Independent.
(A) Analysis of the susceptibility to R. solanacearum and P. cucumerina of irx1-6 double mutants with defects in the SA (irx1-6 NahG), ET (irx1-6 ein2-5), or JA (irx1-6 coi1-1) signal transduction pathway. Average disease ratings (±sd) of the indicated genotypes at 10 d after inoculation are represented. The disease rating varies between 0 (no symptoms) and 4 (dead plants). At least 10 plants per genotype were tested. Asterisks indicate values significantly different (P < 0.01, t test) from those of wild-type plants.
(B) Disease symptoms of P. cucumerina–inoculated plants at 12 d after inoculation.
(C) Expression analysis of SA and ET/JA marker genes in irx1-6 double mutants. RNA gel blot of PR1 and PDF1.2 defense genes in wild-type plants, single mutants, NahG transgenic plants, and double mutants. Total RNA (2.5 μg/lane) was extracted from plants collected at the indicated days after inoculation (dpi) with P. cucumerina (Pc) or treated with water (mock [M]). Blots were hybridized with the PR1, PDF1.2, and Pc ITS probes. β-Tubulin (β-TUB) hybridization was included as a loading control. Three independent experiments were performed and gave similar results.
In agreement with the infection data, the SA-responsive gene PR1 and the ET/JA-associated gene PDF1.2 were not constitutively expressed in uninfected irx plants (Figures 1B and 4C). Upon infection with the pathogens, the steady state levels of these genes were weakly increased in irx plants (Figures 1B and 4C). In the case of the R. solanacearum–inoculated plants, PR1 and PDF1.2 expression levels were similar to those found in the resistant Nd-1 plants (data not shown). Moreover, after fungal infection, the expression of PDF1.2 and PR1 in irx1-6 NahG, irx1-6 coi1-1, and irx1-6 ein2-5 plants was similar or lower than that observed in the NahG, coi1-1, and ein2-5 mutants (Figure 4C). These data demonstrated that these signaling pathways did not contribute significantly to the reduced susceptibility of the irx mutants to pathogens.
ABA-Responsive, Defense-Related Genes Are Expressed Constitutively in the irx1-6 and irx5-5 Mutants
To further investigate the mechanisms controlling irx-mediated resistance, comparative gene expression analysis of noninoculated wild-type, irx5-5, and irx1-6 plants was performed. Of the 22,000 genes tested, 301 genes were constitutively upregulated and 265 genes were constitutively downregulated in both irx1-6 and irx5-5 mutants compared with wild-type plants (see Supplemental Tables 1 and 2 online). Most of these differentially expressed genes seemed to be specific to the secondary cell wall irx mutants, as quantitative RT-PCR analysis revealed that their expression in primary cell wall ixr mutants was similar to that in wild-type plants (see Supplemental Figure 2 online; data not shown). A significant number of genes encoding cell wall–related proteins were differentially regulated in the irx mutants (see Supplemental Tables 1 and 2 online), as expected from the substantial changes in secondary cell wall structure and composition caused by the irx mutations (Taylor et al., 2003; Somerville et al., 2004; Brown et al., 2005; Persson et al., 2005).
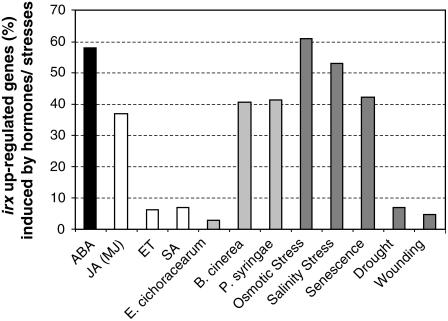
Among the constitutively irx upregulated genes, a reduced number (6 to 7%) of ET- or SA-dependent defense-related genes was identified (Figure 5), which was in agreement with the genetic and RNA gel blot expression data (Figure 4). A noticeably greater percentage (37%) of constitutively upregulated genes were found to be JA-regulated (Figure 5). However, these genes may not be essential for irx-mediated resistance, as the phenotype of the irx1-6 mutant was not affected by the JA-insensitive coi1 and jar1 mutations (Figures 4A and 4B; data not shown). Interestingly, the majority (58%) of the genes strongly activated in the irx mutants were ABA-regulated (Figure 5; see Supplemental Table 1 online). Among those, we identified genes encoding key components of the ABA biosynthetic (e.g., NCED3) and signaling (e.g., ABI1, ABI2, and ABI4) pathways (Shinozaki et al., 2003) as well as defense-related proteins (e.g., Lipid Transfer Proteins [LTPs] and ATR1 [García-Olmedo et al., 1998; Grubb and Abel, 2006]). The constitutive activation of ABA signaling in the irx mutants was in agreement with the recent demonstration that disruption of the CESA8/IRX1 gene in the lew2-1/irx1-6 allele resulted in an increased accumulation of ABA in planta and an enhanced tolerance to drought and osmotic stresses (Chen et al., 2005). Consistently, a significant number of the irx upregulated genes also responded to these abiotic stresses (Figure 5).
Figure 5.
Expression of the irx Upregulated Genes in Response to Defense Signaling Compounds and to Abiotic and Biotic Stresses.
Expression of the 301 genes upregulated in the irx5-5 and irx1-6 genes was analyzed using the Genevestigator Meta-Analyzer tools (www.genevestigator.ethz.ch/at/). The corresponding percentages were determined by selecting upregulated genes showing fold (normalized) values > 2.
Previous studies indicate that ABA functions as a negative modulator of the ET/JA-dependent defense response and as a positive regulator of JA-dependent defensive genes (Anderson et al., 2004; Lorenzo et al., 2004). In agreement with these data, some ET-associated genes were downregulated in the irx mutants compared with wild-type plants (see Supplemental Table 2 online).
Many constitutively upregulated genes in the irx mutants were defense-related genes, which responded to infection by different pathogens (e.g., B. cinerea and Pseudomonas syringae) (Figure 5). Among these pathogen-responsive genes, some encoded antimicrobial peptides belonging to four distinct families (e.g., LTPs, thionins [THs], snakin/GASA, and pEARLY), PR proteins, enzymes involved in the synthesis and activation of antimicrobial secondary metabolites (e.g., CYP79B2 and CYP79B3), as well as transcriptional regulators (e.g., ATR1) of this pathway (Table 2).
Table 2.
Defensive Genes Constitutively Upregulated in the irx5-5 and irx1-6 Mutants
| Gene Description | Locus | Normalized irx5-5/irx1-6a | References |
|---|---|---|---|
| Thaumatin, pathogenesis-related protein | At1g20030 | 9.4 | Hu and Reddy (1997) |
| Subtilase protein family | At1g20160 | 4.0 | Jorda et al. (1999) |
| Myrosinase binding protein | At1g52000 | 3.9 | Kliebenstein et al. (2005) |
| Myrosinase binding protein (MBP2) | At1g52030 | 2.9 | Capella et al. (2001) |
| Myrosinase-associated protein | At1g54020 | 11.9 | Kliebenstein et al. (2005) |
| Terpene synthase/cyclase protein | At1g61120 | 4.8 | Kappers et al. (2005) |
| pEARLY peptide | At1g62510 | 16.1 | Weyman et al. (2005) |
| Flavin monooxygenase (FMO) | At1g62540 | 4.8 | Bartsch et al. (2006) |
| Flavin monooxygenase (FMO) | At1g62570 | 6.6 | Bartsch et al. (2006) |
| Antimicrobial peptide thionin (THI2.1) | At1g72260 | 5.5 | Molina et al. (1993a); Epple et al. (1997) |
| Cytochrome P450 CYP79B3 | At2g22330 | 3.5 | Kliebenstein et al. (2005); Grubb and Abel (2006) |
| Flavin adenine dinucleotide binding domain–containing protein | At2g34810 | 6.0 | Bartsch et al. (2006) |
| Protease inhibitor/antimicrobial | At2g37870 | 14.9 | García-Olmedo et al. (1998) |
| Lipid transfer protein (LTP), putative | |||
| Jacalin lectin family protein | At3g16450 | 4.2 | Kliebenstein et al. (2005) |
| Antimicrobial peptide LTP | At3g53980 | 19.2 | García-Olmedo et al. (1998) |
| Strictosidine synthase | At3g57010 | 7.5 | Facchini and St-Pierre (2005) |
| C-S lyase (CORI3) | At4g23600 | 2.9 | Grubb and Abel (2006) |
| Antimicrobial peptide LTP | At4g33550 | 7.8 | García-Olmedo et al. (1998) |
| ELI3-2 defensive protein | At4g37990 | 5.5 | Williamson et al. (1995) |
| Cytochrome P450 CYP79B2 | At4g39950 | 2.3 | Kliebenstein et al. (2005); Grubb and Abel (2006) |
| Anthranilate synthase, α-subunit (ASA1) | At5g05730 | 3.2 | Niyogi and Fink (1992) |
| Gibberellin-regulated protein 4 (GASA4) | At5g15230 | 2.4 | Berrocal-Lobo et al. (2002b) |
| Antimicrobial peptide LTP4 | At5g59310 | 11.1 | Segura et al. (1993) |
| Antimicrobial peptide LTP3 | At5g59320 | 13.2 | Segura et al. (1993) |
| Transcriptional factor ATR1 (MYB34) | At5g60890 | 4.3 | Celenza et al. (2005) |
Normalized, average fold ratios (irx5-5/irx1-6 mutants versus wild-type plants). Genes were selected using values of P < 0.01 and raw data in the mutants >70 (see Methods).
ABA Signaling Function in irx-Mediated Resistance
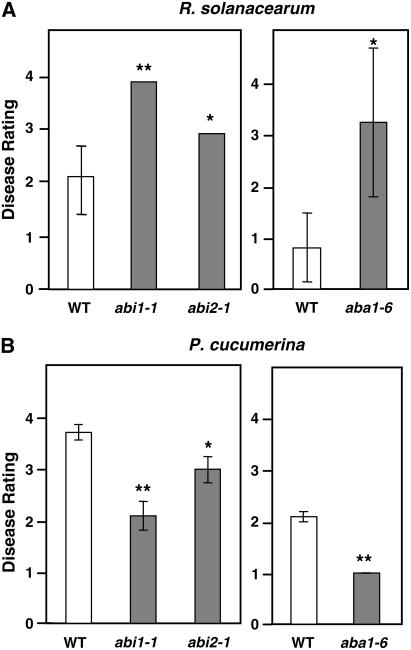
We further investigated the role of ABA signaling in the resistance to P. cucumerina and R. solanacearum by testing abi1-1 and abi2-1, two ABA-insensitive mutants impaired in genes upregulated in irx1-6/irx5-5 plants (see Supplemental Figure 2 online), as well as aba1-6, a mutant defective in ABA biosynthesis (Niyogi et al., 1998). These mutants displayed a higher susceptibility toward the bacterial pathogen (Figure 6A), which further corroborates the involvement of ABA signaling in the control of wilt disease development caused by R. solanacearum. By contrast, the disease symptoms caused by the necrotrophic fungus were reduced in the abi1-1, abi2-1, and aba1-6 mutants compared with those observed in wild-type plants (Figure 6B). The reduced susceptibility to P. cucumerina of abi1-1, abi2-1, and aba1-6 mutants was in agreement with the previously described reduced susceptibility of Arabidopsis ABA-insensitive and ABA-deficient mutants to the vascular pathogen Fusarium oxysporum and of ABA-deficient tomato (Solanum lycopersicum) plants and Arabidopsis ABA-insensitive mutants to the necrotroph B. cinerea (Audenaert et al., 2002; Anderson et al., 2004; AbuQamar et al., 2006).
Figure 6.
ABA Signaling Affects Arabidopsis Resistance to R. solanacearum and P. cucumerina.
The resistance of abi1-1, abi2-1, and aba1-6 mutants to R. solanacearum (A) and P. cucumerina (B) was evaluated. Average disease ratings (±sd) of wild-type plants (Ler and Col-0), abi1-1 and abi2-1 mutants (Ler background), and the aba1-6 mutant (Col-0 background) after inoculation with R. solanacearum GMI1000 (5 days after inoculation) or P. cucumerina (12 days after inoculation) are shown. The disease rating varies between 0 (no symptoms) and 4 (dead plants). At least 10 plants per genotype were tested. Asterisks indicate values significantly different (** P < 0.01, * P < 0.05, t test) from those of wild-type plants.
To corroborate the function of ABA signaling in irx-mediated resistance, we disabled ABA biosynthesis and ABA signaling in the irx1-1 and irx3-1 mutants by generating irx1-1 abi1-1 and irx3-1 aba3-2 double mutants. A small number of dwarf plants with pronounced developmental abnormalities, corresponding to the double homozygous mutants, were identified under high-humidity growth conditions (see Supplemental Figure 3 online; data not shown). However, these irx1-1 abi1-1 and irx3-1 aba3-2 double mutants died 2 to 3 weeks after sowing, making it impossible to test their resistance to pathogens. These results suggested that ABA signaling played a significant role in the regulation of the irx developmental phenotype.
Function of Secondary Metabolites in irx-Mediated Resistance
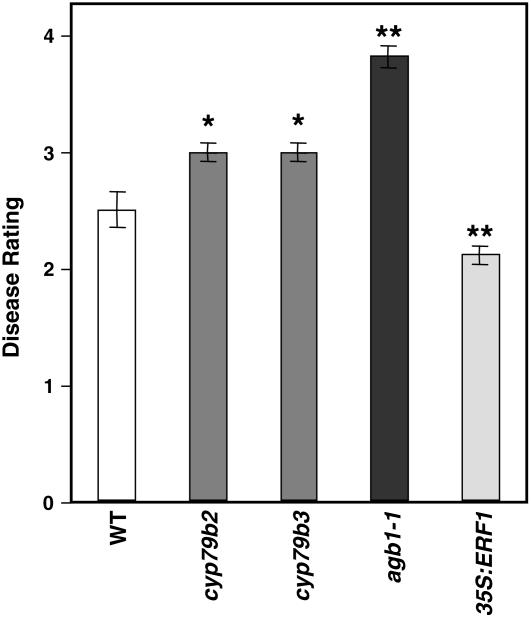
A significant number of defensive genes were constitutively upregulated in the irx/cesa mutants, including some encoding antimicrobial peptides (e.g., LTPs and THs) (Table 2; see Supplemental Figure 2 online) and enzymes involved in the synthesis and activation of secondary metabolites. Secondary metabolites, such as glucosinolates and camalexin, act as antimicrobials (Grubb and Abel, 2006) that affect the interaction between Arabidopsis and various pathogens, including the necrotrophic fungus B. cinerea (Kliebenstein et al., 2005). The synthesis of these metabolites is regulated by several enzymes (e.g., CYP79B2 and CYP79B3) and transcriptional factors (e.g., ATR1) whose corresponding genes were constitutively upregulated in the irx mutants (Table 2). To further determine the relevance of the secondary metabolites in irx-mediated resistance, we analyzed the susceptibility to P. cucumerina of the defective cyp79b2 and cyp79b3 mutants and found that they were slightly, although significantly, more susceptible to the fungus than wild-type plants (Figure 7). The susceptibility of cyp79b2 and cyp79b3 was lower than that of the highly susceptible agb1-1 mutant, which is altered in the β-subunit of the heterotrimeric G protein (Figure 7) (Llorente et al., 2005). Based on these results, the simplest hypothesis to explain irx-mediated resistance is that the secondary metabolites accumulated in the irx mutants may create a hostile and antimicrobial-enriched environment for the pathogen, hindering its multiplication.
Figure 7.
Secondary Metabolites Are Required for Arabidopsis Resistance to the Necrotrophic Fungus P. cucumerina.
Average disease ratings (±sd) of wild-type plants and of the cyp79b2 and cyp79b3 mutants, which are impaired in secondary metabolite biosynthesis, at 10 d after inoculation with P. cucumerina are shown. The disease rating varies between 0 (no symptoms) and 4 (dead plants). At least 10 plants per genotype were tested. The agb1-1 mutant and the transgenic 35S:ERF1, which show enhanced susceptibility and resistance to P. cucumerina, respectively, were included for comparison (Berrocal-Lobo et al., 2002a; Llorente et al., 2005). Asterisks indicate values significantly different (** P < 0.01, * P < 0.05, t test) from those of wild-type plants.
DISCUSSION
The role of the cell wall in resistance and/or disease development has been highlighted by several studies (Schulze-Lefert, 2004; Somerville et al., 2004). Apart from the previously mentioned ixr1/cev1, pmr5, and pmr6 mutants, which do not support the growth of two powdery mildew species (Ellis et al., 2002; Vogel et al., 2002, 2004), inactivation of a cell surface arabinogalactan protein, RAT1, and of a putative glycosyltransferase, RAT4, confers enhanced resistance to Agrobacterium tumefaciens infection (Zhu et al., 2003). In this study, we demonstrated that the irx mutants, impaired in the CESA proteins required for the synthesis of secondary cell wall cellulose, show an enhanced resistance to fungal (P. cucumerina, B. cinerea, and powdery mildew) and bacterial (R. solanacearum) pathogens. The reduced susceptibility of irx/cesa mutants to these pathogens was SA-, ET-, and JA-independent, like that of pmr5 and pmr6 mutants (Vogel et al., 2002, 2004). By contrast, the enhanced resistance to powdery mildew of the primary cell wall mutant cev1/ixr1, which is impaired in the CESA3 protein, depended on a concomitant activation of the JA and ET pathways (Ellis et al., 2002). Interestingly, the susceptibility to P. cucumerina and R. solanacearum of CESA primary cell wall mutants, including cev1/ixr1, and of pmr5 and pmr6 mutants was similar to that of wild-type plants (Figures 1 and 3). These results illustrate the high level of specificity and complexity of the resistance associated with the impairment of primary and secondary cell wall integrity caused by the irx, ixr, and pmr mutations.
Most of the cell wall mutants present phenotypic alterations, which reveal the importance of the corresponding genes for plant development (Somerville et al., 2004). Developmental alterations have also been described for the majority of Arabidopsis mutants exhibiting a constitutive activation of disease resistance (Glazebrook, 2001). In the pmr6 and pmr5 mutants, a pectin component appears to be altered, and both mutants are reduced in stature compared with wild-type plants (Vogel et al., 2002, 2004). Similarly, mutations in the CESA/IRX genes cause significant modifications of the structure and noncellulosic carbohydrate composition of the cell wall, which lead to widespread morphological alterations, including the collapse of xylem vessels, that might affect pathogen attachment and spread within the plants (Taylor et al., 2003; Brown et al., 2005). This possibility is rather unlikely, because (1) microscopic examination showed that pathogen attachment was not impaired in irx mutants (data not shown); (2) bacteria inoculated by dipping wounded roots in a microbial suspension could be detected within minutes in the aerial parts of irx mutants (Figure 1D); (3) the induction of bacterial hrp genes, which is required for disease development (Aldon et al., 2000), was unaffected in the irx mutants (data not shown); and (4) the resistance to P. cucumerina of the gpx1 mutant, which presents severe alterations in xylem vessels (Turner and Hall, 2000), did not differ from that of wild-type plants (data not shown).
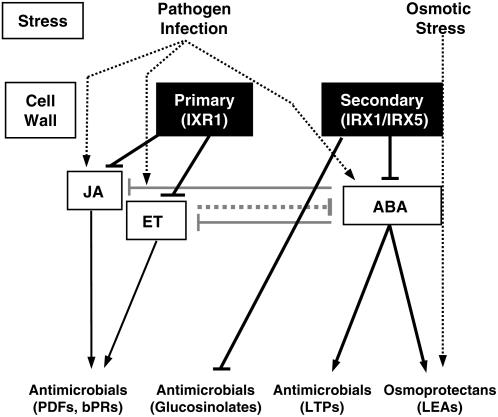
Disruption of the CESA8/IRX1 gene in the irx1-6/ern1/lew2-1 allele leads to increased endogenous ABA levels in planta and to enhanced tolerance to drought and osmotic stresses (Chen et al., 2005) (Figure 8). Consistent with these data, transcriptomic analyses revealed that a majority (58%) of the constitutively upregulated genes in the irx mutants were ABA-responsive (Figure 5). Among the proteins encoded by these ABA-regulated genes, we identified enzymes and regulators of the ABA biosynthetic and signaling pathways (e.g., NECD3, ABI1, ABI2, and ABI4) (Shinozaki et al., 2003) as well as antimicrobial, defense-related peptides (e.g., LTPs) (Molina et al., 1993b) and transcriptional regulators of secondary metabolite biosynthesis (e.g., ATR1) (Grubb and Abel, 2006). ABA plays a significant role in the control of plant development and the adaptive response to osmotic stresses (Shinozaki et al., 2003). Also, ABA modulates disease resistance by interfering with biotic stress signaling. Thus, ABA can act as a negative regulator of the ET/JA-dependent defense response and as a positive modulator of JA-dependent defensive genes (Anderson et al., 2004; Lorenzo et al., 2004). In agreement with these data, some ET-associated genes were constitutively downregulated in the irx mutants (see Supplemental Table 2 online), and upon infection, the induction of ET/JA-regulated genes, such as PDF1.2, was weaker in the irx mutants than in wild-type plants (Figures 1B and 4C).
Figure 8.
Scheme of the Interconnections between Alteration of Cell Wall Integrity and Hormone Signaling Pathways in the Regulation of Arabidopsis Resistance to Pathogens and Osmotic Stress.
Pathogen infection and abiotic stresses lead to the activation of signaling pathways (ET, JA, or ABA) that regulate the expression of different groups of antimicrobials and osmoprotectant compounds. Antagonistic interactions between the ET/JA and ABA signaling pathways have been described (Anderson et al., 2004; Lorenzo et al., 2004). Alteration of primary cell walls caused by mutations (ixr1) in the IXR1 protein leads to the constitutive activation of the ET/JA pathway, which controls the expression of antimicrobial proteins, such as defensins (PDFs) and basic PR proteins (bPRs) (Ellis et al., 2002). Modifications of the secondary cell walls caused by the impairment of IRX1 and IRX5 proteins, as occurs in the irx/cesa mutants, triggers ABA signaling and may lead to the accumulation of antimicrobial peptides (e.g., LTPs), secondary metabolites (e.g., glucosinolates), and osmoprotectants (e.g., late embryogenesis–associated proteins [LEAs]). Arrows indicate activation, and broken arrows indicate negative regulation.
The involvement of ABA in primed callose production at the site of pathogen penetration is one of the few examples of a positive function of this hormone in defense and may be the basis for the described β-aminobutyric acid–induced resistance (Ton and Mauch-Mani, 2004; Mauch-Mani and Mauch, 2005; Ton et al., 2005). However, callose deposition at the site of P. cucumerina infection was similar in irx1-6 mutant and wild-type plants, which excludes priming for callose production as a mediator of irx-reduced susceptibility (data not shown). We now provide strong evidence for a direct involvement of ABA signaling in the control of Arabidopsis resistance to R. solanacearum. This ABA function was supported by different observations: (1) the ABA-insensitive mutants abi1-1 and abi2-1, and the ABA-deficient mutant aba1-6, were more susceptible to the bacterium than were wild-type plants (Figure 6); (2) the constitutive expression in the irx mutants of some ABA signaling regulators, including ABI1-1 and ABI2-1, was associated with an increased resistance to the bacterium (Figure 1, Table 2); and (3) the ein2-1 mutant, a phenotypic suppressor of abi1-1, was shown to be more tolerant to virulent strains of the bacterium (Baudoin et al., 2000; Hirsch et al., 2002) (Figure 4A).
ABA was recently implicated as a regulator of defense responses to necrotrophic pathogens. Thus, ABA-deficient mutants exhibited reduced susceptibility to the necrotroph B. cinerea and the vascular pathogen F. oxysporum (Audenaert et al., 2002; Anderson et al., 2004) and ABA-sensitive mutants showed increased susceptibility to B. cinerea (AbuQamar et al., 2006). In agreement with these results, we previously demonstrated that the ABA-hypersensitive mutant agb1-1 (Pandey et al., 2006) was more susceptible than wild-type plants to these fungi and P. cucumerina (Llorente et al., 2005). We now demonstrate that the constitutive activation of ABA signaling in the irx mutants, or the impairment of the ABA pathway, as it occurs in the abi and aba mutants, led to an enhanced resistance to P. cucumerina via the activation of separate pathways, which seem to regulate distinct sets of defense genes (Figure 8). In the irx mutants, ABA-regulated genes encoding antimicrobial peptides (e.g., LTPs and THs) (Molina et al., 1993b) or regulators of secondary metabolite biosynthesis (e.g., ATR1) (Grubb and Abel, 2006) were constitutively expressed. The accumulation of these antimicrobial peptides/metabolites in these mutants may explain their enhanced resistance to pathogens (Figure 8). Additionally, we have observed that in the abi and aba mutants some ET/JA-regulated, defense-related genes (e.g., PDF1.2) were overexpressed (data not shown), in agreement with the proposed function of ABA as a negative modulator of the ET/JA-dependent defense response (Anderson et al., 2004; Lorenzo et al., 2004). This observation may explain the reduced susceptibility of these abi and aba mutants to P. cucumerina (Figure 8). Indeed, we have demonstrated previously that the constitutive activation of the ET and JA pathways by overexpression of the ERF1 transcriptional factor was sufficient to confer enhanced resistance to necrotrophs (Figures 1A and 7) (Berrocal-Lobo et al., 2002a).
The previous observations implicating ABA as a regulator of defense responses, those reported here, and the recent demonstration that stomata have an important role in defense against the bacterium P. syringae (Melotto et al., 2006) suggest that the ABA pathway, in addition to mediating the adaptative response to osmotic stress, may play a significant role in the fine-tuned regulation of the plant immune system. Examples of positive and negative crosstalk between biotic and abiotic stresses are emerging (Park et al., 2001; Mengiste et al., 2003). Interestingly, the enhanced resistance to biotic and abiotic stresses mediated by irx/cesa mutations was JA- and ET-independent.
Based on the transcriptomic analysis, the simplest hypothesis to explain irx-mediated resistance is that the antimicrobial compounds that might accumulate in these mutants (e.g., peptides, PR proteins, and secondary metabolites) create a hostile environment for pathogens, whose progression within the plant is hindered. Secondary metabolites have been suggested to constitute one of the determinants of nonhost and basal resistance to several pathogens, including P. cucumerina (Lipka et al., 2005; Stein et al., 2006), and to affect the interaction between Arabidopsis and the necrotrophic fungus B. cinerea (Kliebenstein et al., 2005). Also, the defensive functions of some plant antimicrobial peptides encoded by genes upregulated in the irx mutants are well established (Table 2) (García-Olmedo et al., 1998). Indeed, THs and LTPs, including those encoded by the irx-upregulated LTP3 and LTP4 genes (Table 2), possess in vitro antibiotic activity, and their constitutive overexpression in Arabidopsis increases plant tolerance to various pathogens (Molina et al., 1993a, 1993b; Segura et al., 1993; Epple et al., 1997; Molina and García-Olmedo, 1997; Titarenko et al., 1997). Our hypothesis is supported by the demonstration of an enhanced susceptibility to P. cucumerina of the cyp79b2 and cyp79b3 mutants, which are impaired in the accumulation of antimicrobial secondary metabolites (Figure 8). Moreover, we purified and tested the in vitro antibiotic activity of Arabidopsis LTPs encoded by genes upregulated in the irx mutants and found that they were active against P. cucumerina and R. solanacearum GMI1000 (data not shown). The activity of these LTPs was, as described previously for other pathogens, essentially equivalent to that of other plant LTPs (e.g., EC50 values for P. cucumerina of 3.2 ± 0.7 μM for barley [Hordeum vulgare] LTPs [LTP3 + LTP4] and 6.2 ± 0.9 μM for Arabidopsis LTPs [Molina et al., 1993b; Segura et al., 1993]).
In yeast, there is a mechanism controlling cell wall integrity that is regulated by the WSC genes (for cell wall integrity and stress response), which encode integral membrane proteins acting as surface sensors responding to environmental stress (Philip and Levin, 2001; Merchan et al., 2004). Our data suggest that a similar sensing system to that of yeast may exist in plants, because modifications of the integrity of the primary or secondary cell wall activate different signaling pathways, at least partially ABA-dependent in the irx mutants, ET/JA-dependent in the ixr mutants, and still uncharacterized in the pmr5 and pmr6 mutants (Figure 8). Although the precise functions of most genes exhibiting altered expression in the irx mutants remain to be determined, some may encode putative sensors/receptors of cell wall integrity (see Supplemental Table 1 online). The characterization of plant sensors involved in the detection of pathogen-induced perturbations of cell wall integrity, and the identification of their corresponding ligands and downstream effectors, are future challenges in the plant resistance field.
METHODS
Plant Lines and Growth Conditions
Arabidopsis thaliana accessions used in this study were Col-0, Landsberg erecta (Ler), Col-5 (a glabrous [gl] derivative of Col-0), Nd-1, and Kas-1. The primary and secondary cell wall mutants ixr1-1, ixr2-1, rsw2-1, irx1-1, irx3-1, and irx5-4 analyzed here have been described previously (Fagard et al., 2000; Scheible et al., 2001; Desprez et al., 2002; Ellis et al., 2002; Taylor et al., 2003; Brown et al., 2005; Persson et al., 2005). The ABA mutants aba1-6 (Col-0 background), abi1-1, abi2-1, and aba3-2 (Ler background) analyzed in this study have been characterized previously (Niyogi et al., 1998; Gosti et al., 1999; Xiong et al., 2001). Plants were grown in growth chambers at 20 to 22°C with a 10-h photoperiod and a light intensity of ∼150 μE·m−2·s−1, as described previously (Berrocal-Lobo et al., 2002a; Deslandes et al., 2003). For infection experiments with Erysiphe cichoracearum (Golovinomyces cichoracearum), plants were grown in growth chambers at 22°C with a 14-h photoperiod of ∼125 μE·m−2·s−1 in the 400- to 700-nm range.
Ralstonia solanacearum GMI1000 is a wild-type strain isolated from tomato (Solanum lycopersicum). R. solanacearum was grown at 28°C in B broth medium (Boucher et al., 1985). The ascomycete fungus Plectosphaerella cucumerina and the necrotroph Botrytis cinerea were grown at 28°C on potato (Solanum tuberosum) dextrose agar and their spores harvested as reported (Berrocal-Lobo et al., 2002a; Llorente et al., 2005). Pseudomonas syringae pv tomato DC3000 and P. syringae pv tomato DC3000 (AvrRPM1) were grown on nutrient broth at 28°C as described previously (Berrocal-Lobo et al., 2002a). Powdery mildew (E. cichoracearum UCSC1) was cultured and applied to Arabidopsis as described (Vogel and Somerville, 2000). Hyaloperonospora parasitica (Peronospora parasitica) isolates Noco-2 and Emwa1 were maintained on the genetically susceptible Arabidopsis accessions Col-0 and Ler, respectively, as described (Dangl et al., 1992). Xanthomonas campestris pv campestris bacteria were grown as described previously (Lummerzheim et al., 2004).
Pathogenicity Assays
The pathogens used in this study were P. cucumerina, P. syringae pv tomato DC3000 (AvrRPM1), and P. syringae pv tomato DC3000 (Berrocal-Lobo et al., 2002a), B. cinerea (Llorente et al., 2005), R. solanacearum GMI1000 (Deslandes et al., 1998), the isolates Noco2 and Emwa1 of H. parasitica (Dangl et al., 1992), E. cichoracearum UCSC1 (Vogel and Somerville, 2000), and the strains 8004 and 147 of X. campestris pv campestris (Lummerzheim et al., 2004).
Plant infection with R. solanacearum was done as reported by inoculating roots of 4-week-old plants with the bacterial suspension (Deslandes et al., 2003). Bacterial in planta growth curves were measured as described (Deslandes et al., 1998). Infection with P. cucumerina was performed by spraying 4-week-old plants with a spore suspension (2 × 106 spores/mL) of the fungus. After inoculation, plants were kept under the same growth conditions and the average disease rating (±sd) was measured at different times (days after inoculation) as reported (Llorente et al., 2005). Disease rating was as follows: 0, no symptoms, 1, 1 to 3 leaves showing some necrosis; 2, 4 to 8 leaves with some necrosis; 3, 8 to 12 leaves showing necrosis; 4, all leaves showing profuse necrosis or decayed/dead plant. At least 10 plants per genotype were inoculated in each of two to three experiments, and the disease rating means and sd were estimated at different times.
Plant inoculation with B. cinerea and disease resistance analysis were done as described (Llorente et al., 2005). Arabidopsis plants were dip-inoculated with P. syringae pv tomato DC3000 (AvRPM1) or P. syringae pv tomato DC3000, and in planta bacterial growth was determined at different times (Berrocal-Lobo et al., 2002a). Disease resistance analysis of H. parasitica was performed by spraying 2-week-old plants with a conidiophore suspension (4 × 104 spores/mL) of the isolates Noco2 or Emwa1, and the degree of infection was quantified as described (Llorente et al., 2005). For E. cichoracearum infections, 3-week-old Arabidopsis plants were inoculated as described above, phenotypes were monitored visually at 4, 5, 6, and 7 d after inoculation, and the number of conidiophores per colony was determined at the end of the experiment (Vogel and Somerville, 2000). For X. campestris infection, plants were kept at high humidity for 12 h before inoculation, and then the abaxial side of the leaves was injected with a bacterial suspension of 2 × 107 to 108 colony-forming units/mL using a blunt syringe (Lummerzheim et al., 2004). At least 10 plants per genotype were inoculated in each of two to three experiments performed.
Statistical analysis of the pathogenicity experiments was performed using a two-tailed Student's t test.
In Vitro Inhibition Tests
Arabidopsis LTPs and barley (Hordeum vulgare) LTPs (a mixture of LTP3 and LTP4) used for the in vitro inhibition assays were purified as described previously (Molina et al., 1993a; Segura et al., 1993). Pathogen inhibition was done on microtiter plates as reported (Molina et al., 1993b) by mixing 33 μL of the corresponding peptide solution with 66 μL of medium containing the fungal spores or bacteria (104 colony-forming units or spores/mL) to give the desired peptide concentrations. The experiment was repeated at least two times.
Microscopic Analysis
Four-week-old irx1-6 and wild-type plants were mock-inoculated or challenged, as indicated above, with a spore suspension of P. cucumerina. Callose staining at different times (0, 1, 3, and 4 d after inoculation) was performed as reported previously (Llorente et al., 2005). At least 10 plants per genotype were analyzed, and the experiment was repeated three times.
Identification and Cloning of the nws2/irx5-5 Gene
Thirty-five thousand T-DNA–tagged Col-5 plants were inoculated with the virulent GMI1000 bacterium, and plants that did not develop wilt symptoms were selected and allowed to set seed. Cosegregation analysis in the F2 population resulting from the nws2 backcross indicated that the resistant phenotype was tightly linked to the T-DNA insertion. The cloning of the nws2/irx5-5 gene was performed by inverse PCR as described (http://www.darmouth.edu/tjack/cloning_flanking_DNA_from_.html). Primers used to amplify the right border were as follows: 5′-TCGGGCCTAACTTTTGGTG-3′ (adjacent right border) and 5′-AGTGCCAAGCTTGCATGC-3′ (HindIII site in the polylinker). Genomic DNA (200 ng) was digested with various enzymes (XbaI, SalI, BamHI, and PstI). For each digestion, 20 ng was used in a ligation reaction. PCR was performed using the following conditions: 94°C for 20 s, 54°C for 30 s, and 68°C for 10 min (35 cycles). An 8-kb fragment was obtained using, as a template, the self-ligated XbaI fragments.
Isolation and Map-Based Cloning of the ern1/irx1-6 Mutant
Approximately 80,000 10-d-old plants of an ethyl methanesulfonate M2 mutagenized population of Arabidopsis Col-0 were sprayed with 2 × 106 spores/mL P. cucumerina fungus for the identification of ern mutants, which showed reduced disease symptoms compared with Col-0 wild-type plants. This screen yielded the recessive ern1-1 mutant. The ERN1 gene was mapped using an ern1-1 (Col-0) × Ler F2 population of 430 individuals using standard PCR-based marker techniques (Lukowitz et al., 2000). New markers within the fine-mapping interval on chromosome IV were generated using the CEREON database of Col and Ler polymorphisms (http://www.arabidopsis.org/browse/Cereon/index.jsp). The derived oligonucleotide sequences used for mapping were as follows: F15J5A, 5′-TCTAAGCGGGTCGGGTCGATTC-3′ and 5′-AATCAGACCGGACCGGACCGG-3′; F28J12, 5′-CAAATCCATACATCTCTCCCTCTC-3′ and 5′-GATGGAGTAGATAGAGAAAGGGAG-3′; F28A21A, 5′-AGGCATTCTTGCTAAGGGC-3′ and 5′-AATTCACGTGGGACTTGGG-3′; F28A21B, 5′-AAAAGTTACGAGTCAGTTACATC-3′ and 5′-GTTGGTGATGAATGATGATG-3′; F28A21C, 5′-CTTTGAGGTTAGCAAAAACTCAC-3′ and 5′-CATTGAACTCTTTTCACATGGCAG-3′; and T16H5, 5′-ATTCTAGCTTTTGAGAGGTCAG-3′ and 5′-CCCAATCCAAATCAAATTCC-3′.
Generation and Selection of Double Mutants
The mutant alleles used for the construction of double mutants with irx1-6 were coi1-1 (Feys et al., 1994), npr1-1 (Cao et al., 1994), ein2-5 (Roman et al., 1995), NahG (Lawton et al., 1995), jar1-1 (Staswick et al., 1992), sid2-1 (Wildermuth et al., 2001), abi1-1 (Gosti et al., 1999), and aba3-2 (Xiong et al., 2001). All of the mutant lines were in the Col-0 background except coi1-1, which was in the Col-6 background, and abi1-1 and aba3-2, which were in the Ler background. We generated the double mutants by standard genetic crosses, following the mutations in the mutant alleles (npr1-1, sid2-1, coi1-1, jar1-1, aba3-2, and abi1-1) or the transgene (NahG) by PCR-based methods, or by selection on Murashige and Skoog plates containing 10 μM 1-aminocyclopropane-1-carboxylic acid, the ET precursor (ein2-5 [Roman et al., 1995]), 50 μM JA (coi1-1 and jar1-1 [Feys et al., 1994]), or 1 μM ABA (abi1-1 [Gosti et al., 1999]). The NahG, coi1-1, sid2-1, npr1-1, and jar1-1 oligonucleotides used for PCR-based detection have been described (Stein et al., 2006). Genotyping of the irx1-6 mutation in all of the double mutants was confirmed by PCR amplification and sequencing of the mutation (oligonucleotides 5′-GGTTAATAGGAGACCATTTAG-3′ and 5′-TCCATCCAAATCTCAATCCC-3′). As described previously (Brown et al., 2005; Chen et al., 2005), the homozygous irx1-6 and irx5-5 plants were sterile; therefore, for the disease resistance and expression analysis, heterozygous populations were sown on soil and the corresponding irx1-6 and irx5-5 single or double mutants that showed a clear distinguishable phenotype were selected for the analyses. For the study of irx1-6 coi1-1 plants, seeds of heterozygous irx1-6 coi1-1 and coi1-1 plants were sown on plates containing 50 μM JA for coi1-1 selection (Feys et al., 1994). Plants were then transferred to soil for the analysis, and the irx1-6 coi1-1 double mutants were selected according to their phenotype. For the selection of irx3-1 aba3-2 plants, a cleaved amplified polymorphism was used to identify the irx3-1 mutation (5′-CGGGAATCTTGGAATTGAGATT-3′ and 5′-GATCAGCAGTGTTGTCCATTTG-3′ and MseI digestion), and the aba3-2 mutation was confirmed by DNA sequencing (5′-CTCAAGTCGCTTACACCTTCTG-3′ and 5′-GGGATAACTCGAGATACTTTG-3′). Genotyping of the irx1-1 abi1-1 double mutant was confirmed by PCR amplification of abi1-1 (PCR amplification with oligonucleotides 5′-GCCTTTGTATGGTTTTACTTC-3′ and 5′-CAGCCACGTATCACCATCG-3′ followed by NcoI digestion) and sequencing of irx1-1 (oligonucleotides 5′-GGTTAATAGGAGACCATTTAG-3′ and 5′-TCCATCCAAATCTCAATCCC-3′).
Expression Analysis
Total RNA isolation from plants challenged or not with R. solanacearum and RNA gel blotting were performed according to the protocol of Hirsch et al. (2002). RNA extraction of P. cucumerina–infected or mock-inoculated plants and RNA gel blot analysis of these RNAs were done as described (Llorente et al., 2005). The hybridization probes (PDF1.2, PR1, Pc ITS1, and β-TUB) used in this study have been described (Llorente et al. 2005).
For quantitative RT-PCR analyses, 5 μg of total RNA was treated with DNase and then reverse-transcribed for 90 min at 42°C in a 20-μL reaction volume using the SuperScript II First-Strand Synthesis System kit (Invitrogen) and oligo(dT). Oligonucleotides used for cDNA amplification were designed with Primer Express (version 2.0; Applied Biosystems): At1g18710, 5′-GAGCTGCAGATTAAGGTGGCTT-3′ and 5′-CCGAGAACAGCATGAAGTTGG-3′; At2g39800, 5′-ATGCAGTCTGTGTGTGCACTCC-3′ and 5′-ACCATGAGTACTGTGCCAAGGC-3′; At2g46680, 5′-CGATGATGTCAGCAGAGCCAT-3′ and 5′-CAGTGACTTGATCTGCTCGTCG-3′; At3g1440, 5′-ACATCTTTACGGCGATAACCG-3′ and 5′-TTCCATGTCTTCTCGTCGTGA-3′; At4g26080, 5′-TGTTTTCCCGTCTCACATCTTC-3′ and 5′-CCGGTTTATGGTCAACGGAT-3′; At4g39950, 5′-GCCGACCCACTTTGCTTTAAA-3′ and 5′-GCACAACCTCTTTTCCCGGTA-3′; At5g57050, 5′-TCAGTGCGGCGAGTAAAAGAA-3′ and 5′-TCCGTTTCCGAGCCAAATC-3′; and At3g18780, 5′-GACCAGCTCTTCCATCGAGAA-3′ and 5′-GCAGCTTCCATTCCCACAA-3′.
Real-time quantitative PCR amplification and detection was performed in the 7300 real-time PCR system (Applied Biosystems). Reactions were done in a final volume of 20 μL and contained 10 μL of 2× SYBR Green Master Mix (Applied Biosystems), 0.5 μM of each primer, and 8 ng of cDNA. PCR conditions were as follows: 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. At the end of each experiment, a dissociation stage (95°C for 15 s, 60°C for 30 s, and 95°C for 15 s) was performed to ensure that only single products were formed. Actin2 (At3g18780) expression was used to normalize the transcript level in each sample. Data analysis was performed using the sequence detector software (version 1.2.2; Applied Biosystems).
Microarray Experiments and Analysis
Leaves from uninfected 4-week-old homozygous irx5-5 and irx1-6 plants and their corresponding heterozygous counterparts, which were fully susceptible to both R. solanacearum and P. cucumerina and showed a wild-type phenotype, were collected for microarray analysis. Three biological replicates from irx5-5 and wild-type plants and two replicates from irx1-6 and wild-type plants were harvested for microarray analysis. Total RNA was extracted using the NucleoSpin RNAII kit (Macherey-Nagel). Probes were synthesized from the RNA samples and hybridized to the Affymetrix GeneChip (ATHI, 22 K) according to the procedures provided by the manufacturer. The array images were analyzed with the Affymetrix Microarray Suite 5.0 with the target intensity set to 500. The expression levels of the genes were analyzed with GeneSpring 7.2 software (Silicon Genetics), and the chip-to-chip signal variation was minimized by normalizing signal intensities to the averaged intensity values of wild-type plants using the expression levels of the top 50th percentile of probe sets. Differentially expressed genes in both the irx5-5 (three replicates) and irx1-6 (two replicates) mutants relative to wild-type samples (five replicates) were identified using one-way analysis of variance and a Benjamini and Hochberg multiple testing correction (GeneSpring 7.2), as described previously (Stein et al., 2006). Genes were considered differentially expressed at P ≤ 0.01. Upregulated and downregulated genes were selected using normalized values (fold) > 2 or < 0.5 relative to wild-type plants, respectively.
Accession Numbers
The data obtained by the microarray hybridizations have been submitted to NASCArrays (reference number /NASCARRAYS-368/). Arabidopsis Genome Initiative locus identifiers for the genes impaired in the mutants characterized in this article are as follows: CESA8/IRX1/ERN1, At4g18780; CESA4/IRX5/NWS2, At5g44030.
Supplemental Data
The following materials are available in the online version of this article
Supplemental Figure 1. Cloning of the ERN1/IRX1 and NWS2/IRX5 Genes.
Supplemental Figure 2. Quantitative RT-PCR Analysis of Genes Differentially Upregulated or Downregulated in Secondary Cell Wall irx Mutants.
Supplemental Figure 3. Developmental Phenotype of the irx3-1 aba3-2 Double Mutant.
Supplemental Table 1. Genes Constitutively Upregulated in the irx5-5 and irx1-6 Mutants.
Supplemental Table 2. Genes Constitutively Downregulated in the irx5-5 and irx1-6 Mutants.
Supplementary Material
Acknowledgments
We thank Gemma López (Centro de Biotecnología y Genómica de Plantas, Universidad Politecnica de Madrid) for technical assistance and F. García-Olmedo and F. García-Arenal (Centro de Biotecnología y Genómica de Plantas) and X. Dong (Duke University) for critical reading of the manuscript. C.H.-B., A.S.-V., and C.S.-R. were recipients of Ph.D. fellowships from the Ministerio de Educación y Ciencia (MEC) of Spain. F.L. and M.B.-L. were postdoctoral fellows of the Comunidad de Madrid and MEC (Spain), respectively. This work was supported by MEC (Grant BIO2003-4424 to A.M.), by Genoplante (Grant AF2001060 to Y.M.), and by the National Science Foundation (Grant 0114783 to S.S.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Yves Marco (yves.marco@inra.toulouse.fr) and Antonio Molina (antonio.molina@upm.es).
Online version contains Web-only data.
References
- AbuQamar, S., Chen, X., Dhawan, R., Bluhm, B., Salmeron, J., Lam, S., Dietrich, R.A., and Mengiste, T. (2006). Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J. 48 28–44. [DOI] [PubMed] [Google Scholar]
- Aldon, D., Brito, B., Boucher, C., and Genin, S. (2000). A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 10 2304–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.P., Badruzsaufari, E., Schenk, P.M., Manners, J.M., Desmond, O.J., Ehlert, C., Maclean, D.J., Ebert, P.R., and Kazan, K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert, K., De Meyer, G.B., and Hofte, M.M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch, M., Gobbato, E., Bednarek, P., Debey, S., Schultze, J.L., Bautor, J., and Parker, J.E. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudoin, N., Serizet, C., Gosti, F., and Giraudat, J. (2000). Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo, M., Molina, A., and Solano, R. (2002. a). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29 23–32. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo, M., Segura, A., Moreno, M., Lopez, G., Garcia-Olmedo, F., and Molina, A. (2002. b). Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 128 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher, C.A., Barberis, P.A., Trigalet, A.P., and Demery, D.A.J. (1985). Transposon mutagenesis of Pseudomonas solanacearum: Isolation of Tn5-induced virulent mutants. J. Gen. Microbiol. 131 2449–2457. [Google Scholar]
- Brown, D.M., Zeef, L.A., Ellis, J., Goodacre, R., and Turner, S.R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella, A.N., Menossi, M., Arruda, P., and Benedetti, C.E. (2001). COI1 affects myrosinase activity and controls the expression of two flower-specific myrosinase-binding protein homologues in Arabidopsis. Planta 213 691–699. [DOI] [PubMed] [Google Scholar]
- Carpita, N.C., and McCann, M. (2000). The cell wall. In Biochemistry and Molecular Biology of Plants, B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 52–108.
- Celenza, J.L., Quiel, J.A., Smolen, G.A., Merrikh, H., Silvestro, A.R., Normanly, J., and Bender, J. (2005). The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 137 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Hong, X., Zhang, H., Wang, Y., Li, X., Zhu, J.K., and Gong, Z. (2005). Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 43 273–283. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., Ritter, C., Gibbon, M.J., Mur, L.A., Wood, J.R., Gross, S., Mansfield, J., and Taylor, J.D. (1992). Functional homologs of the Arabidopsis RPM1 disease resistance gene in bean and pea. Plant Cell 4 1359–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L., Olivier, J., Peeters, N., Feng, D.X., Khounlotham, M., Boucher, C., Somssich, I., Genin, S., and Marco, Y. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L., Pileur, F., Liaubet, L., Camut, S., Can, C., Williams, K., Holub, E, Beynon, J., Arlat, M., and Marco, Y. (1998). Genetic characterization of RRS1, a recessive locus in Arabidopsis thaliana that confers resistance to the bacterial soilborne pathogen Ralstonia solanacearum. Mol. Plant Microbe Interact. 11 659–667. [DOI] [PubMed] [Google Scholar]
- Desprez, T., Vernhettes, S., Fagard, M., Refregier, G., Desnos, T., Aletti, E., Py, H., Pelletier, S., and Hofte, H. (2002). Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 128 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C., Karafyllidis, I., Wasternack, C., and Turner, J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple, P., Apel, K., and Bohlmann, H. (1997). Overexpression of an endogenous thionin enhances resistance of Arabidopsis against Fusarium oxysporum. Plant Cell 9 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini, P.J., and St-Pierre, B. (2005). Synthesis and trafficking of alkaloid biosynthetic enzymes. Curr. Opin. Plant Biol. 8 657–666. [DOI] [PubMed] [Google Scholar]
- Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., McCann, M., Rayon, C., Vernhettes, S., and Hofte, H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12 2409–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Olmedo, F., Molina, A., Alamillo, J.M., and Rodriguez-Palenzuela, P. (1998). Plant defense peptides. Biopolymers 47 479–491. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis. Curr. Opin. Plant Biol. 4 301–308. [DOI] [PubMed] [Google Scholar]
- Godiard, L., Sauviac, L., Torii, K.U., Grenon, O., Mangin, B., Grimsley, N.L., and Marco, Y. (2004). ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 36 353–365. [DOI] [PubMed] [Google Scholar]
- Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb, C.D., and Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11 89–100. [DOI] [PubMed] [Google Scholar]
- Hirsch, J., Deslandes, L., Feng, D.X., Balagué, C., and Marco, Y. (2002). Delayed symptom development in ein2-1, an Arabidopsis ethylene-insensitive mutant, in response to bacterial wilt caused by Ralstonia solanacearum. Phytopathology 92 1142–1148. [DOI] [PubMed] [Google Scholar]
- Hu, X., and Reddy, A.S. (1997). Cloning and expression of a PR5-like protein from Arabidopsis: Inhibition of fungal growth by bacterially expressed protein. Plant Mol. Biol. 34 949–959. [DOI] [PubMed] [Google Scholar]
- Jorda, L., Coego, A., Conejero, V., and Vera, P. (1999). A genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J. Biol. Chem. 274 2360–2365. [DOI] [PubMed] [Google Scholar]
- Kappers, I.F., Aharoni, A., van Herpen, T.W., Luckerhoff, L.L., Dicke, M., and Bouwmeester, H.J. (2005). Genetic engineering of terpenoid metabolism attracts bodyguards to Arabidopsis. Science 309 2070–2072. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Kroymann, J., and Mitchell-Olds, T. (2005). The glucosinolate-myrosinase system in an ecological and evolutionary context. Curr. Opin. Plant Biol. 8 264–271. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Rowe, H.C., and Denby, K.J. (2005). Secondary metabolites influence Arabidopsis/Botrytis interactions: Variation in host production and pathogen sensitivity. Plant J. 44 25–36. [DOI] [PubMed] [Google Scholar]
- Lawton, K., Weymann, K., Friedrich, L., Vernooij, B., Uknes, S., and Ryals, J. (1995). Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol. Plant Microbe Interact. 8 863–870. [DOI] [PubMed] [Google Scholar]
- Lipka, V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 1180–1183. [DOI] [PubMed] [Google Scholar]
- Llorente, F., Alonso-Blanco, C., Sanchez-Rodriguez, C., Jorda, L., and Molina, A. (2005). ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 43 165–180. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O., Chico, J.M., Sanchez-Serrano, J.J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Gillmor, C.S., and Scheible, W.R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lummerzheim, M., Kroj, T., Ferreira, M., Tronchet, M., Godard, F., Van Montagu, M., and Roby, D. (2004). An Arabidopsis mutant with altered hypersensitive response to Xanthomonas campestris pv. campestris, hxc1, displays a complex pathophenotype. Mol. Plant Pathol. 5 453–464. [DOI] [PubMed] [Google Scholar]
- Mauch-Mani, B., and Mauch, F. (2005). The role of abscisic acid in plant-pathogen interactions. Curr. Opin. Plant Biol. 8 409–414. [DOI] [PubMed] [Google Scholar]
- Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126 969–980. [DOI] [PubMed] [Google Scholar]
- Mengiste, T., Chen, X., Salmeron, J., and Dietrich, R. (2003). The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15 2551–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchan, S., Bernal, D., Serrano, R., and Yenush, L. (2004). Response of the Saccharomyces cerevisiae Mpk1 mitogen-activated protein kinase pathway to increases in internal turgor pressure caused by loss of Ppz protein phosphatases. Eukaryot. Cell 3 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, A., Ahl-Goy, P., Fraile, A., Sanchez-Monge, R., and Garcia-Olmedo, F. (1993. a). Inhibition of bacterial and fungal plant pathogens by thionins of types I and II. Plant Sci. 92 169–177. [Google Scholar]
- Molina, A., and García-Olmedo, F. (1997). Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. Plant J. 12 669–675. [DOI] [PubMed] [Google Scholar]
- Molina, A., Segura, A., and García-Olmedo, F. (1993. b). Lipid transfer proteins (nsLTPs) from barley and maize leaves are potent inhibitors of bacterial and fungal plant pathogens. FEBS Lett. 316 119–122. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K., and Fink, G.R. (1992). Two anthranilate synthase genes in Arabidopsis: Defense-related regulation of the tryptophan pathway. Plant Cell 4 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi, K.K., Grossman, A.R., and Bjorkman, O. (1998). Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S., Chen, J.G., Jones, A.M., and Assmann, S.M. (2006). G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol. 141 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Park, J.M., Park, C.-H., Lee, S.-B., Hamd, B.-K., Shin, R., and Paek, K.-H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, S., Wei, H., Milne, J., Page, G.P., and Somerville, C.R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 102 8633–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip, B., and Levin, D.E. (2001). Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman, G., Lubarsky, B., Kieber, J.J., Rothenberg, M., and Ecker, J.R. (1995). Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics 139 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible, W.R., Eshed, R., Richmond, T., Delmer, D., and Somervillle, C.R. (2001). Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis ixr1 mutants. Proc. Natl. Acad. Sci. USA 98 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert, P. (2004). Knocking on the heaven's wall: Pathogenesis of and resistance to biotrophic fungi at the cell wall. Curr. Opin. Plant Biol. 7 377–383. [DOI] [PubMed] [Google Scholar]
- Segura, A., Moreno, M., and Garcia-Olmedo, F. (1993). Purification and antipathogenic activity of lipid transfer proteins (LTPs) from the leaves of Arabidopsis and spinach. FEBS Lett. 332 243–246. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6 410–417. [DOI] [PubMed] [Google Scholar]
- Somerville, C.R., et al. (2004). Toward a systems approach to understanding plant cell walls. Science 306 2206–2211. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, M., Dittgen, J., Sanchez-Rodriguez, C., Hou, B.H., Molina, A., Schulze-Lefert, P., Lipka, V., and Somerville, S. (2006). Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Howells, R.M., Huttly, A.K., Vickers, K., and Turner, S.R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko, E., Lopez-Solanilla, E., Garcia-Olmedo, F., and Rodriguez-Palenzuela, P. (1997). Mutants of Ralstonia (Pseudomonas) solanacearum sensitive to antimicrobial peptides are altered in their lipopolysaccharide structure and are avirulent in tobacco. J. Bacteriol. 179 6699–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, J., Jakab, G., Toquin, V., Flors, V., Iavicoli, A., Maeder, M.N., Metraux, J.P., and Mauch-Mani, B. (2005). Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell 17 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, J., and Mauch-Mani, B. (2004). Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant J. 38 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S., and Hall, M. (2000). The gapped xylem mutant identifies a common regulatory step in secondary cell wall deposition. Plant J. 24 477–488. [DOI] [PubMed] [Google Scholar]
- Vogel, J., and Somerville, S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 97 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J.P., Raab, T.K., Schiff, C., and Somerville, S.C. (2002). PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14 2095–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J.P., Raab, T.K., Somerville, C.R., and Somerville, S.C. (2004). Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 40 968–978. [DOI] [PubMed] [Google Scholar]
- Weyman, P.D., Pan, Z., Feng, Q., Gilchrist, D.G., and Bostock, R.M. (2005). A circadian rhythm-regulated tomato gene is induced by arachidonic acid and Phytophthora infestans infection. Plant Physiol. 140 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565. [DOI] [PubMed] [Google Scholar]
- Williamson, J.D., Stoop, J.M., Massel, M.O., Conkling, M.A., and Pharr, D.M. (1995). Sequence analysis of a mannitol dehydrogenase cDNA from plants reveals a function for the pathogenesis-related protein ELI3. Proc. Natl. Acad. Sci. USA 92 7148–7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Ishitani, M., Lee, H., and Zhu, J.K. (2001). The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13 2063–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., et al. (2003). Identification of Arabidopsis rat mutants. Plant Physiol. 132 494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.