Abstract
In most eukaryotes, the prospective chromosomal positions of meiotic crossovers are marked during meiotic prophase by protein complexes called late recombination nodules (LNs). In tomato (Solanum lycopersicum), a cytological recombination map has been constructed based on LN positions. We demonstrate that the mismatch repair protein MLH1 occurs in LNs. We determined the positions of MLH1 foci along the 12 tomato chromosome pairs (bivalents) during meiotic prophase and compared the map of MLH1 focus positions with that of LN positions. On all 12 bivalents, the number of MLH1 foci was ∼70% of the number of LNs. Bivalents with zero MLH1 foci were rare, which argues against random failure of detecting MLH1 in the LNs. We inferred that there are two types of LNs, MLH1-positive and MLH1-negative LNs, and that each bivalent gets an obligate MLH1-positive LN. The two LN types are differently distributed along the bivalents. Furthermore, cytological interference among MLH1 foci was much stronger than interference among LNs, implying that MLH1 marks the positions of a subset of strongly interfering crossovers. Based on the distances between MLH1 foci or LNs, we propose that MLH1-positive and MLH1-negative LNs stem from the same population of weakly interfering precursors.
INTRODUCTION
During meiosis, reciprocal exchanges (crossovers [COs]) between homologous chromosomes (homologs) occur at a high frequency. In many organisms, ultrastructurally detectable electron-dense protein complexes named late recombination nodules (LNs) mark the chromosomal positions of COs during the pachytene stage of meiotic prophase (Anderson and Stack, 2005). LNs are associated with zipper-like structures that closely appose homologs during meiotic prophase: the synaptonemal complexes (SCs). SCs consist of two axial elements (AEs), one along each homolog, and numerous transverse filaments, which connect the AEs of homologs along their entire length (reviewed in Page and Hawley, 2004). In organisms with a favorable cytology, including tomato (Solanum lycopersicum), it has been possible to construct detailed cytological recombination maps based on LN positions along SCs in electron micrographs of spread meiocytes (reviewed in Sherman and Stack, 1995; Anderson and Stack, 2005). Such LN maps are invaluable because they provide high-resolution genome-wide information about meiotic crossing over, whereas polymorphic genetic markers are not required. In a later stage of meiotic prophase (diakinesis), COs can be recognized as chiasmata, so in principle it is also possible to construct a cytological recombination map based on chiasma positions. However, in most species, chromosome condensation precludes the precise location of chiasmata along bivalents. Nevertheless, analyses of chiasma positions in tomato have been useful for validation of the LN-based recombination map because at the level of chromosome arms, the occurrence of LNs corresponds closely with the occurrence of chiasmata (reviewed in Havekes et al., 1994; Sherman and Stack, 1995; Anderson and Stack, 2005), which indicates that LNs mark all COs in tomato.
Because the ultrastructural analysis of LN positions is labor-intensive and LNs are not easily recognized in all organisms, Anderson et al. (1999) and Froenicke et al. (2002) followed another approach for the construction of a cytological CO map of the male mouse, namely, immunofluorescent double labeling of spermatocytes using antibodies against SCs and a protein component of LNs, MLH1 (Moens et al., 2002; Marcon and Moens, 2003). MLH1 is one of the eukaryotic homologs of Escherichia coli MutL. In meiosis, heterodimers of MLH1 with another MutL homolog, MLH3, are essential for wild-type levels of crossing over (Baker et al., 1996; Wang et al., 1999; Lipkin et al., 2002; Jackson et al., 2006). In mouse, Froenicke et al. (2002) found that the occurrence of immunofluorescent MLH1 foci correlated closely with the occurrence of COs as detected genetically (Broman et al., 2002), which implies that MLH1 foci represent virtually all prospective CO positions in this species. However, it is doubtful whether this correlation exists in all organisms. First, cytological CO mapping is based on snapshots in time; if the life span of the cytological CO marker does not entirely overlap with the time window of observation, part of the CO positions will be missed. Second, some species have more than one pathway for meiotic crossing over. In yeast, two major pathways have been proposed (reviewed in Hollingsworth and Brill, 2004; Whitby, 2005); one pathway depends on a group of proteins that are collectively indicated as ZMM proteins and yields approximately two-thirds of the yeast meiotic COs, the class I COs (Chua and Roeder, 1998; Agarwal and Roeder, 2000; Börner et al., 2004). These COs depend largely, though not entirely, on Mlh1 (Argueso et al., 2004). The other major pathway does not require ZMM proteins but depends on the Mus81/Mms4 endonuclease (de los Santos et al., 2003). COs produced by this pathway, named class II COs, do not depend on Mlh1 (Argueso et al., 2004). It is therefore uncertain whether MLH1 marks all class I COs in all organisms, whereas it seems highly unlikely that it marks class II COs (discussed in Hoffmann and Borts, 2004).
In Arabidopsis thaliana, two genes homologous to yeast ZMM genes have been identified, namely, MSH4 (Higgins et al., 2004) and MER3/RCK (Chen et al., 2005; Mercier et al., 2005). Mutants for these genes form ∼15% of the wild-type level of COs. If these residual 15% COs correspond to yeast class II COs, they would not be expected to be marked by MLH1 or MLH3. Furthermore, it has been suggested that some class I COs are still formed in the absence of MLH3 (Jackson et al., 2006). It is thus doubtful whether MLH3 (and MLH1) mark all meiotic COs in Arabidopsis, but unfortunately this cannot be analyzed because it is technically not feasible to perform a systematic comparison of the positions of MLH1 or MLH3 foci with the positions of LNs or COs in this species. One of the few species where this can be done is tomato, which has a favorable meiotic cytology, whereas recombination maps based on LN positions (Sherman and Stack, 1995) and on genetic analyses (Tanksley et al., 1992) have already been made. In this study, we demonstrate by immunogold labeling that MLH1 forms part of the tomato LNs. Subsequently, using immunofluorescent labeling, we determined the positions of MLH1 foci along all 12 tomato SCs in spread pollen mother cells (PMCs). Antibodies recognizing AEs, MLH1, and centromeric regions were not yet available for tomato and were developed during the course of this study. Using these antibodies, we found that MLH1 foci mark only 70% of all LNs in wild-type tomato.
In most analyzed eukaryotes, including yeast and tomato, meiotic COs are more evenly spaced than expected if they were randomly distributed along the chromosomes, a phenomenon called (positive) CO interference (reviewed in Hillers, 2004). It is now generally assumed that yeast class I COs display interference, whereas class II COs don't (discussed by de los Santos et al., 2003). We found that interference among tomato MLH1 foci was much stronger than interference among LNs, which is consistent with the idea that MLH1 foci specifically mark those LNs, and thus prospective CO positions, that correspond to yeast class I COs.
RESULTS
Immunocytological Detection of MLH1 Foci, SCs, and Kinetochore Regions in Spread Tomato PMCs
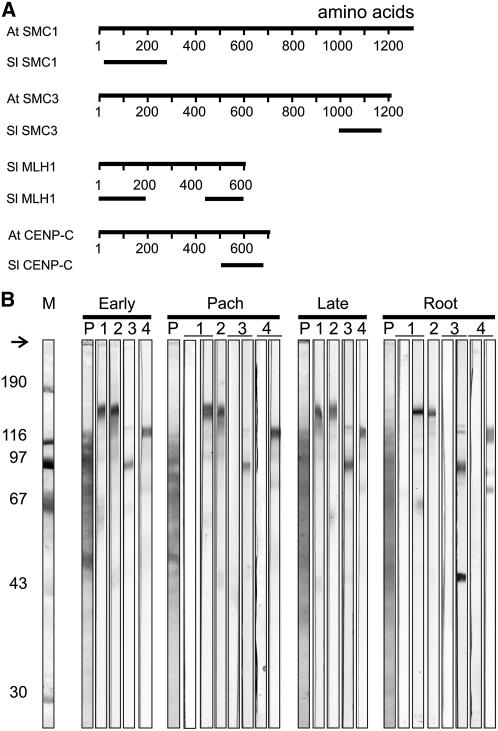
For the construction of a map of MLH1 foci along tomato SCs, we needed to perform immunofluorescence triple labeling of MLH1, SCs, and kinetochoric regions in spread pachytene PMCs of tomato. For this purpose, we prepared antisera against tomato MLH1 (Sl MLH1), Sl SMC1, Sl SMC3 and Sl CENPC. SMC1 and SMC3 are components of the cohesin complex, which ensures sister chromatid cohesion in mitosis and meiosis (reviewed in Nasmyth, 2001). In various organisms, including Arabidopsis, SMC1 and SMC3 lie along the AEs of SCs during meiotic prophase (Klein et al., 1999; Eijpe et al., 2000; Revenkova et al., 2004; Lam et al., 2005b). CENPC is an inner kinetochore protein (Dawe et al., 1999, and references therein). Affinity-purified antibodies from the sera raised against Sl MLH1, Sl SMC1, Sl SMC3, and Sl CENPC recognized single bands of the expected electrophoretic mobilities on immunoblots of tomato anthers (Figure 1). On blots of tomato roots, the α Sl MLH1 antibodies from serum 8, but not from serum 621, also recognized an unknown Mr 45,000 peptide (Figure 1B), which we have not characterized because it was not detected in anther extracts.
Figure 1.
Antibodies Used in This Study.
(A) Positions of peptides that were used for immunization. For each protein, the top line represents the predicted protein encoded by the full-length Arabidopsis (At) or tomato (Sl) cDNA, whereas the bottom line(s) represents the positions of the peptides of the tomato protein that were used for immunization.
(B) Characterization of the antibodies. Immunoblot strips carrying molecular weight markers (M), proteins from tomato anthers containing PMCs in early meiotic prophase (Early), pachytene (Pach), or late meiotic prophase (Late), or proteins from tomato root tips (Root) were incubated in Ponceau S (P) or in affinity-purified antibodies from sera elicited against peptides shown in (A): lane 1, anti-Sl SMC1 (serum 5); left strip, preimmune serum; right strip, anti-SMC1 antibodies; lane 2, anti-Sl SMC3 (serum 623), anti-SMC3 antibodies; lane 3, anti-Sl MLH1 C-terminal peptide (serum 8); left strip: preimmune serum; right strip, anti-MLH1 antibodies; lane 4, anti-Sl CENPC (serum 20); left strip, preimmune serum; right strip, anti-CENPC antibodies. The arrow indicates the top of the gel, and the numbers to the left represent the molecular masses in kilodaltons of the marker proteins.
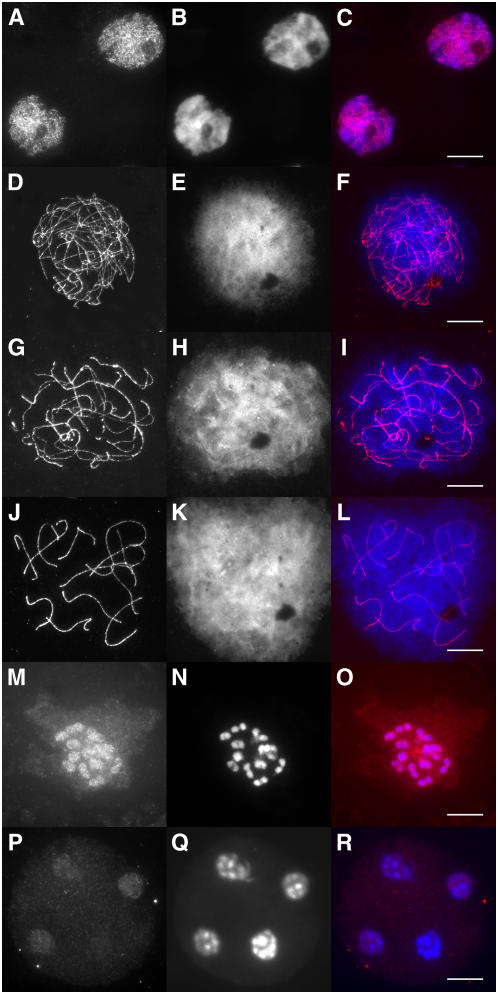
In immunofluorescence experiments, the affinity-purified α Sl SMC1 and α Sl SMC3 antibodies labeled the AEs of SCs from leptotene till diplotene (shown for Sl SMC1 in Figures 2 and 3) and produced little signal in the remainder of the nucleus. In metaphase I, part of the Sl SMC1 and Sl SMC3 proteins remained associated with the chromosomes, and part was found diffusely distributed throughout the PMC (Figure 2O). Telophase II nuclei still displayed a weak, diffuse Sl SMC1 or Sl SMC3 signal (Figure 2R). This pattern strongly resembled that reported previously for SMC3 (Lam et al., 2005b) in Arabidopsis. The affinity-purified α Sl MLH1 antibodies specifically recognized distinct foci on the SCs in immunofluorescence experiments (Figures 3A and 3E; see Supplemental Figure 1 online), whereas in immunogold labeling experiments, the α Sl MLH1 antibodies labeled LNs specifically, indicating that MLH1 foci correspond with LNs (Figure 4). In some late zygotene cells, part of the bivalents displayed MLH1 foci (data not shown), whereas in pachytene PMCs, most or all bivalents displayed at least one MLH1 focus, irrespective of the pachytene sub stage (see Supplemental Figure 1 online). Sl CENPC was not detectable before early pachytene (cf. Dawe et al., 1999). The intensity of the Sl CENPC signal increased as pachytene progressed (see Supplemental Figure 1 online) and reached its maximum in late pachytene/early diplotene.
Figure 2.
Immunolocalization of Sl SMC1 throughout Meiosis in Tomato.
Squashes ([A] to [C] and [M] to [R]) or spreads ([D] to [L]) of tomato PMCs were labeled with α Sl SMC1 ([A], [D], [G], [J], [M], and [P]) and counterstained with 4',6-diamidino-2-phenylindole (DAPI) ([B], [E], [H], [K], [N], and [Q]); the merged images are shown in ([C], [F], [I], [L], [O], and [R]). Leptotene ([A] to [C]), early zygotene ([D] to [F]), late zygotene ([G] to [I]) pachytene ([J] to [L]), metaphase I ([M] to [O]), and telophase II ([P] to [R]). Bars = 10 μm.
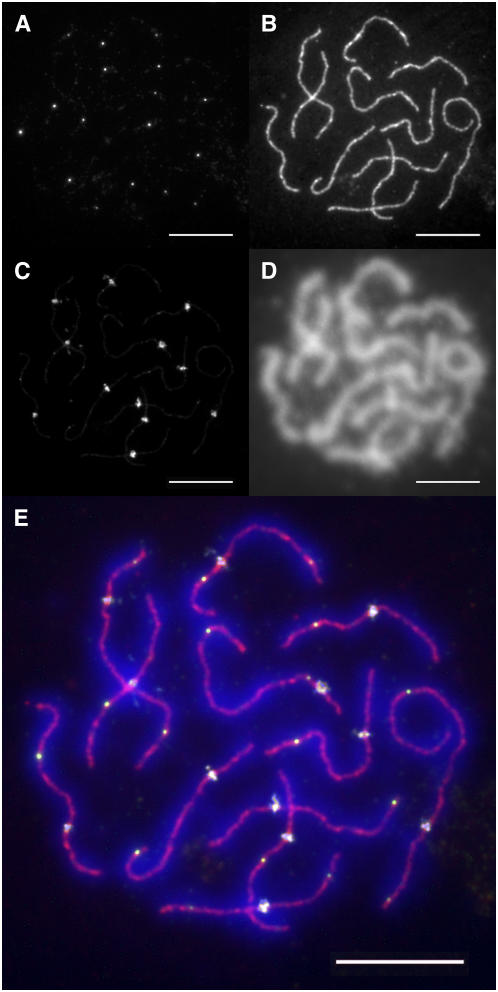
Figure 3.
Immunofluorescence Triple Labeling of Spread Tomato PMC Nuclei.
Spread PMC nuclei were successively incubated in antibodies recognizing MLH1 (A), SMC1 (B), and CENPC (C) as described in Methods and then counterstained with DAPI (D). (E) shows the merged images of the four labels. Bars = 10 μm.
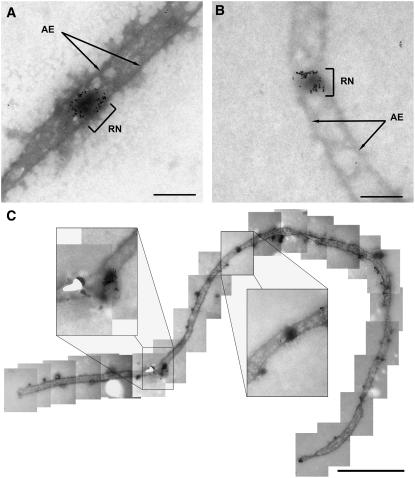
Figure 4.
Immunogold Labeling of MLH1 in LNs of Tomato.
Shown are details of spread pachytene tomato PMCs after immunogold labeling of MLH1 and counterstaining with uranyl acetate.
(A) and (B) Segments of SCs carrying an MLH1-positive LN (RN). Bars = 200 nm.
(C) Overview of an SC carrying an MLH1-positive and an MLH1-negative LN. The insets shows these LNs at a higher magnification. Note the terminal knobs, which are characteristic of pachytene SCs (Stack and Anderson, 1986a). For a high-resolution assembly of the entire SC, see Supplemental Figure 2 online. Bar = 1 μm.
In MLH1/SMC1/CENPC triple labeling experiments, the Sl CENPC signal did not exceed the Sl SMC1 signal until mid pachytene (Figures 3C and 3E; see Supplemental Figure 1 online). Because the CENPC signal was essential for distinguishing between the two arms of most chromosomes and determining the arm length ratio, we only analyzed MLH1 foci in mid and late pachytene PMCs (see below).
Relating Tomato SCs to Specific Chromosomes
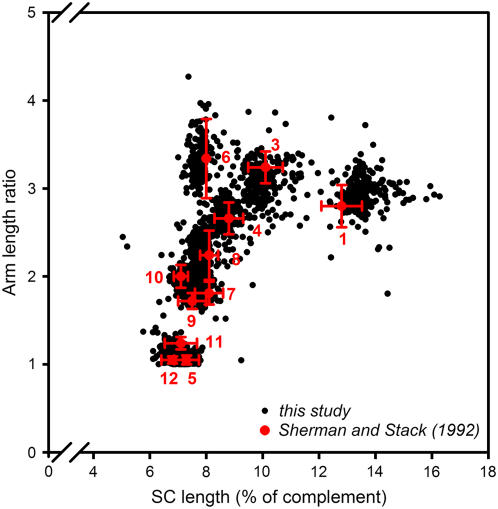
Tomato SCs can be related to specific chromosomes by their length and arm length ratio. In an ultrastructural study of spread PMCs from a series of plants that were each trisomic for one of the tomato chromosomes, Sherman and Stack (1992) determined the length and arm length ratio of all 12 tomato SCs. By these two criteria, SCs of chromosomes 1 to 4, 6, and 8 can be identified individually (Figure 5), and two groups of chromosomes can be distinguished, one consisting of chromosomes 5, 11, and 12 and one of chromosomes 7, 9, and 10 (Figure 5). Sherman and Stack (1992) differentiated further between SCs of the same group by comparing the lengths and arm length ratios of SCs within the same PMC nucleus. Based on their lengths and arm length ratios, SCs analyzed by immunofluorescence (this study) clustered similarly as ultrastructurally analyzed SCs do (Sherman and Stack, 1992) (Figure 5), and we could therefore relate SCs to specific chromosomes using the same criteria as had been used by Sherman and Stack (1992). We consider the distinction between SCs of the same group and the distinction between the long and short arms of chromosomes 5 and 12 as tentative.
Figure 5.
SC Length versus Arm Length Ratio in Spread Tomato PMCs.
The black dots represent the results from 200 PMC nuclei of which the complete set of 12 SCs could be analyzed by immunofluorescence labeling (this study), with each dot representing an individual SC. The red symbols and numbers refer to data from Sherman and Stack (1992). The red dots represent the average SC length versus the average arm length ratio of chromosomes 1 to 12 (except chromosome 2), as identified by ultrastructural analysis of spread PMCs from trisomic tomato plants (red numbers). The red error bars represent 1 sd, in both directions, of the SC lengths and arm length ratios as determined by Sherman and Stack (1992). Chromosome 2 is not represented in this figure because its arm length ratio cannot be determined reliably: the AEs of its short arms stay asynapsed and are often broken. However, because of these features, SC 2 is easily identified. The SC length is expressed as a percentage of the total SC complement length (excluding the short arms of SC 2) of the relevant PMC nucleus to account for variation in length in individual PMCs (e.g., due to variation in spreading conditions).
MLH1 Foci Represent a Specific Subset of Tomato LNs
Tomato PMCs contained on average 15.05 MLH1 foci per nucleus (Table 1), which is only 69% of the average number of LNs per nucleus (Table 1). We found such a difference between the number of MLH1 foci and the number of LNs for each of the 12 bivalents (Table 1). Furthermore, we found that PMCs that had been immunogold-labeled for MLH1 displayed immunogold-negative LNs besides immunogold-positive LNs (Figure 4C; see Supplemental Figure 2 online), but we were unable to determine the proportion of MLH1-negative LNs in the immunogold preparations because the low contrast of the uranyl acetate counterstain did not allow the unambiguous identification of every individual LN. We considered two possible explanations for these observations. (1) There is one type of LN, but we failed to detect MLH1 in part of the LNs for technical reasons and/or because MLH1 is not present in LNs during the entire time window, as defined by our selection of PMCs for analysis (mid to late pachytene); or (2) there are two types of LNs: MLH1-positive LNs and MLH1-negative LNs. To distinguish between these possibilities, we calculated for each chromosome the expected percentage of SCs without MLH1 foci if we would fail at random to detect MLH1 in part of the LNs, and/or, equivalently, if MLH1 foci would be present in LNs only part of the time. For instance, Sherman and Stack (1995) detected 2.48 LNs on SC 1, whereas we detected on average only 1.63 MLH1 foci on this SC. If we would fail at random to detect MLH1 in LNs, the probability of displaying MLH1 upon immunofluorescence labeling is thus 1.63/2.48 = 0.66 for every LN on SC 1. Sherman and Stack (1995) found that the number of LNs per SC ranged between 1 and 4 for all chromosomes. On SC 1, 15% had one LN, 43% had two LNs, 36% had three LNs, and 6% had four LNs. The expected percentage of SC 1 with no MLH1 foci is thus [(1 − 0.66) × 0.15 + (1 − 0.66)2 × 0.43 + (1 − 0.66)3 × 0.36 + (1 − 0.66)4 × 0.06] ×100 = 11.8%. If MLH1 foci are not present in LNs during the entire time window as defined by our selection of SCs for analysis (mid to late pachytene; see Methods), it would have a similar effect on the observed percentage of zero-focus SCs as random failure of detection of MLH1 in part of the LNs. Because we found for all chromosomes a much lower percentage of SCs with zero MLH1 foci than expected (Table 1; see Supplemental Figure 1 online), we infer that there are two types of LNs in tomato PMCs: MLH1-positive LNs, constituting ∼70% of all LNs, and MLH1-negative LNs, representing the remaining 30% of the LNs.
Table 1.
Numbers of MLH1 Foci and LNs on Tomato SCs
| SCs without MLH1 Foci (%)
|
||||||
|---|---|---|---|---|---|---|
| SC | No. of SCs | MLH1 foci/SC (±sd) | LNs/SCa | MLH1 Foci/ LNs (R) | Observed | Expectedb |
| 1 | 199 | 1.63 ± 0.60 | 2.48 | 0.66 | 0 | 11.8 |
| 2 | 201 | 1.31 ± 0.47 | 2.08 | 0.63 | 0 | 20.9 |
| 3 | 202 | 1.50 ± 0.51 | 2.10 | 0.71 | 0 | 11.9 |
| 4 | 200 | 1.32 ± 0.48 | 1.89 | 0.70 | 0 | 14.2 |
| 5 | 203 | 1.20 ± 0.44 | 1.67 | 0.72 | 0.99 | 16.9 |
| 6 | 200 | 1.08 ± 0.29 | 1.73 | 0.62 | 0.50 | 23.4 |
| 7 | 199 | 1.22 ± 0.44 | 1.77 | 0.69 | 1.01 | 17.2 |
| 8 | 200 | 1.11 ± 0.32 | 1.68 | 0.66 | 0.50 | 21.0 |
| 9 | 199 | 1.18 ± 0.41 | 1.58 | 0.74 | 1.01 | 14.0 |
| 10 | 199 | 1.17 ± 0.40 | 1.66 | 0.71 | 1.01 | 16.5 |
| 11 | 197 | 1.16 ± 0.39 | 1.66 | 0.70 | 1.02 | 17.0 |
| 12 | 203 | 1.18 ± 0.42 | 1.59 | 0.74 | 1.48 | 13.7 |
| Total | 15.05 ± 1.94 | 21.89 | 0.69 | |||
All data about LNs were taken from Sherman and Stack (1995).
Expected percentages of SCs with zero MLH1 foci if the difference between the number of MLH1 foci/SC and the number of LNs/SC would be due to random failure of detecting MLH1 in LNs. Because the number of LNs per SC ranges between 1 and 4 for all tomato SCs (Sherman and Stack, 1995), the expected percentage of, for instance, SC 1 with zero MLH1 foci is 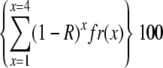 , where x is the number of LNs on an SC, fr(x) is the relative frequency of SC 1 with x foci, and R is the ratio MLH1 foci/LNs for SC 1 (0.66). The values for fr(x) were derived from Table 4 in Sherman and Stack (1995). See also text.
, where x is the number of LNs on an SC, fr(x) is the relative frequency of SC 1 with x foci, and R is the ratio MLH1 foci/LNs for SC 1 (0.66). The values for fr(x) were derived from Table 4 in Sherman and Stack (1995). See also text.
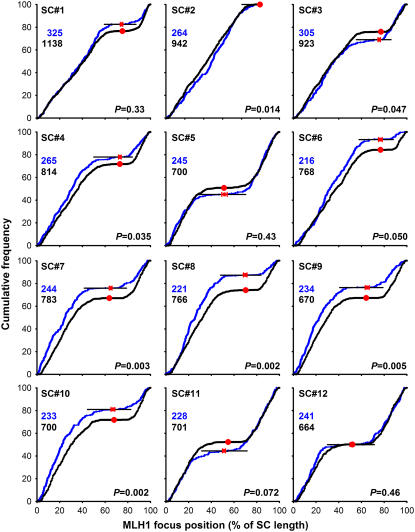
MLH1-Positive and MLH1-Negative LNs Are Distributed Differently along the SCs
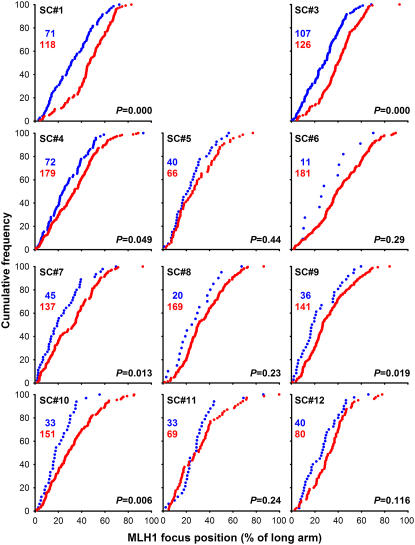
Figure 6 shows a comparison of the positions of MLH1 foci on the 12 tomato SCs with the positions of LNs as determined by Sherman and Stack (1995). The MLH1 focus map has in common with the LN map that the pericentromeric regions are virtually focus-free, as was to be expected if the MLH1 foci represent a subset of the LNs. Presumably, these regions lack a common precursor of MLH1-positive and -negative LNs because early recombination nodules (ENs), which are ultrastructurally recognizable complexes containing recombination proteins (Anderson et al., 1997), of which a small proportion is thought to develop into LNs (reviewed in Anderson and Stack, 2005), are also scarce in pericentromeric regions (Stack and Anderson, 1986b). Recombination suppression in pericentromeric regions has been observed in a variety of plant species (Choo, 1998).
Figure 6.
Positions of MLH1 Foci and LNs on Tomato SCs.
Shown are the cumulative frequencies of foci or LNs as a function of the distance to the telomeric end of the long arm of the SC. The distance is expressed as percentage of the length of the SC on which the focus or LN was located. The graphs show for every position on the SC the percentage of MLH1 foci (or LNs) that is found on the SC stretch between that position and the telomeric end of the long arm. The cumulative frequencies of MLH1 foci (this study) are shown in blue and cumulative frequencies of the LNs (Sherman and Stack, 1995) in black. A uniform distribution of foci or LNs along a given SC would yield a straight line from the bottom left to the top right corner of each graph. Focus or LN-poor regions show up as horizontal stretches in the cumulative curves: the black horizontal lines mark the focus-poor regions around the centromeres. Focus-rich regions are visible as steep parts of the curves, for instance, the subtelomeric region of the long arm of SC 7. The red dots indicate the positions of the centromeres; for the MLH1 data set (this study), horizontal error bars indicate 1 sd (in both directions) of the centromere positions. The numbers of MLH1 foci or LNs on which the curves are based are indicated in the top left corners of the panels. For each chromosome, the P value represents the probability that the MLH1 focus positions (this study) and the LN positions (Sherman and Stack, 1995) stem from the same distribution (Kolmogorov-Smirnov two-sample test).
However, there are also striking differences between the two maps. For SCs 2, 3, 4, 7, 8, 9, and 10, the distribution of MLH1 foci along the SC differed significantly (P < 0.05) from that of LNs, whereas for SC 6, the result was borderline (P = 0.05). For most of these SCs, namely, SCs 4, 6, 7, 8, 9, and 10, the difference between the cumulative frequency of MLH1 foci and that of LNs was greatest in the centromeric region, with a higher cumulative frequency for MLH1 foci than for LNs. This means that on these SCs, a higher percentage of the MLH1 foci than of the LNs is on the long arm. The reverse is found on SC 3. Furthermore, the cumulative curves for MLH1 foci are steep in the subtelomeric regions of the long arms of some SCs, particularly of SCs 7, 8, 9, and 10, indicating that MLH1 foci tend to localize there, whereas this is not obvious for LNs (Sherman and Stack, 1995). Finally, the cumulative curves for MLH1 foci are less smooth than those for LNs. These differences suggest that the rules determining the MLH1 focus positions do not apply to all LNs, which is consistent with the idea that MLH1 foci mark a specific subset of LNs. We emphasize that these inferences are based on comparisons of two data sets from different laboratories. Although we have taken all possible precautions to avoid systematic differences between the data sets (we used the same tomato strain and the same spreading technique as had been used in Sherman and Stack [1995]; see Methods), these inferences should be considered with caution.
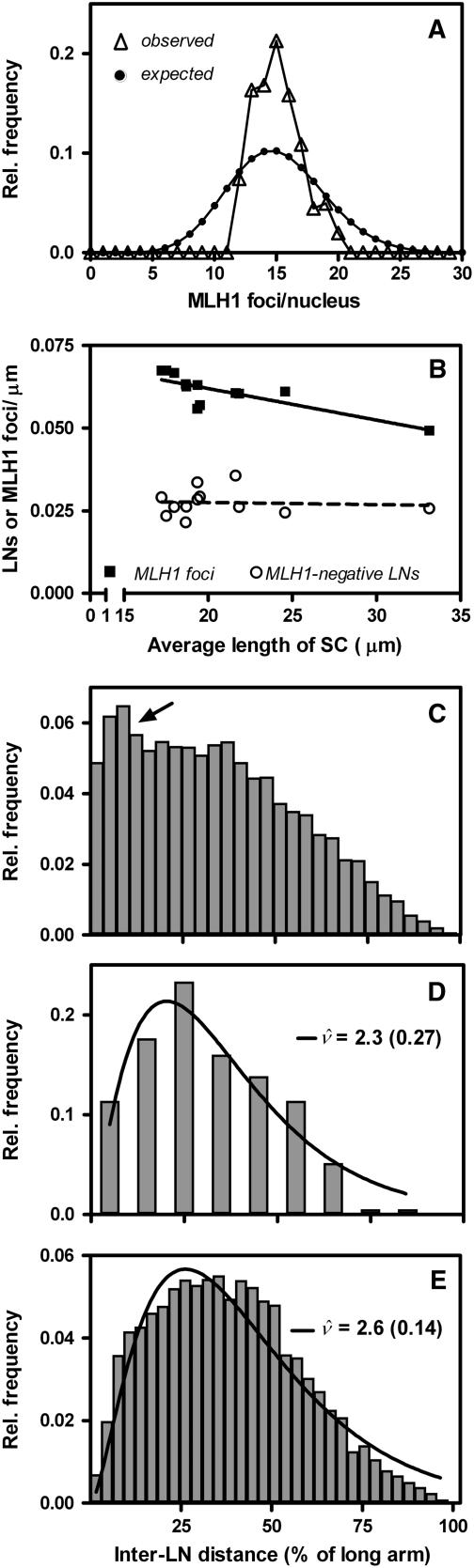
Interference among MLH1 Foci Is Much Stronger Than Interference among LNs
The MLH1-positive LNs probably correspond to yeast class I COs (de los Santos et al., 2003) because yeast Mlh1 is specifically involved in the formation of this CO type (Argueso et al., 2004). It is generally assumed that class I COs display interference and class II COs don't (de los Santos et al., 2003). CO interference manifests itself as a more even spacing of COs along the bivalents than expected if COs would be placed at random (reviewed in Hillers, 2004). It is correlated with two other features of CO distribution, namely, a much narrower frequency distribution of the numbers of COs per nucleus than expected if COs were distributed randomly among nuclei and the occurrence of at least one CO on each bivalent (the obligate CO). The nonrandom distribution of COs among nuclei, the even spacing, and the obligate CO are possibly three manifestations of the same mechanism because they are usually lost together (reviewed in Hillers, 2004; Jones and Franklin, 2006). MLH1 foci showed all three features. First, the frequency distribution of the numbers of MLH1 foci/nucleus was much narrower than the Poisson distribution that was to be expected if MLH1 foci were distributed randomly among nuclei (P < 0.001; χ2 test) (Figure 7A). Second, MLH1 foci displayed strong (cytological) interference: Using the interference parameter ν of the gamma model as a measure for the strength of interference (i.e., the evenness of spacing; not the average interfocus distance; McPeek and Speed, 1995; de Boer et al., 2006), we estimated the strength of interference among MLH1 foci on the long arms of tomato chromosomes 1 and 2 and compared it with the strength of interference among LNs (Table 2). Interference among MLH1 foci was much stronger than interference among LNs, which provides further support for the idea that MLH1 foci represent a specific subset of LNs. Third, the rarity of SCs without MLH1 foci (Table 1) is most likely a cytological manifestation of the obligate CO (i.e., each SC gets an obligate MLH1 focus, which should yield the obligate CO). We analyzed for chromosome 2 whether the low frequency of SCs with zero MLH1 foci follows directly from the observed strength of interference and average number of foci per bivalent. We assumed that the MLH1 foci are distributed uniformly along the bivalent and that the gamma model applies for the spacing of MLH1 foci (which is roughly correct; see Figure 6, Table 2). Using computer simulation, we then estimated the expected proportion of bivalents with zero, one, and two foci based on the corrected ν estimate, rounded to an integer value (ν = 5; Table 2) and the observed average number of foci per bivalent (1.31; Table 1). We found a significant difference between the observed and expected distribution of the numbers of foci per bivalent, with more bivalents with zero MLH1 foci expected than observed (P < 0.001, χ2 test; see Supplemental Table 1 online). The obligate MLH1 focus is thus not merely a consequence of the even spacing of foci (cf. Jones and Franklin, 2006). Another indication that additional assumptions are required to account for the low frequency of SCs without MLH1 foci is that the density (foci/μm) of MLH1 foci is higher on short than on long SCs (Figure 7B). This is generally considered a consequence of some mechanism that ensures at least one CO (or MLH1 focus) on each bivalent, irrespective of its length. Apparently, such a mechanism only has an effect on MLH1-positive LNs (MLH1 foci) because the density of MLH1-negative LNs was not negatively correlated with SC length (Figure 7B). This last inference should be considered with caution because we calculated the density of MLH1-negative LNs by subtracting the number of MLH1 foci (this study) from the total number of LNs as determined by Sherman and Stack (1995).
Figure 7.
Analysis of the Distributions of MLH1 Foci and LNs among Nuclei and among and along SCs.
(A) Observed and expected numbers of MLH1 foci per PMC nucleus. Open triangles represent the observed relative frequencies of MLH1 foci per PMC nucleus. Closed circles represent the frequency distribution expected if MLH1 foci were distributed randomly among PMC nuclei (i.e., a Poisson distribution for on average 15.05 MLH1 foci/nucleus).
(B) Density of MLH1-positive LNs (MLH1 foci) and MLH1-negative LNs (i.e., total LNs minus MLH1 foci) as a function of the SC length. The density of MLH1 foci (i.e., foci/μm) is higher on short than on long SCs (negative correlation; r2 = 0.63; P = 0.002); the density of MLH1-negative LNs is not correlated with SC length (r2 = 0.005; P = 0.82).
(C) to (E) Observed and expected frequency distributions of inter-LN distances on the long arm of SC 1.
(C) Frequency distribution of inter-LN distances expected if LNs would consist of a mixture of strongly interfering MLH1-positive LNs and randomly placed MLH1-negative LNs. The distribution was obtained by simulation, assuming the following: average number of MLH1-positive LNs, 1.32 (this study); average number of MLH1-negative LNs, 0.58 (= the difference between the observed average number of MLH1 foci [this study) and the average number of LNs on the long arm of SC 1 reported in Sherman and Stack [1995]); interference parameter ν for MLH1-positive LNs, 6 (the corrected ν value for MLH1 foci on the long arm of SC 1 rounded to an integer value; this study; Table 2); and random placement of MLH1-negative LNs (i.e., no interference among MLH1-negative LNs or among MLH1-negative and MLH1-positive LNs). The arrow indicates the peak of small inter-LN distances that is expected based on these assumptions.
(D) Frequency distribution of inter-LN distances on the long arm of SC 1 as observed by Sherman and Stack (1995).
(E) Frequency distribution of inter-LN distances expected if MLH1-positive and MLH1-negative LNs would both originate from the same population of weakly interfering recombination intermediates. We assumed the following: on average, 1.32 MLH1-positive and 0.58 MLH1-negative LNs per long arm of SC 1 (see above); interference parameter ν for MLH1-positive LNs, 6 (see above); recruitment of both MLH1-positive and MLH1-negative LNs from a population of intermediates displaying weak interference, corresponding to ν = 2 (ν estimate based on data from Anderson et al. [2001]; see Figure 8); and the MLH1-positive LNs are recruited in such a way that a spacing corresponding to ν = 6 is generated, whereas the MLH1-negative LNs are taken at random from the ν = 2 intermediates.
The bars represent the observed (D) or expected ([C] and [E]) relative frequencies of inter-LN distances, the curves represent the best fits to the gamma distribution, and the numbers represent the ν values (with se) for which the best fits to the gamma distribution were obtained. Because the frequency distribution in (E), and supposedly also in (D), consists of a mixture of two populations of LNs, it doesn't fit well to the gamma distribution. Note that the ν estimates for all LNs (MLH1-positive plus MLH1-negative) are much lower (ν = 2.3 for the Sherman and Stack [1995] data and ν = 2.6 for the simulation) than for MLH1-positive LNs only (ν = 6).
Table 2.
Interference among MLH1 Foci and among LNs of Tomato
| MLH1 Foci
|
LNsa
|
||||
|---|---|---|---|---|---|
| Chromosome (Long Arm) | No. of Intervals | ν (se)b | Pc | Corr. νd | ν (se)e |
| 1 | 91 | 7.9 (1.2) | 0.07 | 5.6 | 2.3 (0.27) |
| 2 | 85 | 6.9 (1.0) | 0.03 | 4.9 | 2.9 (0.15) |
All data about LNs were taken from Sherman and Stack (1995).
Maximum likelihood estimate of the interference parameter ν in the gamma model (with estimated se).
Estimated P value. P is the probability of finding an as-bad or worse fit of the observed interfocus distances to the gamma distribution due to sampling error.
Maximum likelihood estimate of ν corrected for the limited range of observable interfocus distances.
Estimate of the interference parameter ν in the gamma model (with estimated se), determined by the least squares method.
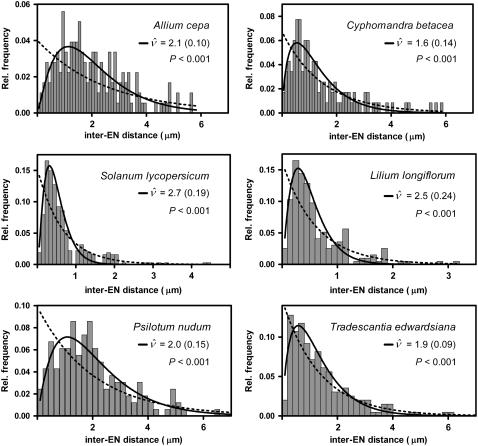
Whether MLH1-negative LNs display interference could not be analyzed directly. However, LNs cannot be considered a mixture of strongly interfering MLH1-positive LNs and randomly placed MLH1-negative LNs because the expected distribution of inter-LN distances based on this assumption shows a peak of small inter-LN distances (arrow in Figure 7C; for details, see legend of Figure 7), which is lacking from the observed distribution on all SCs, as is shown for the long arm of SC 1 in Figure 7D (for the other chromosomes, this can be inferred from Figure 17 and Table 10 in Sherman and Stack, 1995). One possible explanation could be that both MLH1-positive and MLH1-negative LNs stem from the same population of weakly interfering intermediates. In mouse, weak interference among protein complexes representing intermediate events in meiotic recombination has been observed (de Boer et al., 2006). Based on the relative frequencies of inter-EN distances (Anderson et al., 2001), we estimated that tomato ENs, which are candidate precursors of LNs (see above), display a low level of interference, corresponding to a ν-value of ∼2 (Figure 8). The distribution of inter-LN distances expected if both MLH1-positive and MLH1-negative LNs would stem from such a population of weakly interfering intermediates does not have a peak of small interfocus distances (Figure 7E; for details, see legend of Figure 7). However, other explanations for the paucity of small inter-LN distances are conceivable. For instance, Fung et al. (2004) proposed that class II COs would experience interference from class I COs but would not exert interference themselves. Under those circumstances, MLH1-negative LNs should display negative interference (discussed in de Boer and Heyting, 2006), whereas small inter-LN distances are still likely to occur between MLH1-negative LNs but not between MLH1-positive LNs or between adjacent MLH-negative LNs and MLH1-positive LNs. Testing these predictions must await the development of antibodies that recognize specifically all MLH1-negative LNs, or all LNs, so that these three types of inter-LN distances can be distinguished.
Figure 8.
Weak Interference among ENs in Six Plant Species.
The bars show the relative frequencies of inter-EN distances in six plant species, as published by Anderson et al. (2001). The continuous curves show the best fit of the observed relative frequencies of inter-EN distances to the gamma distribution. The ν values represent the estimates of the interference parameter ν (with se) for which the best fit was obtained. The dotted curves show the exponential distributions that were to be expected for the observed average inter-EN distances (Anderson et al., 2001) if there were no interference (i.e., interference parameter ν =1). The P values refer to the comparison of the fit to the gamma distribution with the fit to the exponential distribution (F test). P is the probability that the difference in sum-of-squares for the gamma model and the exponential distribution is as large as observed or larger if the exponential distribution is correct.
Interference among MLH1 Foci across Centromeres
It has been a controversial issue whether interference acts across the centromere (discussed in Colombo and Jones, 1997), although convincing evidence for chiasma interference across centromeres has been presented in various species (Colombo and Jones, 1997; Broman and Weber, 2000; Esch, 2005; Drouaud et al., 2006). In tomato, Sherman and Stack (1995) noted that there must be some control on LN numbers across centromeres because they found at least one LN on every chromosome, but not on every chromosome arm. The same was true for MLH1 foci: almost every chromosome had at least one focus, but there was not an MLH1 focus on every chromosome arm, as is evident from the average number of foci/bivalent, which was <2 for all chromosomes (Table 1). Because MLH1 foci are excluded from pericentromeric regions (Figure 6), factors other than interference codetermine the interfocus distances spanning the centromeres. Therefore, we could not estimate the strength of interference across centromeres from the distribution of interfocus distances. However, we found that the presence of a focus on one arm influenced the positions of foci on the other arm. For instance, the position of a single focus on the long arm is on average more distal if there is also a focus on the short arm than if there is no focus on the short arm (Figure 9). For six of the 11 analyzed SCs, this difference was significant at the 5% level (P values in Figure 9), which indicates positive interference among MLH1 foci across the centromeres of these SCs.
Figure 9.
Interference across the Centromere.
Shown are the cumulative frequencies of MLH1 foci on the long arm of the SC as a function of the distance to the telomeric end of the long arm of the SC. The distance is expressed as a percentage of the length of the long arm of the SC on which the focus or LN was. The curves show for every position on the SC the percentage of MLH1 foci (or LNs) that is found on the SC stretch between that position and the telomeric end of the long arm. The positions of MLH1 foci on the long arms of SCs with one MLH1 focus on the long arm and one MLH1 focus on the short arm are shown in blue. The positions of MLH1 foci on the long arms of SCs with one MLH1 focus on the long arm and no focus on the short arm are shown in red. The numbers of MLH1 foci or LNs on which the curves are based are indicated in the top left corners of the panels. For each chromosome, the P value represents the probability that the MLH1 focus positions represented in blue and the MLH1 focus positions represented in red stem from the same distribution (Kolmogorov-Smirnov two-sample test).
DISCUSSION
We have generated antibodies against tomato MLH1 and against AEs and centromeres of tomato to construct a map of MLH1 focus locations along individually identified pachytene SCs in spread tomato PMCs. The immunocytochemical studies performed with these antibodies (Figures 2 to 4) confirmed and extended observations made in other organisms regarding the localization of MLH1 (Moens et al., 2002; Marcon and Moens, 2003; Higgins et al., 2005), SMC1 and SMC3 (Lam et al., 2005b), and CENPC (Dawe et al., 1999) in meiotic prophase meiocytes (reviewed for plants in Hamant et al., 2006). The antibodies add to an extensive list of attributes (reviewed in Stack and Anderson, 1986b) that make tomato a unique experimental organism for cytological studies of meiosis. In this study, we have taken advantage of this when comparing our MLH1 focus map with an existing ultrastructural map of LN locations along tomato SCs.
MLH1-Positive and MLH1-Negative LNs
We inferred from the differences between LNs and MLH1 foci with respect to their numbers and distributions among and along SCs that there are two types of LNs: MLH1-positive and MLH1-negative LNs. Both types most likely mark CO sites because there is a close correspondence between the presence of LNs and the occurrence of chiasmata not only in tomato (Havekes et al., 1994; Sherman and Stack, 1995) but also in various other species (reviewed in Zickler and Kleckner, 1999; Anderson and Stack, 2005). All MLH1-positive LNs in tomato are likely to correspond to yeast class I COs because in yeast meiosis Mlh1 contributes specifically to class I COs (Argueso et al., 2004). For reasons discussed below, it seems likely that MLH1-positive LNs (MLH1 foci) mark all class I COs in wild-type tomato. In other words, in wild-type tomato, there is probably a 1:1 correlation between the presence of an MLH1 focus and the occurrence of a class I CO. It is possible that the MLH1-negative LNs correspond to yeast class II COs, but this still needs to be confirmed, for instance, by the detection of tomato proteins homologous to yeast Mus81 or Mms4 in MLH1-negative LNs. Although the two proposed CO types in tomato are thought to be formed by different sets of recombination proteins, they are marked by morphologically similar, if not indistinguishable, LNs (Figure 4C; Sherman and Stack, 1995). Possibly, the ultrastructural appearance of tomato LNs is not primarily determined by recombination proteins, but, for instance, by protein complexes that crosswise link nonsister chromatid axes.
Interference among MLH1 Foci and among LNs
The correspondence of MLH1-positive LNs with yeast class I COs is not only indicated by the presence of MLH1 but also by the strong interference among MLH1 foci. Tomato has two types of CO, of which the MLH1-marked type displays strong interference. A similar situation likely exists in Arabidopsis. The Arabidopsis mutants mer3 (also known as rck) and msh4, which are each deficient for another ZMM protein, still form 15 to 30% of the wild-type level of COs, which are randomly distributed among nuclei and might correspond to yeast class II COs (Higgins et al., 2004; Chen et al., 2005; Mercier et al., 2005). Furthermore, Copenhaver et al. (2002) analyzed in wild-type Arabidopsis whether the distribution of genetic inter-CO distances was consistent with the coexistence of an interfering and a noninterfering CO type. They estimated from genetic data that ∼80% of the COs in wild-type Arabidopsis would belong to a strongly interfering CO type; (genetic) interference among these COs would be as strong as or stronger than (cytological) interference among MLH1-positive LNs in tomato (Table 2; Copenhaver et al., 2002). Twenty percent would belong to a noninterfering CO type, which fits well with the amount of COs found in Arabidopsis ZMM mutants. These estimates were based on the assumption that the noninterfering COs were placed at random on a background of interfering COs (i.e., noninterfering COs would not influence the position of any other CO, and interfering COs would only influence the position of other interfering COs). However, in tomato, the observed distribution of inter-LN distances (Figure 7D; Sherman and Stack, 1995) is not consistent with this assumption because it lacks a peak of small inter-LN distances that should occur if noninterfering COs were scattered randomly among strongly interfering COs (Figure 7C). We propose therefore that both CO types in tomato are derived from common precursors that display already weak interference (Figure 8). That would imply that interference is imposed in two steps and that the strong interference among class I–type (MLH1-marked) COs is imposed after the weak interference among precursors has been established. This has implications for the development of gene-mapping algorithms and the recognition of possible genotyping errors in gene mapping studies (discussed in Housworth and Stahl, 2003). If there were no interference among the recombination intermediates from which both types of COs are derived, tight double COs are expected to occur occasionally, and recombinants that appear to result from tight double COs are thus not necessarily due to genotyping errors. However, if there is already some interference among these intermediates, tight double COs are less likely to occur.
Copenhaver et al. (2002) and Lam et al. (2005a) inferred that the proportion of noninterfering COs would be different on individual Arabidopsis chromosomes, with few noninterfering COs on the two nucleolus-organizing regions (NORs) bearing chromosomes. They speculate that the noninterfering COs are a by-product of recombinational interactions that serve homolog recognition and that such interactions are not required on the two NOR-bearing chromosomes because NORs would serve as homolog recognition sites. However, in tomato, the proportion of MLH1-negative LNs was similar on all tomato chromosomes, including the NOR-bearing chromosome 2. Likewise, Higgins et al. (2004) concluded that MSH4-independent chiasmata in Arabidopsis were distributed randomly among all chromosomes, including the two NOR-bearing chromosomes.
MLH1-Positive LNs Provide the Obligate COs
In several species (reviewed in Zickler and Kleckner, 1999), including tomato (Sherman and Stack, 1995), the density of LNs is higher on short than on long chromosomes, and this is considered a consequence of the formation of at least one LN on each bivalent (the obligate LN). It now turns out that tomato PMCs specifically control the occurrence of at least one MLH1-positive LN (MLH1 focus) rather than one LN on each SC (Table 1, Figure 7B), and this control does not detectably affect the distribution of MLH1-negative LNs (Figure 7B). Because MLH1-positive LNs display strong interference, this represents yet another correlation between the phenomena of interference and the obligate CO (or MLH1 focus). As has been discussed by Jones and Franklin (2006), this correlation can be due to a dependence of both phenomena on a common component or to a mechanistic link between he two phenomena. A mechanistic link (if any) could be partly due to a sequential determination of MLH1 focus positions, so that the first (prospective) MLH1 focus on an SC will not experience interference.
Because SCs without MLH1 focus are rare (Table 1), virtually all obligate CO positions are marked by MLH1. This implies that MLH1 foci mark virtually all class I CO positions in wild-type tomato, unless obligate COs have a higher probability of being marked by MLH1 than other class I COs, which seems unlikely. At first sight, this contrasts with observations in yeast mlh1 and Arabidopsis mlh3 mutants, which suggest that part of the prospective class I COs are resolved as COs in the absence of Mlh1 and At MLH3, respectively (Argueso et al., 2004; Jackson et al., 2006). However, it is possible that these mutants resolve some prospective class I COs (i.e., ZMM-dependent CO precursors) into COs via pathways that are not normally used for this purpose (Hoffmann and Borts, 2004; discussed in Franklin et al., 2006). The cytological labeling of MLH1 in tomato suggests that in a wild-type situation, MLH1 is involved in all class I COs.
The obligate MLH1-positive LN (obligate MLH1 focus) should yield the obligate CO, which is thought to ensure proper chromosome disjunction at meiotic metaphase I. Whether (COs resulting from) MLH1-negative LNs can ensure chromosome disjunction remains to be investigated.
A Provisional Map of MLH1 Focus Positions
The preparation of antibodies against SCs and centromeres of tomato has paved the way for the construction of an immunocytochemical recombination map, and in this article, we present a provisional map of MLH1 focus positions along tomato SCs (Figure 6). As discussed above, we could make several inferences from a comparison of this map with an existing ultrastructural map of LN positions, thanks to a remarkably good correspondence between the two maps (Figure 5). As we have noted above, we consider the distinction between SCs of the same group (Figure 5) and the distinction between the long and short arms of chromosomes 5 and 12 tentative, and the provisional MLH1 focus map (Figure 6) can therefore be further improved by combining the immunofluorescence labeling with fluorescence in situ hybridization so that all chromosomes and chromosome arms can be identified unequivocally (cf. Froenicke et al., 2002).
The immunocytological map will become even more useful if suitable immunocytochemical markers of MLH1-negative LNs and/or all LNs become available because this will enable us to construct an immunofluorescence recombination map of tomato that includes all COs. That will allow us to (1) consolidate in a single investigation all inferences that in this study were based on a comparison of two data sets (immunofluorescence and electron microscopy) from different laboratories; (2) develop realistic gene-mapping algorithms, which might improve the efficiency of crosses; (3) identify CO-poor or CO-free regions (e.g., in hybrids); (4) analyze both MLH1-negative and MLH1-positive LNs and the interaction (if any) between the two types of LNs in wild-type and mutant backgrounds; and (5) study the regulation of the numbers and positions of class I and class II COs in higher plants.
METHODS
Plants
In all experiments, we used plants of the same line of diploid tomato (Solanum lycopersicum var cherry) as had been used by Sherman and Stack (1992, 1995). Seeds from this line (kindly provided by S.M. Stack and L.K. Anderson) were sown bimonthly, and plants were grown to maturity in a climate chamber under a 16-h-light/8-h-dark rhythm and a relative humidity of 70%. For each experiment, we sampled flower buds from at least six individuals. Plants were discarded when they were 4 months old.
Antibodies
For the production of antibodies against tomato proteins, we identified ESTs of tomato by BLAST searches of the SOL Genomics Network EST libraries (http://www.sgn.cornell.edu/tools/blast/simple.pl) based on known amino acid sequences of homologous proteins in Arabidopsis thaliana and other species. Then, using primers derived from the identified ESTs, we performed PCR on cDNA preparations from various tomato tissues and cloned the thus obtained cDNA fragments in the pGEMT-easy vector (Promega) and then in pET28 (Novagen). After inspection of the inserts in the pET28 constructs by sequencing, the encoded peptides were produced in Escherichia coli from the pET28 constructs, purified on a Ni2+-NTA resin (Qiagen), and used for immunization as described (Offenberg et al., 1991). This yielded rabbit sera 5 (anti-tomato SMC1 or α Sl SMC1), 623 (α Sl SMC3), 621 (α Sl MLH1, N-terminal peptide), 8 (α Sl MLH1, C-terminal peptide), and 20 (α Sl CENPC) and rat sera 501 and 502 (α Sl CENPC). For immunocytology and immunoblotting, we affinity-purified antibodies from these sera on columns that carried the peptides that we had used for immunization. Most immunofluorescence experiments were performed with α Sl SMC1 (serum 5), α Sl MLH1 (serum 8), and α Sl CENPC (serum 20).
Immunocytochemistry
The progression of meiotic prophase in tomato PMCs is correlated with the size of the flower buds. After collecting buds of the appropriate size, we squashed PMCs from one anther of each bud in 2% aceto-carmine in 45% acetic acid and verified the stage of meiotic prophase as described (Havekes et al., 1994). We then spread the PMCs from the other four anthers by the hypotonic bursting technique (Stack, 1982; Sherman and Stack, 1992) and performed immunocytochemical labeling as described (de Vries et al., 2005) using affinity-purified antibodies from the above-mentioned antisera. In most experiments, we used antibodies from rabbit serum 8 (α Sl MLH1, C-terminal peptide) for labeling MLH1 foci, rabbit serum 5 (α Sl SMC1) for labeling AEs of SCs, and rabbit serum 20 (α Sl CENPC) for labeling kinetochore regions. Because these three sera had been elicited in the same species, rabbit, we performed immunofluorescent triple labeling as follows. First, we incubated the slides in α Sl MLH1 and visualized binding of the α Sl MLH1 antibodies using fluorescein isothiocyanate–conjugated Fab' fragments of goat-anti-rabbit antibodies (Jackson Immunoresearch). Then, we incubated the slides in, successively, α Sl SMC1 and Texas Red–conjugated goat-anti-rabbit antibodies and mounted them in Vectashield (Vector Laboratories) containing 2 μg/mL DAPI. The Sl MLH1 (green), Sl SMC1 (red), and DAPI (blue) images of pachytene nuclei were then micrographed through a Zeiss Axiovert epifluorescence microscope equipped with a ×100 Plan Apo objective (numerical aperture of 1.32), a computer-controlled stage, a Zeiss Axiocam MRc CCD camera, and Zeiss Axiovision rel. 4.4 software, and the positions of the micrographed nuclei on the slides were stored in a database for the next round of observations. Subsequently, we removed the cover slips and successively exposed the slides to α Sl CENPC and Alexa 488–conjugated goat-anti-rabbit antibodies (Jackson), mounted them again in Vectashield containing 2 μg/mL DAPI, and recorded the green (Sl CENPC, Sl SMC1, and Sl MLH1) and blue (DAPI) images of the same nuclei that had been micrographed before. All antibody incubations were preceded by blocking incubations and followed by washes as described (Heyting and Dietrich, 1991). The blocking and antibody incubations were performed in PBS (140 mM NaCl and 10 mM sodium phosphate, pH 7.4) containing 3% BSA, 0.1% Triton X-100, and 0.01% sodium azide; washes were performed in PBS. We processed and combined the images using Adobe Photoshop and the public domain ImageJ program (developed at the U.S. National Institutes of Health by Wayne Rasband and available on the Internet at http://rsb.info.nih.gov/ij/).
For immunogold labeling and ultrastructural analysis, we spread tomato PMCs by the above-described procedure on slides that had been coated with polystyrene and glow discharged as follows: brand new slides were wiped once with a Kim wipe tissue and dipped for 10 s in a solution consisting of 0.6% polystyrene-co-acrylonitrile in 1,2-dichloroethane. The excess of plastic solution was drained, and the slides were allowed to air-dry vertically in a tube rack above a filter paper. The plastic-coated slides were glow discharged under argon at 0.1 Torr and 3 A for 5 min. After spreading PMCs on these slides, we wiped the back of the slides with a wet tissue to remove the plastic film and sealed the edges of the slides with clear nail polish to prevent unwanted lifting of the plastic during the subsequent incubation steps. For immunogold labeling, we blocked the slides for 30 min at room temperature on drops of filter-sterilized blocking buffer containing 0.05% NaN3. Then, we incubated the slides for 1 h at 37°C, 48 h at 4°C, and again 1 h at 37°C on drops containing affinity-purified rabbit-α Sl MLH1 antibodies in blocking buffer. After three washes of 5 min each on drops of PBS, the slides were incubated for 90 min at 37°C on drops of 6-nm gold-conjugated goat anti-rabbit IgG in blocking buffer. The slides were washed three times for 5 min on drops of PBS and then fixed for 10 min at room temperature on drops of 2% paraformaldehyde in PBS. Then followed three washes of 5 min each on drops of MilliQ water, and the slides were allowed to dry thoroughly overnight at room temperature. The immunogold-labeled slides were stained with 5% aqueous uranyl acetate for 5 min at room temperature in the dark, washed four times for 5 min on drops of MilliQ water, and then allowed to thoroughly dry on filter paper. We selected well-spread cells by phase contrast microscopy and scored the plastic around these cells with a razor blade. Then, we detached the plastic film fragments supporting the selected PMCs from the glass slide by applying 1% hydrofluoric acid on the scored edges and floated the film fragments off the slides by dipping the slides gently into a beaker containing filter-sterilized MilliQ water. The 100-μm mesh nickel grids were then placed onto the plastic rafts, and the plastic-coated grids were collected onto Parafilm and then allowed to dry thoroughly overnight on filter paper. We observed the grids in a JEM 1200 EX II transmission electron microscope (JEOL) operating at 80 keV and captured images using a Keenview 12-bit CCD TEM camera using the iTEM software package (SiS)
Image and Data Analysis
We only analyzed PMC spreads that fulfilled the following criteria: (1) the signal-to-background ratio for both MLH1 and SMC1 was sufficiently high to allow unambiguous identification of the MLH1 foci and tracing of all SCs from one end to the other; (2) the cells displayed full synapsis, which is indicative of the pachytene stage; and (3) the kinetochores, as visualized by CENPC labeling, were clear and well-delimited, which is characteristic of mid to late pachytene SCs (see Supplemental Figure 1 online; Stack and Anderson, 1986a). For the generation of the provisional MLH1 focus map, we only analyzed PMC spreads with complete sets of 12 SCs; if one or more chromosomes in a given set could not be identified based on the relative length and arm length ratio, the entire set was discarded. For counting the number of MLH1 foci/SCs (Table 1) and interference measurements (Table 2), we also analyzed SCs in incomplete sets, provided that the SCs could be identified unambiguously. The average length of each of the 12 tomato chromosomes was calculated from all available complete sets.
For measurements of the lengths of SCs and AEs and the positions of foci on SCs, we prepared a macro for the public domain image analysis program Object Image, which is an extended version of NIH Image by N. Vischer (University of Amsterdam) and is available at http://simon.bio.uva.nl/object-image.html. The macro allows batch measurement of SC lengths and positions of MLH1 foci and centromeres for several nuclei. For the construction of the map of MLH1 focus positions, we analyzed only nuclei with complete sets of SCs. The data were collected in a spreadsheet, which was exported into Microsoft Excel for further analysis.
We estimated the strength of interference among MLH1 foci or LNs on the long arms of chromosomes 1 and 2 using the interference parameter ν of the gamma model as a measure for the strength of interference (de Boer et al., 2006). For MLH1 foci, we obtained a first estimate of ν by fitting the observed interfocus distances to the gamma distribution by the maximum likelihood method using the GENSTAT software package (VSN International). Subsequently, using computer simulations, we applied a correction for the limited range of interfocus distances that we can observe, as described (de Boer et al., 2006); this correction leads to slightly lower estimates of ν. For LNs, we estimated the ν values by fitting the relative frequencies of inter-LN distances on the long arms of chromosomes 1 and 2 (Table 10 in Sherman and Stack, 1995) to the gamma distribution by the least squares method using the GraphPad Prism 4 program (GraphPad Software); by the same procedure, we estimated the strength of interference among ENs of tomato from the relative frequencies of inter-EN distances reported by Anderson et al. (2001). We determined the expected distribution of the numbers of foci per bivalent of chromosome 2 for the estimated ν value and the observed average number of MLH1 foci on SC 2 by simulating the positions of MLH1 foci on 10,000 chromosomes using a macro in Microsoft Excel. The macro is available upon request from the corresponding author.
Accession Numbers
Sequences data of the tomato cDNA fragments can be found in the GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) data library under accession numbers EF071926 (Sl CENPC1), EF071927 (Sl MLH1), EF071928 (Sl SMC3), and EF071929 (Sl SMC3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Immunofluorescence Triple Labeling of Spread Pollen Mother Cells in Early, Mid, and Late Pachytene.
Supplemental Figure 2. Immunogold Labeling of MLH1 in Late Recombination Nodules of Tomato.
Supplemental Table 1. Observed versus Expected Distribution of the Numbers of Foci per SC of Chromosome 2.
Supplementary Material
Acknowledgments
We thank S.M. Stack and L.K. Anderson (Colorado State University) for instructing F.G.P.L. in the techniques of spreading and labeling tomato PMCs and for providing us with seeds of the tomato line used by them in previous studies. We also thank E. Holub and the animal and plant facilities at Wageningen University for expert technical assistance and E. de Boer for useful comments.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Franck G.P. Lhuissier (franck.lhuissier@keygene.com) and Christa Heyting (christa.heyting@wur.nl).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Agarwal, S., and Roeder, G.S. (2000). Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102 245–255. [DOI] [PubMed] [Google Scholar]
- Anderson, L.K., Hooker, K.D., and Stack, S.M. (2001). The distribution of early recombination nodules on zygotene bivalents from plants. Genetics 159 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L.K., Offenberg, H.H., Verkuijlen, W.C., and Heyting, C. (1997). RecA-like proteins make part of early meiotic nodules in lily. Proc. Natl. Acad. Sci. USA 94 6868–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L.K., Reeves, A., Webb, L.M., and Ashley, T. (1999). Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L.K., and Stack, S.M. (2005). Recombination nodules in plants. Cytogenet. Genome Res. 109 198–204. [DOI] [PubMed] [Google Scholar]
- Argueso, J.L., Wanat, J., Gemici, Z., and Alani, E. (2004). Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, S.M., Plug, A.W., Prolla, T.A., Bronner, C.E., Harris, A.C., Yao, X., Christie, D.-M., Monell, C., Arnheim, N., Bradley, A., Ashley, T., and Liskay, R.M. (1996). Involvement of mouse MLH1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13 336–342. [DOI] [PubMed] [Google Scholar]
- Börner, G.V., Kleckner, N., and Hunter, N. (2004). Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117 29–45. [DOI] [PubMed] [Google Scholar]
- Broman, K.W., Rowe, L.B., Churchill, G.A., and Paigen, K. (2002). Crossover interference in the mouse. Genetics 160 1123–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman, K.W., and Weber, J.L. (2000). Characterization of human crossover interference. Am. J. Hum. Genet. 66 1911–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Zhang, W., Timofejeva, L., Gerardin, Y., and Ma, H. (2005). The Arabidopsis ROCK-N-ROLLERS gene encodes a homolog of the yeast ATP-dependent DNA helicase MER3 and is required for normal meiotic crossover formation. Plant J. 43 321–334. [DOI] [PubMed] [Google Scholar]
- Choo, K.H. (1998). Why is the centromere so cold? Genome Res. 8 81–82. [DOI] [PubMed] [Google Scholar]
- Chua, P.R., and Roeder, G.S. (1998). Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93 349–359. [DOI] [PubMed] [Google Scholar]
- Colombo, P.C., and Jones, G.H. (1997). Chiasma interference is blind to the centromere. Heredity 79 214–227. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P., Housworth, E.A., and Stahl, F.W. (2002). Crossover interference in Arabidopsis. Genetics 160 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R.K., Reed, L.M., Yu, H.G., Muszynski, M.G., and Hiatt, E.N. (1999). A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer, E., and Heyting, C. (2006). The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma 115 220–234. [DOI] [PubMed] [Google Scholar]
- de Boer, E., Stam, P., Dietrich, A.J.J., Pastink, A., and Heyting, C. (2006). Two levels of interference in mouse meiotic recombination. Proc. Natl. Acad. Sci. USA 103 9607–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos, T., Hunter, N., Lee, C., Larkin, B., Loidl, J., and Hollingsworth, N.M. (2003). The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, F.A.T., de Boer, E., van den Bosch, M., Baarends, W.M., Ooms, M., Yuan, L., Liu, J.-G., Heyting, C., and Pastink, A. (2005). Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19 1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud, J., Camilleri, C., Bourguignon, P.Y., Canaguier, A., Berard, A., Vezon, D., Giancola, S., Brunel, D., Colot, V., Prum, B., Quesneville, H., and Mezard, C. (2006). Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res. 16 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijpe, M., Heyting, C., Gross, B., and Jessberger, R. (2000). Association of mammalian SMC1 and SMC3 proteins with meiotic chromosomes and synaptonemal complexes. J. Cell Sci. 113 673–682. [DOI] [PubMed] [Google Scholar]
- Esch, E. (2005). Estimation of gametic frequencies from F2 populations using the EM algorithm and its application in the analysis of crossover interference in rice. Theor. Appl. Genet. 111 100–109. [DOI] [PubMed] [Google Scholar]
- Franklin, F.C., Higgins, J.D., Sanchez-Moran, E., Armstrong, S.J., Osman, K.E., Jackson, N., and Jones, G.H. (2006). Control of meiotic recombination in Arabidopsis: Role of the MutL and MutS homologues. Biochem. Soc. Trans. 34 542–544. [DOI] [PubMed] [Google Scholar]
- Froenicke, L., Anderson, L.K., Wienberg, J., and Ashley, T. (2002). Male mouse recombination maps for each autosome identified by chromosome painting. Am. J. Hum. Genet. 71 1353–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, J.C., Rockmill, B., Odell, M., and Roeder, G.S. (2004). Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116 795–802. [DOI] [PubMed] [Google Scholar]
- Hamant, O., Ma, H., and Cande, W.Z. (2006). Genetics of meiotic prophase I in plants. Annu. Rev. Plant Biol. 57 267–302. [DOI] [PubMed] [Google Scholar]
- Havekes, F.W.J., de Jong, J.H., Heyting, C., and Ramanna, M.S. (1994). Synapsis and chiasma formation in four meiotic mutants of tomato (Lycopersicon esculentum). Chromosome Res. 2 315–325. [DOI] [PubMed] [Google Scholar]
- Heyting, C., and Dietrich, A.J. (1991). Meiotic chromosome preparation and protein labeling. Methods Cell Biol. 35 177–202. [DOI] [PubMed] [Google Scholar]
- Higgins, J.D., Armstrong, S.J., Franklin, F.C.H., and Jones, G.H. (2004). The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev. 18 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, J.D., Sanchez-Moran, E., Armstrong, S.J., Jones, G.H., and Franklin, F.C. (2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers, K.J. (2004). Crossover interference. Curr. Biol. 14 R1036–R1037. [DOI] [PubMed] [Google Scholar]
- Hoffmann, E.R., and Borts, R.H. (2004). Meiotic recombination intermediates and mismatch repair proteins. Cytogenet. Genome Res. 107 232–248. [DOI] [PubMed] [Google Scholar]
- Hollingsworth, N.M., and Brill, S.J. (2004). The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes Dev. 18 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housworth, E.A., and Stahl, F.W. (2003). Crossover interference in humans. Am. J. Hum. Genet. 73 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, N., Sanchez-Moran, E., Buckling, E., Armstrong, S.J., Jones, G.H., and Franklin, F.C.H. (2006). Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J. 15 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G.H., and Franklin, F.C. (2006). Meiotic crossing-over: Obligation and interference. Cell 126 246–248. [DOI] [PubMed] [Google Scholar]
- Klein, F., Mahr, P., Galova, M., Buonomo, S.B., Michaelis, C., Nairz, K., and Nasmyth, K. (1999). A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98 91–103. [DOI] [PubMed] [Google Scholar]
- Lam, S.Y., Horn, S.R., Radford, S.J., Housworth, E.A., Stahl, F.W., and Copenhaver, G.P. (2005. a). Crossover interference on nucleolus organizing region-bearing chromosomes in Arabidopsis. Genetics 170 807–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, W.S., Yang, X., and Makaroff, C.A. (2005. b). Characterization of Arabidopsis thaliana SMC1 and SMC3: Evidence that AtSMC3 may function beyond chromosome cohesion. J. Cell Sci. 118 3037–3048. [DOI] [PubMed] [Google Scholar]
- Lipkin, S.M., et al. (2002). Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31 385–390. [DOI] [PubMed] [Google Scholar]
- Marcon, E., and Moens, P. (2003). MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 165 2283–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek, M.S., and Speed, T.P. (1995). Modeling interference in genetic recombination. Genetics 139 1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier, R., Jolivet, S., Vezon, D., Huppe, E., Chelysheva, L., Giovanni, M., Nogue, F., Doutriaux, M.P., Horlow, C., Grelon, M., and Mezard, C. (2005). Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3, whereas the other one is not. Curr. Biol. 15 692–701. [DOI] [PubMed] [Google Scholar]
- Moens, P.B., Kolas, N.K., Tarsounas, M., Marcon, E., Cohen, P.E., and Spyropoulos, B. (2002). The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115 1611–1622. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. (2001). Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35 673–745. [DOI] [PubMed] [Google Scholar]
- Offenberg, H.H., Dietrich, A.J.J., and Heyting, C. (1991). Tissue distribution of two major components of synaptonemal complexes of the rat. Chromosoma 101 83–91. [DOI] [PubMed] [Google Scholar]
- Page, S.L., and Hawley, R.S. (2004). The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20 525–558. [DOI] [PubMed] [Google Scholar]
- Revenkova, E., Eijpe, M., Heyting, C., Hodges, C., Hunt, P.A., Liebe, B., Scherthan, H., and Jessberger, R. (2004). Cohesin SMC1β is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 6 555–562. [DOI] [PubMed] [Google Scholar]
- Sherman, J.D., and Stack, S.M. (1992). Two-dimensional spreads of synaptonemal complexes from solanaceous plants. V. Tomato (Lycopersicon esculentum) karyotype and idiogram. Genome 35 907–915.1473725 [Google Scholar]
- Sherman, J.D., and Stack, S.M. (1995). Two-dimensional spreads of synaptonemal complexes from solanaceous plants. IV. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141 683–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack, S. (1982). Two-dimensional spreads of synaptonermal complexes from solanaceous plants. I. The technique. Stain Technol. 57 265–272. [DOI] [PubMed] [Google Scholar]
- Stack, S.M., and Anderson, L.K. (1986. a). Two-dimensional spreads of synaptonemal complexes from solanaceous plants. II. Synapsis in Lycopersicon esculentum (tomato). Am. J. Bot. 73 264–281. [Google Scholar]
- Stack, S.M., and Anderson, L.K. (1986. b). Two-dimensional spreads of synaptonemal complexes from solanaceous plants. III. Recombination nodules and crossing over in Lycopersicon esculentum (tomato). Chromosoma 94 253–256. [Google Scholar]
- Tanksley, S.D., et al. (1992). High density molecular linkage maps of the tomato and potato genomes. Genetics 132 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T.F., Kleckner, N., and Hunter, N. (1999). Functional specificity of MutL homologs in yeast: Evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. USA 96 13914–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby, M.C. (2005). Making crossovers during meiosis. Biochem. Soc. Trans. 33 1451–1455. [DOI] [PubMed] [Google Scholar]
- Zickler, D., and Kleckner, N. (1999). Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33 603–754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.