Abstract
Arabidopsis thaliana seedling growth with pure oxylipins resulted in root waving, loss of root apical dominance, and decreased root elongation. 9-Hydroxyoctadecatrienoic acid (9-HOT) was a potent inducer of root waving. Studies with noxy2 (for nonresponding to oxylipins2), a new 9-HOT–insensitive mutant, and coronatine insensitive1-1 (jasmonate-insensitive) revealed at least three signaling cascades mediating the oxylipin actions. Treatment with 9-HOT resulted in a reduction in lateral roots and an increase in stage V primordia. Roots showed strong 9-lipoxygenase (9-LOX) activity, and root primordia expressed 9-LOX genes. These results, along with findings that noxy2 and mutants with defective 9-LOX activity showed increased numbers of lateral roots, suggest that 9-HOT, or a closely related 9-LOX product, is an endogenous modulator of lateral root formation. Histochemical and molecular analyses revealed that 9-HOT activated events common to development and defense responses. A subset of 9-HOT–responding root genes was also induced in leaves after 9-HOT treatment or pathogen inoculation. The results that noxy2 displayed altered root development, enhanced susceptibility to Pseudomonas, and reduced the activation of 9-HOT–responding genes are consistent with mechanistic links among these processes. The nature of the changes detected suggests that oxylipins from the 9-LOX pathway function in cell wall modifications required for lateral root development and pathogen arrest.
INTRODUCTION
The capacity of plants to survive adverse conditions and reach reproductive maturity critically depends on their ability to continuously adapt to changes in the environment. Therefore, plants have evolved an array of intricate regulatory mechanisms that involve the generation of signaling molecules mediating the activation of required adaptive responses. In particular, the activation of pathogen-specific defense mechanisms upon microbial infection, as well as the acquisition of structural and physiological adjustments to environmental changes, permit survival, plant development, and reproduction.
A common group of metabolites playing a fundamental role in the physiological and pathological responses of plants and vertebrates are lipid derivatives generated by oxygenation and further transformation of fatty acids. In mammals, eicosanoids, including prostaglandins and leukotrienes, are generated mainly by the oxygenation of arachidonic acid, and their receptor-mediated roles in various processes, such as the immune response, fever, pain, and inflammation, are well understood. By contrast, information regarding the roles of octadecanoids derived from linoleic and linolenic acids in plants is more limited.
The biosynthesis of plant oxylipins is initiated by the action of lipoxygenases (9-LOX and 13-LOX) or α-dioxygenase (α-DOX), which catalyze the oxygenation of predominantly linoleic acid (18:2) and linolenic acid (18:3) into reactive hydroperoxides. Secondary transformations of these hydroperoxides are catalyzed by allene oxide synthases, divinyl ether synthases, hydroperoxide lyases, peroxygenases, and epoxy alcohol synthase. The oxygenated derivatives thus formed include the phytohormone jasmonic acid (JA) as well as reactive oxylipins possessing epoxide, conjugated carbonyl, or aldehyde functionalities (reviewed in Blée, 2002; Howe and Schilmiller, 2002; Liavonchanka and Feussner, 2006). A schematic representation of the oxylipin metabolic pathways and of the products synthesized is provided in Supplemental Figure 1 online.
LOX-derived oxylipins are involved in physiological processes of plants, such as growth and fertility (Creelman and Mullet, 1997; Sanders et al., 2000; Stintzi and Browse, 2000) and mechanotransduction (Stelmach et al., 1998). In addition, oxylipins have important roles in the adaptation of plants to adverse growth conditions (Staswick et al., 1992; Creelman and Mullet, 1995; Armengaud et al., 2004) and in the defense reactions that take place as a consequence of the infection of plants with bacterial and fungal pathogens. Numerous studies aimed at defining the action of oxylipins have shown that the expression of genes encoding the enzymes initiating the synthesis of oxylipins is specifically induced upon inoculation with plant pathogens (Melan et al., 1993; Sanz et al., 1998; Jalloul et al., 2002; Turner et al., 2002). Moreover, alterations in the synthesis of oxylipins in mutants and transgenic lines have been shown to modify the plant response to pathogen infection (Rancé et al., 1998; Vijayan et al., 1998; De León et al., 2002; Farmer et al., 2003).
Analytical approaches have identified the different types of oxylipins produced in plants (Mueller et al., 2006). Furthermore, improvements of synthetic and biosynthetic methodologies provide increasing quantities and numbers of oxylipins for use in biological work (Prost et al., 2005). It is known that oxylipins have antimicrobial effects, stimulate plant defense gene expression, and regulate plant cell death. In vitro studies using pure derivatives showed that oxylipins impair the growth of some plant microbial pathogens, including bacteria, oomycetes, and fungi (Croft et al., 1993; Weber et al., 1999; Prost et al., 2005). On the other hand, many 13-LOX–derived compounds, including the hormone JA, 12-oxophytodienoic acid (OPDA), 13-hydroxyoctadecatrienoic acid (13-HOT), and 13-hydroperoxide lyase–derived C6 aldehydes, are regulators of plant defense gene expression (Bate and Rothstein, 1998; Weichert et al., 1999; Alméras et al., 2003). Also, electrophilic ketodienes (KODs) and ketotrienes (KOTs), derived from linoleic and linolenic acids, respectively, were shown to induce plant defense genes (Vollenweider et al., 2000). Importantly, JA has been shown to be a key defense molecule that interacts with additional defense pathways to control resistance to necrotrophic pathogens (Penninckx et al., 1996; Glazebrook, 2005). Activation of the oxylipin pathways can also lead to the production of molecules regulating plant cell death. Thus, apart from being precursors of numerous other oxylipins, LOX-generated 9- and 13-hydroperoxides have been shown to affect plant cell viability, and a role of hydroperoxides in regulating localized cell death during the hypersensitive reaction has been suggested (Rustérucci et al., 1999). Similarly, fatty acid KODs and KOTs were shown to display cell death–promoting activities (Vollenweider et al., 2000). Also, the α-DOX pathway contributes to plant defense against bacterial infection by controlling the development of the hypersensitive reaction (De León et al., 2002; Hamberg et al., 2003).
Despite increasing experimental evidence, the mechanisms by which oxylipins exert their activity remain essentially unknown. JA is an exception because a number of the components mediating its signaling have been defined recently (for a recent review, see Lorenzo and Solano, 2005). Among those, the F-box protein CORONATINE INSENSITIVE (COI) plays a central role (Xie et al., 1998).
Here, we used a collection of pure oxylipins synthesized through the 13-LOX, 9-LOX, and α-DOX pathways and an in vitro seedling assay to study the functionality of these compounds in physiological and pathological processes. Systematic analyses of plants grown in the presence of oxylipins showed three different phenotypic alterations: root waving with lateral root arrest, growth arrest with loss of root apical dominance, and overall decrease of root elongation. Detailed characterization of the waving response to 9-HOT, a 9-LOX derivative, suggested a role of this oxylipin in both the formation of lateral roots and the defense against pathogens. Moreover, studies of mutants that do not develop the root-waving morphology allowed the definition of the signaling pathways that mediate the cellular responses to oxylipins.
RESULTS
Oxylipins Induce Distinct Developmental Alterations in Roots of Arabidopsis Seedlings
Functional analyses using a set of pure and well-characterized oxylipins (see Supplemental Figure 1 online) demonstrated striking effects on root development. As shown in Figure 1, we found that 10 of the oxylipins tested, all possessing hydroxyl and/or keto functionalities (9-HOT, 13-HOT, 13- and 9-hydroxyoctadecadienoic [13- and 9-HOD], 2-hydroxyoctadecenoic acid [2-HOE], 9-ketooctadecatrienoic acid [9-KOT], 13- and 9-ketooctadienoic acid [9- and 13-KOD], 9-hydroxy-10-ketooctadecenoic acid [9,10-KHOE], and 13-hydroxy-12-ketooctadecadienoic [12,13-KHOD]) induced waving of Arabidopsis thaliana roots with growth arrest of lateral roots. In addition, another group of six of oxylipins (9-oxononanoic acid [9-oxo-C9], 12-oxo-12:1(E) [traumatin], colnelenic acid, colneleic acid, JA, and OPDA) was found to arrest root growth. Among these, two phenotypes were observed: loss of apical dominance, with the development of increased numbers of lateral and adventitious roots (induced by 9-oxo-C9, traumatin, colnelenic acid, and colneleic acid [i.e., short-chain oxoacids or potential precursors of such compounds]); and a general decrease in root elongation, resulting in the formation of shorter roots than in control plants (induced by the cyclic oxylipins JA and OPDA). By contrast, growing plants in the presence of the remaining 28 oxylipins, including hydroperoxides, aldehydes, epoxides, epoxy alcohols, diols, and triols (see Supplemental Figure 1 online) produced no visible root phenotype. Concentrations of oxylipins tested in these analyses are described in Methods, and optimal amounts to induce the phenotypes described are shown in Figure 1.
Figure 1.
Phenotypic Alterations Provoked in Root of Arabidopsis Seedlings by Distinct Oxylipins.
Seedlings were grown vertically on the surface of hard agar plates. Shown are 10-d-old seedlings grown in control MS medium and in medium containing different concentrations of oxylipins. Three types of phenotypes were noted: root waving with lateral root arrest, caused by 2-HOE, 13-HOT, 13-HOD, 13-KOD, 12,13-KHOD, 9-HOT, 9-HOD, 9-KOT, 9-KOD, and 9,10-KHOE; growth arrest with loss of apical dominance, caused by 9-oxo-C9, 12-oxo-12:1, colnelenic acid (Cln), and colneleic acid (CL); and overall root shortening, caused by OPDA and JA. The last panel in the middle row shows an Arabidopsis seedling grown for 6 d in MS medium and then transferred for 4 additional days to a 9-HOT–containing MS medium. Optimal concentrations inducing the phenotypes described are indicated. Complete names of oxylipins are shown in Supplemental Figure 1 online.
The group of oxylipins producing root waving are biosynthesized from fatty acid hydroperoxides by allene oxide synthase (9,10-KHOE and 12,13-KHOD), reductase/peroxidase (hydroxy acids), or LOX (keto acids). Concentrations needed for activity ranged from 25 to 75 μM, although 9-HOT was active even at 15 μM (see Supplemental Figure 2 online). Additional results with seedlings grown for 6 d on Murashige and Skoog (MS) medium and then transferred to 9-HOT–containing plates revealed that the waving phenotype was locally restricted to the root zone grown in the presence of inducer (Figure 1).
By contrast, oxylipins producing a root growth–inhibiting effect with loss of apical dominance and increase of lateral roots are biosynthesized from hydroperoxides by either hydroperoxide lyase or divinyl ether synthase. The strongest activity was observed with 9-oxo-C9 (active at 15 μM), an oxylipin produced from linole(n)ic acid 9-hydroperoxides by hydroperoxide lyase. The third phenotype, overall decreased root elongation, was induced by OPDA and JA at concentrations of 2 and 10 μM, respectively. Biosynthesis of these oxylipins takes place from linolenic acid 13-hydroperoxide and involves allene oxide synthase and allene oxide cyclase. Therefore, the three distinct morphological changes are induced by distinct sets of derivatives that are formed from different parts of the oxylipin biosynthetic pathway.
9-HOT Causes Waving of Roots and Inhibits the Emergence of Lateral Roots
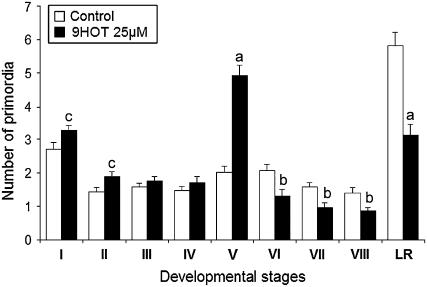
The root-waving phenotype was accompanied by an obvious reduction in the number of lateral roots. Lateral root development proceeds through a number of discrete stages designated I to VIII (Malamy and Benfey, 1997). As seen in Figure 2, the profile of primordia of seedlings grown in the presence of 25 μM 9-HOT was shifted toward an increased abundance of early stages (I to V) and a reduced abundance of later stages (VI to VIII and lateral roots). The strongest effect was noted in the number of stage V primordia, which was 2.5- to 3-fold higher in 9-HOT–treated roots compared with control roots. A similar distribution of primordia was observed in roots grown in the presence of 15 or 35 μM 9-HOT, and the abundance of primordia at stage V was consistently higher than that in control roots (percentages of stage V primordia of total primordia: 14, 27, and 10% for roots grown in the presence of 15 and 35 μM 9-HOT or in the absence of 9-HOT, respectively) (see Supplemental Figure 2 online). This finding suggests that 9-HOT treatment results in a blockage of the lateral root development pathway at stage V, whereas early stages of lateral root development are little affected. The dose-dependent response observed is consistent with oxylipins playing a physiological role in the development but not in the initiation of the lateral root primordia (LRP).
Figure 2.
Analyses of LRP in Columbia Wild-Type Plants.
Number of primordia found in roots of 10-d-old Arabidopsis seedlings grown on MS medium and on 9-HOT–containing medium. Eight developmental stages and emergent lateral roots were counted. A representative example of the developmental stages examined is shown in Supplemental Figure 3 online. Means and se are shown. Letters above the bars indicate statistically significant differences between seedlings grown on MS medium and 9-HOT–containing medium (t test; P < 0.001 [a], 0.001 < P < 0.01 [b], 0.001 < P < 0.05 [c]). The data presented are results obtained in five independent experiments.
LOX1, LOX3, and LOX5 Genes Are Expressed in LRP
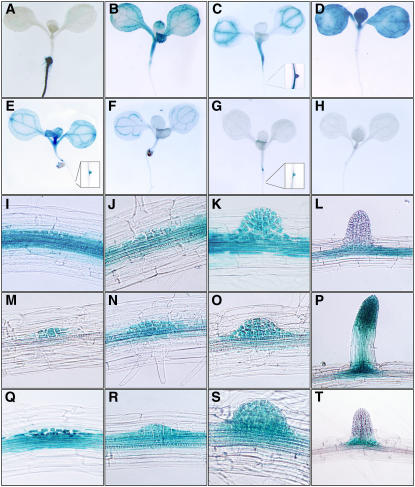
To evaluate the functional role of oxylipins during the development of lateral roots, the expression of LOX and α-DOX genes was examined by means of promoter β-glucuronidase (GUS) constructs (see Methods for a detailed description). Analysis of gene transcription revealed expression in leaves of transgenic seedlings with α-DOX2, LOX1, LOX2, LOX3, and LOX4 reporter genes and in roots of plants containing α-DOX1, LOX1, LOX3, and LOX5 constructs, whereas no GUS activity was observed in seedlings containing the LOX6 reporter gene (Figures 3A to 3H). GUS staining in roots of the α-DOX1 lines was localized in the epidermal cells, whereas plants expressing the LOX1, LOX3, and LOX5 promoters showed staining in LRP (Figure 3). In plants harboring the LOX1:GUS construct, GUS activity was detected in the pericycle cells and in the LRP from stages I to VII, from which expression declined at the time of lateral root emergence (Figures 3I to 3L). Expression directed by LOX3 and LOX5 was primarily restricted to the LRP, in which case blue staining was detected from the first pericycle divisions. Notably, whereas expression in LOX5:GUS plants diminished before root emergence, GUS activity was maintained in lateral roots in LOX3 plants (Figures 3M to 3T).
Figure 3.
Expression Analyses of α-DOX and LOX Genes Encoding the Enzymes Initiating the Synthesis of Oxylipins.
Histological examination of GUS activity in seedlings of transgenic lines containing promoter GUS constructs is shown.
(A) to (H) GUS staining of seedlings from α-DOX1:GUS (A), α-DOX2:GUS (B), LOX1:GUS (C), LOX2:GUS (D), LOX3:GUS (E), LOX4:GUS (F), LOX5:GUS (G), and LOX6:GUS (H). Details of the GUS activity found in root primordia are shown for LOX1:GUS (C), LOX3:GUS (E), and LOX5:GUS (G).
(I) to (T) Selected phases during the formation of LRP in plants with reporters: LOX1, (I) to (L); LOX3, (M) to (P); and LOX5, (Q) to (T).
The pattern of gene expression characterized is consistent with a role of LOX1, LOX3, and LOX5 in the developmental process leading to the formation of lateral roots. Based on their putative amino acid sequences, LOX1 and LOX5 have been proposed to be 9-LOXs, whereas LOX3 is likely to encode a 13-LOX isoform (Liavonchanka and Feussner, 2006). These results suggest a role of both 9-LOX– and 13-LOX–generated oxylipins in the development of LRP and also suggest that the 9-LOX products could be primarily associated with the development process preceding root emergence.
Oxygenation of Linolenic Acid in Roots of Arabidopsis
Because the exogenous application of 9-HOT alters root development and the LOX1 and LOX5 genes are expressed in roots, the 9-LOX activity of roots was examined by incubating linolenic acid with root homogenates from wild-type seedlings. Results from such analyses revealed that linolenic acid (300 μM) was completely metabolized into polar products when incubated with a 1:10 (w/v) homogenate of wild-type Arabidopsis roots. LOX products detected by gas chromatography–mass spectrometry (GC-MS) included 9-HOT and 9-KOT as well as epoxy alcohols and trihydroxy acids (see Supplemental Figure 3 online). The latter two derivatives can be formed as nonspecific degradation products of hydroperoxides accumulating because of low activity of specific hydroperoxide-metabolizing enzymes. That an accumulation of 9-HPOT indeed took place was proven by a separate incubation in which addition of the hydroperoxide reductant SnCl2 inhibited the appearance of degradation products and at the same time strongly enhanced the recovery of the hydroperoxide reduction product 9-HOT. Additional compounds detected in these studies were the α-DOX products 8,11,14-heptadecatrienal and 2-HOT as well as an α-DOX–9-LOX double oxygenation product tentatively identified as 2-hydroxy-9-KOT. Interestingly, 13-LOX products were not observed in these incubations. The 9-LOX activity found in root homogenates is consistent with a role of this oxylipin pathway in root development, in which the 9-LOX derivatives produced might regulate the emergence of lateral roots.
Mutation of LOX1 and LOX5 Increases the Number of Lateral Roots
Given that LOX1 and LOX5 are expressed in LRP and that treatment of wild-type plants with 9-HOT represses lateral root development, we predicted that plants lacking LOX1 and LOX5 function should develop more lateral roots than wild-type plants. Therefore, we examined the phenotypes of plants lacking 9-LOX function for defective lateral root phenotypes. Homozygous T-DNA insertion mutants that lacked LOX1 (lox1-1) and LOX5 (lox5-1) function developed more emergent (stage VIII) and lateral roots than wild-type plants at 10 d, as predicted (Figure 4), whereas no significant differences were observed at the remaining developmental stages (I to VII) (see Supplemental Figure 4 online). Analyses of two additional T-DNA insertion lines, lox1-2 and lox5-2, further supported our results showing that the lack of LOX1 and LOX5 function led to an increment in the number of lateral roots (Figure 4). In addition, a moderate increase in the length of the primary root was observed in the lox1 and lox5 mutants examined, which thus maintained the overall density of primordia as in roots of wild-type plants (Figure 4). This finding suggests that the effect of the lox1 and lox5 mutations is not to increase lateral root initiation but to promote the number of lateral roots. Therefore, these analyses indicated that LOX1 and LOX5 may function as regulators of root development by controlling the emergence of lateral roots through the production of 9-HOT.
Figure 4.
Analyses of LRP in Columbia Wild-Type Plants and in lox1 and lox5 Mutants.
(A) Scheme of LOX1 and LOX5 genomic structures with exons represented as black boxes. The positions of the T-DNA insertions are indicated.
(B) RT-PCR of RNA isolated from leaves of the genotypes indicated. Gene At1g43170 encoding the 60S ribosomal protein L3 (RPL3A) was used to normalize transcript levels in each sample. Gene-specific primer sets used for the evaluation of RNA are shown in Supplemental Table 2 online.
(C) Number of emergent (VIII) and lateral roots (LR) found in roots of 10-d-old wild-type Arabidopsis seedlings and lox1-1, lox1-2, lox5-1, and lox5-2 mutants grown on MS medium. Means and se are shown. The data presented are results obtained in three independent experiments. Letters above the bars indicate statistically significant differences between the corresponding mutants and wild-type plants (t test; P < 0.001 [a], 0.001 < P < 0.01 [b], 0.001 < P < 0.05 [c]).
(D) Number of total root primordia, length of the primary root, and lateral root density in 10-d-old wild-type Arabidopsis, lox1-1, lox1-2, lox5-1, and lox5-2 seedlings grown on MS medium. Means and se of measurements of 20 seedlings are shown.
9-HOT Induces the Formation of Polysaccharide Deposits and the Production of Reactive Oxygen Species in Waving Roots and in Leaves
Further examination using specific dyes revealed that the root-waving response induced by 9-HOT was accompanied by the accumulation of callose, a high-molecular-weight β-1,3-glucan that plays a role in development and in the response of plants to biotic and abiotic stress (Verma and Hong, 2001). As shown in Figure 5, aniline blue staining revealed deposits of callose that were frequently located along the concave face of the waves. Visualization of root transverse sections revealed that callose deposits were present in two or three closely located epidermal cells per section (Figure 5B), clearly visible from the root surface as cellular vesicles (Figures 5C to 5E). In addition to aniline blue staining, accumulation of callose in the 9-HOT–treated roots was determined using a specific anti-callose antibody. Both markers (aniline blue and anti-callose antibodies) stained the same cellular structures, unambiguously establishing the identity as callose (see Supplemental Figure 5 online). In contrast with 9-HOT–treated roots, no callose deposits were detected in roots of control untreated plants (see Supplemental Figure 5 online).
Figure 5.
Histological Examination of 9-HOT–Induced Waved Roots.
(A), (D), (G), and (J) Fluorescence visualization of callose deposits observed in 9 HOT roots stained with aniline blue. All bars = 50 μm.
(B) Transverse section of an aniline blue–stained root.
(C) Nomarski image of a 9-HOT–treated root.
(E) Merging of images (C) and (D).
(F) Transmitted light microscopic visualization of root sections stained with ruthenium red. Pectin deposits are indicated by arrowheads.
(H) Merging of images (F) and (G).
(I) Transmitted light microscopic visualization of ROS in roots stained with NBT.
(K) Merging of images (I) and (J).
In addition to β-1,3-glucan, the use of ruthenium red revealed the presence of pectin colocalizing with the callose, showing that these deposits were infiltrated by additional polysaccharides (Figures 5F to 5H). Furthermore, staining of waved roots with nitroblue tetrazolium (NBT) revealed a strong production of reactive oxygen species (ROS), visualized as blue formazan deposits formed by the reduction of NBT in the presence of superoxide ions (Figure 5I). The accumulation of ROS was not restricted to the callose-containing vesicles; instead, abundant NBT precipitates were found dispersed along the roots (Figures 5J and 5K), with staining as its most intense in the meristematic and elongation zones.
Challenging leaves with 9-HOT resulted in callose accumulation and ROS production, as it did in roots. Detection of ROS was maximal at 30 min after treatment, whereas the formation of callose deposits started at ∼8 h after treatment and increased to its highest accumulation at 24 h (Figure 6). Moreover, ROS were observed homogeneously distributed through the leaf area infiltrated, whereas callose was found dispersed, forming small deposits. A weak accumulation of callose and production of ROS were found in leaves treated with the 9-HOT precursor linolenic acid, whereas both responses were nearly undetectable in untreated leaves or in leaves infiltrated with water, used as controls in these experiments.
Figure 6.
Analyses of Callose Accumulation and ROS Production in Leaves of 4-Week-Old Arabidopsis Plants.
Representative examples of treated leaves are shown.
(A) Aniline blue staining of nontreated leaves and leaves infiltrated with water, 9-HOT, or linolenic acid.
(B) NBT staining in leaf samples treated as described for (A).
Identification of Genes Responding to 9-HOT Treatment
To gain a further understanding of the cellular responses activated in roots when growing in the presence of 9-HOT, changes in gene expression were examined by microarray analysis. Transcriptional profiles of RNA samples from roots after treatment during 3 and 5 d with 25 μM 9-HOT were compared with those of roots growing in the absence of 9-HOT for the same periods (see plant treatments in Methods for a detailed description). For each time point, three separate microarrays were hybridized using RNA extracted from three independent biological replicates. Complete microarray data sets obtained were analyzed, and a list of differentially expressed genes is shown in Supplemental Table 1 online. Details of computational methods to process gene expression data are described in Methods. Of the 26,173 genes represented on the microarray (Galbraith et al., 2004), 178 showed altered expression, with genes induced by 9-HOT (cluster 1, containing 34 genes) and genes repressed by 9-HOT (cluster 2, containing 144 genes). Moreover, three subclusters, 2A, 2B, and 2C, were defined according to the grade and the timing of suppression (Figure 7A). Genes from clusters 1 and 2A, upregulated and downregulated, respectively, were selected for further examination by semiquantitative RT-PCR using specific primers and RNA samples from control and 9-HOT–treated roots (Figure 7B; see Supplemental Table 2 online). Furthermore, because the 9-LOX oxylipin pathway is activated in leaves responding to bacterial infection (Hamberg et al., 2003), gene expression was examined in leaves subjected to 9-HOT treatment as well as in leaves of plants responding to the avirulent bacterium Pseudomonas syringae pv tomato (Pst) DC3000 avrRpm1 and to the virulent strain Pst DC3000.
Figure 7.
Identification of Genes Responding to 9-HOT and Analyses of Gene Expression.
(A) Cluster analysis of genes changing their expression in roots treated with 9-HOT. Cluster 1 contains upregulated genes. Clusters 2A, 2B, and 2C are highly, slightly, and moderately downregulated genes, respectively. A complete list is provided in Supplemental Table 1 online.
(B) List of genes selected from microarray data for further characterization. Shown are locus designations, descriptions of putative encoded proteins, and differential expression ratios between roots grown in the absence of 9-HOT and roots grown for 3 or 5 d in 9-HOT–containing medium.
(C) RT-PCR analysis of genes in control and 9-HOT–treated roots, leaves subjected to 9-HOT treatment, and leaves responding to inoculation with the avirulent bacterium Pst DC3000 avrRpm1 or the virulent strain Pst DC3000. Gene At1g43170 encoding RPL3A was used to normalize transcript levels in each sample.
As shown in Figure 7C, significant levels of transcripts were detected for most genes (10 of 11) in control roots that varied according to the microarray data in the 9-HOT–treated samples. The nature of the genes suggested that 9-HOT treatment altered the expression of genes encoding proteins involved in oxidative stress and modifications of the cell wall (Figure 7B). Basal gene expression was much lower in leaves. Approximately 30% of the upregulated genes in roots were also activated in 9-HOT–treated leaves, and this proportion increased to 57% in leaves responding to bacterial inoculation. Remaining root upregulated genes were either not expressed in leaves (At5g22140 and At5g10230) or did not alter its expression during these treatments (see At1g05560 in 9-HOT infiltration and in bacterial inoculation). Notably, a significant proportion of the genes downregulated in roots were induced in 9-HOT leaves (25%) and in bacteria-treated leaves (75%). An explanation for this expression pattern may be that the differences in basal expression observed in roots versus leaves are likely to reflect a variation in the production of endogenous signal(s) mediating expression in these two tissue types. Also, additional regulatory pathways might interact differentially in roots and leaves with the 9-HOT response. Further analyses of gene expression are being performed to investigate these possibilities.
Isolation of Mutants Not Responding to 9-HOT
The root-waving phenotype characterized here was used to screen for mutants that are insensitive to 9-HOT. From a screen of ∼20,000 M2 seeds, 18 putative mutants, designated noxy (for nonresponding to oxylipins), failed to induce root waving, and one of them, noxy2, was selected for further characterization in this article (Figure 8). The genetic nature of the noxy2 mutation was examined in F1 and F2 populations generated from a backcross with wild-type plants. Results from these analyses indicated that the noxy2 mutation segregated as a single recessive trait. For mapping, noxy2 was outcrossed to the C-24 ecotype. Molecular markers polymorphic between these two ecotypes defined the position of noxy2 on chromosome 5, flanked by simple sequence length polymorphisms nga249 and ca72. Generation of additional markers based on the information from the Marker Tracker and MASC single nucleotide polymorphism databases delimited NOXY2 to a 400-kb region between markers N5-3597960 and MASC04596. As shown in Figure 8A, the noxy2 mutant failed to form root waves when grown on 9-HOT–containing plates. Analyses of callose accumulation in response to 9-HOT showed that plugs were almost absent. Furthermore, ROS production in roots, as determined by NBT staining, was diminished compared with that in wild-type plants (Figure 8A). Finally, the noxy2 mutant showed a significant increase in the formation of lateral roots (Figure 8B).
Figure 8.
Characterization of the 9-HOT–Insensitive Mutant noxy2.
(A) Wild-type and noxy2 seedlings grown in 9-HOT–containing medium. Shown are transmitted light visualization of seedlings, fluorescence visualization of roots stained with aniline blue, and transmitted light visualization of roots stained with NBT.
(B) Number of LRP in roots of wild-type plants and noxy-2 mutants grown on MS medium. Means and se of measurements of 20 seedlings are shown. Letters above the bars indicate statistically significant differences between the corresponding mutants and wild-type plants (t test; P < 0.001 [a] and 0.001 < P < 0.05 [c]).
(C) Lesions developed in leaves of wild-type and noxy2 plants after bacterial inoculation. Shown are representative examples of symptoms developed at 48 h after infiltration of a suspension of Pst DC3000 avrRpm1 (106 cfu/mL) or Pst DC3000 (105 cfu/mL).
(D) Bacterial growth in wild-type and noxy2 plants. Means and se obtained in three independent experiments are shown.
(E) Analyses of defense gene expression in wild-type and noxy2 plants at different intervals after bacterial inoculation. Blots were hybridized to riboprobes for genes encoding PR1, PR2, and PR4. Hybridization against an 18S rRNA radioactive probe was used as a loading control. Shown are representative examples of results obtained with RNA from three independent experiments.
(F) RT-PCR analysis of 9-HOT–responsive genes in leaves of wild-type and noxy2 plants at different intervals after bacterial inoculation. Gene At1g43170 encoding RPL3A was used to normalize transcript levels in each sample. Shown are representative examples of results obtained with RNA from three independent experiments.
NOXY2 Contributes to Plant Defense
Because roots of noxy2 were insensitive to 9-HOT and the 9-LOX oxylipin pathway is activated in leaves responding to bacterial infection (Hamberg et al., 2003), plants lacking NOXY2 function might respond differently than wild-type plants to pathogen infection. Therefore, we examined the response of noxy2 to the bacterial strains Pst DC3000 avrRPM1 (avirulent) and Pst DC3000 (virulent). As seen in Figure 8, control plants treated with Pst DC3000 avrRPM1 showed a typical hypersensitive reaction necrotic response in the area inoculated. This response varied significantly in noxy2 mutants. In this line, leaves developed chlorotic symptoms of variable size that were most prominent between leaf veins. Symptoms described appeared simultaneously in both types of plants and were clearly distinguished at 48 h after inoculation with a bacterial suspension of 106 colony-forming units (cfu)/mL (Figure 8C). Significant differences in symptom development were also apparent after using a low-dose inoculum (105 cfu/mL) of Pst DC3000. A light yellow area was observed in wild-type plants at 48 h after bacterial inoculation, whereas a more intense chlorotic zone was formed in noxy2 mutants (Figure 8C). As infection progressed, control plants developed a necrotic zone surrounded by a chlorotic halo, characteristic of Pst DC3000, whereas chlorotic symptoms were primarily observed in noxy2 plants. The variation in the formation of symptoms diminished when plants were treated with a high-dose inoculum (107 cfu/mL).
Further investigation of NOXY2 function was performed by measuring in planta bacterial growth and expression of both well-known defense markers and the 9-HOT–responsive genes identified in this study (Figure 7). As shown in Figure 8D, higher growth rates were found in noxy2 mutants. The growth of Pst DC3000 avrRPM1 was increased ∼5-fold in noxy2 relative to control plants, whereas the growth of Pst DC3000 increased 10-fold in noxy2 mutants with respect to wild-type plants. Enhanced bacterial growth in noxy2 mutants was accompanied by a delayed and reduced accumulation of transcripts from the salicylic acid–responsive genes PR1 and PR2. Also, the accumulation of transcripts from the 9-HOT–responsive genes (At5g64120, A1g09530, At2g30750, and At4g14630) was decreased in noxy2 mutants, in which lower levels of RNA corresponded to leaves inoculated with the virulent bacterium. As with these genes, expression of the JA and ethylene marker gene PR4 was preferentially reduced in noxy2 plants after Pst DC3000 inoculation (Figure 8E). Finally, examination of At1g05560 and At1g02920 (upregulated in 9-HOT–treated roots but whose expression did not vary in leaves of control plants subjected to 9-HOT or bacterial inoculation) did not show any apparent variation in infected noxy2 leaves with respect to control plants. These analyses suggest a positive role of NOXY2 in basal defense of plants, likely through the regulation of signals required for the full expression of resistance against biotrophic bacteria. Moreover, our results suggest the participation of NOXY2 and of the 9-LOX pathways in signaling defense responses to limit pathogen growth.
Different Signaling Pathways Involved in the Action of Oxylipins
Given that noxy mutants failed to form root waves when grown on 9-HOT, the set of oxylipins used in this study in wild-type Arabidopsis (see Supplemental Figure 1 online) was used on the noxy2 plants to obtain information about the signaling pathways involved. Additionally, the JA-insensitive coi1-1 mutant (Xie et al., 1998) was included in these analyses (Figure 9). Results from these studies showed that the noxy2 plants were insensitive to the 10 waving-inducing oxylipins, whereas the coi1-1 mutant responded as wild-type plants to all of these molecules. On the other hand, the noxy2 mutant responded similarly to wild-type plants to the six root growth–arresting oxylipins. As expected, the coi1-1 mutants were insensitive to the presence of the two cyclic oxylipins used, JA and its precursor OPDA. However, a root growth–arresting phenotype similar to that observed with wild-type plants was induced by 9-oxo-C9, traumatin, colnelenic acid, or colneleic acid in coi1-1 mutants. Results in Figure 9 show the responses of wild-type plants, noxy2, and coi1-1 mutants to treatment with 9-HOT, 9-oxo-C9, and JA. Also, a scheme of the synthetic pathways leading to active oxylipins is shown. This suggests that the morphological changes that are induced by the three sets of oxylipins are activated through three distinct signaling cascades.
Figure 9.
Root Phenotypes of Wild-Type, noxy2, and coi1-1 Plants Responding to Oxylipins.
Ten-day-old seedlings grown on MS medium and in the presence of 9-HOT (25 μM), 9-oxo-C9 (15 μM), or JA (10 μM). The scheme at right shows the branches leading to the synthesis of active oxylipins and the phenotypes induced. AOC, allene oxide cyclase; AOS, allene oxide synthase; DES, divinyl ether synthase; HPL, hydroperoxide lyase; LOX, lipoxygenase; PR, peroxygenase; RD, reductase.
DISCUSSION
The plant oxylipin metabolome constitutes a large number of structurally diverse compounds formed by the oxygenation of fatty acids. The importance of oxylipins such as JA and its derivatives in physiological and pathological processes in plants is being gradually revealed. Here, we have studied the involvement of oxylipins in root growth and development, an area that to date has received limited attention. Three kinds of phenotypic alterations were observed in Arabidopsis germinating in the presence of oxylipins, and structure–activity relationships were readily apparent. Thus, waving of roots accompanied by inhibition of the growth of lateral roots was induced by hydroxy, keto, and keto-hydroxy oxylipins, whereas root growth arrest with a loss of apical dominance was seen in the presence of divinyl ethers and short-chain ω-oxoacids. Inhibition of overall root elongation was induced by the cyclic oxylipins JA and OPDA. Consequently, our results suggest that the metabolism of LOX-generated hydroperoxides by different secondary enzymatic branches results in oxylipins having distinct actions on roots.
Special attention was devoted to the phenotype showing root waving and inhibition of lateral roots. Of 44 oxylipins tested, the 9-LOX derivative 9-HOT was the most potent inducer of this phenotype (Figure 1; see Supplemental Figure 2 online). Interestingly, the formation of lateral roots was blocked at stage V, whereas a minor alteration was observed at the early stages of development (Figure 2), indicating that 9-HOT exerted its effect on the growth of existing lateral roots but not on their initiation. LOX1 and LOX5 genes encoding 9-LOXs are expressed in the LRP (Figure 3), and 9-LOX activity was found in roots of wild-type seedlings (see Supplemental Figure 3 online). Moreover, seedlings of insertion mutants lacking LOX1 or LOX5 activity as well as the noxy2 mutant insensitive to 9-HOT all displayed an increased emergence of lateral roots (Figures 4 and 8). Together, these findings strongly indicate that 9-HOT generated locally by 9-LOX activity is causally involved in the regulation of lateral root growth, although further studies are needed to conclusively identify the endogenous 9-LOX–derived mediator.
It is known that auxins are required to initiate the formation of lateral roots as well as to continue further development to reach the postemergence stage (Celenza et al., 1995). Also, abscisic acid has been shown to regulate an auxin-independent point in lateral root development controlling meristem activation (De Smet et al., 2003). Results from microarray data (from this study and from available microarray databases [GENEVESTIGATOR; http://www.genevestigator.ethz.ch/]) suggest that 9-HOT might interfere with lateral root emergence by affecting abscisic acid signaling. Thus, whereas a low proportion (<5%) of the genes changing their expression in 9-HOT–treated roots are regulated by auxins, 30% of the root genes upregulated by 9-HOT are induced by abscisic acid (see Supplemental Table 3 online).
Histochemical characterization of the changes induced by 9-HOT treatment indicated that this oxylipin modulates root development through the modification of the cell wall. This conclusion was based on the finding that treatment with 9-HOT led to the formation of polysaccharide deposits, composed of callose and pectin, and to the production of ROS, namely superoxide ion. Callose (β-1,3-glucan) and pectin (complex galacturonans) are components of the cell wall, and their deposition and subsequent modification allow wild-type growth and development (Micheli, 2001; Enns et al., 2005). The production of ROS also plays a developmental role by modifying cross-linking in the cell wall, causing the scission of hemicellulose molecules and the activation of calcium channels required during growth (Schopfer, 2001; Foreman et al., 2003; Carol et al., 2005). Further evidence to support the role of 9-HOT in the modification of the cell wall comes from the changes in gene expression observed in 9-HOT–treated roots. 9-HOT treatment causes transcriptional changes of genes that encode cell wall–modifying products such as pectinesterase, Hyp-rich protein, UDP-glucose transferase, and annexin, of which the latter two have been proposed to form a complex with callose synthase (Cassab, 1998; Micheli, 2001; Verma and Hong, 2001) as well as of genes that are likely to affect the level of ROS, such as peroxidase, annexin, germin-like protein (homologous with oxalate oxidase), and glucosyl oxidase (a flavin adenine dinucleotide binding domain–containing protein) (Gidrol et al., 1996; Custers et al., 2004; Tamás et al., 2005; Rouet et al., 2006).
The molecular events that are controlled by 9-HOT during development are remarkably similar to those that occur in plants as part of the active defense to control pathogen infection (Dangl and Jones, 2001). The deposition of callose as focal deposits was traditionally regarded as a barrier that reinforces the cell wall (Smart et al., 1986), although recent studies suggest that it might play a signaling role to regulate the salicylic acid pathway (Jacobs et al., 2003; Nishimura et al., 2003). As with callose, the relevance of pectin in plant defense has been highlighted by recent results showing that its alteration in the pmr5 mutant confers resistance to certain biotrophic fungi (Vogel et al., 2004). Moreover, production of ROS is a nearly ubiquitous response to pathogen invasion, causing the reinforcement of the cell wall through the immobilization of proteins and also acting as signaling molecules to activate further defenses (Bradley et al., 1992; Lamb and Dixon, 1997; Govrin and Levine, 2000).
The finding that treatment of leaves with 9-HOT provoked an accumulation of callose as well as the production of ROS (Figure 6) is in agreement with a role of this oxylipin in plant defense. Further support for such involvement was provided by the observation that a subset of the 9-HOT root-responsive genes show enhanced expression both in leaves treated with 9-HOT and in leaves inoculated with Pseudomonas (Figure 7C). Additional support came from studies of the 9-HOT–insensitive noxy2 mutant that showed increased bacterial growth and reduced activation of 9-HOT–responsive and salicylic acid–inducible genes than in wild-type plants, indicating that NOXY2 is required for full activation of resistance and that 9-HOT might be involved in this response (Figure 8). Although these findings strongly indicated that the 9-LOX oxylipin pathway plays a role in defense against pathogens, definite experimental evidence for this is still lacking. Because of the functional redundancy of multiple LOX genes and of interactions between the LOX and α-DOX pathways (Hamberg et al., 2003), studies of the role of the 9-LOX pathway in plant defense will required double or multiple mutant LOX/DOX plants rather than mutants of single enzymes. This work is under way in our laboratory.
The fact that the noxy2 mutant was defective in both plant defense and root development indicates that both processes share common signaling components. As discussed above, the nature of the changes activated by 9-HOT in both situations suggests that the participation of this oxylipin, and thus of the 9-LOX pathway, operates mainly through the modification of the cell wall. In addition, as both cell wall components and ROS are known to signal stress responses in plants (Grant and Loake, 2000; Ellis et al., 2002; Nishimura et al., 2003), it is possible that the changes in these cellular products contribute to generate other signal molecules acting as mediators of the responses activated by 9-HOT. Further examination will be required to ascertain this point; however, results shown here suggest that the modifications effected through participation of the 9-LOX and NOXY2 proteins might help to restrain pathogen invasion and to facilitate the proper emergence of lateral roots through the cell layers of the parent root.
Finally, studies with 9-HOT–insensitive (noxy2) and JA-insensitive (coi1-1) mutants revealed that the signaling mechanisms that mediate the response to 9-HOT (as well as to the additionally identified root-waving–inducing oxylipins) are activated through a JA-independent pathway. Furthermore, we have found that this pathway differs from that used by oxoacids (9-oxo-C9 and 12-oxo-12:1(E) and divinyl ethers [colnelenic acid and colneleic acid]) to arrest root growth and the loss of apical dominance, and that the later response is also activated independently of JA. These results indicate that the functional specialization of the different oxylipins has been accompanied by the diversification of their signaling pathways. On the other hand, the insensitivity of noxy2 to all waving-inducing oxylipins identified (produced through 13-LOX, 9-LOX, and α-DOX activities) might reflect a partial functional redundancy of the different oxylipin biosynthetic pathways.
The results of this study indicate that, in addition to JA, other oxylipins contribute to the adaptation of plants to their environment. This would include defense against pathogens and the modulation of root architecture, an important response for the accommodation of plants to different growth situations. Also, the modification of root architecture could form part of the defense mechanisms of plants to the attack of root pathogens. In this context, the complexity of the oxylipin signaling network might enhance the adaptative flexibility of plants facing a multitude of biotic and abiotic stresses.
METHODS
Plant Material, Mutant Characterization, and Growth Conditions
Arabidopsis thaliana wild type, transgenic lines, and coi1-1 mutants used in this study were derived from ecotype Columbia. Insertion mutants used were identified using the SIGnAL T-DNA Express Arabidopsis gene mapping tool (http://signal.salk.edu/). SALK lines SALK_059431(lox1-1), SALK_000058 (lox1-2), SALK_044826 (lox5-1), and SALK_038475 (lox5-2) were distributed by the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info). Homozygous insertion mutants were identified by PCR using T-DNA and gene-specific primer sets as described on the T-DNA Express homepage. To evaluate whether the isolated mutants were mRNA null mutants, semiquantitative RT-PCR was performed using gene-specific primers (sequences of primer sets used in lox1 and lox5 mutants are shown in Supplemental Table 2 online).
Sterilized seeds were vernalized for 3 d at 4°C and grown in vertically oriented square Petri dishes (120 mm × 120 mm; Deltalab) containing 1× MS medium, pH 6.0, 1.5% (w/v) sucrose, and 1.5% (w/v) agar (Bacto Agar; Becton-Dickinson). Metacrylate platforms designed to guarantee verticality were used to hold the plates (see Supplemental Figure 6 online). Oxylipins were added to molten medium (50°C) at the indicated concentrations and then poured onto plates. Freshly prepared plates were always used to avoid product breakdown or instability. Phenotypes were generally observed at 10 d after seed germination. An M2 population from ethyl methanesulfonate–mutagenized seeds was obtained from Lehle Seeds and used to screen for mutants not responding to oxylipins. Growth conditions were 16 h of light, 8 h of dark, and 22°C. For pathology assays, seeds were sown on soil and vernalized for 4 d at 4°C. Ten days after germination, seedlings were transferred to individual pots and grown in chambers (22°C, 70% RH, 250 μE·m−2·s−1 fluorescent illumination) under a 14-h-light/10-h-dark photoperiod. Plants were treated and examined 4 weeks after seed germination.
Oxylipins
The oxylipins used were purchased from Larodan Fine Chemicals. The chemical and stereochemical purity of all compounds was checked by gas chromatography, straight-phase high-pressure liquid chromatography, and mass spectrometry and found to be in accord with the supplier's specifications (i.e., >97%). Large-scale preparation of 9-HOT was performed by incubating linolenic acid with tomato (Solanum lycopersicum) fruit LOX (Matthew et al., 1977) followed by sodium borohydride reduction and isolation by straight-phase high-pressure liquid chromatography. This provided >97% pure material (up to 1 g) in yields of ∼35%. A range of concentrations, from 15 to 75 μM, was tested to evaluate the effect on root development of all available products. Lesser amounts of products were also examined if required. Optimal concentrations provoking clear and reproducible phenotypes are shown in Figure 1 and were used for further analyses.
Incubation of Arabidopsis Roots with Linolenic Acid
Roots (500 mg) from 10-d-old Arabidopsis seedlings were homogenized at 0°C in 0.1 M potassium phosphate buffer, pH 6.7 (5 mL), containing linolenic acid (300 μM). The homogenate was incubated at 23°C for 30 min and subsequently extracted with diethyl ether. The material obtained was methyl-esterified and trimethylsilylated and analyzed by GC-MS using a Hewlett-Packard model 5970 mass selective detector attached to a Hewlett-Packard model 5890 gas chromatograph. In other experiments, the incubations were interrupted after 10 min by the addition of ethanol (5 mL) containing SnCl2 (100 mg). After standing for 5 min, material was isolated by solvent extraction, derivatized, and analyzed by GC-MS as described above.
Histology and Histochemistry
Clarification of roots for counting root primordia was performed as described by Malamy and Benfey (1997). Primordia from ∼20 seedlings were counted in each experiment for each of the plants examined. Reported data are means and se of the results obtained in at least three independent experiments. Data was statistically analyzed by t test (P < 0.001 [a], 0.001 < P < 0.01 [b], and 0.001 < P < 0.05 [c]) using the GraphPad Prism version 4 computer program. For the detection of callose, plant tissues were stained with a 0.1-mg/mL solution of aniline blue fluorochrome (Sirofluor; Biosupplies) during 30 min in the dark. Samples were washed, mounted in 50% glycerol on glass microscope slides, and examined with a Leica DMR fluorescence microscope. For transverse sections, samples were fixed and embedded in Historesin according to the protocol described by Di Laurenzio et al. (1996). Pectin was visualized by staining root sections with an aqueous red ruthenium solution (0.05%) for 30 min, followed by water destaining. Callose was visualized by aniline blue staining or by incubation with a 200:1 dilution of anti-callose antibodies in PBS (Biosupplies). Nitroblue tetrazolium (Roche Diagnostics) was used to stain for the site of superoxide production, as described by Carol et al. (2005). Staining of callose and superoxide production in leaves was performed as described for roots. Leaves of 4-week-old plants were vacuum-infiltrated with the appropriate solution (water, 50 μM 9-HOT, or 50 μM linolenic acid), excised from the plants, and stained at different times after infiltration. Leaves were destained by washing in an ethanol solution before visualization.
Construction of Transgenic Lines and Analyses of GUS Activity
Genomic sequences extending to ∼1 kb from the translational start site of the Arabidopsis α-DOX1, α-DOX2, LOX1, and LOX2 genes, characterized previously (Melan et al., 1993; Bell et al., 1995; De León et al., 2002; Hamberg et al., 2005), and from the four additional LOX genomic sequences, LOX3, LOX4, LOX5, and LOX6, listed in the databases were amplified by PCR from wild-type Columbia using Expand High Fidelity polymerase (Roche). Forward and reverse primers used in each case are shown in Supplemental Table 2 online. The resulting PCR fragments were inserted into the plasmid pGEM-T Easy vector system I (Promega) and sequenced to ensure correct amplification. Promoters were fused to the coding region of the GUS gene present in the plasmid pBI101.2, which confers resistance to kanamycin in planta, introduced into Agrobacterium tumefaciens, and transferred into Columbia wild-type plants. Examination of GUS activity in transgenic seedlings was performed as described by Malamy and Benfey (1997).
Plant Treatment and RNA Isolation
From microarray analyses, 6-d-old seedlings grown on MS medium were transferred to fresh plates and grown for 3 or 5 additional days in the absence or presence of 9-HOT (25 μM). Root fragments grown after transferring to fresh plates (∼20 and ∼30 mm from the tip at 3 and 5 d, respectively) were excised and used to compare gene expression between controls (roots grown on MS medium) and 9-HOT–treated seedlings. Root fragments from ∼400 seedlings were collected for each independent sample. Total RNA was isolated from three independent biological replicates. For chemical treatment, 4-week-old Arabidopsis plants were vacuum-infiltrated with 50 μM 9-HOT, and leaves were excised from the plant at the times indicated. Bacterial inoculation was performed by injecting a bacterial suspension of 106 cfu/mL Pseudomonas syringae pv tomato DC3000 avrRpm1 or P. syringae pv tomato DC3000 into the abaxial side of rosette leaves of 4-week-old plants. Plant tissues were immediately frozen in liquid nitrogen after harvest. Total RNA was isolated according to Logemann et al. (1987) and then using the RNeasy mini kit (Qiagen). RNA samples were examined as described below.
Microarray Analysis
Gene expression of roots grown on MS medium versus MS medium + 9-HOT was compared using Superamine Telechem slides of the Qiagen-Operon Arabidopsis version 3.0 obtained from David Galbraith (Arizona University). The whole-genome oligonucleotide set represents 26,173 protein-coding genes and 28,964 transcripts (Galbraith et al., 2004). More information about printing and the oligonucleotide set can be found at http://ag.arizona.edu/microarray/. In these experiments, 6-d-old seedlings grown on MS medium were transferred to fresh plates and grown for 3 or 5 additional days in the absence or presence of 9-HOT. RNA was prepared from the root fragments grown after transferring to fresh plates (∼20 and ∼30 mm from the tip at 3 and 5 d, respectively) and used to compare gene expression between controls (roots grown on MS medium) and 9-HOT–treated seedlings. Three independent hybridizations were performed for each comparison using RNAs obtained from three independent experiments, and the results were examined as described below. RNA was quantified using a Nanodrop ND-1000 UV-Vis spectrophotometer (Nanodrop Technology) and assessed using an Agilent 2100 bioanalyzer (Agilent Technology). One microgram of total RNA from each sample was amplified and aminoallyl-labeled using the MessageAmp II aRNA kit (Ambion) and 5-(3-aminoallyl)-2′-deoxyuridine-5′-triphosphate (Ambion) according to the manufacturer's instructions. A quantity (7.5 μg) of aminoallyl-labeled aRNA were resuspended in 0.1 M Na2CO3, pH 9.0, and labeled with either Cy3 or Cy5 Mono NHS ester (Cy Dye Postlabeling Reactive Dye Pack; Amersham). Samples were purified according to the manufacturer's instructions for Megaclear (Ambion), and Cy3 and Cy5 incorporation was measured using 1 μL of the probe in the Nanodrop spectrophotometer. Hybridization and washing of microarrays are described in the Supplemental Methods online.
Microarray Data Analysis
Background correction and normalization of the expression data were performed using the LIMMA software package (Smyth and Speed, 2003). To avoid exaggerated variability of log ratios in low-intensity spots, the normexp method in LIMMA was used to adjust the local median background estimates. Log ratio values were scaled as described by Storey (2003) to have similar distribution across arrays and consistency among arrays. The rank products method was used to determine genes differentially expressed. For each chip, probes were sorted by their normalized expression ratio in two lists of ascending and descending order. Rank products were calculated for each gene according to Breitling et al. (2004) and compared with the root primordia of 5000 random permutations of the same data to assign E-values. The multiple testing problem inherent to microarray experiments was corrected using the false discovery rate (FDR): we divided the E-value of each gene by its position in the list of changed transcripts (Storey, 2003). An FDR of 5% means that only 5% or less of the genes up to this position are expected to be observed by chance (false positives), the remaining 95% being genes that are indeed significantly affected (true positives). Significantly upregulated and downregulated genes obtained in Arabidopsis roots after 3 and 5 d of 9-HOT treatment (at FDR of 5%), represented in red and green, are listed in Supplemental Table 1 online in ascending order of FDR. Additionally, fold change and M values, representing differential expression ratios and their logarithmic forms, respectively, are listed for each gene. Hierarchical clustering of both significantly upregulated and downregulated genes was performed using The Institute for Genomic Research Multiexperiment Viewer version 3.0.3 using Euclidean distance and the average linkage clustering (Saeed et al., 2003). Annotation of Arabidopsis genes based on the probe set identifiers was obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org).
Analyses of Gene Expression
RT-PCR was performed with a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems) using the Titan One Tube RT-PCR system (Roche Applied Science) as specified by the manufacturer. Total RNA was treated with DNase TURBO DNA-free (Ambion) to remove contaminating DNA. A quantity (100 ng) of this RNA was used in each one-step RT-PCR procedure. Primers used and the lengths of amplification products are described in Supplemental Table 2 online. Gene At1g43170 encoding RPL3A was used as an internal standard. For RNA gel blots, RNA (5 μg/lane) was analyzed on agarose-formaldehyde gels, transferred to Hybond N membranes, and hybridized to single-stranded riboprobes according to standard procedures (Sambrook et al., 1989). Radioactive probes were prepared for PR1, PR2 (Uknes et al., 1992), and PR4 (EST clone 119C17, obtained from the Nottingham Arabidopsis Stock Centre). The amount of loaded RNA was verified by the addition of ethidium bromide to the samples and photography under UV light after electrophoresis, followed by hybridization to 18S rRNA (Ruiz-García et al., 1997). Blots shown are representative examples of the results obtained in three independent experiments.
Mutant Isolation
A screen of ∼20,000 M2 seeds from an ethyl methanesulfonate–mutagenized population was performed. Seeds were grown on vertical MS plates for 4 d and then transferred to 9-HOT (25 μM)–containing MS plates. Seedlings failing to show a root-waving phenotype in the presence of 9-HOT were selected as putative mutants and were designated noxy. Progeny was retested for the same phenotype and crossed to wild-type Columbia plants to perform segregation analyses. Crosses were performed for complementation analyses. For mapping purposes, noxy mutants were crossed to the wild type of the C24 ecotype and F2 mutants were selected.
Analyses of Symptom Development and Bacterial in Vivo Growth Curves
Bacterial symptoms in the wild type and noxy2 mutants were visually examined after infecting leaves of 4-week-old plants with a suspension of 105 or 106 cfu/mL Pst DC3000 avrRpm1 or Pst DC3000. For each genotype, a minimum of 20 plants was examined in three independent experiments. Bacterial growth was evaluated by infecting leaves with a bacterial suspension of 105 cfu/mL. Discs of 0.6 cm2 were excised from each infected leaf using a core borer, pooled in triplicate, and homogenized in sterile water using a plastic pestle. Eight replicates were used for each time interval examined. Bacterial populations were determined by plating appropriate dilutions from each sample in King's medium. Reported data are means and se of the values obtained in three independent experiments.
Accession Numbers
The accession numbers for the genes discussed in this article are as follows: αDOX1 (At3g01420), αDOX2 (At1g73680), LOX1 (At1g55020), LOX2 (At3g45140), LOX3 (At1g17420), LOX4 (At1g72520), LOX5 (At3g22400), LOX6 (At1g67560), PR1 (At2g14610), PR2 (At3g57260), PR4 (At3g04720), and RPL3A (At1g43170). Arabidopsis Genome Initiative numbers for differentially expressed genes are given in Figure 7B and in Supplemental Table 1 online. Microarray data from this article have been deposited with the ArrayExpress data library (http://www.ebi.ac.uk/arrayexpress/) under accession number E-ATMX-14.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Genes Differentially Expressed by 9-HOT Treatment in Roots.
Supplemental Table 2. Sets of Primers Used to Examine Gene Expression and to Amplify Promoters of α-DOX and LOX Genes.
Supplemental Table 3. Genes Upregulated in Roots of Control Wild-Type Plants Treated with 9-HOT (Genes Shown in Boldface Were Found to be Induced by Abscisic Acid [Fold Change > 2] in Available Microarray Databases).
Supplemental Figure 1. Major Pathways in the Oxygenation of Fatty Acids.
Supplemental Figure 2. Dose-Dependent Response of the 9-HOT–Induced Root Waving.
Supplemental Figure 3. Oxygenation of Linolenic Acid in Roots of Arabidopsis Plants.
Supplemental Figure 4. Analyses of LRP in Columbia Wild-Type Plants and in LOX1 and LOX5 Mutants.
Supplemental Figure 5. Histological Examination of 9-HOT–Induced Waved Roots.
Supplemental Figure 6. Growth of Seedlings on Vertically Oriented Square Plates.
Supplemental Methods. Microarray Hybridization.
Supplementary Material
Acknowledgments
We thank G. Bannenberg, J. Paz-Ares, and R. Solano for critical reading of the manuscript; A.P. Calvo, M. Pernas, and S. Takeda for help with histochemical analyses; I. Poveda for expert photography; R. Piqueras and M. Peinado for help with in vitro plant growth; and G. Hamberg for assistance during the preparation of the oxylipins used. The T-DNA insertion lines used in these studies were from the SALK collection and were obtained from the Nottingham Arabidopsis Stock Centre. Microarray analyses were performed at the genomic facilities of the Centro Nacional de Biotecnología (http://www.cnb.uam.es/∼genomica/). This work was supported by Grant BIO2003-02338 to C.C. from the Ministry of Education and Science, by Grant GEN2003-20218 to C.C. from the Trilateral Project, by Grant 2001-2553 to M.H. from the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning, and by Grant QLK5-CT-2001-02445 from the European Union. T.V. and J.V. were supported by fellowships from the Ministry of Education and Science.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Carmen Castresana (ccastresana@cnb.uam.es).
Online version contains Web-only data.
References
- Alméras, E., Stolz, S., Vollenweider, S., Reymond, P., Mène-Saffrané, L., and Farmer, E.E. (2003). Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J. 34 205–216. [DOI] [PubMed] [Google Scholar]
- Armengaud, P., Breitling, R., and Amtmann, A. (2004). The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 136 2556–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate, N.J., and Rothstein, S.J. (1998). C6-volatiles derived from the lipoxygenase pathway induce a subset of defense-related genes. Plant J. 16 561–569. [DOI] [PubMed] [Google Scholar]
- Bell, E., Creelman, R.A., and Mullet, J.E. (1995). A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 92 8675–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blée, E. (2002). Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 7 315–321. [DOI] [PubMed] [Google Scholar]
- Bradley, D.J., Kjellbom, P., and Lamb, C.J. (1992). Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: A novel, rapid defense response. Cell 70 21–30. [DOI] [PubMed] [Google Scholar]
- Breitling, R., Armengaud, P., Amtmann, A., and Herzyk, P. (2004). Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573 83–92. [DOI] [PubMed] [Google Scholar]
- Carol, R.J., Takeda, S., Linstead, P., Durrant, M.C., Kakesova, H., Derbyshire, P., Drea, S., Zarsky, V., and Dolan, L. (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438 1013–1016. [DOI] [PubMed] [Google Scholar]
- Cassab, G.I. (1998). Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 281–309. [DOI] [PubMed] [Google Scholar]
- Celenza, J.L., Jr., Grisafi, P.L., and Fink, G.R. (1995). A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 9 2131–2142. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1995). Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 92 4114–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 355–381. [DOI] [PubMed] [Google Scholar]
- Croft, K., Jüttner, F., and Slusarenko, A.J. (1993). Volatile products of the lipoxygenase pathway evolved from Phaseolus vulgaris (L.) leaves inoculated with Pseudomonas syringae pv phaseolicola. Plant Physiol. 101 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custers, J.H., Harrison, S.J., Sela-Buurlage, M.B., van Deventer, E., Lageweg, W., Howe, P.W., van der Meijs, P.J., Ponstein, A.S., Simons, B.H., Melchers, L.S., and Stuiver, M.H. (2004). Isolation and characterisation of a class of carbohydrate oxidases from higher plants, with a role in active defence. Plant J. 39 147–160. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- De Smet, I., Signora, L., Beeckman, T., Inzé, D., Foyer, C.H., and Zhang, H. (2003). An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 33 543–555. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio, L., Wysocka-Diller, J., Malamy, J.E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M.G., Feldmann, K.A., and Benfey, P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86 423–433. [DOI] [PubMed] [Google Scholar]
- Ellis, C., Karafyllidis, I., Wasternack, C., and Turner, J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enns, L.C., Kanaoka, M.M., Torii, K.U., Comai, L., Okada, K., and Cleland, R.E. (2005). Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol. Biol. 58 333–349. [DOI] [PubMed] [Google Scholar]
- Farmer, E.E., Alméras, E., and Krishnamurthy, V. (2003). Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol. 6 372–378. [DOI] [PubMed] [Google Scholar]
- Foreman, J., Demidchik, V., Bothwell, J.H., Mylona, P., Miedema, H., Torres, M.A., Linstead, P., Costa, S., Brownlee, C., Jones, J.D., Davies, J.M., and Dolan, L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422 442–446. [DOI] [PubMed] [Google Scholar]
- Galbraith, D.W., Elumalai, R., and Gong, F.C. (2004). Integrative flow cytometric and microarray approaches for use in transcriptional profiling. Methods Mol. Biol. 263 259–280. [DOI] [PubMed] [Google Scholar]
- Gidrol, X., Sabelli, P.A., Fern, Y.S., and Kush, A.K. (1996). Annexin-like protein from Arabidopsis thaliana rescues ΔoxyR mutant of Escherichia coli from H2O2 stress. Proc. Natl. Acad. Sci. USA 93 11268–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43 205–227. [DOI] [PubMed] [Google Scholar]
- Govrin, E.M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10 751–757. [DOI] [PubMed] [Google Scholar]
- Grant, J.J., and Loake, G.J. (2000). Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol. 124 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberg, M., Ponce de Leon, I., Rodriguez, M.J., and Castresana, C. (2005). α-Dioxygenases. Biochem. Biophys. Res. Commun. 338 169–174. [DOI] [PubMed] [Google Scholar]
- Hamberg, M., Sanz, A., Rodriguez, M.J., Calvo, A.P., and Castresana, C. (2003). Activation of the fatty acid α-dioxygenase pathway during bacterial infection of tobacco leaves. Formation of oxylipins protecting against cell death. J. Biol. Chem. 278 51796–51805. [DOI] [PubMed] [Google Scholar]
- Howe, G.A., and Schilmiller, A.L. (2002). Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 5 230–236. [DOI] [PubMed] [Google Scholar]
- Jacobs, A.K., Lipka, V., Burton, R.A., Panstruga, R., Strizhov, N., Schulze-Lefert, P., and Fincher, G.B. (2003). An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15 2503–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalloul, A., Montillet, J.L., Assigbetsé, K., Agnel, J.P., Delannoy, E., Triantaphylidès, C., Daniel, J.F., Marmey, P., Geiger, J.P., and Nicole, M. (2002). Lipid peroxidation in cotton: Xanthomonas interactions and the role of lipoxygenases during the hypersensitive reaction. Plant J. 32 1–12. [DOI] [PubMed] [Google Scholar]
- Lamb, C., and Dixon, R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 251–275. [DOI] [PubMed] [Google Scholar]
- Liavonchanka, A., and Feussner, I. (2006). Lipoxygenases: Occurrence, functions and catalysis. J. Plant Physiol. 163 348–357. [DOI] [PubMed] [Google Scholar]
- Logemann, J., Schell, J., and Willmitzer, L. (1987). Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163 16–20. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O., and Solano, R. (2005). Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 8 532–540. [DOI] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124 33–44. [DOI] [PubMed] [Google Scholar]
- Matthew, J.A., Chan, H.W., and Galliard, T. (1977). A simple method for the preparation of pure 9-d-hydroperoxide of linoleic acid and methyl linoleate based on the positional specificity of lipoxygenase in tomato fruit. Lipids 12 324–326. [DOI] [PubMed] [Google Scholar]
- Melan, M.A., Dong, X., Endara, M.E., Davis, K.R., Ausubel, F.M., and Peterman, T.K. (1993). An Arabidopsis thaliana lipoxygenase gene can be induced by pathogens, abscisic acid, and methyl jasmonate. Plant Physiol. 101 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli, F. (2001). Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6 414–419. [DOI] [PubMed] [Google Scholar]
- Mueller, M.J., Mène-Saffrané, L., Grun, C., Karg, K., and Farmer, E.E. (2006). Oxylipin analysis methods. Plant J. 45 472–489. [DOI] [PubMed] [Google Scholar]
- Nishimura, M.T., Stein, M., Hou, B.H., Vogel, J.P., Edwards, H., and Somerville, S.C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 969–972. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A., Eggermont, K., Terras, F.R., Thomma, B.P., De Samblanx, G.W., Buchala, A., Métraux, J.P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De León, I.P., Sanz, A., Hamberg, M., and Castresana, C. (2002). Involvement of the Arabidopsis α-DOX1 fatty acid dioxygenase in protection against oxidative stress and cell death. Plant J. 29 61–72. [DOI] [PubMed] [Google Scholar]
- Prost, I., et al. (2005). Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 139 1902–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancé, I., Fournier, J., and Esquerré-Tugayé, M.T. (1998). The incompatible interaction between Phytophthora parasitica var. nicotianae race 0 and tobacco is suppressed in transgenic plants expressing antisense lipoxygenase sequences. Proc. Natl. Acad. Sci. USA 95 6554–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet, M.A., Mathieu, Y., Barbier-Brygoo, H., and Laurière, C. (2006). Characterization of active oxygen-producing proteins in response to hypo-osmolarity in tobacco and Arabidopsis cell suspensions: Identification of a cell wall peroxidase. J. Exp. Bot. 57 1323–1332. [DOI] [PubMed] [Google Scholar]
- Ruiz-García, L., Madueño, F., Wilkinson, M., Haughn, G., Salinas, J., and Martínez-Zapater, J.M. (1997). Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9 1921–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci, C., Montillet, J.L., Agnel, J.P., Battesti, C., Alonso, B., Knoll, A., Bessoule, J.J., Etienne, P., Suty, L., Blein, J.P., and Triantaphylidès, C. (1999). Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J. Biol. Chem. 274 36446–36455. [DOI] [PubMed] [Google Scholar]
- Saeed, A.I., et al. (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34 374–378. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritisch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sanders, P.M., Lee, P.Y., Biesgen, C., Boone, J.D., Beals, T.P., Weiler, E.W., and Goldberg, R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz, A., Moreno, J.I., and Castresana, C. (1998). PIOX, a new pathogen-induced oxygenase with homology to animal cyclooxygenase. Plant Cell 10 1523–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer, P. (2001). Hydroxyl radical-induced cell-wall loosening in vitro and in vivo: Implications for the control of elongation growth. Plant J. 28 679–688. [DOI] [PubMed] [Google Scholar]
- Smart, M.G., Aist, J.R., and Israel, H.W. (1986). Structure and function of wall appositions. I. General histochemistry of papillae in barley (Hordeum vulgare) coleoptiles attacked by Erysiphe graminis f. sp. hordei. Can. J. Bot. 64 793–801. [Google Scholar]
- Smyth, G.K., and Speed, T. (2003). Normalization of cDNA microarray data. Methods 31 265–273. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelmach, B.A., Müller, A., Hennig, P., Laudert, D., Andert, L., and Weiler, E.W. (1998). Quantitation of the octadecanoid 12-oxophytodienoic acid, a signalling compound in plant mechanotransduction. Phytochemistry 47 539–546. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J.D. (2003). The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Statist. 31 2013–2035. [Google Scholar]
- Tamás, L., Budíková, S., Huttová, J., Mistrík, I., Šimonovičová, M., and Široká, B. (2005). Aluminum-induced cell death of barley-root border cells is correlated with peroxidase- and oxalate oxidase-mediated hydrogen peroxide production. Plant Cell Rep. 24 189–194. [DOI] [PubMed] [Google Scholar]
- Turner, J.G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14 (suppl.): S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., Chandler, D., Slusarenko, A., Ward, E., and Ryals, J. (1992). Acquired resistance in Arabidopsis. Plant Cell 4 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, D.P., and Hong, Z. (2001). Plant callose synthase complexes. Plant Mol. Biol. 47 693–701. [DOI] [PubMed] [Google Scholar]
- Vijayan, P., Shockey, J., Lévesque, C.A., Cook, R.J., and Browse, J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J.P., Raab, T.K., Somerville, C.R., and Somerville, S.C. (2004). Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 40 968–978. [DOI] [PubMed] [Google Scholar]
- Vollenweider, S., Weber, H., Stolz, S., Chételat, A., and Farmer, E.E. (2000). Fatty acid ketodienes and fatty acid ketotrienes: Michael addition acceptors that accumulate in wounded and diseased Arabidopsis leaves. Plant J. 24 467–476. [DOI] [PubMed] [Google Scholar]
- Weber, H., Chételat, A., Caldelari, D., and Farmer, E.E. (1999). Divinyl ether fatty acid synthesis in late blight-diseased potato leaves. Plant Cell 11 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert, H., Stenzel, I., Berndt, E., Wasternack, C., and Feussner, I. (1999). Metabolic profiling of oxylipins upon salicylate treatment in barley leaves—Preferential induction of the reductase pathway by salicylate. FEBS Lett. 464 133–137. [DOI] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.