Abstract
To translate basic research findings into clinical practice, it is essential that information about mutations and variations in the human genome are communicated easily and unequivocally. Unfortunately, there has been much confusion regarding the description of genetic sequence variants. This is largely because research articles that first report novel sequence variants do not often use standard nomenclature, and the final genomic sequence is compiled over many separate entries. In this article, we discuss issues crucial to clear communication, using examples of genes that are commonly assayed in clinical laboratories. Although molecular diagnostics is a dynamic field, this should not inhibit the need for and movement toward consensus nomenclature for accurate reporting among laboratories. Our aim is to alert laboratory scientists and other health care professionals to the important issues and provide a foundation for further discussions that will ultimately lead to solutions.
The complexity and inherent variation of the human genome sequence have placed unprecedented demands on bioinformatics resources to assure organized data management.1 Genomic data are continuously translated into clinical molecular tests, and laboratory reports are generated for patient management and clinical and epidemiological studies. The consistent use of uniform nomenclature in the management of DNA sequence data is especially critical for concise communication of diagnostic testing and genetic risk assessment. Just as standards were established early in the Human Genome Project for uniform documentation and collation of sequence data, conventions for standardized nomenclature of variant sequences—mutations and polymorphisms—have been developed and promulgated.2,3,4,5 Although in this article we use the term “mutation” to imply a deleterious genetic sequence variation, our discussion here is relevant to all small genetic sequence variations, whether neutral or deleterious. Despite the nominal acceptance of these standards, clinical mutation testing and screening for major genetic disorders still suffer from the use of nonstandard and variable mutation nomenclature. In fact, colloquial designations for mutations of clinical importance are used so broadly that many geneticists and molecular diagnosticians are probably unaware that they are nonstandard. This may cause confusion when cross-referencing between the original literature and modern databases.
In an effort to clarify the nomenclature recommendations of the Human Genome Variation Society (HGVS), we first briefly illustrate how to name a particular sequence variant (either novel or known) using standard nomenclature. Recommendations for methods of interpreting sequence variants, whether deleterious or neutral, have been reviewed elsewhere.6 We next raise issues of standard and nonstandard nomenclature in a limited number of examples of genes that have been commonly used for molecular diagnostics. It is noteworthy that nomenclature problems exist not only for germline mutations and polymorphisms but also for somatic alterations in genes associated with cancer.
Standard Nomenclature for Genes and Mutations
Figures 1and 2 exemplify how to number nucleotides and name mutations or variants, respectively, according to the standard nomenclature recommendations of the HGVS (http://www.HGVS.org/mutnomen/). These numbering examples are based on coding DNA reference sequences and protein-level amino acid sequences. “Coding DNA reference sequence” refers to a cDNA-derived sequence containing the full length of all coding regions and noncoding untranslated regions [5′ untranslated region (UTR) and 3′-UTR]; splice variants may lack one or more of the coding exons. Nucleotide numbering is in relation to the translation initiation codon, starting with number 1 at the A of the ATG. Standard mutation nomenclature based on coding DNA reference sequences and protein-level amino acid sequences requires prefixes “c.” and “p.,” respectively, as in Figure 2. Standard nomenclature based on genomic DNA reference sequences and RNA reference sequences is not shown. “Genomic DNA reference sequence” simply indicates any human DNA sequence in the database that is not based on a cDNA sequence. Standard mutation nomenclature based on a “genomic DNA reference sequence” requires a prefix “g.” and numbering starts with number 1 for the first nucleotide in the file.
Figure 1.
Example of nucleotide numbering based on a coding DNA sequence. Exonic sequences are numbered sequentially from the initiation codon to the stop codon. Untranslated sequences in the 5′- and 3′-UTRs, as well as in intronic sequences, are numbered in relation to the coding exonic sequences as shown. Note that lengths of DNA sequence are arbitrary.
Figure 2.
Example of standard mutation nomenclature based on a coding DNA sequence. Note that the amino acid change for “c.1A>T” is described as “p.0?” because amino acid changes secondary to codon 1 mutations are frequently unpredictable. In this example, c.1A>T cannot be described as “p.Met1Leu” because it either creates no protein or creates a different protein starting from a cryptic translation initiation site. One may describe the amino acid sequence change as “p.0” if there is experimental proof that no protein forms.
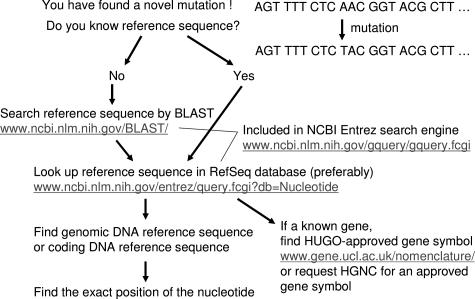
Figure 3 illustrates the process for finding a reference sequence that describes a novel mutation or for searching for the sequence surrounding a particular mutation. As shown in Figure 3, it is essential to find and use the gene symbol approved by the Human Genome Organisation (HUGO) Gene Nomenclature Committee (HGNC; http://www.gene.ucl.ac.uk/nomenclature/index.html).7,8 A major problem has been the highly variable use of gene nomenclature in the literature, producing multiple symbols and names for one and the same gene9,10 or one gene/protein symbol that stands for completely different genes or proteins.11,12,13,14 Up to one third of human genes may have been affected by the homonym problem,15 mainly because of the nonuse of HGNC-approved official gene symbols.
Figure 3.
How to find a DNA reference sequence and HGNC-approved gene symbol. BLAST, Basic Local Alignment Search Tool; HUGO, Human Genome Organisation; NCBI, National Center for Biotechnology Information.
In addition to the use of the HGNC-approved gene symbol, one needs to find the most appropriate reference sequence for a novel mutation. The most appropriate reference sequence may be a coding DNA sequence based on full-length mRNA or a genomic DNA reference sequence. Even if one finds the mutation based on a reference sequence, it may not be the most updated or the most appropriate reference sequence. For example, the reference sequence that has been used to identify a novel exonic mutation might comprise the sequence of only one exon of the gene. In this case, it is appropriate to search for a coding DNA reference sequence based on full-length cDNA.
Nomenclature for CFTR Mutations: An Example
The cystic fibrosis transmembrane conductance regulator (CFTR) gene (Online Mendelian Inheritance of Man no. 602421) is the gene that, when mutated, causes cystic fibrosis (Online Mendelian Inheritance of Man no. 219700). Although there is a single major deletion mutation of phenylalanine at codon 508, over 1000 different CFTR mutations and variants have been described (http://www.genet.sickkids.on.ca/cftr/). As in other human disease-related genes of interest, the nomenclature for the CFTR mutations and variants has been a persistent problem. The description of CFTR mutations associated with pathogenic changes started well before any mutation nomenclature recommendations were proposed. Consequently, published and commonly used designations for many CFTR mutations have been at variance with the evolving standard nomenclature guidelines (see http://www.HGVS.org/mutnomen/).2,3 Genetics clinics and diagnostic laboratories primarily use these variant or colloquial descriptions.
Table 1 lists the CFTR mutations included in the American College of Medical Genetics-recommended carrier screening panel16 by standard nomenclature and colloquial nomenclature side by side. To describe a single nucleotide substitution based on a coding DNA reference sequence using the standard nomenclature, one must describe it with 1) the GenBank accession number and version number of the coding DNA (or cDNA) reference sequence used, followed by 2) a colon “:”; 3) prefix “c.”; 4) the nucleotide number; 5) a wild-type nucleotide; 6) the symbol “>” (indicating a change); and 7) a mutant nucleotide. For example, in the nomenclature of the CFTR mutation “NM_000492.3:c.350G>A” (ie, p.Arg117His), “NM_000492.3” indicates the GenBank cDNA reference sequence used, c. indicates that the nucleotide number “350” is based on coding DNA sequence (see Figure 1), and “G>A” indicates that the nucleotide substitution is G to A.
Table 1.
Standard and Colloquial Nomenclature for CFTR Mutations and Variants
| DNA sequence change* | Amino acid change (three-letter code) | Commonly used colloquial nomenclature | Site of mutation (exon/intron number)§ | Type of mutation |
|---|---|---|---|---|
| c.254G>A | p.Gly85Glu | G85E | Exon 3 | Missense |
| c.350G>A | p.Arg117His | R117H | Exon 4 | Missense |
| c.443T>C† | p.Ile148Thr | I148T | Exon 4 | Missense |
| c.489+1G>T (AJ574942.1:g.240G>T) | 621+1G>T | Intron 4 | Splice site | |
| c.579+1G>T (AJ574943.1:g.261G>T) | 711+1G>T | Intron 5 | Splice site | |
| c.948delT† | p.Phe316LeufsX12 | 1078delT | Exon 7 (no. 8) | Frameshift |
| c.1000C>T | p.Arg334Trp | R334W | Exon 7 (no. 8) | Missense |
| c.1040G>C | p.Arg347Pro | R347P | Exon 7 (no. 8) | Missense |
| c.1210−12T(5_9) (AJ574948.1:g.152T (5_9)) | 5T/7T/9T polymorphism | Intron 8 (no. 9) | Splice site | |
| c.1210−12[5]‡ (AJ574948.1:g.152T[5]‡) | 5T | |||
| c.1210−12T[9]‡ (AJ574948.1:g.152T[9]‡) | 9T | |||
| c.1364C>A | p.Ala455Glu | A455E | Exon 9 (no. 10) | Missense |
| c.1519_1521delATC | p.Ile507del | Delta I507 | Exon 10 (no. 11) | In-frame deletion |
| c.1521_1523delCTT | p.Phe508del | Delta F508 | Exon 10 (no. 11) | In-frame deletion |
| c.1585−1G>A (AJ574980.1:g.116G>A) | 1717−1G>A | Intron 10 (no. 11) | Splice site | |
| c.1624G>T | p.Gly542X | G542X | Exon 11 (no. 12) | Nonsense |
| c.1652G>A | p.Gly551Asp | G551D | Exon 11 (no. 12) | Missense |
| c.1657C>T | p.Arg553X | R553X | Exon 11 (no. 12) | Nonsense |
| c.1679G>C | p.Arg560Thr | R560T | Exon 11 (no. 12) | Missense |
| c.1766+1G>A (AJ574983.1:g.179G>A) | 1898+1G>A | Intron 12 (no. 13) | Splice site | |
| c.2052delA | p.Lys684AsnfsX38 | 2184delA | Exon 13 (no. 14) | Frameshift |
| c.2657+5G>A (AJ574995.1:g.216G>A) | 2789+5G>A | Intron 14b (no. 16) | Splice site | |
| c.2988+1G>T (AJ575003.1:g.305G>T) | 3120+1G>T | Intron 16 (no. 18) | Splice site | |
| c.3437delC | p.Ala1146ValfsX2 | 3569delC | Exon 18 (no. 21) | Frameshift |
| c.3484C>T | p.Arg1162X | R1162X | Exon 19 (no. 22) | Nonsense |
| c.3718−2477C>T (AY848832.1:g.40725C>T) | 3849+10kbC>T | Intron 19 (no. 22) | Other | |
| c.3846G>A | p.Trp1282X | W1282X | Exon 20 (no. 23) | Nonsense |
| c.3909C>G | p.Asn1303Lys | N1303K | Exon 21 (no. 24) | Missense |
If not specified, nucleotide numbering is based on DNA reference sequence NM_000492.3. Note that the version number of this reference sequence may be frequently updated.
These two mutations in the initial 25-mutation panel have been excluded in the recent American College of Medical Genetics recommendations.19
Common repeat polymorphisms should be described as the first nucleotide number, followed by a repeated nucleotide(s), and the number of repeats in square brackets.
Conventional CFTR exon/intron numbering includes exons 6a and 6b, exons 14a and 14b, and exons 17a and 17b; for exon/intron numbers in parentheses, these exon pairs are simply numbered sequentially, without modifiers such as ′6a′ and ′6b.′
Coding DNA Reference Sequence
Problems in colloquial CFTR mutation nomenclature reside mainly in the numbering of nucleotide positions. Although the colloquial notations of CFTR mutations are also based on the GenBank cDNA reference sequence NM_000492.3, the colloquial notations use nucleotide numbering with the A of the ATG initiation codon at the nucleotide number 133. One can retrieve the coding DNA sequence (“CDS”) for CFTR simply by clicking on the CDS link in GenBank NM_000492.3; this opens a window in which the nucleotide numbering starts with +1 at the A of the ATG initiation codon, thus eliminating 132 nucleotides from the 5′-UTR. There is only one coding DNA reference sequence for a given GenBank accession number, so that one can describe nucleotide positions unequivocally.
Nomenclature for Intronic Variants
Another important issue is the choice of proper nomenclature for intronic variants. To describe an intronic variant clearly and unequivocally, one should use a genomic reference sequence that contains uninterrupted genomic DNA sequence including introns. In contrast, a coding DNA reference sequence does not contain intronic sequences. It is desirable to describe an intronic variant by nomenclature based on not only a genomic DNA reference sequence but also a coding DNA reference sequence. This is because a genomic reference sequence cannot describe the relation to an adjacent exon as can nomenclature based on a coding DNA reference sequence in the form of “c.###+#G>T” or “c.###−#A>C” (where ### and # represent integers; see Figure 1). The relation to an adjacent exon is often clinically important information because it may indicate a predicted pathogenic effect of the variant. For example, there is a CFTR mutation commonly named “621+1G>T” (ie, AJ574942.1:g.240G>T and NM_000492.3:c.489+1G>T using the standard nomenclature). The nomenclature “AJ574942.1:g.240G>T” can provide precise information on the mutated locus and adjacent nucleotides in the intron, whereas the nomenclature “NM_000492.3:c.489+1G>T” provides information on the relation to the adjacent exon (ie, one base after the 489th coding nucleotide at the end of the exon). To describe an intronic mutation such as NM_000492.3:c.489+1G>T based on a coding DNA reference sequence, the distance of a mutated intronic nucleotide to the closest exonic nucleotide is used (see Figure 1). Note that the intronic sequence itself is not present in the coding DNA reference sequence NM_000492.3. Using the standard nomenclature, one can call this mutation neither “c.621+1G>T” because this numbering “621” is not based on a coding DNA reference sequence nor “g.621+1G>T” because a g description cannot be based on a cDNA sequence and a g description cannot contain nucleotide numbering such as “621+1.” One may simply describe this variant as “c.489+1G>T”; however, to prevent confusion when mutations in different genes and/or different transcript variants are described, the prefix “NM_000492.3:” is required to indicate the reference sequence (the version number of any reference sequence may be updated frequently). We described the major intronic mutations of CFTR using both genomic reference sequences and the coding DNA reference sequence (NM_000492.3) in Table 1.
Nomenclature for Nucleotide Repeat Sequence
Nomenclature for nucleotide repeat sequences in the literature and clinical practice is currently in a state of confusion. HGVS has recently updated recommendations to minimize confusion. According to the recommendations, a known common polymorphism of a nucleotide repeat sequence should be described as starting with the first nucleotide number of a repeat sequence, followed by a repeated nucleotide unit (eg, T) or repeated nucleotides (eg, CA) and the number of repeats in square brackets (eg, [5]). Thus, the common polymorphism of CFTR, so called the “5T intron 8 polymorphism” should be described as “c.1210–12T[5] (AJ574948.1:g.152T[5]).” To describe polymorphisms collectively in the same locus, a range of repeat numbers is indicated by underscore, for example, [5_9].
To describe a unique mutation (or variant) of a nucleotide repeat sequence, one should use “dup” or “del” as for other mutations, and nucleotide numbering is based on the most 3′ end of a repeat sequence. For example, if one finds a complete duplication or deletion of the “CTFR 7T intron 8” sequence, one should describe the former as “c.1210−12_1210−6dup” and the latter as “c.1210−12_1210−6del.” However, it is not always possible to determine whether a given variant is common or unique, and any current unique variants may become common variants in the near future. Additional issues include large expansions observed in some trinucleotide expansions such as in FMR1 (the fragile X mental retardation 1 gene) or FXN (the frataxin gene). Nucleotide repeat expansions may have interruptions or mutations within the expanded sequence (eg, AGG among CGG repeats in FMR1), which sometimes have clinical significance even in relatively small expansions. All of these issues need to be resolved.
Description at DNA Level versus Amino Acid Level
As shown in Table 1, genetic sequence changes occur at the DNA level, and we usually identify mutations at the DNA level in a clinical genetic testing. Descriptions at the amino acid level are usually inferred with no experimental proof and are not unequivocal because amino acid codes are degenerate. For example, the most common CFTR mutation, p.Phe508del, due to DNA sequence change c.1521_1523delCTT, can be caused by other DNA sequence changes, including c.1522_1524delTTT. In addition, DNA sequence changes may have unforeseen effects, impairing gene function through other mechanisms such as influencing RNA stability or splicing (disrupting an exonic splice enhancer, activating a cryptic splice site or creating a new splice site). There is also a risk that some one-letter amino acid codes (such as A, C, G, and T) may be confused with nucleotide code when a variant is rare or unfamiliar to health care providers. Nonetheless, specific amino acid changes should be included if such amino acid changes have been experimentally demonstrated, because those amino acid changes likely indicate pathogenic mechanisms of the mutations and provide clinically important information.
Other Commonly Tested Gene Variants
We have found that, even in a gene that is commonly tested in clinical practice, it can be difficult to trace some mutations. An example is the common thrombophilic variant of the prothrombin (F2) gene “20210G>A,” which should be described as F2 AF478696.1:g.21538G>A (c.*97G>A). The “c.*97G>A” notation indicates that this variant is present 97 nucleotides downstream of the stop codon. The designation of “20210” appears to be based on a historical reference sequence. Other examples of standard and nonstandard colloquial nomenclature of genes and variants are listed in Table 2.
Table 2.
Standard and Colloquial Nomenclature of Common Gene Variants
| Standard nomenclature | Colloquial nomenclature | Associated disease |
|---|---|---|
| F2 AF478696.1: g.21538G>A (c.* 97G>A)† | Prothrombin G20210A (or 20210G>A) | Venous thrombosis |
| F5 NM_000130.3: c.1601G>A (p.Arg534Gln) | Factor V 1691G>A (R506Q) | Venous thrombosis |
| MTHFR NM_005957.3: c.665C>T (p.Ala222Val) | MTHFR C677T (or 677C>T) | Homocystinemia |
| HFENM_000410.3: c.845G>A (p.Cys282Tyr) | HFE C282Y | Hemochromatosis |
| HFE NM_000410.3: c.187C>G (p.His63Asp) | HFE H63D | Hemochromatosis |
The symbol
is used for 3′-UTR region. The nucleotide number indicates the distance from the end of the stop codon, in this example, the 97th nucleotide after the stop codon. See Figure 1.
Practical and Educational Issues
Use of correct nomenclature in the literature as well as in laboratory reports is desirable because clinical diagnosis and decision-making are based on data in the literature regarding a specific mutation detected in a proband or affected family member. In a timely editorial, den Dunnen and Paalman addressed the importance of adherence to correct nomenclature in detail.4 Some of our co-authors proposed the use of standard nomenclature for small intragenic mutations of SMN1 (Online Mendelian Inheritance of Man no. 600354), the disease gene for spinal muscular atrophy (SMA; Online Mendelian Inheritance of Man no. 253300 for SMA type I, no. 253550 for SMA type II, and no. 253400 for SMA type III).17,18 Notations for SMN1 mutations have been sufficiently ambiguous and inconsistent, such that one mutation has been reported as two different mutations.17 The HGVS recommends including traditional descriptions, initially, between parentheses or in a second column in a summary table (http://www.HGVS.org/mutnomen/); for example, CFTR NM_000492.3:c.489+1G>T (621+1G>T), NM_000492.3:c.3437delC (3569delC), NM_000492.3:c.1521_1523del (delF508), etc.
Adopting standard nomenclature should help remove confusion. The HGVS recommendations have now been widely accepted, and an increasing number of journals demand that authors follow the recommendations. Colloquial mutation nomenclature is in wide use, however, by clinical laboratories, clinical geneticists, genetic counselors, and diagnostic reagent manufacturers and has set a “standard” in these fields. Consequently, the fact that not all journals or medical societies insist on consistent use of HGVS-recommended nomenclature may cause additional confusion; for example, a novel mutation in CFTR might be described in one report by standard nomenclature and in another report by the traditional description. Therefore, the use of HGVS standard nomenclature can be accompanied by the colloquial term in parentheses. To avoid confusion in the future, the HGVS standard nomenclature should be used for newly discovered CFTR mutations as well as those in other genes of clinical or research interest. Many reports describe new mutations only in terms of the amino acid change, despite the degeneracy of the amino acid code. It is conceivable that future therapies targeted to specific alterations in DNA would be different for different mutations that cause the same amino acid change. Mutation nomenclature should be unequivocal and should be described at the DNA level as discussed in the previous section.
Summary
We have raised issues of standard and nonstandard nomenclature of gene variants and mutations, using a limited number of commonly tested genes as examples, particularly CFTR. Similar issues and problems exist for many other genes. The confusion surrounding nomenclature has potential far-reaching impact, and care needs to be taken to communicate accurately. As we move forward in defining nomenclature rules, it is important for us (laboratory scientists) to educate ourselves and other health care professionals so that standard nomenclature of gene variants and mutations is used uniformly across a wide variety of medical specialties.
Note:
Standardized report format, including standard nomenclature for gene sequence variants, is important in laboratory medicine and clinical practice. College of American Pathologists Molecular Pathology Committee has been compiling comprehensive recommendations for molecular diagnostics laboratories (ML Gulley, RM Braziel, KC Halling, ED Hsi, JA Kant, MN Nikiforova, JA Nowak, S Ogino, A Oliveira, HF Polesky, L Silverman, RR Tubbs, VM Van Deerlin, GV Vance, J Versalovic, for the College of American Pathologists Molecular Pathology Resource Committee; Clinical Laboratory Reports in Molecular Pathology, revised version submited to Arch Pathol Lab Med).
Acknowledgments
We thank Wayne Grody, S. Terence Dunn, Marsha Speevak, Daniel Farkas, Victoria Pratt, Jeffrey Kant, and the Council, Publications Committee and Genetics Subdivision of the Association for Molecular Pathology for helpful discussion and critical reading of the manuscript.
Footnotes
The 2005 Association for Molecular Pathology Training and Education Committee consists of Deborah Payne (Chair), Mary C. Lowery Nordberg, Jerald Z. Gong, Amy E. Krafft, Shuji Ogino, Timothy S. Uphoff, Peter Donahue, Jennifer Hunt, and Gladys Garrison.
Standard of practice is not being defined by this article, and there may be alternatives.
References
- Horaitis O, Cotton RG. The challenge of documenting mutation across the genome: the human genome variation society approach. Hum Mutat. 2004;23:447–452. doi: 10.1002/humu.20038. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE. Recommendations for a nomenclature system for human gene mutations. Nomenclature Working Group. Hum Mutat. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Paalman MH. Standardizing mutation nomenclature: why bother? Hum Mutat. 2003;22:181–182. doi: 10.1002/humu.10262. [DOI] [PubMed] [Google Scholar]
- Ad Hoc Committee on Mutation Nomenclature Update on nomenclature for human gene mutations. Hum Mutat. 1996;8:197–202. doi: 10.1002/humu.1380080302. [DOI] [PubMed] [Google Scholar]
- ACMG Laboratory Practice Committee Working Group ACMG recommendations for standards for interpretation of sequence variations. Genet Med. 2000;2:302–303. [Google Scholar]
- Wain HM, Lush MJ, Ducluzeau F, Khodiyar VK, Povey S. Genew: the Human Gene Nomenclature Database, 2004 updates. Nucleic Acids Res. 2004;32:D255–D257. doi: 10.1093/nar/gkh072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain HM, Bruford EA, Lovering RC, Lush MJ, Wright MW, Povey S. Guidelines for human gene nomenclature. Genomics. 2002;79:464–470. doi: 10.1006/geno.2002.6748. [DOI] [PubMed] [Google Scholar]
- Obstacles of nomenclature. Nature. 1997;389:1. doi: 10.1038/37816. [DOI] [PubMed] [Google Scholar]
- Wain H, White J, Povey S. The changing challenges of nomenclature. Cytogenet Cell Genet. 1999;86:162–164. doi: 10.1159/000015372. [DOI] [PubMed] [Google Scholar]
- Gopalan B, Litvak A, Sharma S, Mhashilkar AM, Chada S, Ramesh R. Activation of the Fas-FasL signaling pathway by MDA-7/IL-24 kills human ovarian cancer cells. Cancer Res. 2005;65:3017–3024. doi: 10.1158/0008-5472.CAN-04-3758. [DOI] [PubMed] [Google Scholar]
- Korner A, Ma L, Franks PW, Kiess W, Baier LJ, Stumvoll M, Kovacs P: Sex-specific effect of the Val1483Ile polymorphism in the fatty acid synthase gene (FAS) on body mass index and lipid profile in Caucasian children. Int J Obes (Lond) (in press) [DOI] [PubMed] [Google Scholar]
- Li Y, Raffo AJ, Drew L, Mao Y, Tran A, Petrylak DP, Fine RL. Fas-mediated apoptosis is dependent on wild-type p53 status in human cancer cells expressing a temperature-sensitive p53 mutant alanine-143. Cancer Res. 2003;63:1527–1533. [PubMed] [Google Scholar]
- Bertram J, Peacock JW, Tan C, Mui AL, Chung SW, Gleave ME, Dedhar S, Cox ME, Ong CJ. Inhibition of the phosphatidylinositol 3′-kinase pathway promotes autocrine Fas-induced death of phosphatase and tensin homologue-deficient prostate cancer cells. Cancer Res. 2006;66:4781–4788. doi: 10.1158/0008-5472.CAN-05-3173. [DOI] [PubMed] [Google Scholar]
- Schijvenaars B, Mons B, Weeber M, Schuemie M, van Mulligen E, Wain H, Kors J. Thesaurus-based disambiguation of gene symbols. BMC Bioinformatics. 2005;6:149. doi: 10.1186/1471-2105-6-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grody WW, Cutting GR, Klinger KW, Richards CS, Watson MS, Desnick RJ. Laboratory standards and guidelines for population-based cystic fibrosis carrier screening. Genet Med. 2001;3:149–154. doi: 10.1097/00125817-200103000-00010. [DOI] [PubMed] [Google Scholar]
- Ogino S, Wilson RB. Importance of standard nomenclature for SMN1 small intragenic (“subtle”) mutations. Hum Mutat. 2004;23:392–393. doi: 10.1002/humu.20013. [DOI] [PubMed] [Google Scholar]
- Ogino S, Wilson RB. Spinal muscular atrophy: molecular genetics and diagnostics. Expert Rev Mol Diagn. 2004;4:15–29. doi: 10.1586/14737159.4.1.15. [DOI] [PubMed] [Google Scholar]
- Watson MS, Cutting GR, Desnick RJ, Driscoll DA, Klinger K, Mennuti M, Palomaki GE, Popovich BW, Pratt VM, Rohlfs EM, Strom CM, Richards CS, Witt DR, Grody WW. Cystic fibrosis population carrier screening: 2004 revision of American College of Medical Genetics mutation panel. Genet Med. 2004;6:387–391. doi: 10.1097/01.GIM.0000139506.11694.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]